Abstract
p63, a p53 family member, is critical for proper skin and limb development and directly regulates gene expression in the ectoderm. Mice lacking p63 exhibit skin and craniofacial defects including cleft palate. In humans p63 mutations are associated with several distinct developmental syndromes. p63 sterile-α-motif domain, AEC (ankyloblepharon-ectodermal dysplasia-clefting)-associated mutations are associated with a high prevalence of orofacial clefting disorders, which are less common in EEC (ectrodactyly-ectodermal dysplasia-clefting) patients with DNA binding domain p63 mutations. However, the mechanisms by which these mutations differentially influence p63 function remain unclear, and interactions with other proteins implicated in craniofacial development have not been identified. Here, we show that AEC p63 mutations affect the ability of the p63 protein to interact with special AT-rich binding protein-2 (SATB2), which has recently also been implicated in the development of cleft palate. p63 and SATB2 are co-expressed early in development in the ectoderm of the first and second branchial arches, two essential sites where signaling is required for craniofacial patterning. SATB2 attenuates p63-mediated gene expression of perp (p53 apoptosis effector related to PMP-22), a critical downstream target gene during development, and specifically decreases p63 perp promoter binding. Interestingly, AEC but not EEC p63 mutations affect the ability of p63 to interact with SATB2 and the inhibitory effects of SATB2 on p63 transactivation of perp are most pronounced for AEC-associated p63 mutations. Our findings reveal a novel gain-of-function property of AEC-causing p63 mutations and identify SATB2 as the first p63 binding partner that differentially influences AEC and EEC p63 mutant proteins.
Keywords: Craniofacial Development, DNA-binding Protein, Genetic Diseases, Molecular Biology, p63, SATB2, Ectoderm, perp
Introduction
p63, a member of the p53 family of transcription factors, encodes multiple isoforms that arise from alternative splicing and transcription from two distinct promoters (1, 2). The latter mechanism generates transactivating (TA)4 or dominant negative (ΔN) p63 proteins (1, 2). TAp63 proteins possess typical p53-like properties whereas ΔNp63 are incapable of inducing characteristic p53 target genes and can also inhibit TAp53 family members (1, 2).
In addition to inhibiting TAp53 family proteins, ΔNp63 can also activate transcription of a distinct subset of genes, including integrin-β4, dlx3/5/6, p57Kip2, and claudin-1 (3–7). These genes have been implicated in early embryonic patterning processes and ectodermal development. Consistent with these observations, p63−/− mice exhibit malformations in ectoderm-derived structures such as the skin and limbs that are not found in mice lacking the other p53 family members (8, 9). Furthermore, a majority of mice specifically lacking TAp63 die during embryonic development, and any remaining pups that are born exhibit multiple defects (3). A significant proportion of TAp63−/− animals develop blisters and ulcerations due to impaired wound healing (3). Hair follicle defects and cysts were also detected in the bladder, kidney, and stomach of TAp63−/− mice (3). Together, this suggests that TA- and ΔNp63α-mediated gene activation is critically important during development.
Recently, perp (p53 apoptosis effector related to PMP-22) has been shown to be an important downstream target gene of p63 during development (10). Originally identified as an apoptotic effector of p53, perp encodes a transmembrane protein that is involved in the formation of desmosomes that mediate cellular connections in stratified epithelia (10–13). Consistent with its role in apoptosis and tumor suppression, specific ablation of perp in the epidermis predisposes mutant mice to carcinoma development (14). However, the expression of perp is substantially diminished in the ectoderm of branchial arches of p63−/− murine embryos as early as embryonic day 10 (E10) (10). perp−/− mice exhibit extensive blistering reminiscent of the skin lacerations detected in p63−/− as well as TAp63−/− mice, supporting the notion that during embryonic development, perp is a p63 target gene (3, 8–10).
In humans, germ line p63 mutations are associated with a group of autosomal dominant, ectodermal disorders that are characterized by abnormalities similar to those observed in p63−/− mice (15). A genotype-phenotype correlation for p63 mutations has been reported (15). p63 mutations that map to the DNA binding domain are associated with EEC (ectrodactyly-ectodermal dysplasia-clefting) syndrome, whereas mutations in the SAM (sterile-α-motif) domain, which is only found in the longest α isoforms of p63, are associated with AEC (ankyloblepharon-ectodermal dysplasia-clefting) syndrome (16, 17). Notably, 80% of AEC patients exhibit orofacial clefting, whereas only 20% of EEC patients display this defect (15). However, the unique mechanisms underlying these disorders are poorly understood. Studies utilizing overexpression of p63 mutant proteins have demonstrated that EEC mutants are unable to activate known p53 family target genes, but AEC mutants retain transcriptional activity for a subset of genes, including the ability to induce perp (18). Because AEC p63α mutations are associated with significant craniofacial defects despite intact transcriptional activity, it is likely that mutation-induced changes in p63 transcriptional activity alone cannot account for distinct AEC and EEC phenotypes and that additional factors exist that likely influence AEC mutant proteins (16, 18).
