Background: P450 catalytic cycles start with Fe(III) in vitro, but the resting form in vivo is unknown.
Results: Significant Fe(II) P450 is present in intact cells even after vigorous aeration.
Conclusion: The P450 form and cell environment determine the proportion of Fe(II) P450 in vivo.
Significance: Fe(II) in the resting state is unexpected due to the potential for futile cycling.
Keywords: Cytochrome P450, Gating, Heme, Oxidation-Reduction, Reactive Oxygen Species (ROS), difference Spectroscopy, Heme Redox State, Heme-Thiolate Protein, Resting State
Abstract
Cytochrome P450 enzymes (P450s) are exceptionally versatile monooxygenases, mediating hydroxylations of unactivated C–H bonds, epoxidations, dealkylations, and N- and S-oxidations as well as other less common reactions. In the conventional view of the catalytic cycle, based upon studies of P450s in vitro, substrate binding to the Fe(III) resting state facilitates the first 1-electron reduction of the heme. However, the resting state of P450s in vivo has not been examined. In the present study, whole cell difference spectroscopy of bacterial (CYP101A1 and CYP176A1, i.e. P450cam and P450cin) and mammalian (CYP1A2, CYP2C9, CYP2A6, CYP2C19, and CYP3A4) P450s expressed within intact Escherichia coli revealed that both Fe(III) and Fe(II) forms of the enzyme are present in the absence of substrates. The relevance of this finding was supported by similar observations of Fe(II) P450 heme in intact rat hepatocytes. Electron paramagnetic resonance (EPR) spectroscopy of the bacterial forms in intact cells showed that a proportion of the P450 in cells was in an EPR-silent form in the native state consistent with the presence of Fe(II) P450. Coexpression of suitable cognate electron donors increased the degree of endogenous reduction to over 80%. A significant proportion of intracellular P450 remained in the Fe(II) form after vigorous aeration of cells. The addition of substrates increased the proportion of Fe(II) heme, suggesting a kinetic gate to heme reduction in the absence of substrate. In summary, these observations suggest that the resting state of P450s should be regarded as a mixture of Fe(III) and Fe(II) forms in both aerobic and oxygen-limited conditions.
Introduction
The heme-thiolate enzymes in the cytochrome P450 gene superfamily carry out an unequalled variety of functions by virtue of the chemistry effected at the heme center and the evolutionary versatility of the P450 fold. More than 12,000 different forms are currently known (48), the vast majority of which are believed to share a common catalytic mechanism. Most P450s act as monooxygenases, requiring one or more redox partners to transfer electrons from a pyridine-containing coenzyme. Monooxygenation consumes 1 equivalent each of molecular oxygen and NAD(P)H, with the production of 1 equivalent of water in the typical reaction. By this general reaction, a number of common (aliphatic and aromatic C-hydroxylation, heteroatom dealkylation and oxidation) and uncommon (group migration, C–C bond cleavage, ring expansion and contraction, phenolic coupling) reactions are mediated (1–3).
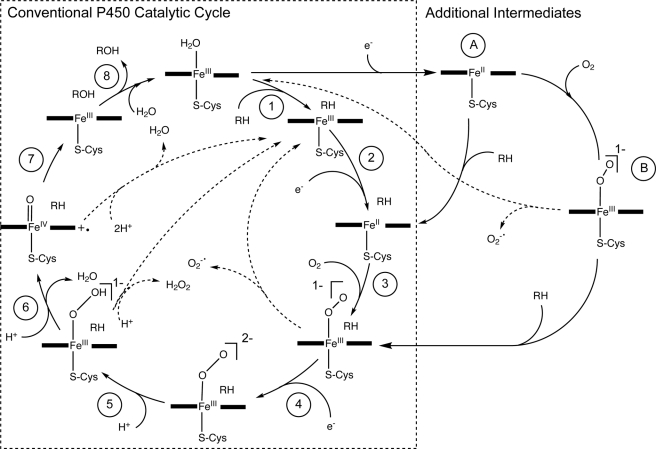
In the conventional representation of the P450 catalytic cycle (Fig. 1, steps 1–8) (4–7), the first step is substrate binding to the oxidized ferric heme resting form of the enzyme, with conversion of the iron from low to high spin and concurrent loss of a water ligand (step 1). Results from the prototypical bacterial form, CYP101A1 (P450cam), suggest that substrate binding potentiates the subsequent single electron transfer from the cognate redox partner to the heme (step 2). This generates the ferrous form, and rapid coordination of dioxygen follows (step 3), setting in motion steps 4–8. Uncoupling side reactions, i.e. the consumption of electrons and oxygen without substrate oxidation, can occur at three steps (Fig. 1). The observation that substrate binding to the ferric form accelerates delivery of the first electron has led to the proposal that the cycle is thermodynamically gated at the first reduction step in the absence of substrate (4, 8) to protect the cell from the release of damaging reactive oxygen species.
