Abstract
When growing in its native habitat, Thlaspi goesingense can hyperaccumulate 1.2% of its shoot dry weight as nickel. We reported previously that both constitutively elevated activity of serine acetyltransferase (SAT) and concentration of glutathione (GSH) are involved in the ability of T. goesingense to tolerate nickel. A feature of SAT is its feedback inhibition by l-cysteine. To understand the role of this regulation of SAT by Cys on GSH-mediated nickel tolerance in T. goesingense, we characterized the enzymatic properties of SATs from T. goesingense. We demonstrate that all three isoforms of SAT in T. goesingense are insensitive to inhibition by Cys. Further, two amino acids (proline and alanine) in the C-terminal region of the cytosolic SAT (SAT-c) from T. goesingense are responsible for converting the enzyme from a Cys-sensitive to a Cys-insensitive form. Furthermore, the Cys-insensitive isoform of SAT-c confers elevated resistance to nickel when expressed in Escherichia coli and Arabidopsis thaliana, supporting a role for altered regulation of SAT by Cys in nickel tolerance in T. goesingense.
Keywords: Allosteric Regulation, Amino Acid, Antioxidants, Arabidopsis, Nickel, Cysteine, Glutathione, Hyperaccumulation, Serine Acetyltransferase, Tolerance
Introduction
Hyperaccumulators can absorb and store high concentration of metal(loid)s in their aerial tissues without showing symptoms of toxicity (1). Nickel hyperaccumulators have been defined as plants with nickel concentrations higher than 1,000 μg of nickel/g in their aerial dry weight (2). The genus Thlaspi (Brassicaceae) contains species that can hyperaccumulate nickel, and various Thlaspi species have been used to understand the mechanism of nickel hyperaccumulation. Among Thlaspi nickel hyperaccumulators, Thlaspi goesingense occurs on Austrian nickel-rich ultramafic (serpentine) soils (3), where it has been reported to accumulate 5,476 ± 974 μg/g nickel in shoot dry biomass (4). This high nickel concentration in T. goesingense is more than 20-fold higher than the nonaccumulator Silene cucubalus, which contained 252 ± 146 μg/g nickel in shoot dry biomass, although these two species were collected from the same site (4).
Glutathione (GSH) is proposed to be an important antioxidant compound in reducing oxidative stress induced by the high concentration of nickel accumulated by the nickel hyperaccumulator T. goesingense (5). Nickel concentrations in the shoot are strongly correlated with GSH and its precursors l-cysteine (Cys) and O-acetylserine (OAS)2 in various Thlaspi hyperaccumulators and nonaccumulators (5, 6). Furthermore, the elevated OAS, Cys, and GSH contents in T. goesingense are probably due to elevated serine acetyltransferase (SAT; EC 2.3.1.30) activity (5). By expressing the gene encoding the mitochondrially targeted SAT from T. goesingense (TgSAT-m) in the nonaccumulator Arabidopsis thaliana, Freeman et al. (5) showed that elevated GSH content reduces nickel-induced oxidative stress and enhances nickel resistance. X-ray absorption spectroscopy showed no significant presence of Ni-S complexes in either transgenic A. thaliana expressing TgSAT-m (5) or in hydroponically or natural field-grown T. goesingense (5, 7), confirming that elevated GSH does not play a role in binding nickel directly.
SAT is involved in the biosynthesis of OAS from acetyl-CoA and serine, and Cys is synthesized by O-acetylserine (thiol) lyase (OAS-TL; EC 4.2.99.8) from OAS and sulfide. SAT and OAS-TL form the cysteine synthase complex, an important regulatory complex (8, 9). Cys is a precursor for GSH biosynthesis through the action of γ-glutamylcysteine synthetase (EC 6.3.2.2) and glutathione synthetase (EC 6.3.2.3) (10). SAT is known as the first rate-limiting enzyme in Cys synthesis (11–13), because SAT is at 100–400-fold lower concentrations than OAS-TL in cells (14–17). There are at least three known compartmental isoforms of SAT in plants, localized in the cytosol, chloroplast, and mitochondria, as observed in multiple species, such as A. thaliana (18, 19) and Citrullus vulgaris (20, 21). In A. thaliana, there are three cytosolic SATs, one mitochondrial SAT, and a chloroplast SAT (22).
One of the features of SAT is that at least one SAT isoform is sensitive to feedback inhibition by Cys. The sensitivity of SAT to inhibition by Cys differs between plant species and subcellular compartments. The sensitivity of SAT to Cys has been mainly reported in cytosolic SAT isoforms in various species (18, 20, 23–25). On the other hand, SAT insensitive to Cys has been reported for the plastidic and mitochondrial SAT isoforms (18, 22). It has been proposed that Cys feedback inhibition of SAT is part of the sensor system that maintains the Cys concentration in various subcellular compartments (26). SAT has two important domains: a protein-protein interaction domain for the interaction of SAT/SAT and SAT/OAS-TL and a domain for Cys-dependent regulation. It is known that several amino acids in the C-terminal region of SAT play important roles in both Cys and OAS-TL binding (19, 27). The N-terminal region of SAT is also involved in Cys inhibition, although this region does not have a catalytic domain (19).
We hypothesize that an increased Cys insensitivity of SAT in T. goesingense is a possible mechanism to account for at least part of the increased OAS production observed in T. goesingense and which we have shown leads to elevated GSH and nickel resistance (5). To test this hypothesis, we characterized the enzymatic properties of the cytosolic (SAT-c), plastid (SAT-p), and mitochondrial (SAT-m) isoforms of SAT from T. goesingense. Furthermore, we heterologously expressed SAT with contrasting Cys sensitivities from T. goesingense in A. thaliana and Escherichia coli to assess the effect of overproduction of Cys-insensitive and Cys-sensitive SAT isoforms on nickel resistance.
EXPERIMENTAL PROCEDURES
Bacterial Strains
Several E. coli strains were used. For general cloning, TOP10F′ (F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG) and DH5α (supE44 ΔlacU169 (phi80 lacZ ΔM15) hsdR17 recA1 endA1 gryA96 thi-1 relA1) were used. For recombinant protein expression experiments, Rosetta (DE3) pLysS (F− ompT hsdSB (rB− mB−) gal dcm lacY1 (DE3) pLysSRARE6, Cmr, Novagen) was used. E. coli strain JM15 (cysE tfr), which lacks SAT activity and requires Cys for growth, was kindly provided by the Yale E. coli Genetic Stock Center and used for functional complementation with the various SAT constructs. For plant transformation, disarmed Agrobacterium tumefaciens GV3101 was used. All pGreen constructs were transformed with pSoup plasmid DNA into Agrobacterium using electroporation.
Expression Vector Constructions
To clone the SAT coding region from A. thaliana and T. goesingense into the E. coli expression vector pET32(a)+ (Novagen), full-length open reading frame specific primers for PCR were designed. AtSAT-c (AtSerat1;1, U30298), TgSAT-m (AY618468), TgSAT-p (AY618469), and TgSAT-c (AY618470) were used as the template for PCR amplification. For the generation of recombinant protein pET32-AtSAT-c, the 5′ primer was 5′-CCATGGAAATGCCACCGGCCGGAGAACT-3′ (NcoI), and 3′ primer was 5′-GAATTCCAACTTTATATGATGTAATCTG-3′ (EcoRI). The NcoI and EcoRI-digested fragment was inserted into NcoI and EcoRI-linearized pET32(a)+ expression vector. For the pET32-TgSAT-m, 5′ primer was 5′-TTACCTGCCTATCATGTTCCCGGTCACAATGC-3′ (BspMI), and 3′ primer was 5′-TTAAGCTTTCAAATTACATAATCCGAC-3′ (HindIII). For the pET32-TgSAT-p, 5′ primer was 5′-TTGAAGACAACATGGCACCGTGCATCG-3′ (BbsI), and 3′ primer was 5′-TTAAGCTTTTAGATAACGTAAT CAGAC-3′ (HindIII). For the pET32-TgSAT-c, 5′ primer was 5-TTACCTGCTAATCATGCCGCCTGCCG-3′ (BspMI), and 3′ primer was 5′-TTGAATTCTCAAATAATGTAATCTGACC-3′ (EcoRI). All PCR products were directly ligated into the pGEM-T-easy vector (Promega) after purification of PCR products using a PCR product purification kit (Invitrogen). The resultant plasmids were fragmented using a pair of restriction enzymes: BspMI and HindIII for TgSAT-m, BbsI and HindIII for TgSAT-p, and BspMI and EcoRI for TgSAT-c. Fragments of the three SAT genes were ligated into linearized pE32(a)+. All constructs were confirmed by DNA sequencing using the T7 and S-tag sequences in pET32(a)+ for primer design.