SATB2 (special AT-rich binding protein-2), a transcription factor that binds DNA in nuclear matrix attachment regions (19) influences gene expression both by orchestrating chromatin structure and by functioning as a transcriptional co-factor (20–22). SATB2−/− mice have defects in bone development and osteoblast differentiation (22). In addition, satb2−/− mice and humans with loss-of-function satb2 mutations develop craniofacial abnormalities including orofacial clefting (22–25). Previously, we have shown that SATB2 and ΔNp63α form an endogenous molecular complex in vivo, regulating proapoptotic target gene transcriptional activation and survival of cancer cells (26). Here, we show that unlike EEC TAp63α, AEC TAp63α-mediated transactivation function is inhibited by SATB2 and specifically attenuates the transactivation of perp, a gene involved in ectodermal development (10). This report provides the first evidence for a p63α-binding protein that differentially influences AEC and EEC mutant proteins.
EXPERIMENTAL PROCEDURES
Hematoxylin and Eosin Staining
p63+/− animals were mated, and pregnant females were killed at E17.5. Following dissection, p63+/+ and p63−/− embryos were decapitated, and the heads were fixed in 4% paraformaldehyde overnight at 4 °C. Specimens were washed with 1× PBS twice for 30 min each, dehydrated in ethanol series, and washed with xylene. Heads were then embedded in wax, and 5-μm sections were taken. Slides were then heated at 60 °C overnight.
Staining was performed by washing the slides in xylene and hydrating in ethanol series. Samples were then stained with hematoxylin for 10 min, 0.5% HCl in 70% ethanol for 10 s, and 1% lithium carbonate for 5 s with water washes in between each step. Slides were incubated in 30%, 50%, and 70% ethanol for 1 min each and then stained with 0.5% eosin solution in 70% ethanol for 5 min. Specimens were dehydrated in ethanol series, washed with xylene, and mounted with Permount.
Whole Mount in Situ Hybridization
Timed pregnant CD1 mice were purchased from Jackson Laboratories (Bar Harbor, ME). Following dissection, embryos were fixed in 4% paraformaldehyde overnight at 4 °C. The next day, embryos were washed three times with 1× PBT (PBS with 0.1% Tween 20) and then 25%, 50%, and 75% methanol/PBT and methanol for 10 min each at 4 °C. The embryos were then bleached with 4:1 methanol/30% H2O2 for 6 h at room temperature in the dark, washed in methanol/PBT again, digested with 20 μg/ml proteinase K, and refixed in a solution of 0.2% glutaraldehyde, 4% paraformaldehyde solution in PBS for 20 min at room temperature. Probe hybridization was performed by using 2.0 μg/ml sense and antisense probes at 63 °C overnight with rocking in hybridization buffer (50% formamide, 0.75 m NaCl, 1×PE (10 mm PIPES, pH 6.8, 0.1 mm EDTA), 100 μg/ml tRNA, 0.05% heparin, and 1% SDS). Embryos were washed in WB1 (300 mm NaCl, 1× PE, and 1% SDS) twice for 30 min at 63 °C, WB1.5 (50 mm NaCl, 1× PE, and 0.1% SDS) twice for 30 min at 50 °C, and treated with RNase A for 1 h at 37 °C. Following RNase A digestion, embryos were washed with WB2 (50% formamide, 300 mm NaCl, 1× PE, and 1% SDS) for 30 min at 50 °C, WB3 (50% formamide, 150 mm NaCl, 1× PE, and 0.1% Tween 20) for 30 min at 50 °C, and WB4 (500 mm NaCl, 1× PE, and 0.1% Tween 20) twice for 5 min at room temperature and once for 20 min at 70 °C. Blocking was carried out for 1 h at room temperature (TBST, 2 mm levamisole, 10% heat- inactivated FBS) and incubated with anti-digoxigenin alkaline phosphatase antibody (1:3000) overnight at 4 °C. Embryos were washed with TBST containing 2 mm levamisole at room temperature several times and then twice with NTMT (100 mm Tris, pH 9.5, 100 mm NaCl, 50 mm MgCl2, and 0.1% Tween 20) with 2 mm levamisole for 20 min each. Color development was started by added BM purple at room temperature and stopped with three washes of PBT containing 1 mm EDTA, pH 8. Specimens were washed in methanol/PBT series, refixed in 0.1% glutaraldehyde, 4% paraformaldehyde solution, and stored in PBT supplemented with 0.5 mm EDTA at 4 °C.
Sense and antisense probes were prepared by cloning the DNA binding domain of murine p63 into the pGem-T-Easy vector (Promega, Madison, WI). The constructs were linearized by digestion with SphI. 1 μg of template was used in an in vitro transcription reaction with Sp6 RNA polymerase according to the manufacturer's instructions (Promega). Ethanol-precipitated RNA probes were dissolved in 50% formamide/Diethylpyrocarbonate water solution.
Confocal Immunofluorescence
E10 embryos were dissected, fixed in 4% paraformaldehyde overnight at 4 °C, and dehydrated in 30% sucrose overnight at 4 °C. Embryos were embedded in OCT embedding medium, and 16-μm transverse sections were taken using the cryostat. Sections were dried at 37 °C for 10 min, fixed with 4% paraformaldehyde for 15 min, and treated with 10% BSA in 0.3% Triton-X/PBS solution for 1 h. Blocking and immunostaining were performed using the MOM kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions, counterstained with DAPI, and mounted using Permafluor.
Cells and Plasmids
SCC9, HEK293, and H1299 cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT) at 5% CO2 at 37 °C. NIH 3T3 cells were cultured in DMEM with 10% heat-inactivated fetal calf serum (HyClone) under the same atmospheric conditions as above.