FIGURE 1.
The conventional P450 catalytic cycle (steps 1–8) (4–7), with additional intermediates (A and B) resulting from reduction of ferric to ferrous heme in the absence of substrate. Dotted arrows represent uncoupling reactions.
This conventional view of the catalytic cycle, although drawn primarily from experiments with soluble P450s that are easily studied, such as CYP101A1, has also been applied to other systems (9). However, evidence has steadily accumulated that contradicts aspects of this model. In particular, for P450s involved in the metabolism of xenobiotics that typically show relatively poor affinity for their substrates, reduction of the ferric P450 appears to proceed to a greater or lesser extent in the absence of a substrate (10). First electron transfer is not necessarily accelerated by substrate binding and depends to some degree on the environment of the enzyme (10). Additionally, substrate binding to these forms does not necessarily induce the displacement of the water ligand to the heme, leading to a spin state shift.
The resting state of P450s is typically portrayed as the ferric form in representations and descriptions of the P450 catalytic cycle (5–7, 11–14). To our knowledge, no experimental evidence exists that addresses the in vivo oxidation state of P450s. However, the reduction of P450s in vitro in the absence of substrate suggests that P450s may exist in both Fe(III) and Fe(II) forms within the cell. The heme chromophore provides a convenient reporter of the redox state of the enzyme within the cell. The peak at ∼450 nm in the ferrous-CO versus ferrous difference spectrum differentiates P450s and a small number of other heme-thiolate proteins from other hemoproteins (15). The spectral characterization of P450s has typically been performed on P450s after extraction from the normal intracellular environment. However, sensitive methodology for quantifying P450s within intact bacterial cells (16, 17) now provides a system for investigating the physiological redox state of P450s in whole cells. The aim of the present study was to test the hypothesis that P450s are partially reduced in their resting state within the intact cell and to explore the role of substrate and cellular redox state in regulating the redox state of the heme in P450s from both bacterial and mammalian sources.
EXPERIMENTAL PROCEDURES
Materials
Culture media were obtained from BD Biosciences (North Ryde, Australia). Cineole and d-camphor were purchased from Sigma-Aldrich (Castle Hill, Australia), and n-butylisocyanide (nBIC)2 was from Thermo Fisher Scientific (Richlands, Australia). All other chemicals were obtained from local suppliers at the highest quality available. The Escherichia coli strain DH5α F′IQ was obtained from Invitrogen (Mulgrave, Australia). Male Sprague-Dawley rat cryopreserved hepatocytes were obtained from Celsis In Vitro Technologies (Chicago, IL). Livers were obtained from male Sprague-Dawley rats (Central Animal Breeding Facility of the University of Queensland) under procedures approved by University of Queensland ethics committees, and microsomes were prepared as described previously (18).
Expression of Recombinant P450s in E. coli
CYP1A2, CYP2A6, CYP2A13, CYP2C9, CYP2C19, CYP3A4, CYP176A1 (P450cin), and CYP101A1 were expressed in E. coli according to the general procedures outlined previously (19–24). Bacterial chaperones were coexpressed using the pGro7 plasmid to augment P450 yields (22, 25, 26). CYP176A1 and CYP101A1 were purified as described previously (24). P450s were expressed without any recombinant redox partners (monocistronic format) as well as with the cognate redox partners (bi- or tricistronic format): human NADPH cytochrome P450 reductase (hCPR) for all human P450s, putidaredoxin plus putidaredoxin reductase for CYP101A1, and cindoxin (24) plus E. coli flavodoxin reductase (FdR, a substitute for cindoxin reductase) for CYP176A1. Starter cultures prepared from single isolated colonies were used to inoculate 100-μl cultures in 96-well flat-bottomed microplates or 1-ml cultures in 24-well plates. Cultures were induced with isopropyl-1-thio-β-d-galactopyranoside and arabinose at 5 h to initiate expression, and then microplates were sealed (microaerobic cultures) with Breathe-Easy gas-permeable membranes (Diversified Biotech, Boston, MA) or covered with the microplate lid (aerobic cultures) and incubated at 25 °C for a further 67 h before harvest unless stated otherwise.