To produce the domain-swapped constructs between TgSAT-c and AtSAT-c (At-N/Tg-C and Tg-N/At-C), pET32-AtSAT-c and pET32-TgSAT-c were used as templates for digestion and ligation. The unique BglII restriction site, in both the pET32(a)+ vector and SAT coding region, was used to prepare the domain-swapped constructs. After treatment of both pET32-AtSAT-c and TgSAT-c with BglII, two fragments from each reaction were eluted from 0.8% agarose gel using a gel elution kit. For the At-N/Tg-C construct, an ∼400-bp fragment from pET32-AtSAT-c was ligated into BglII-linearized pET32-TgSAT-c, and vice versa for the Tg-N/At-C. All constructs were confirmed by DNA sequencing.
For site-directed mutagenesis of SAT, a combination of 5′ gene-specific primer harboring the point mutation and a 3′ primer with an EcoRI site was used. For the C268P point mutation, the 5′ primer 5′-GACGTGCCTCCTCGAGGTACTGCGGTTG-3′ (BgmBI) was used. For the G270A point mutation, the 5′ primer 5′-GACGTGCCTTGTCGAGCTACTGCGGTTG-3′ (BgmBI) was used. For the C268P/G270A point mutation, the 5′ primer 5′-GACGTGCCTCCTCGAGCTACTGCGGTTG-3′ (BgmBI) was used. The common 3′ gene-specific primer was 5′-GAATTCCAACTTTATATGATGTAATCTG (EcoRI). The BgmBI and EcoRI-treated fragments of each of the PCR products were ligated into pET32-ATSAT-c, which was linearized with BgmBI and EcoRI.
Expression and Purification of Recombinant Proteins
The resultant cDNAs in the pET32 vector, which were in frame with the N-terminal thioredoxin, S-tag, and His6 tag, were transformed into the Rosetta DE3 pLysS E. coli strain (Calbiochem-Novabiochem). For protein expression, cells were grown at 30 °C in liquid Luria-Bertani (LB) medium containing 100 μg/ml ampicillin. At an optical density of 0.6 measured at 600 nm, isopropyl 1-thio-β-d-galactopyranoside (1 mm) was added, and the culture was grown for 10 h at 30 °C. For purification of recombinant protein, bacterial cells were pelleted at 10,000 × g for 10 min, resuspended in BugBuster (Novagen), and incubated for 30 min at room temperature for the disruption of the E. coli cells. After clarification by centrifugation at 10,000 × g for 25 min, recombinant proteins were affinity-purified using TALON cobalt resin (Clontech) according to the manufacturer's instructions. Protein concentration was determined in each fraction using bicinchoninic acid colorimetric detection and quantification assay (BCA protein assay kit; Thermo Fisher Scientific Inc., Rockford, IL) using a bovine serum albumin standard curve. Recombinant proteins were freshly purified for each experiment.
SAT Assay with Recombinant SAT Proteins
The activity of recombinant SAT proteins was assayed according to Noji et al. (18). The reaction mixture consisted of 50 mm Tris-HCl (pH 8.0), 0.1 mm acetyl-CoA, 1 mm serine, and a known amount of the purified recombinant SAT protein. Assays were carried out in a final reaction volume of 0.1 ml at room temperature. The reaction was initiated by the addition of serine, and the decrease in acetyl-CoA was monitored spectrophotometrically by absorption at 232 nm. The nmol of acetyl-CoA cleaved during the assay were calculated using a molar extinction coefficient of 4,500 m−1 cm−1 (18).
Nickel Resistance in E. coli
For the measurement of nickel resistance in E. coli expressing various SAT constructs, strains were grown in liquid LB medium. All SAT cDNAs tested, including three SAT cDNAs from T. goesingense, SAT-c from A. thaliana, domain-swapped SAT constructs, and point-mutated SAT constructs, were expressed in cysE− E. coli from the pET32(a)+ vector. Nickel resistance was measured in liquid LB medium containing ampicillin (100 μg/ml) and isopropyl 1-thio-β-d-galactopyranoside (1 mm), with or without nickel acetate (2 mm) at 30 °C in a shaking incubator at 300 r.p.m. for 24 h, and growth was measured as optical density at 600 nm (A600). In this assay, a starter culture containing various strains hosting the SAT constructs was initiated in LB medium containing ampicillin (100 μg/ml) and subcultured at an A600 of 0.6 three consecutive times. After a final A600 of 0.6 was achieved, 200 μl of culture was subcultured into 20 ml of LB medium containing ampicillin (100 μg/ml) and isopropyl 1-thio-β-d-galactopyranoside (1 mm), with or without nickel acetate (2 mm). After 24 h, A600 was measured, and cultured cells were harvested at 10,000 × g for 15 min at 4 °C. Pelleted cells were washed twice by resuspension in 0.9% NaCl and centrifuged at 10,000 × g for 5 min at 4 °C. Collected cells were immediately frozen in liquid nitrogen and stored at −80 °C.
Determination of Cys and GSH Concentrations
For determination of Cys and GSH concentrations in E. coli, we used a modified procedure of Saby et al. (28). In brief, frozen samples were resuspended with vigorous vortexing for 5 min in 0.1 m HCl containing 2 mm disodium EDTA (4 °C) and sonicated twice for 5 min in an ultrasound bath (Prolabo 670/H; power, 9) at 4 °C. Suspensions were incubated for 10 min in an ice bath and centrifuged at 10,000 × g for 5 min. The resulting supernatants were immediately frozen in liquid nitrogen and fast thawed in a water bath at 30 °C three times and centrifuged at 10,000 × g for 5 min, and the supernatants were transferred to new tubes. For the determination of Cys and GSH in plants, extraction was performed according to Tsakraklides et al. (29). Shoot tissue (minimum 0.05 g, fresh weight) was harvested, weighed, and frozen immediately in liquid nitrogen. Frozen tissue was ground to a fine powder in a mortar and pestle at 4 °C. The fine powder was resuspended with extraction buffer containing 0.1 m HCl, 1 mm EDTA, and N-acetyl cysteine as an internal standard. After grinding and vortexing, samples were centrifuged at 14,000 × g in a refrigerated centrifuge at 4 °C for 15 min. The supernatant was then transferred to a new tube.
For the derivatization of samples from E. coli and plants, 75 μl of sample in 0.1 m HCl and 1 mm EDTA was added into analysis buffer containing a final concentration of 75 mm sodium borate (pH 9.5–10) and 4 mm DTT, which was vortexed for 3 min, and incubated for 10 min at room temperature. After incubation, 12 mm bromobimane was added and incubated for 15 min at room temperature in the dark. The final volume of the reaction mixture was 100 μl, and the reaction was terminated with the same volume of 20% (v/v) acetic acid. Derivatized samples were analyzed using the AccQ Tag amino acid analysis system (Waters Corp., Milford, MA) using a Waters HPLC system consisting of a Waters Separation module 2695 with a Waters 2475 fluorescence detector.
Construction of Plant Transformation Vectors
For the heterologous expression studies, SAT coding regions were cloned into the pGreen 0299 plant transformation vector. For the heterologous expression of TgSAT-c, the 5′ primer was 5′- GCTCTAGAATGCCGCCTGCCGGAGAA-3′ (XbaI), and the 3′ primer was 5′-GGAATTCTCAAATAATGTAATCTGACC-3′ (EcoRI). The XbaI and EcoRI-digested fragment was inserted into XbaI and EcoRI-linearized pGreen vector. For the overexpression of AtSAT-c and the three point-mutated constructs, the 5′ primer was 5′-CGGGATCCATGCCACCGGCCGGAGAAC-3′ (BamHI), and the 3′ primer was 5′-GGAATTCTTATATGATGTAATCTGACCA-3′ (EcoRI). AtSAT-c, C268P, G270A, and C268P/G270A pET32(a)+ constructs were used as the template for PCR amplification. The BamHI and EcoRI-digested fragment was inserted into BamHI and EcoRI-linearized pGreen vector.