Plasmids encoding wild-type TA and ΔNp63α as well as T7-SATB2 have been described previously (26). AEC- and EEC-associated p63α mutants were generated using a QuikChange Site-directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. The perp luciferase (pPerp-luc-PS) as well as the mutant (mutD) reporter plasmids were kind gifts from Dr. Laura Attardi.
Transfections and Lentivirus Infections
HEK293 and H1299 cells were transfected using calcium phosphate and polyethylenimine methods, respectively, as described previously (26). NIH 3T3 cells were transiently transfected using FuGENE6 transfection reagent (Roche Applied Science) according to the manufacturer's instructions. Production of lentivirus-expressing shGFP and two different shRNAs targeting SATB2 as well as infection of SCC9 cells were performed as described previously (26).
Chromatin Immunoprecipitations (ChIPs), Immunoprecipitations, Immunoblotting, and Luciferase Assays
ChIPs, immunoprecipitations, immunoblotting, and luciferase assays were performed as described previously (26).
Quantitative Reverse Transcription PCR
Quantitative RT-PCR was performed as described previously. The sequences for primers used for perp and gapdh are as follows: perp-forward, 5′-tcagagcctcatggagtacg-3′; perp-reverse, 5′-ccagggagatgatctggaac-3′; gapdh-forward, 5′-ctcaagggcatcctgggcta-3′; gapdh-reverse, 5′-ggaggagtgggtgtcgctgt-3′.
RESULTS
p63−/− Mice Develop Clefting and Craniofacial Defects
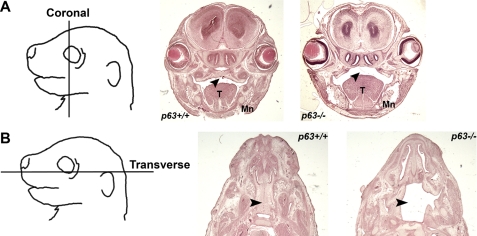
Human p63 mutations are associated with craniofacial clefting (15). In p63−/− E17.5 embryos, craniofacial structures including the tongue and mandible were misshaped and hypoplastic (Fig. 1A). Importantly, the secondary palate in p63−/− embryos is absent (black arrowheads) (Fig. 1, A and B). This is consistent with previous reports showing that loss of p63 affects multiple signaling pathways implicated in craniofacial development including FGF, BMP, and Notch pathways (27, 28). In contrast, the primary palate in p63-knock-out embryos was morphologically normal (data not shown). This suggests that, similar to the developmental role of p63 in humans, p63 plays an essential role in craniofacial patterning and cleft palatogenesis in mice.
FIGURE 1.
Cleft palate is observed in p63-knock-out mice. Heads of E17.5 p63+/+ or p63−/− embryos were dissected, and coronal (A) or transverse (B) sections were cut and stained with hematoxylin and eosin. T, tongue; Mn, mandible. Black arrowheads denote the secondary palate.
p63α and SATB2 Are Co-expressed in the Ectoderm of the Branchial Arches
The severe craniofacial malformations in p63−/− mice suggest that p63 may play a role in early patterning. Therefore, we focused on determining the specific expression of p63 from E9 to E12 because cell-cell signaling is critical as early as mid-gestation for craniofacial patterning (2, 29). A p63 probe that recognizes all p63 isoforms detected p63 transcripts as early as E10 in the first and second branchial arches (Fig. 2A, red arrows), which are transient primitive organizing centers for craniofacial development (29). In addition, p63 mRNA can also be detected in the apical ectodermal ridge, a patterning center for developing limbs (Fig. 2A, red arrowheads) (8, 9). Expression of p63 becomes increasingly prominent in the brain as well as the epidermis starting at E11 and E12, respectively (Fig. 2, C and D, red arrowheads) (30, 31). Furthermore, p63α protein, the longest p63 C-terminal isoform that has the SAM domain, is present in the nuclei of cells located in the outer ectoderm of the first branchial arch (Fig. 2, B–D).
FIGURE 2.
p63α and SATB2 are co-expressed during development. A, whole mount in situ hybridization of wild-type embryos at E9, E10, E11, and E12. The red arrows and arrowheads at E10 denote the branchial arches and the limb buds, respectively. At E11, the red arrowheads highlight the neural staining. B–D, transverse sections of E10 embryos across the branchial arches (B; red arrowheads) stained with anti-p63 (C) or anti-p63α (D) antibodies and analyzed by confocal immunofluorescence. E, similar sections co-stained with anti-pan-p63 and anti-SATB2-CT antibodies. F, NIH 3T3 cells transfected with TA or ΔNp63α. ChIP (IP) analysis was performed on the perp promoter (perp-1). The β-actin gene promoter was used as a negative control. G, murine newborn P3 brains subjected to ChIP analysis using the indicated antibodies. PCR was performed interrogating three sites of the perp promoter. H, co-localization of SATB2 and p63α onto the perp promoter (perp-1 site) analyzed by ChIP/re-ChIP using the indicated antibodies. Scale bars, 10 μm.