Fe(II)-CO versus Fe(II) Difference Spectroscopy in Whole Cells
The P450 contents of bacterial cells were quantified by Fe(II)-CO versus Fe(II) difference spectroscopy as described previously (17). Bacterial cultures were resuspended in 0.5 volume of whole cell assay buffer (100 mm potassium phosphate, 6 mm magnesium acetate, 10 mm D-glucose, pH 7.4) prior to spectral analysis. Unless otherwise stated, spectral measurements were performed on resuspensions that had been left unstirred for 30–60 min. For measurement of total P450 concentrations, freshly prepared sodium dithionite was added to 10 mg/ml, and cells were incubated for 90 min without agitation to reduce the total P450 pool (dithionite reducing treatment). Dithionite was omitted for measurement of endogenously reduced P450. Following a 5-s mix to resuspend cells, a baseline spectrum was recorded at 2 nm intervals from 400–500 nm in a SpectraMax M2 microplate reader. Resuspensions were then gassed with CO for 5 min at 1 atm prior to measurement of Fe(II)-CO versus Fe(II) difference spectra. Other conditions were as indicated in the results for individual experiments.
Cryopreserved rat hepatocytes were thawed according to procedures recommended by the supplier and resuspended in sterile phosphate-buffered saline (100 mm potassium phosphate, pH 7.4, containing 750 mm NaCl; 2.5 × 106 cells/ml). Fe(II)-CO versus Fe(II) difference spectra were obtained on undiluted resuspensions as described above.
Measurement of Ligand Binding Spectra in Whole Cells
For ligand binding spectra, 10 μm nBIC, cineole, or D-camphor was added to bacterial cultures prepared as described above. Difference spectra were obtained with reference to baseline spectra recorded on the same samples prior to the addition of ligand. For some experiments, cells were either reduced by the addition of 10 mg/ml freshly prepared sodium dithionite in whole cell assay buffer (reducing treatment) or oxidized by vigorous agitation in air in a VorTemp mixer for 10 min at 1000 rpm in the presence of 10 mg/ml freshly prepared sodium dithionite (oxidizing treatment).
Check for Endogenous Ligands in E. coli Cell Lysate by Ligand Binding Spectroscopy
Difference spectroscopy was performed on purified bacterial P450s and isolated bacterial membranes containing recombinant P450s to investigate the presence of endogenous ligands. Bacterial membranes containing recombinant human P450s were prepared using established methods (22). Ligand binding spectra were obtained using purified CYP101A1 (9 μm), purified CYP176A1 (10 μm), or bacterial membranes containing 0.5 μm recombinant human P450s, incubated with and without a 5-fold dilution of E. coli cell lysate, that had been prepared from a 10-ml culture of E. coli transformed with the empty expression vector pCW, lysed by sonication, and clarified by centrifugation at 20,000 × g for 30 min.
EPR Spectroscopy
Continuous wave X-band EPR spectra were recorded with a Bruker BioSpin ELEXSYS E580 EPR spectrometer fitted with a super high Q-cavity. Magnetic field and microwave frequency calibration were achieved with a Bruker BioSpin ER 036M teslameter and a Bruker BioSpin microwave frequency counter, respectively. Temperatures (8.00 ± 0.02 K) were controlled using a flow-through cryostat (ESR910) in conjunction with an Oxford ITC503 temperature controller. Spectrometer tuning, signal averaging, and subsequent spectral comparisons were performed with the Xepr (version 2.6) software from Bruker BioSpin.