Plant Cultivation and Transformation
For plant growth in soil, seeds were planted in Scotts (Marysville, OH) Redi-earth artificial soil mix and cold-treated at 4 °C for 5 days to synchronize germination. Seeds were germinated in a climate-controlled room at 19–24 °C with 10 h of photosynthetically active light at an intensity of 90–150 μmol m−2 s−1. A. thaliana was transformed using Agrobacterium by floral dipping (30), and transformed seedlings were selected on glyphosate-ammonium (Pestanal, Sigma-Aldrich). To check segregation ratios in the T2 and T3 generation, at least 200 seeds were sown onto the surface of sterilized 2.22 g/liter MS medium (Caisson Laboratory, Inc.), containing 1× MS vitamin stock (Caisson Laboratory, Inc.), 100 μm glyphosate-ammonium, and 1.2% agar (Sigma-Aldrich) in 100 × 15-mm polystyrene Petri dishes. The T2 lines, showing an ∼75% survival rate (single T-DNA insertion), were selected. T3 seeds collected from the individual T2 plants were again selected on 100 μm glyphosate-ammonium containing MS medium, and only lines showing 100% survival were selected (homozygous for single insertion). Homozygous plant lines for the SAT transgene in the T3 generation were used in all experiments presented.
Transient Expression of GFP Fusion Constructs in A. thaliana Protoplasts
For the protoplast transient expression, the SAT coding regions were amplified from the pET32(a)+ constructs using PCR with 5′ primer 5′-GCTCTAGAATGTTCCCGGTCACAAGTC-3′ (XbaI) and 3′ primer 5′-TCCCCCGGGAATTACATAATCCGACCA-3′ (SmaI) for TgSAT-m, 5′ primer 5′-GCTCTAGAATGGCACCGTGCATCGACA-3′ (XbaI) and 3′ primer 5′- TCCCCCGGGGATAACGTAATCAGACC-3′ (SmaI) for TgSAT-p, 5′ primer 5′-GCTCTAGAATGCCGCCTGCCGGAGAA-3′ (XbaI) and 3′ primer 5′-TCCCCCGGGAATAATGTAATCTGACC-3′ (SmaI) for TgSAT-c, and 5′ primer 5′-CGGGATCCATGCCACCGGCCGGAGAAC-3′ (BamHI) and 3′ primer 5′-TCCCCCGGGTATGATGTAATCTGACCA-3′ (SmaI) for AtSAT-c and point mutation constructs. All PCR products were confirmed by sequencing and were inserted into the XbaI and SmaI or BamHI and SmaI sites of the plasmid pUC-sGFP to create chimeric GFP fusion constructs under the control of the CaMV 35S (Cauliflower mosaic virus 35S) promoter. For the protoplast isolation and plasmid transformation, leaf tissues from 5-week-old plants were collected and incubated with 30 ml of enzyme solution containing 0.25% Marcerozyme R-10 (Yokult Honsha Co., Ltd., Tokyo, Japan), 1% Cellulase R10 (Yokult Honsha Co., Ltd.), 500 mm mannitol, 1 mm CaCl2, and 5 mm MES-KOH (pH 5.6) for 8 h with gentle agitation (50–75 r.p.m) at 22 °C in dark conditions. Incubated mixtures were filtered through a 70-μm nylon mesh and were washed with the same volume of W5 solution containing 154 mm NaCl, 125 ml CaCl2, 5 mm KCl, 5 mm glucose, and 1.5 mm MES. After centrifuging at 46 × g for 5 min, protoplasts were resuspended in fresh W5 solution and overlaid onto a 20-ml 21% (w/w) sucrose cushion and centrifuged at 100 × g for 10 min. The intact protoplasts were transferred to tubes containing 40 ml of W5 solution and incubated at 4 °C at least 5 h before plasmid transformation. Polyethylene glycol-mediated transformation was used for transformation of sGFP fusion constructs into A. thaliana protoplast (31). Images were captured with a Zeiss Axioplan fluorescence microscope (Carl Zeiss Co., Jena, Germany) using the XF116 (exciter, 474AF20; dichroic, 500 DRLP; emitter, 605DF50) filter set for GFP.
Stable Expression of GFP Fusion Constructs in A. thaliana
For the stable expression of GFP fusion constructs, the sGFP coding region was amplified with 5′ primer 5′-TCCCCCGGGATGGTGAGCAAGGGCGAGGAG-3′ (SmaI) and 3′ primer 5′-CGGAATTCTTAGGACTTGTACAGC-3′ (EcoRI). The PCR product was confirmed by sequencing and inserted into the SmaI and EcoRI sites of the plasmid pGreen 0299 plant transformation vector under the control of the CaMV 35S promoter to make pGreen-sGFP constructs. All PCR-amplified SAT cDNAs from the pET32(a)+ vector constructs were fused in frame to sGFP. A. thaliana was transformed using Agrobacterium by floral dipping (30), and transformed seedlings were selected on glyphosate-ammonium. Images were acquired using a Zeiss LSM 710 laser-scanning microscope. The fixed 488 nm was used to excite the GFP, and the fluorescence emitted between 492 and 537 nm was collected. Chlorophyll was simultaneously excited using the 633 nm, and the emission was collected between 647 and 721 nm. Mitotracker was excited using 594 nm, and emission was collected between 562 and 620 nm. A transmitted light image was also collected for reference. Images were processed using the Zeiss imaging software and Photoshop (Adobe, San Jose, CA).
Nickel Resistance in Plants
The measurement of nickel resistance of A. thaliana heterologously expressing SAT was performed on agar plates according to Freeman et al. (5). Surface-sterilized seeds were sown onto the surface of half-strength MS medium (Caisson Laboratory, Inc.) containing 1.2% agar (Sigma-Aldrich) as concentric circles 5 mm apart around a central filter paper disc (6-mm diameter; BD Biosciences) soaked with 50 μl of 100 mm nickel acetate. After 13 days of growth at 19–24 °C with 10 h of 90–150 μmol m−2 s−1 photosynthetically active light, both upright seedlings with secondary leaves and seedlings with hooked roots that represent root growth to the bottom of the agar were recorded for each ring. For Cys and GSH analysis, seedlings were collected, and Cys and GSH were measured using a Waters HPLC system as described previously.
Statistical Analysis
Statistical analyses were performed using Office Excel 2007 software from Microsoft. One-way analysis of variance was done to test the equality of three or more means. Student's t tests were done to statistically compare pairs of means. Statistically significant differences were determined at a 5% level of probability for all comparisons. IC50 was calculated using software as described by Brooks (33).
RESULTS
Cys Sensitivity of SAT
We had previously cloned TgSAT-m (AY618468), TgSAT-p (AY618469), and TgSAT-c (AY618470) from T. goesingense using a nickel sensitivity suppression screen in E. coli (5, 34). The predicted subcellular localization of these proteins (TgSAT-c, TgSAT-m, and TgSAT-p) was determined by stable expression of translational fusions of SAT with GFP in A. thaliana (supplemental Fig. 1). As predicted from the targeting sequences, TgSAT-m was found to be localized to the mitochondria. The GFP signal observed from TgSAT-p-GFP was found to co-localize with autofluorescence from chlorophyll A. The GFP signal from AtSAT-c-GFP localized to the cytoplasm as previously reported by Kawashima et al. (22). The GFP signal from TgSAT-c-GFP also localized to the cytosol as predicted from the homology to AtSAT-c. These data were collected from at least 10 independent A. thaliana transgenic plant lines stably expressing each GFP-fused construct and are consistent with data obtained from transient expression assays in A. thaliana protoplasts (supplemental Fig. 2).