We have shown previously that SATB2 forms a molecular complex with ΔNp63α and augments its function to promote chemoresistance in head and neck squamous cell carcinoma (26). Interestingly, heterozygous satb2 loss-of-function mutations in humans that are associated with craniofacial patterning abnormalities, including a cleft secondary palate, are also observed in satb2−/− mice (22–25). The similarities between SATB2- and p63-deficient phenotypes suggest that these proteins may also function in the same pathways during development. We detected nuclear SATB2 expression in the ectoderm by confocal immunofluorescence analysis (Fig. 2E). The SATB2 staining pattern overlapped with anti-p63 staining (Fig. 2E). An essential gene regulated by p63 during ectoderm development is perp (10, 18). ChIP analysis showed that overexpressed TA and ΔNp63α (Fig. 2F) as well as endogenous SATB2 (Fig. 2G) can be detected on multiple sites of the perp promoter (Fig. 2G). Sequential ChIP/reChIP analysis confirmed that these two proteins are co-localized onto the perp promoter in vivo (Fig. 2H). These data suggest that SATB2 and p63 may co-regulate perp expression.
SATB2 Decreases Wild-type TAp63α Binding to the perp Promoter and Inhibits p63-mediated perp Transactivation
Because SATB2 increases the DNA binding capability of ΔNp63α to the promoters of apoptotic target genes in cancer cells (26), we predicted that SATB2 may have a similar effect on the localization of p63α on the perp promoter. Real-time ChIP analysis performed on NIH 3T3 cells transiently transfected with TAp63α or TAp63α together with T7-SATB2 showed that T7-SATB2 increased DNA binding of TAp63α onto the noxa promoter, another apoptotic target gene of the p53 family. Surprisingly, T7-SATB2 had the opposite effect on perp promoter occupancy because less TAp63α was detected on the perp promoter in the presence of T7-SATB2 compared with TAp63α alone (Fig. 3A). These results indicate that SATB2 differentially influences the ability of p63α to interact with target gene promoters.
FIGURE 3.
SATB2 inhibits p63α-mediated perp activation. A, NIH 3T3 cells were transiently transfected with the indicated plasmids and quantitative real-time ChIP analysis was performed on the perp promoter (perp-1 site). Values were normalized to the amount of input DNA that was present. Real-time ChIP was also performed on mock-transfected cells as a control. B, a luciferase reporter assay using the murine perp promoter was performed in H1299 cells transfected with TA or ΔNp63α. A mutant reporter (MutD) with a point mutation in the perp promoter was used as a negative control. C, increasing amounts of T7-SATB2 were co-transfected with TAp63α into H1299 cells, and a perp luciferase reporter assay was performed. D, SCC9 cells were infected with lentivirus-expressing and shRNA targeting GFP or two different shRNAs directed against SATB2. Total RNA was extracted, and quantitative RT-PCR was performed using primers specific for perp. Values were normalized to gapdh housekeeper. IB, immunoblotting. E, SCC9 cells stably infected with lentivirus-expressing shGFP or shSATB2–2 were lysed, and Perp expression was analyzed by immunoblotting.
We next asked whether these changes in perp promoter binding had any effect on perp induction. As expected, expression of either TA or ΔNp63α increased wild-type but not mutant perp promoter-driven luciferase activity (Fig. 3B). (Fig. 3B). Importantly, TAp63α-mediated reporter transactivation was diminished in a dose-dependent manner by SATB2 (Fig. 3C). SATB2 had a similar effect on ΔNp63α-mediated perp reporter activity (data not shown). Furthermore, lentivirus-mediated knockdown of endogenous SATB2 increased endogenous perp transcript levels (Fig. 3D) and Perp protein (Fig. 3E). These results suggest that SATB2 binding to TAp63α attenuates the transcription of perp.
SATB2 Differentially Regulates Human Disease-associated AEC and EEC p63α Mutations
Because AEC-associated p63α mutations (Fig. 4A) and the loss of SATB2 have been linked to craniofacial defects, we asked whether SATB2 differentially interacts with and affects mutant p63α proteins. Compared with wild-type p63α, a panel of AEC-associated p63α point mutants as well as a truncated p63α lacking the SAM domain showed markedly increased binding to SATB2 (Fig. 4, B and C). Furthermore, EEC-associated p63α harboring DNA binding domain mutations bound to SATB2 at a level similar to wild-type p63α (Fig. 4D). Disruption of the p63 oligomerization domain (OD) by truncation (Fig. 4C, ΔCT mutant) or substitution of isoleucine 378 to proline (I378P) that abolishes p63 oligomerization (Fig. 4E) completely abrogated the interaction with SATB2 (Fig. 4F). These results suggest that a functional OD is required for p63 binding to SATB2 and that AEC p63α mutations in the SAM domain specifically affect SATB2 binding. We also tested whether SATB2 formed a complex with TAp63β and TAp63γ, two other C-terminal p63 isoforms that lack the SAM domain but retain the same OD (1, 2). T7-tagged SATB2 failed to co-precipitate with 3×FLAG-tagged TAp63β or TAp63γ, suggesting that SATB2 is a unique p63α-binding protein (Fig. 4G) and that the SAM domain is not sufficient for SATB2 binding.
FIGURE 4.