Microaerobic cultures of cells expressing CYP101A1 and CYP176A1 were prepared as described above and harvested at 72 h. Half of each culture was added immediately to a Wilmad 707-SQ EPR sample tube and snap-frozen in liquid nitrogen (“native” sample). An identical aliquot was incubated with sodium dithionite (10 mg/ml final concentration) in a microplate and subjected to a 10-min agitation at 1000 rpm in a VorTemp microplate shaker in air prior to freezing in the sample tube (“oxidized” sample). All samples were briefly degassed with argon to remove dissolved oxygen and oxygen present above the frozen sample, to avoid resonances from triplet state oxygen and poor baselines. Identical sample tubes were used to measure the EPR spectra of the native and oxidized whole cell environments, and the sample tubes were placed into an identical position in the liquid helium flow-through quartz assembly within the super high Q-cavity. This was judged by the lack of change in microwave frequency and detector current between sample changes. Background EPR spectra of E. coli cells lacking any P450 were also recorded and subtracted from the experimental EPR spectra of CYP101A1 and CYP176A1 in whole cells to remove signals from other paramagnetic centers (e.g. Cu(II), Mn(II), and high spin rhombic Fe(III)) not associated with P450s. Computer simulation of the EPR spectra was performed using the XSophe-Sophe-XeprView computer simulation software suite (version 1.1.4) running on a PC utilizing Mandriva 2009.1 as the Linux operating system (27).
RESULTS
P450 Reduction State in Resting Whole Cells without Added Oxygen or Redox Partners
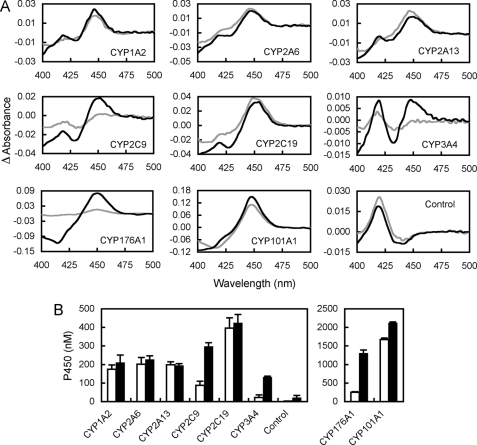
Of the bacterial (CYP101A1 and CYP176A1) and human (CYP1A2, CYP2C9, CYP2A6, CYP2A13, and CYP3A4) P450s tested in whole cells, all but CYP3A4 showed a significant absorbance peak at 450 nm Fe(II)-CO versus Fe(II) difference spectrum in the absence of exogenous reductant (Fig. 2), indicating naturally present, CO-binding, ferrous P450 (subsequently referred to as endogenously reduced P450). Dithionite reducing treatment increased the peak size to different extents depending upon the P450 form under examination. The dithionite-reducible P450 peak was assumed to represent the total amount of P450 in the cell. The ratio of the peak size observed in the absence to that in the presence of dithionite (reducing treatment) represents the proportion of total P450 that is endogenously reduced within the intact bacterial cell.
FIGURE 2.
Fe(II)-CO versus Fe(II) difference spectra of recombinant P450 monocistronic expression cultures. A, spectra were recorded with (black) and without (gray) the addition of external reductant (10 mg/ml sodium dithionite, 90-min contact time). Spectra from three independent experiments were averaged to produce the data shown in each case. B, quantitative analysis of spectra presented in A. Histograms show mean ± S.D. of ferrous P450 concentrations calculated from n = 3 spectra from 72-h microaerobic cultures. Filled bars represent ferrous P450 after reduction by dithionite (total P450), and open bars represent endogenously reduced P450. The control spectrum is from bacteria expressing no P450.
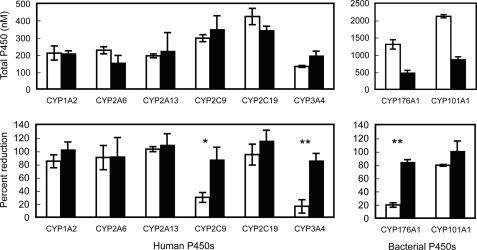
Effect of Coexpressed Cognate Redox Partners on P450 Whole Cell Reduction State
Coexpression of the cognate electron donors (hCPR for human P450s; putidaredoxin plus putidaredoxin reductase for CYP101A1; or cindoxin plus endogenous E. coli FdR for CYP176A1) in bi- or tricistronic cultures with the respective P450s consistently increased the proportion of endogenously reduced P450 detected by Fe(II)-CO versus Fe(II) difference spectroscopy (Fig. 3). However, this increase was only statistically significant for those forms that were not already mostly endogenously reduced in the monocistronic system (CYP2C9, CYP3A4, and CYP176A1, where the reduction percentage increased from <40% to >80%; Fig. 3). All P450s could be considered predominantly reduced (>80%) in the presence of their cognate redox partners. Coexpression of appropriate redox partners had little effect on total P450 expression levels for the recombinant human forms. However, the yields of the more highly expressed bacterial P450s were significantly decreased (Fig. 3).