To characterize the enzymatic properties of SAT from T. goesingense, we prepared four SAT constructs containing TgSAT-m, TgSAT-p, and TgSAT-c, and AtSAT-c from A. thaliana, as a control. We expressed the constructs in E. coli, purified the recombinant enzymes, and determined Cys inhibition of the SAT activity. AtSAT-c from A. thaliana was used as a positive control for the Cys inhibition experiments because this SAT isoform is known to be Cys-sensitive (18). In our experiments, the cytosolic AtSAT-c from A. thaliana was found to be inhibited by Cys with an inhibitory concentration (IC50) = 3.3 μm (Fig. 1A and Table 1), which compares well with the published value of IC50 = 1.8 μm (18). Both the TgSAT-m and TgSAT-p isoforms of SAT were not inhibited by Cys up to 100 μm (Fig. 1A), as we would expect for the chloroplast and mitochondria isoforms from A. thaliana (18). Surprisingly, TgSAT-c was found to be significantly less sensitive to Cys up to 100 μm than its orthologue from A. thaliana (Fig. 1A and Table 1).
FIGURE 1.
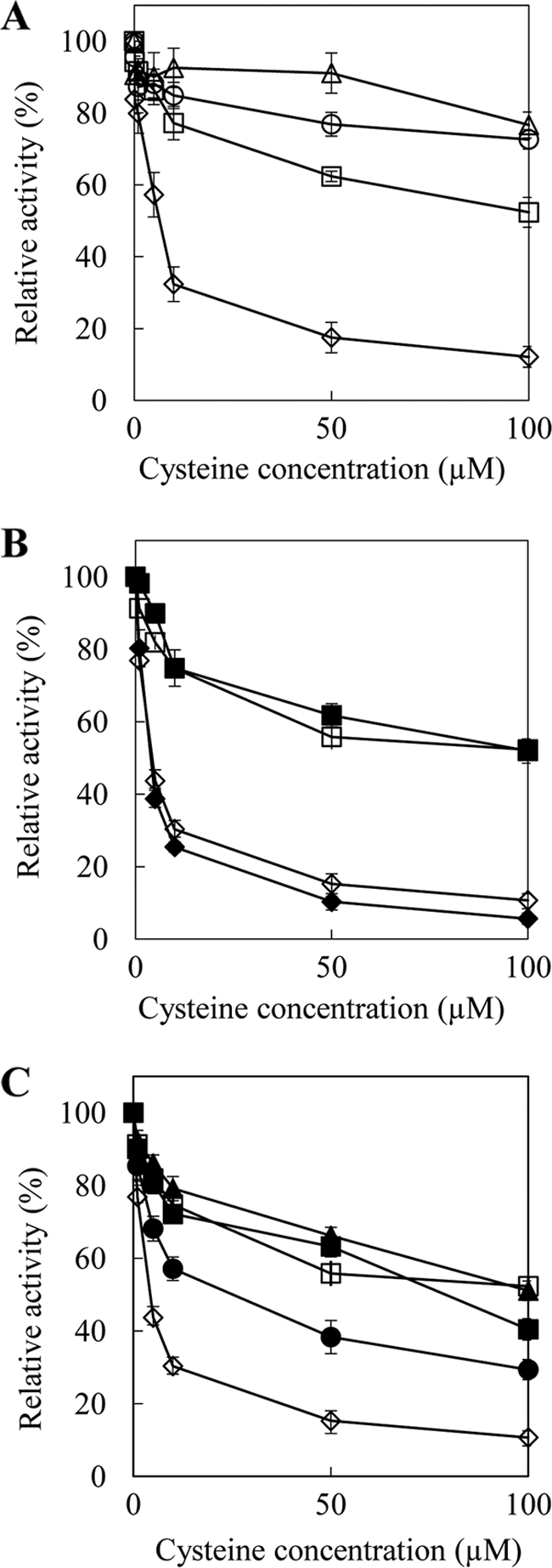
Inhibition of SAT activity by Cys. A, SAT activity was determined in the presence of various concentrations of Cys. Open diamonds, AtSAT-c; open triangles, TgSAT-m; open circles, TgSAT-p; open squares, TgSAT-c. B, Cys inhibition of domain-swapped SATs. At-N/Tg-C represents domain swapping between the N terminus of AtSAT-c and the C terminus of TgSAT-c, and Tg-N/At-C represents domain swapping between the N terminus of TgSAT-c and the C terminus of AtSAT-c. Open diamonds, AtSAT-c; closed diamonds, Tg-N/At-C; open squares, TgSAT-c; closed squares, At-N/Tg-C. C, Cys inhibition of point-mutated AtSAT-c. Open diamonds, AtSAT-c; open squares, TgSAT-c; closed squares, C268P; closed circles, G270A; closed triangles, C268P/G270A. Data represent the average ± S.E. (error bars) of three replicate protein samples with three independent assays each (n = 9). All assays were done in the presence of 1 mm l-serine and 0.1 mm acetyl-CoA.
TABLE 1.
Differences in Cys sensitivity and IC50 of various SATs
Data represent the average of three replicate protein samples with three independent assays each. IC50 represents the Cys concentration that inhibits 50% of the SAT activity.
| Isoforms | Inhibition by l-Cys | IC50 |
|---|---|---|
| μm | ||
| AtSAT-c | Sensitive | 3.3a |
| TgSAT-m | Insensitive | >100b |
| TgSAT-p | Insensitive | >100b |
| TgSAT-c | Insensitive | >100b |
| Tg-N/At-C | Sensitive | 3.4a |
| At-N/Tg-C | Insensitive | >100b |
| C268P | Sensitive | 83.9c |
| G270A | Sensitive | 17.7d |
| C268P/G270A | Insensitive | >100b |
a,b,c,d Values followed by different letters within a treatment are significantly different (one-way analysis of variance, p < 0.05, n = 9).
C-terminal Domain of TgSAT-c Is Important for Resistance to Inhibition by Cys
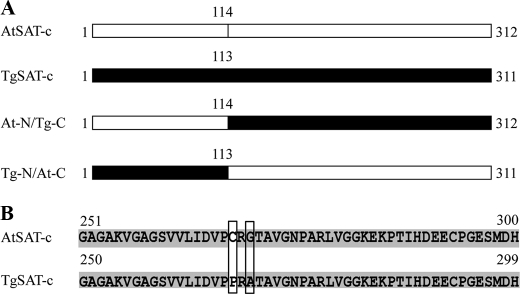
To examine which region of TgSAT-c is responsible for its Cys insensitivity when compared with AtSAT-c, we generated two chimeric proteins in which we swapped the N- and C-terminal domains of the A. thaliana and T. goesingense proteins. These chimeric proteins contained the N-terminal region of AtSAT-c and the C-terminal region of TgSAT-c and vice versa, swapped at amino acid 113 of TgSAT-c or 114 of AtSAT-c (Fig. 2A). The N-terminal region of AtSAT-c was fused with the C-terminal region of TgSAT-c and named At-N/Tg-C. In a separate construct, the N-terminal domain of TgSAT-c was fused to the C-terminal domain of AtSAT-c and named Tg-N/At-C. After expression and purification of the recombinant chimeric proteins, we examined the Cys inhibition of the enzymes. We observed that Tg-N/At-C SAT activity was inhibited by Cys with IC50 = 3.4 μm, the same IC50 as we observed for AtSAT-c (Fig. 1, A and B, and Table 1). The At-N/Tg-C enzyme showed the same insensitivity to Cys as TgSAT-c, with an IC50 of >100 μm (Fig. 1B and Table 1).
FIGURE 2.
A, schematic diagram of the At-N/Tg-C and Tg-N/At-C constructs. At-N/Tg-C represents domain swapping between the N-terminal 114 amino acids of AtSAT-c and C-terminal 198 amino acids of TgSAT-c, and Tg-N/At-C represents domain swapping between the N-terminal 113 amino acids of TgSAT-c and C-terminal 198 amino acids of AtSAT-c. White and black boxes, AtSAT-c and TgSAT-c, respectively. B, alignment of the predicted C-terminal amino acid sequences of TgSAT-c and AtSAT-c. Gray, identical amino acids; white, non-similar amino acid residues. Boxes indicate the amino acids used for the point-mutated SATs.