AEC-associated, SAM domain, p63α mutations increase binding to SATB2. A, mutations associated with EEC syndrome are found exclusively in the DNA binding domain of p63 whereas those associated with AEC syndrome map to the SAM domain of p63α. A nonsense mutation leading to a premature stop codon upstream of the SAM domain has also been associated with AEC syndrome. B–D, various wild-type or mutant 3×FLAG-ΔNp63α constructs were co-transfected with T7-SATB2, and lysates were immunoprecipitated (IP) with anti-FLAG antibodies. Bound proteins were separated on SDS-PAGE and analyzed by immunoblotting (IB). E, wild-type 3F-ΔNp63α or 3F-ΔNp63α mutants were in vitro translated together with wild-type T7-ΔNp63α and immunoprecipitated with anti-FLAG antibodies. Bound proteins were resolved on SDS-PAGE and immunoblotted with anti-FLAG or anti-T7 antibodies. F and G, HEK293 cells were co-transfected with the indicated expression plasmids. Lysates were immunoprecipitated with control (anti-HA) or anti-FLAG antibodies and blotted with anti-FLAG or anti-T7 antibodies.
TAp63α proteins harboring AEC-associated mutations retain the ability to induce gene expression (Fig. 5A) and perp promoter binding comparable with wild-type levels (Fig. 5B) (18). The steady-state level of TAp63α AEC mutant proteins was lower than that of wild-type TAp63α as published previously (18) and is, at least in part, due to decreased stability of the AEC mutant proteins (data not shown). In contrast, loss of the OD, either by truncation or point mutation (I378P), significantly abrogates reporter induction (Fig. 5C). This is consistent with previous work showing that oligomerization is required for p53 family proteins to induce gene expression (32, 33). Given the increased binding of SATB2 to AEC p63α mutants, we next asked whether SATB2 affects AEC p63α-mediated perp transactivation. Introduction of increasing amounts of SATB2 led to a further decline in reporter activity and, at the highest dose, TAp63α-(G530V) transactivation was inhibited by SATB2 to near basal levels (Fig. 5D). Similar results were obtained using a second AEC TAp63α-(Q536L) mutant (Fig. 5e). Importantly, the expression level of wild-type and AEC mutant TAp63α proteins was similar, indicating that inhibition of transcription activity was not due to SATB2 effects on p63α protein stability. Together, these results suggest that AEC-associated TAp63α proteins possess a novel gain-of-function property. These mutants more readily interact with SATB2 and, in turn, SATB2 exerts a dominant negative effect on AEC-associated TAp63α proteins, rendering these proteins transcriptionally inert.
FIGURE 5.
SATB2 has a dominant negative effect on AEC-p63α mutants. A, a luciferase reporter assay was performed using the perp promoter with the indicated TAp63α mutants in H1299 cells (n = 3, mean ± S.E. (error bars)). B, NIH3T3 cells were transfected with the indicated expression plasmids, and ChIP analysis was performed on the perp-1 site in the perp promoter. C, perp luciferase assay was performed with the indicated TAp63α expression plasmids in H1299 cells (representative experiment, mean ± S.D.). D and E, T7-SATB2 was co-transfected, at increasing amounts, with an expression plasmid encoding TAp63α G530V (D) or TAp63α Q536L and a perp luciferase reporter assay was performed (E). IB, immunoblot.
DISCUSSION
Genetic studies in mice have shown that p63 is indispensible for proper development. Loss of p63 is associated with several developmental abnormalities including skin and hair defects as well as orofacial clefting (15, 27, 34, 35). In humans, p63 mutations have been associated with a spectrum of ectodermal dysplasias that are also characterized by similar defects found in p63−/− mice (36). All of these syndromes are sporadic; although rare familial cases have been documented. Only one defective copy of the p63 gene in the germ line is sufficient to cause sporadic disease, and familial cases all exhibit an autosomal dominant mode of inheritance. This suggests that disease-causing p63 mutations either result from loss-of-function haploinsufficiency or represent gain-of-function mutations.
Interestingly, distinct missense mutations in p63 are associated with different ectodermal dysplasia syndromes (15). Mutations in the DNA binding domain are most often detected in individuals with EEC syndrome whereas mutations in patients with AEC syndrome localize to the SAM domain (16, 17). Phenotypically, AEC patients differ from EEC in that they have a much higher incidence of orofacial clefting and display defects in eyelid morphogenesis and hair growth (15). Although this suggests that EEC and AEC mutations have variable effects on p63α function, the mechanisms by which they interfere with p63α function are unknown. Using protein modeling, EEC mutations found in the p63 DNA binding domain are believed to abrogate DNA binding (16). In support, many of the p63 residues mutated in EEC syndrome are conserved in p53 and are the most common tumor-derived mutations that abolish p53 folding and DNA binding (16, 37). Studies have also shown that AEC mutations completely disrupt folding of the SAM domain, indicating that although its biological functions remain unknown, SAM domains of p63 harboring AEC mutations have significant perturbations in three-dimensional structural (38). Additional molecular analysis has revealed that the SAM domains of p63α and p73α do not mediate oligomerization, which is a function typically associated with SAM domains in other proteins such as the Ephrin family of receptor tyrosine kinases (39). An alternative hypothesis is that the SAM domains in the α variants of p63 and p73 mediate binding to unidentified proteins and that mutations within the SAM domain influence these interactions. However, to date, very few candidate p63-binding proteins have been identified. The E3 ubiquitin ligase Itch degrades p63, and one p63 AEC mutant (I549T) disrupts the Itch binding motif (40). However, the vast majority of EEC- and AEC-associated mutant p63 proteins bind to and are degraded by Itch, suggesting that additional factors exist that can discriminate among various p63α mutant proteins (40, 41).