FIGURE 3.
The effect of coexpression of redox partners on reduction state of human and bacterial P450s in E. coli cells. Expression level and the percentage of reduction of P450s were determined by Fe(II)-CO versus Fe(II) difference spectra on intact cells. Open bars show data from monocistronic cultures; filled bars represent data from cultures containing coexpressed cognate redox partners. Data represent the mean ± S.D. of three independent 72-h microaerobic cultures. Total P450 concentrations were determined from spectra collected after dithionite reducing treatment. Asterisks indicate statistically significant differences in the percentage of reduction between cultures with and without cognate electron donors (paired Student's t test); *, p < 0.05; **, p < 0.01).
Oxidation of P450s in Intact Bacterial Cells
To characterize better the different redox states of the P450 in whole cells, we exploited a serendipitous observation that vigorous aeration of cells in the presence of dithionite decreased the amount of P450 detected by Fe(II)-CO versus Fe(II) difference spectra (supplemental Fig. S1). We speculate that this results from oxidation of the Fe(II) heme by hydrogen peroxide generated intracellularly as aerobic decomposition of dithionite to generate hydrogen peroxide has been demonstrated in solution (28, 29). CYP101A1 cultures treated in this manner (oxidizing treatment) showed an increase in the amplitude of type I ligand binding spectra elicited by the addition of camphor, characteristic of the Fe(III) form, confirming conversion of endogenous Fe(II) P450 to the Fe(III) form by aeration in the presence of dithionite (supplemental Fig. S1). A quantitative assessment of these data is precluded by the fact that the addition of d-camphor for the type I measurement will potentiate reduction of the P450 and turnover of the d-camphor in whole cells.
Ligand Binding Spectra of P450s Expressed in Intact Bacterial Cells
Alkylisocyanides bind to both Fe(II) and Fe(III) forms of P450s, producing visible absorption spectra characteristic of the redox state of the heme. Binding spectra were obtained with nBIC using intact E. coli cells expressing CYP101A1 and CYP176A1 at high concentrations that were (i) reduced with sodium dithionite, (ii) oxidized by vigorous shaking in the presence of sodium dithionite (see above), or (iii) measured after the addition of nBIC alone (native samples) (Fig. 4). Although only a small peak was observed at ∼430 nm in the oxidized samples, characteristic absorbance peaks at ∼455 nm (both enzymes) and ∼430 nm (CYP176A1) were observed in spectra obtained from the reduced and native samples, indicating binding of nBIC to the Fe(II) form (30). The ratio of these peaks differed between the native and reduced spectra for the same P450 form, which is consistent with the pH effect previously observed (30) and the ability of dithionite to decrease the pH (31).
FIGURE 4.
Visible region absorption spectra induced by binding of nBIC to P450s in intact bacterial cells. Difference spectra were recorded in the presence and absence of 10 mm nBIC with E. coli expressing CYP101A1 (A), CYP176A1 (B), or no recombinant proteins (C), subjected to three different treatments: (i) incubation for 10 min with 10 mg/ml sodium dithionite (reduced sample, black lines), (ii) incubation with 10 mg/ml sodium dithionite followed by 10 min of vigorous agitation in air (oxidized sample, pale gray lines), or (iii) no treatment (native, dark gray lines).
Investigation of the Possibility That E. coli Cultures Contain an Unidentified Substrate Responsible for Inducing a Spin Shift and Facilitating Reduction
No type I ligand binding spectrum could be detected that were indicative of any endogenous ligands in the E. coli cell lysate capable of provoking a low to high spin state change in purified CYP101A1, purified CYP176A1, or any of the recombinant human P450s studied here.