Amino Acid Residues That Play a Critical Role in Determining Cys Insensitivity
To address which amino acid(s) of TgSAT-c are important for Cys insensitivity, relative to AtSAT-c, we compared the sequences of TgSAT-c and AtSAT-c. AtSAT-c and TgSAT-c share 94.2% identity and 96.8% similarity. Alignment of the amino acid sequence of the two SAT-c isoforms from A. thaliana and T. goesingense revealed several differences in the sequences. There are a total of 20 amino acid differences between the two proteins, 16 of which are in the N-terminal region and four of which are in the C-terminal region (supplemental Fig. 3). Two differences, Cys → Pro at 268, and Gly → Ala at 270, in the C-terminal region, were noticeable because of the non-conserved nature of the Cys to Pro change and the known importance of the conformation of the SAT C-terminal domain for Cys inhibition (19, 35, 36). To confirm if the Cys to Pro and Gly to Ala are important in conferring Cys insensitivity to TgSAT-c, we engineered three mutated forms of AtSAT-c, in which we replaced Cys with Pro at position 268 (named C268P), Gly with Ala at position 270 (named G270A), and Cys with Pro and Gly with Ala (named C268P/G270A) (Fig. 2B).
We observed that C268P has a 20-fold decrease in its sensitivity to Cys, with an increase in the Cys IC50 from 3.3 to 83.9 μm (Fig. 1C and Table 1), although this single point mutation did not achieve the same level of Cys insensitivity as TgSAT-c, which has an IC50 of >100 μm Cys. G270A was observed to show a 3-fold increase of Cys insensitivity compared with AtSAT-c, with an IC50 value of 17.7 μm (Fig. 1C and Table 1). Further, the C268P/G270A construct, containing both amino acid changes, showed a Cys insensitivity of >100 μm, similar to TgSAT-c (Fig. 1C and Table 1).
Expression of Cys-insensitive SATs in E. coli Confers Increased Nickel Resistance
The functional identity of TgSAT-m, TgSAT-p, TgSAT-c, and AtSAT-c were confirmed by complementation of the Cys auxotrophy of the E. coli CysE− mutant lacking an endogenous SAT. All colonies that carried SAT cDNAs were able to grow in the absence of Cys, whereas controls transformed with empty pET32(a)+ vector were unable to grow without the addition of Cys to the M9 minimal medium.
To address the effect of Cys insensitivity of SAT on Ni2+ resistance in E. coli, we measured E. coli cell growth in the presence or absence of 2 mm Ni2+ after 24 h at 30 °C. These results showed that growth of CysE− E. coli in liquid LB medium containing 2 mm Ni2+ is dependent on the type of SAT being expressed. E. coli transformed with native SAT (CYS E) grown in the presence of Ni2+ showed a 70% inhibition of growth relative to a similar culture in the absence of Ni2+ (Fig. 3A). However, CysE− E. coli expressing TgSAT-p or TgSAT-m showed no inhibition of growth in the presence of 2 mm Ni2+ (Fig. 3A). Significantly, the growth of E. coli in the presence of Ni2+ was found to strongly correlate with the IC50 for Cys of the SAT being expressed. The CysE− mutant expressing the Cys-sensitive CysE SAT (37, 38) showed the largest inhibition of growth, whereas the E. coli expressing the Cys-insensitive TgSAT-m or TgSAT-p showed the lowest inhibition. Cys-insensitive TgSAT-c, including At-N/Tg-C, C268P, and C268P/G270A, showed increased Ni2+ resistance compared with the Cys-sensitive Tg-N/At-C, G270A, and AtSAT-c. Growth of all CysE− E. coli expressing various SATs in liquid LB medium containing no Ni2+ was similar (supplemental Fig. 4). Importantly, the Ni2+ resistance of the various SAT-transformed CysE− E. coli was not related (R2 = 0.38) to the relative level of SAT expression, as determined by immunoblotting (supplemental Fig. 4).
FIGURE 3.
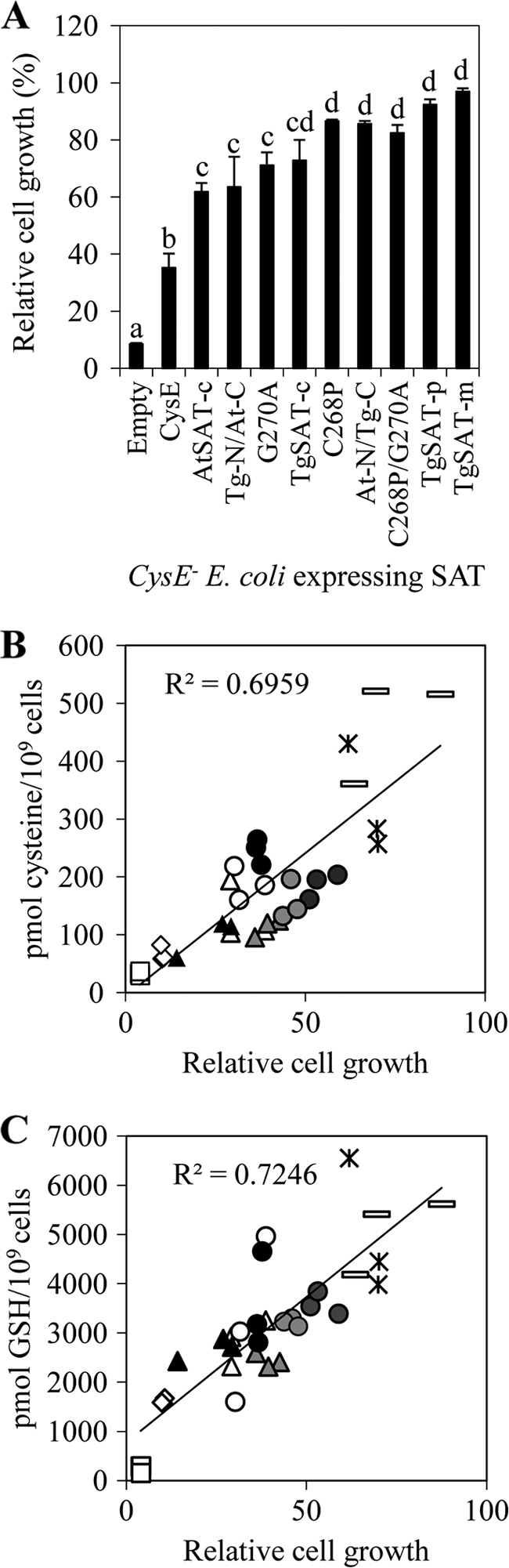
Increased Cys and GSH contents in E. coli confer enhanced nickel resistance. A, relative cell growth of CysE− E. coli expressing various SATs in the presence of 2 mm Ni2+. Letters above the bars represent results of a one-way analysis of variance; the same letters were not significantly different (p > 0.05). Data represent the average ± S.E. (error bars) of three independent samples with three replicates each (n = 9). B, the relationship between the relative cell growth of E. coli expressing various SATs and Cys content in the presence of 2 mm Ni2+. C, the relationship between the relative cell growth of E. coli expressing various SATs and GSH content in the presence of 2 mm Ni2+. B and C, open squares, empty pET32(a) vector; open diamonds, CysE from E. coli; open triangles, AtSAT-c; gray triangles, Tg-N/At-C; closed triangles, G270A; open circles, TgSAT-c; gray circles, C268P; dark gray circles, At-N/Tg-C; closed circles, C268P/G270A; stars, TgSAT-p; open rectangles, TgSAT-m.
To further understand the role of SAT in conferring nickel resistance in CysE− E. coli, we measured levels of Cys and GSH in E. coli cells transformed with the various SAT genes. Increased Cys and GSH levels positively correlated with Ni2+ resistance in CysE− E. coli (Fig. 3, B and C). Furthermore, increases of Cys and GSH levels in CysE− E. coli also correlated with the degree of Cys sensitivity of the SAT being expressed. These positive correlations between the degree of Cys insensitivity of SAT, Cys, and GSH levels in cells and Ni2+ resistance suggest that the degree of Cys insensitivity of SAT is important in determining the level of Ni2+ resistance conferred via elevated GSH biosynthesis.