Here, we show that AEC p63 mutations specifically affect the ability of the p63α protein to interact with SATB2, which has recently also been implicated in the development of cleft palate. This is the first p63-binding partner that differentially influences AEC and EEC p63 mutant proteins. SATB2 and p63α are co-expressed in the ectoderm during embryonic development at a time in which craniofacial patterning is initiated (42). Both SATB2 and p63α co-localize onto the perp promoter, a gene that is a critical developmental downstream target of p63 (10). Compared with wild-type p63α and EEC p63α missense mutations, AEC p63α missense mutations affect the ability of p63α to bind to SATB2. This increased interaction results in enhanced SATB2-mediated inhibition of TAp63α AEC mutant transcriptional activity, rendering them functionally incompetent. This represents a novel gain-of-function property of AEC p63α mutants. These results provide mechanistic insight into the differences between AEC and EEC and may help explain why AEC syndrome clinically present with an autosomal dominant mode of inheritance.
Our mapping experiments revealed that disruption of the p63α OD, either by truncation or point mutation, abrogates the ability of p63α to bind SATB2. This indicates that p63α oligomerization is either required for SATB2 interaction and/or the SATB2 binding motif on p63α is formed by multiple molecules. A similar mechanism of protein-protein interactions has been recently proposed for the p53 OD that constitutes a direct binding interface for two proteins, Cul7 and PARC, and this binding surface is formed by at least two p53 molecules (43). In contrast to the effects of OD disruption, the complete loss of the p63α C terminus downstream of the OD resulted in a dramatic increase in the ability of p63α to interact with SATB2, suggesting that an intact SAM domain may partially block the SATB2 binding site on p63α. One possible explanation for these data is that there is an intramolecular interaction between the SAM and OD of p63α. Consistent with this notion, structural studies carried out on Cep-1 (Caenorhabditis elegans p53, the only known p53 homolog in C. elegans) revealed that a region in its C terminus folds into a domain resembling that of vertebrate p63α (as well as p73α) SAM domain (44). Notably, the putative Cep-1 SAM domain folded back onto its OD to stabilize its dimerization (44). Further, we could not detect an interaction between SATB2 with TAp63β or TAp63γ, which possess the same OD as TAp63α, suggesting that the OD is required but not sufficient for the interaction with SATB2. Notably, p63β and p63γ are proteins that possess unique coding sequences downstream of the OD that are not found in α variants and may influence the overall structure of these proteins (45). In accord, a recent report indicates that under unstressed conditions, TAp63α predominantly exists as dimers and not tetramers as is the case with other p63 isoforms (46). Our data provide additional evidence to indicate that the C terminus of the α isoforms of the p53 family are structurally distinct and function in conjunction with the OD. Further work is required to elucidate the molecular basis for the co-operativity between the OD and SAM domains in mammalian p63α and p73α and how that may be important in human disease as AEC-associated mutations cause significant SAM domain instability and unfolding (47).
Previous work has shown that SATB2 can function as a co-factor to regulate gene expression in osteogenesis (22) and SATB family proteins can act as both co-repressors and co-activators in a context-dependent manner (19, 22, 26). Accordingly, we have demonstrated previously that SATB2 augments the trans-repression function of ΔNp63α in cancer cells by increasing ΔNp63α DNA binding onto the promoters of apoptotic target genes (26). However, our findings indicate that the effects of SATB2 on p63 may be more complex and context-dependent. We unexpectedly found that although SATB2 increased TAp63α binding onto the noxa promoter, it decreased TAp63α DNA binding onto the perp promoter. Thus, in addition to influencing the affinity of DNA binding, SATB2 can also direct DNA binding specificity. This modulatory effect has never been reported previously for any p53 family member and, in part, explains the SATB2-mediated inhibitory effect on wild-type as well as AEC-mutant TAp63α perp transactivation.
How can SATB2 differentially regulate p63α binding onto the perp promoter and proapoptotic genes? The OD is necessary for p63α to interact with SATB2, and it is tempting to speculate that SATB2 may regulate oligomerization of p63α and, thus, DNA binding specificity. Studies have shown that under certain conditions, the OD of p63 and p73 has a greater propensity to exist as dimeric rather than tetrameric complexes more typical of the p53 family (46, 48). It would be of significant interest to determine whether SATB2 influences oligomerization of wild-type and AEC-associated p63α proteins and whether that has an impact on DNA sequence recognition and target gene regulation.
Although changes in p63 DNA recognition may simply be the result of altered oligomerization states, a second possibility that may account for this result is that SATB2 may influence the physical landscape of the genome. This may serve to restrict or permit p63 access to target gene promoters. Previous in vitro studies have shown that the position of p53-REs within a nucleosome affects p53 binding, suggesting that the architecture of packed chromatin can influence p53 family function (49). Furthermore, activation of p53 in vivo resulted in p53-dependent loss of nucleosomal organization surrounding p53-REs in the p21 promoter, indicating that p53 can also elicit changes in chromatin packing (49). Whether this is the case for SATB2 or p63 is unknown and warrants further investigation.
Acknowledgments
We thank members of the Irwin and Kaplan laboratories and Drs. Brent Derry and Alan Davidson for technical assistance and insightful discussions.
This work was supported by Canadian Cancer Society Grant 018054.
- TA
- transactivating
- AEC
- ankyloblepharon-ectodermal dysplasia-clefting
- ΔN
- dominant negative
- E
- embryonic day
- EEC
- ectrodactyly-ectodermal dysplasia-clefting
- OD
- oligomerization domain; perp, p53 apoptosis effector related to PMP-22
- SAM
- sterile-α-motif
- SATB2
- special AT-rich binding protein-2.
REFERENCES
- 1. Melino G., De Laurenzi V., Vousden K. H. (2002) Nat. Rev. Cancer 2, 605–615 [DOI] [PubMed] [Google Scholar]
- 2. Yang A., McKeon F. (2000) Nat. Rev. Mol. Cell Biol. 1, 199–207 [DOI] [PubMed] [Google Scholar]
- 3. Su X., Paris M., Gi Y. J., Tsai K. Y., Cho M. S., Lin Y. L., Biernaskie J. A., Sinha S., Prives C., Pevny L. H., Miller F. D., Flores E. R. (2009) Cell Stem Cell 5, 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carroll D. K., Carroll J. S., Leong C. O., Cheng F., Brown M., Mills A. A., Brugge J. S., Ellisen L. W. (2006) Nat. Cell Biol. 8, 551–561 [DOI] [PubMed] [Google Scholar]
- 5. Lopardo T., Lo Iacono N., Marinari B., Giustizieri M. L., Cyr D. G., Merlo G., Crosti F., Costanzo A., Guerrini L. (2008) PloS One 3, e2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lo Iacono N., Mantero S., Chiarelli A., Garcia E., Mills A. A., Morasso M. I., Costanzo A., Levi G., Guerrini L., Merlo G. R. (2008) Development 135, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 7. Radoja N., Guerrini L., Lo Iacono N., Merlo G. R., Costanzo A., Weinberg W. C., La Mantia G., Calabrò V., Morasso M. I. (2007) Development 134, 13–18 [DOI] [PubMed] [Google Scholar]
- 8. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 9. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 10. Ihrie R. A., Marques M. R., Nguyen B. T., Horner J. S., Papazoglu C., Bronson R. T., Mills A. A., Attardi L. D. (2005) Cell 120, 843–856 [DOI] [PubMed] [Google Scholar]
- 11. Attardi L. D., Reczek E. E., Cosmas C., Demicco E. G., McCurrach M. E., Lowe S. W., Jacks T. (2000) Genes Dev. 14, 704–718 [PMC free article] [PubMed] [Google Scholar]
- 12. Ihrie R. A., Reczek E., Horner J. S., Khachatrian L., Sage J., Jacks T., Attardi L. D. (2003) Curr. Biol. 13, 1985–1990 [DOI] [PubMed] [Google Scholar]
- 13. Reczek E. E., Flores E. R., Tsay A. S., Attardi L. D., Jacks T. (2003) Mol. Cancer Res. 1, 1048–1057 [PubMed] [Google Scholar]
- 14. Beaudry V. G., Jiang D., Dusek R. L., Park E. J., Knezevich S., Ridd K., Vogel H., Bastian B. C., Attardi L. D. (2010) PLoS Genet. 6, e1001168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brunner H. G., Hamel B. C., Van Bokhoven H. (2002) J. Med. Genet. 39, 377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Celli J., Duijf P., Hamel B. C., Bamshad M., Kramer B., Smits A. P., Newbury-Ecob R., Hennekam R. C., Van Buggenhout G., van Haeringen A., Woods C. G., van Essen A. J., de Waal R., Vriend G., Haber D. A., Yang A., McKeon F., Brunner H. G., van Bokhoven H. (1999) Cell 99, 143–153 [DOI] [PubMed] [Google Scholar]
- 17. McGrath J. A., Duijf P. H., Doetsch V., Irvine A. D., de Waal R., Vanmolkot K. R., Wessagowit V., Kelly A., Atherton D. J., Griffiths W. A., Orlow S. J., van Haeringen A., Ausems M. G., Yang A., McKeon F., Bamshad M. A., Brunner H. G., Hamel B. C., van Bokhoven H. (2001) Hum. Mol. Genet. 10, 221–229 [DOI] [PubMed] [Google Scholar]
- 18. Beaudry V. G., Pathak N., Koster M. I., Attardi L. D. (2009) Am. J. Med. Genet. 149A, 1952–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobreva G., Dambacher J., Grosschedl R. (2003) Genes Dev. 17, 3048–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cai S., Han H. J., Kohwi-Shigematsu T. (2003) Nat. Genet. 34, 42–51 [DOI] [PubMed] [Google Scholar]
- 21. Cai S., Lee C. C., Kohwi-Shigematsu T. (2006) Nat. Genet. 38, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 22. Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., Karsenty G., Grosschedl R. (2006) Cell 125, 971–986 [DOI] [PubMed] [Google Scholar]
- 23. Britanova O., Depew M. J., Schwark M., Thomas B. L., Miletich I., Sharpe P., Tarabykin V. (2006) Am. J. Hum. Genet. 79, 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. FitzPatrick D. R., Carr I. M., McLaren L., Leek J. P., Wightman P., Williamson K., Gautier P., McGill N., Hayward C., Firth H., Markham A. F., Fantes J. A., Bonthron D. T. (2003) Hum. Mol. Genet. 12, 2491–2501 [DOI] [PubMed] [Google Scholar]