Expression of Cys-insensitive SATs in A. thaliana Confers Ni2+ Resistance
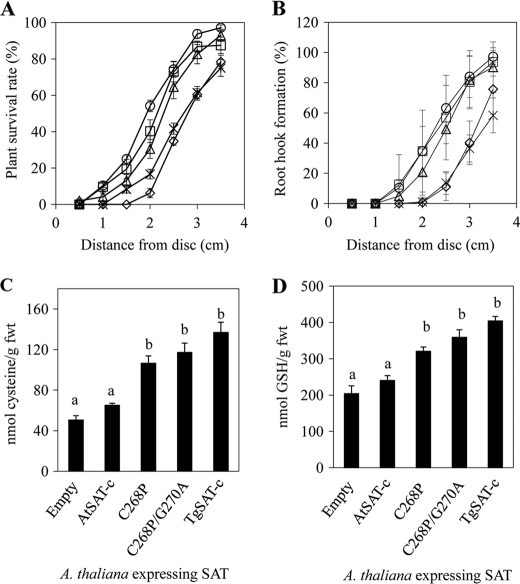
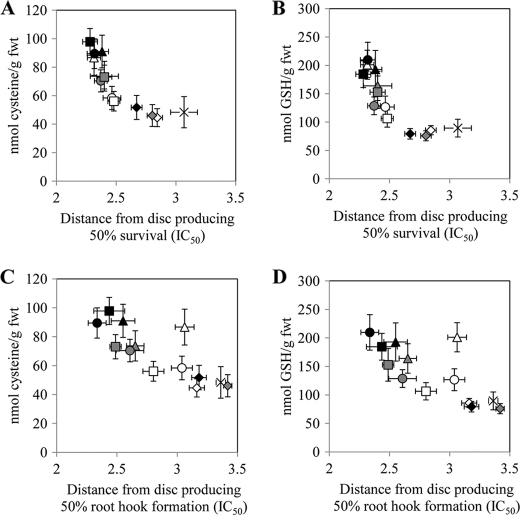
To establish if the degree of Cys insensitivity of SAT affects Ni2+ resistance in plants, we generated four different types of transgenic A. thaliana lines expressing AtSAT-c, C268P, C268P/G270A, and TgSAT-c. Empty vector-transformed A. thaliana plants were used as controls. After identifying homozygous T3 lines for each of the SAT transformation constructs, SAT protein levels were determined using an anti-SAT-specific antibody to identify lines with low, medium, and high levels of accumulation of the various SAT proteins from 15–20 independent lines for each construct (supplemental Fig. 5). Each low, medium, and high expressing class consisted of three independent lines except for the medium accumulating line of C268P, for which only two different lines were characterized. To quantify Ni2+ resistance in these transgenic plants, we used the disc method of Freeman et al. (5). Each transgenic A. thaliana line showed different shoot and root growth patterns in the presence of Ni2+, depending on the type of SAT they were expressing. Growth of all transgenic lines on medium containing no Ni2+ was similar (supplemental Fig. 6). Plants transformed with the Cys-insensitive SAT genes, C268P, C268P/G270A, and TgSAT-c, and accumulating high levels of the SAT proteins showed elevated Ni2+ resistance, measured as either shoot or root growth, compared with the lines accumulating high levels of the Cys-sensitive AtSAT-c (Fig. 4, A and B). Furthermore, Cys and GSH in plants expressing the three Cys-insensitive SAT genes (C268P, C268P/G270A, and TgSAT-c) showed an ∼2-fold increase compared with empty vector- or Cys-sensitive AtSAT-c-overexpressing lines (Fig. 4, C and D). We observed that medium and low SAT-accumulating transgenic lines showed trends similar to that observed for the high SAT-accumulating lines, although the magnitude of the difference from AtSAT-c was less, as we would expect (supplemental Figs. 7 and 8). By determining the distance from the Ni2+-soaked disc that caused a 50% inhibition of growth within a given ring of seedlings, we were able to determine an IC50 for each transgenic line. This IC50 was observed to be strongly correlated with the accumulation of Cys and GSH in these transgenic lines and with the level of Cys sensitivity of the SAT that each line was transformed with (Fig. 5). Furthermore, the degree of nickel resistance conferred by the Cys-insensitive SAT genes (C268P, C268P/G270A, and TgSAT-c) was also directly correlated with the level of accumulation of each of these proteins (Fig. 5). Importantly, however, no trend (R2 = 0.0055) was observed between the level of accumulation of SAT proteins and the degree of nickel resistance when considering all of the various Cys-sensitive and -insensitive forms together. (supplemental Fig. 5).
FIGURE 4.
Nickel resistance of transgenic A. thaliana expressing various SATs. Transgenic plants were grown for 13 days on plates in the presence of 50 μl of 100 mm nickel acetate soaked into a filter paper disc. Data for transgenic lines accumulating high levels of AtSAT-c, C268P, C268P/G270A, and TgSAT-c are presented, and empty vector-transformed lines are used as controls. Both plant growth determined as formation of secondary leaves (A) and root hook (B) and seedling Cys (C) and GSH (D) content are presented from transgenic plants transformed with various SATs. A and B, stars, empty vector-transformed plants; open diamonds, AtSAT-c-transformed plants; open triangles, C268P-transformed plants; open circles, C268P/G270A-transformed plants; open squares, TgSAT-c-transformed plants. Each data point represents an average ± S.E. (error bars) of plants from three high SAT-accumulating lines grown on three independent plates per line. The same letters within graphs represent lines that are not significantly different (p > 0.05, n = 9).
FIGURE 5.
The relationship between Cys content and the distance from the central Ni2+ acetate-soaked disc (50 μl of 100 mm nickel acetate) that allowed 50% survival (IC50) (A), the relationship between GSH content and IC50 (B), the relationship between Cys content and the distance from the central Ni2+ acetate-soaked disc (50 μl of 100 mm nickel acetate) that allowed 50% root hook formation (IC50) (C), and the relationship between GSH content and IC50 (D). Stars, empty vector-transformed plants; diamonds, AtSAT-c-transformed plants; triangles, C268P-transformed plants; circles, C268P/G270A-transformed plants; squares, TgSAT-c-transformed plants. White, low accumulating level of SAT; gray, medium accumulating level of SAT; black, high accumulating level of SAT. All presented data represent an average ± S.E. (error bars) of data from three independent replicate plates.
Using a C268P/G270A-GFP construct, the subcellular localization of C268P/G270A was confirmed to be cytosolic, as predicted from the homology to AtSAT-c (supplemental Figs. 1 and 2). This establishes that the point mutations made in the C-terminal domain of AtSAT-c, which convert it into a Cys-insensitive enzyme, do not affect the localization of AtSAT-c.
Role of Pro and Gly Polymorphisms in SAT-c across the Thlaspi Genus
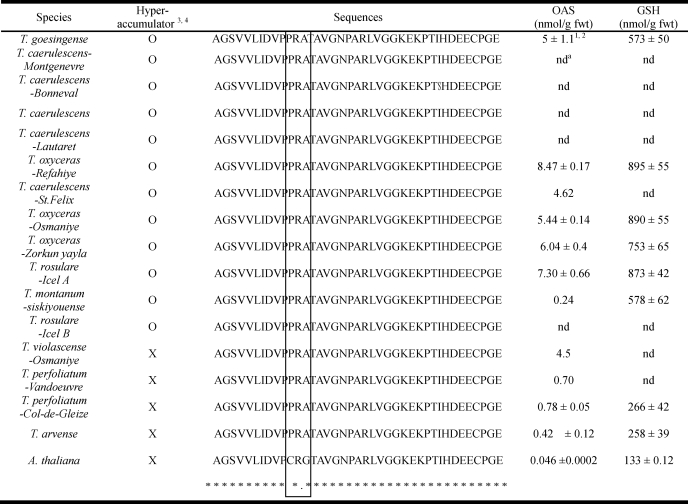
To determine if the Pro and Gly changes in SAT-c, determined to control the Cys insensitivity of TgSAT-c, are common to all Thlaspi species or just to hyperaccumulators within the genus, we sequenced SAT-c from 12 species and accessions of hyperaccumulators and three species of nonaccumulators from the Thlaspi genus. Seeds for the various species and accessions used were collected from France, Italy, Turkey, and Austria (6, 39). The native habitats of most plants were serpentine except for Thlaspi caerulescens (Lautaret) from nonmetalliferous soil, T. caerulescens (St. Felix de Pallieres) from zinc/lead mine waste, and Thlaspi perfoliatum (Col de Gleize) and T. perfoliatum (Vandoeuvre-les-Nancy) from calcareous soils. All Ni2+ hyperaccumulators contained 1,000–7,000 μg/g nickel in shoot dry biomass when growing in their native habitat, and the three nonaccumulators contained a maximum of 300 μg/g nickel in shoot dry biomass (3, 6, 39). Seeds of all species were germinated and grown on 1/2 strength MS medium with no added Ni2+ for 3 weeks. Genomic DNA was isolated from each Thlaspi species and used as a template for PCR amplification. The TgSAT-c-specific primers for the C-terminal domain from amino acid 250 to 314 were designed and used to amplify PCR products that were directly sequenced in order to minimize the possibility of introducing nucleotide changes during cloning procedures. We observed that both the Pro and Gly are conserved in the C-terminal domain of SAT-c from all 12 nickel-hyperaccumulating species and accessions examined (Table 2). The C-terminal domains of all of the hyperaccumulators tested were identical, except for a single amino acid change from isoleucine to serine in T. caerulescens (Bonneval). Interestingly, the three Thlaspi nonaccumulators also have both Pro and Gly residues in the C-terminal domain of SAT-c (Table 2). We conclude that the Pro and Gly changes in SAT are likely to be common to the Thlaspi genus and are not specific just to the hyperaccumulator species. This suggests that cytosolic SATs from Thlaspi species are generally insensitive to Cys, and this is supported by the fact that the nonaccumulators T. perfoliatum, Thlaspi arvense, and Thlaspi violascense also have elevated levels of OAS and GSH compared with A. thaliana (Table 2). However, the concentration of OAS and GSH in nonaccumulators is 4–10-fold lower than observed in the nickel hyperaccumulators (Table 2), suggesting that other mechanisms, such as elevated salicylic acid (30), are also important.
TABLE 2.
Alignment of the predicted C-terminal amino acid sequences of cytosolic SAT from various Thlaspi species, including nickel hyperaccumulator and non-accumulators and A. thaliana
The box indicates the amino acids that are responsible for the different Cys sensitivities of SAT.
a nd, not determined.
1 The OAS and GSH data were taken from Freeman et al. (5).
2 Data represent the average ± S.D. of quantifications of three individual plants per species.
3,4 O and X represent hyperaccumulator and nonaccumulator, respectively.
DISCUSSION
It has been proposed that the Cys sensitivity of AtSAT-c is an important mechanism for the regulation of Cys concentrations in the cytosol of plants (18, 24), where the Cys concentration has been recently estimated to be 300 μm (40). We have previously observed that T. goesingense contains constitutively elevated OAS, Cys, and GSH concentrations compared with closely related nonaccumulators, including A. thaliana (5). Based on these observations, Freeman et al., (5) hypothesized that Cys insensitivity of the cytosolic SAT from T. goesingense leads, at least in part, to the elevated OAS, Cys, and GSH concentration observed in T. goesingense. Here, we show that TgSAT-c is relatively insensitive to Cys up to 100 μm. This contrasts with the orthologous cytosolic SAT from A. thaliana, which shows a 50% inhibition of activity at 3.3 μm Cys. Further, we show that this elevated insensitivity to Cys confers increased resistance to Ni2+ when TgSAT-c is heterologously expressed in E. coli and A. thaliana. This enhanced resistance is conferred through the enhanced accumulation of Cys and GSH.
The Cys-sensitive AtSAT-c and the Cys-insensitive TgSAT-c share 87% identity at the nucleotide level and 94% identity at the amino acid level. Within the N-terminal region of the protein, there is considerable variation, with 16 of the 20 amino acids differing between the proteins. However, our domain swapping experiments (Fig. 1 and Table 1) establish that it is the C terminus of TgSAT-c that plays a critical role in the regulation of the enzyme by Cys. It has been reported that the conformation of the SAT C-terminal domain is important for Cys inhibition because alterations in the structure of this domain can result in changes in the sensitivity to Cys (19, 35, 36). It has also been reported that 20–25 amino acids from the C-terminal region are responsible for the Cys inhibition in E. coli SAT because truncation of these 20 amino acids results in Cys insensitivity without any loss of specific activity of the enzyme (41). In the Cys-sensitive SAT-c from C. vulgaris (WaSATase), a point mutation that changes Met at 280 to Ile was found to completely eliminate the Cys feedback regulation of this SAT (19). In addition, two amino acids (Gly at 277 and His at 282) from WaSATase were also found to be involved in Cys sensitivity because mutation of these residues to glutamine led to an increase in Cys insensitivity from 2.9 μm to 77.8 and 10.1 μm, respectively (19). These amino acids are all located in the C-terminal region of the enzyme, the so-called “allosteric domain.” Inoue et al. (19) concluded that Gly at 277 and His at 282 are essential for SAT activity because both residues are conserved in Cys-sensitive and Cys-insensitive SATs, and they predicted that a particular structure around Met280 and Gly277 may be needed for Cys inhibition, although the exact amino acids involved in this structure were not determined.
Here, we report two newly discovered residues that are specifically responsible for Cys insensitivity but are not required for SAT activity. These two amino acids, Pro at 268 and Ala at 270, are different from previously reported residues. These amino acids were identified by a comparative study of naturally occurring Cys-sensitive and -insensitive isoforms that share 94% identity at the amino acid level and were isolated from closely related species.
Heterologous expression of the Cys-insensitive mitochondrial SAT from T. goesingense (TgSAT-m) has been observed to enhance Ni2+ resistance in A. thaliana (5) and E. coli (34). Data presented here demonstrate that heterologous expression of Cys-insensitive cytosolic SATs in E. coli, including SAT-c from T. goesingense (TgSAT-c), and At-N/Tg-C, C268P, and C268P/G270A also increases Ni2+ resistance when compared with the Cys-sensitive SATs, AtSAT-c and Tg-N/At-C (Fig. 3). This increased Ni2+ resistance strongly correlates with the degree of Cys insensitivity of the SAT expressed and is unrelated to the level at which the enzyme is accumulated in E. coli. Moreover, the Cys insensitivity of the SAT expressed is also positively correlated with both the Cys and GSH contents. Importantly, such effects are also observed in planta. When Cys-insensitive SATs (TgSAT-c, C268P, and C268P/G270A) are heterologously expressed in A. thaliana, we observe increased Ni2+ resistance compared with overexpression of the Cys-sensitive AtSAT-c. This increased Ni2+ resistance also correlates with elevated in planta concentrations of Cys and GSH (Fig. 4). Furthermore, we see no relationship between the degree of nickel resistance and the level of accumulation of the expressed SAT when all Cys-sensitive and -insensitive enzymes are considered together (supplemental Fig. 5).
It has been proposed that OAS is mainly produced in the mitochondria and that Cys is mainly produced in the cytosol, whereas the chloroplast is primarily involved in sulfate reduction (42). Therefore, we predict that a cytosolically localized SAT insensitive to Cys may drive enhanced production of OAS in the cytoplasm, stimulating the production of Cys. Elevated Cys would be expected to give rise to elevated GSH, which would confer increased Ni2+ resistance.
Another possible explanation for the ability of the Cys-insensitive enzymes (TgSAT-c, C268P, and C268P/G270A) to elevate Cys and GSH levels when heterologously expressed in A. thaliana is that their interaction with the endogenous OAS-TL is modified compared with AtSAT-c, causing an altered activity of the SAT·OAS-TL regulatory complex, leading to enhanced Cys biosynthesis. Although plausible, we have no direct evidence for such a mechanism.
Interestingly, both Pro at 268 and Ala at 270 are extremely well conserved in Thlaspi species, occurring in both hyperaccumulators and nonaccumulators (Table 2). However, there are clear quantitative differences in OAS, Cys, and GSH content among Thlaspi nickel hyperaccumulators and nonaccumulators (5, 6). Hyperaccumulators show constitutively elevated steady-state concentrations of OAS and GSH compared with nonaccumulators (Table 2). However, nonaccumulators from the Thlaspi genus, such as T. arvense and T. perfoliatum, also contain at least 10 times more OAS and twice as much GSH as A. thaliana, a nonaccumulator in a related genus. The conservation of the Pro and Ala in the SAT-c from Thlaspi species makes this enzyme insensitive to Cys and may be responsible for the elevated OAS and GSH observed across the Thlaspi genus. Such elevated levels of GSH in the Thlaspi genus may help to preadapt species in this genus for the evolution of the Ni2+ tolerance required for Ni2+ hyperaccumulation. Constitutively elevated salicylic acid has been implicated in the elevated levels of GSH observed in the nickel hyperaccumulator T. goesingense, through activation of SAT activity (32). Together, SA activation of the Cys-insensitive SAT-c may explain the elevated levels of OAS, Cys, and GSH observed in the hyperaccumulator compared with the nonaccumulator Thlaspi species.