- 25. Vieira A. R., Avila J. R., Daack-Hirsch S., Dragan E., Félix T. M., Rahimov F., Harrington J., Schultz R. R., Watanabe Y., Johnson M., Fang J., O'Brien S. E., Orioli I. M., Castilla E. E., Fitzpatrick D. R., Jiang R., Marazita M. L., Murray J. C. (2005) PLoS Genet. 1, e64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chung J., Lau J., Cheng L. S., Grant R. I., Robinson F., Ketela T., Reis P. P., Roche O., Kamel-Reid S., Moffat J., Ohh M., Perez-Ordonez B., Kaplan D. R., Irwin M. S. (2010) EMBO Rep. 11, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomason H. A., Dixon M. J., Dixon J. (2008) Dev. Biol. 321, 273–282 [DOI] [PubMed] [Google Scholar]
- 28. Laurikkala J., Mikkola M. L., James M., Tummers M., Mills A. A., Thesleff I. (2006) Development 133, 1553–1563 [DOI] [PubMed] [Google Scholar]
- 29. Chai Y., Maxson R. E., Jr. (2006) Dev. Dyn. 235, 2353–2375 [DOI] [PubMed] [Google Scholar]
- 30. Dugani C. B., Paquin A., Fujitani M., Kaplan D. R., Miller F. D. (2009) J. Neurosci. 29, 6710–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacobs W. B., Govoni G., Ho D., Atwal J. K., Barnabe-Heider F., Keyes W. M., Mills A. A., Miller F. D., Kaplan D. R. (2005) Neuron 48, 743–756 [DOI] [PubMed] [Google Scholar]
- 32. Irwin M., Marin M. C., Phillips A. C., Seelan R. S., Smith D. I., Liu W., Flores E. R., Tsai K. Y., Jacks T., Vousden K. H., Kaelin W. G., Jr. (2000) Nature 407, 645–648 [DOI] [PubMed] [Google Scholar]
- 33. Davison T. S., Yin P., Nie E., Kay C., Arrowsmith C. H. (1998) Oncogene 17, 651–656 [DOI] [PubMed] [Google Scholar]
- 34. Moretti F., Marinari B., Lo Iacono N., Botti E., Giunta A., Spallone G., Garaffo G., Vernersson-Lindahl E., Merlo G., Mills A. A., Ballarò C., Alemà S., Chimenti S., Guerrini L., Costanzo A. (2010) J. Clin. Invest. 120, 1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomason H. A., Zhou H., Kouwenhoven E. N., Dotto G. P., Restivo G., Nguyen B. C., Little H., Dixon M. J., van Bokhoven H., Dixon J. (2010) J. Clin. Invest. 120, 1561–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rinne T., Brunner H. G., van Bokhoven H. (2007) Cell Cycle 6, 262–268 [DOI] [PubMed] [Google Scholar]
- 37. Soussi T., Ishioka C., Claustres M., Béroud C. (2006) Nat. Rev. Cancer 6, 83–90 [DOI] [PubMed] [Google Scholar]
- 38. Cicero D. O., Falconi M., Candi E., Mele S., Cadot B., Di Venere A., Rufini S., Melino G., Desideri A. (2006) Cell Biochem. Biophys. 44, 475–489 [DOI] [PubMed] [Google Scholar]
- 39. Chi S. W., Ayed A., Arrowsmith C. H. (1999) EMBO J. 18, 4438–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bellomaria A., Barbato G., Melino G., Paci M., Melino S. (2010) Cell Cycle 9, 3730–3739 [PubMed] [Google Scholar]
- 41. Browne G., Cipollone R., Lena A. M., Serra V., Zhou H., van Bokhoven H., Dötsch V., Merico D., Mantovani R., Terrinoni A., Knight R. A., Candi E., Melino G. (2011) J. Cell Sci. 124, 2200–2207 [DOI] [PubMed] [Google Scholar]
- 42. Minoux M., Rijli F. M. (2010) Development 137, 2605–2621 [DOI] [PubMed] [Google Scholar]
- 43. Kaustov L., Lukin J., Lemak A., Duan S., Ho M., Doherty R., Penn L. Z., Arrowsmith C. H. (2007) J. Biol. Chem. 282, 11300–11307 [DOI] [PubMed] [Google Scholar]
- 44. Ou H. D., Löhr F., Vogel V., Mäntele W., Dötsch V. (2007) EMBO J. 26, 3463–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) Molecular cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 46. Deutsch G. B., Zielonka E. M., Coutandin D., Weber T. A., Schäfer B., Hannewald J., Luh L. M., Durst F. G., Ibrahim M., Hoffmann J., Niesen F. H., Sentürk A., Kunkel H., Brutschy B., Schleiff E., Knapp S., Acker-Palmer A., Grez M., McKeon F., Dötsch V. (2011) Cell 144, 566–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sathyamurthy A., Freund S. M., Johnson C. M., Allen M. D., Bycroft M. (2011) FEBS J. 278, 2680–2688 [DOI] [PubMed] [Google Scholar]
- 48. Coutandin D., Löhr F., Niesen F. H., Ikeya T., Weber T. A., Schäfer B., Zielonka E. M., Bullock A. N., Yang A., Güntert P., Knapp S., McKeon F., Ou H. D., Dötsch V. (2009) Cell Death Differ. 16, 1582–1589 [DOI] [PubMed] [Google Scholar]
- 49. Laptenko O., Beckerman R., Freulich E., Prives C. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 10385–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]