In summary, we discovered that two amino acid residues, Pro and Ala, in SAT-c are critically important for the Cys insensitivity of the T. goesingense SAT-c enzyme. Further, we establish that it is the enhanced insensitivity to Cys of T. goesingense SAT-c that allows it to confer elevated Ni2+ resistance in A. thaliana. Taken together, our data support the conclusion that the altered Cys feedback regulation of SAT-c plays an important role in Ni2+ tolerance in the nickel hyperaccumulator T. goesingense.
Supplementary Material
Acknowledgments
We thank John L. Freeman for valuable discussion and technical assistance, Danielle R. Ellis for cloning of three SATs from T. goesingense into expression vectors, and Thomas G. Sors for significant discussions and technical assistance.
This work was supported by National Science Foundation Grant IOS-0419695 (to D. E. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8.
- OAS
- O-acetylserine
- SAT
- serine acetyltransferase
- OAS-TL
- O-acetylserine (thiol) lyase
- SAT-c
- cytosolic SAT
- SAT-p
- plastid SAT
- SAT-m
- mitochondrial SAT
- sGFP
- synthetic GFP.
REFERENCES
- 1. Krämer U. (2010) Annu. Rev. Plant Biol. 61, 517–534 [DOI] [PubMed] [Google Scholar]
- 2. Brooks R., Lee J., Reeves R., Jaffre T. (1977) J. Geochem. Explor. 7, 49–57 [Google Scholar]
- 3. Reeves R., Brooks R. (1983) J. Geochem. Explor. 18, 275–283 [Google Scholar]
- 4. Kramer U., Smith R. D., Wenzel W. W., Raskin I., Salt D. E. (1997) Plant Physiol. 115, 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Freeman J. L., Persans M. W., Nieman K., Albrecht C., Peer W., Pickering I. J., Salt D. E. (2004) Plant Cell 16, 2176–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peer W. A., Mahmoudian M., Freeman J. L., Lahner B., Richards E. L., Reeves R. D., Murphy A. S., Salt D. E. (2006) New Phytol. 172, 248–260 [DOI] [PubMed] [Google Scholar]
- 7. Krämer U., Pickering I. J., Prince R. C., Raskin I., Salt D. E. (2000) Plant Physiol. 122, 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Droux M., Ruffet M. L., Douce R., Job D. (1998) Eur. J. Biochem. 255, 235–245 [DOI] [PubMed] [Google Scholar]
- 9. Feldman-Salit A., Wirtz M., Hell R., Wade R. C. (2009) J. Mol. Biol. 386, 37–59 [DOI] [PubMed] [Google Scholar]
- 10. Rausch T., Gromes R., Liedschulte V., Müller I., Bogs J., Galovic V., Wachter A. (2007) Plant Biol. 9, 565–572 [DOI] [PubMed] [Google Scholar]
- 11. Kredich N. M. (1971) J. Biol. Chem. 246, 3474–3484 [PubMed] [Google Scholar]
- 12. Saito K., Kurosawa M., Tatsuguchi K., Takagi Y., Murakoshi I. (1994) Plant Physiol. 106, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liszewska F., Blaszczyk A., Sirko A. (2001) Acta Biochim. Pol. 48, 647–656 [PubMed] [Google Scholar]
- 14. Ruffet M. L., Droux M., Douce R. (1994) Plant Physiol. 104, 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruffet M. L., Lebrun M., Droux M., Douce R. (1995) Eur. J. Biochem. 227, 500–509 [DOI] [PubMed] [Google Scholar]
- 16. Wirtz M., Droux M. (2005) Photosynth. Res. 86, 345–362 [DOI] [PubMed] [Google Scholar]
- 17. Wirtz M., Hell R. (2006) J. Plant Physiol. 163, 273–286 [DOI] [PubMed] [Google Scholar]
- 18. Noji M., Inoue K., Kimura N., Gouda A., Saito K. (1998) J. Biol. Chem. 273, 32739–32745 [DOI] [PubMed] [Google Scholar]
- 19. Inoue K., Noji M., Saito K. (1999) Eur. J. Biochem. 266, 220–227 [DOI] [PubMed] [Google Scholar]
- 20. Saito K., Yokoyama H., Noji M., Murakoshi I. (1995) J. Biol. Chem. 270, 16321–16326 [DOI] [PubMed] [Google Scholar]
- 21. Saito K., Inoue K., Fukushima R., Noji M. (1997) Gene 189, 57–63 [DOI] [PubMed] [Google Scholar]
- 22. Kawashima C. G., Berkowitz O., Hell R., Noji M., Saito K. (2005) Plant Physiol. 137, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howarth J. R., Roberts M. A., Wray J. L. (1997) Biochim. Biophys. Acta 1350, 123–127 [DOI] [PubMed] [Google Scholar]
- 24. Urano Y., Manabe T., Noji M., Saito K. (2000) Gene 257, 269–277 [DOI] [PubMed] [Google Scholar]
- 25. Chronis D., Krishnan H. B. (2004) Planta 218, 417–426 [DOI] [PubMed] [Google Scholar]
- 26. Noji M., Saito K. (2002) Amino Acids 22, 231–243 [DOI] [PubMed] [Google Scholar]
- 27. Mino K., Hiraoka K., Imamura K., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (2000) Biosci. Biotechnol. Biochem. 64, 1874–1880 [DOI] [PubMed] [Google Scholar]
- 28. Saby S., Leroy P., Block J. C. (1999) Appl. Environ. Microbiol. 65, 5600–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsakraklides G., Martin M., Chalam R., Tarczynski M. C., Schmidt A., Leustek T. (2002) Plant J. 32, 879–889 [DOI] [PubMed] [Google Scholar]
- 30. Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 31. Jin J. B., Kim Y. A., Kim S. J., Lee S. H., Kim D. H., Cheong G. W., Hwang I. (2001) Plant Cell 13, 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freeman J. L., Garcia D., Kim D., Hopf A., Salt D. E. (2005) Plant Physiol. 137, 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brooks S. P. (1992) BioTechniques 13, 906–911 [PubMed] [Google Scholar]
- 34. Freeman J. L., Persans M. W., Nieman K., Salt D. E. (2005) Appl. Environ. Microbiol. 71, 8627–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Francois J. A., Kumaran S., Jez J. M. (2006) Plant Cell 18, 3647–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zubieta C., Arkus K. A., Cahoon R. E., Jez J. M. (2008) J. Biol. Chem. 283, 7561–7567 [DOI] [PubMed] [Google Scholar]
- 37. Denk D., Böck A. (1987) J. Gen. Microbiol. 133, 515–525 [DOI] [PubMed] [Google Scholar]
- 38. Nakamori S., Kobayashi S. I., Kobayashi C., Takagi H. (1998) Appl. Environ. Microbiol. 64, 1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peer W., Mamoudian M., Lahner B., Reeves R., Murphy A., Salt D. E. (2003) New Phytol. 159, 421–430 [DOI] [PubMed] [Google Scholar]
- 40. Krueger S., Niehl A., Lopez Martin M. C., Steinhauser D., Donath A., Hildebrandt T., Romero L. C., Hoefgen R., Gotor C., Hesse H. (2009) Plant Cell Environ. 32, 349–367 [DOI] [PubMed] [Google Scholar]
- 41. Mino K., Yamanoue T., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (1999) Biosci. Biotechnol. Biochem. 63, 168–179 [DOI] [PubMed] [Google Scholar]
- 42. Haas F. H., Heeg C., Queiroz R., Bauer A., Wirtz M., Hell R. (2008) Plant Physiol. 148, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.