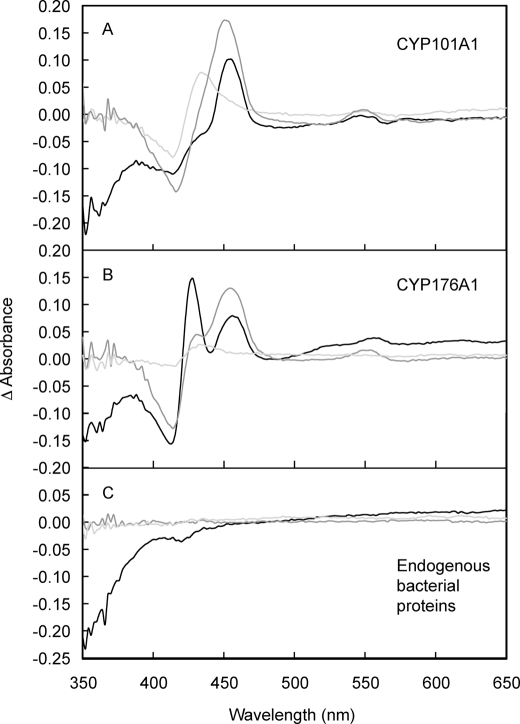
EPR Spectroscopy
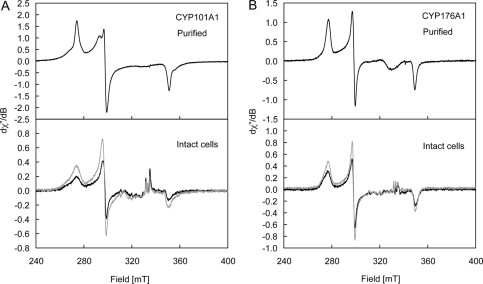
Low temperature (8 K) EPR spectra of purified CYP101A1 and CYP176A1 (Fig. 5, A and B) were measured prior to attempting similar measurements on whole cells. Both enzymes displayed resonances (Fig. 5) attributable to low spin Fe(III) species. X-ray crystallographic structures of the resting CYP101A1 have revealed a ruffled porphyrin ring coordinated axially to a cysteine and an aqua ligand, which is hydrogen-bonded in a cluster of another five water molecules in the substrate-binding pocket (32, 33). Pulsed EPR spectroscopy (34, 35) and model complex studies (36) indicated that polarization within the water cluster produces a hydroxide-like character in the aqua ligand and a low spin state for the Fe(III) ion. The EPR spectrum of purified CYP101A1 (Fig. 5A) was as observed previously (37). A comparison of the g matrices for species A (g1 = 2.4459, g2 = 2.2516, g3 = 1.9072; determined by computer simulation, supplemental Fig. S2) and species B (g1, 2.4856; g2, 2.2745; g3, 1.8853) with known literature values for CYP101A1 (37) indicates that these species correspond to the native enzyme (denoted species A here) and a complex in which the water molecules have been partially displaced by the buffer Tris (32, 33) (species B), respectively. Changing the buffer to HEPES, a non-coordinating buffer, produces a more complicated EPR spectrum (supplemental Fig. S3, A and B) showing a mixture of high spin (geff = 7.77, 3.91, 2.01) and three low spin Fe(III) species. Subtle changes to the low spin Fe(III) resonances (supplemental Fig. S3B) were observed upon the addition of KCl or Tris buffer, which reflect modifications to the water cluster at the active site. In contrast, the EPR spectrum of CYP176A1 shows a single low spin Fe(III) P450 species with g values: g1 = 2.4157, g2 = 2.2450, and g3 = 1.9196 (Fig. 5B, supplemental Fig. S4) (27). Monocistronic cultures of CYP101A1 and CYP176A1 were divided in two with one aliquot snap-frozen immediately and the other identical aliquot subjected to the oxidizing treatment noted above before freezing. A smaller proportion of species B and larger line widths for the native enzyme, species A, were seen in EPR spectra for CYP101A1 from whole cells when compared with the purified enzyme (Fig. 5A). A similar comparison of the EPR spectra for CYP176A1 (Fig. 5B) reveals a small proportion of a second species (low field resonance) in whole cells. A comparison of the EPR spectra for CYP101A1 and CYP176A1 in native and oxidized environments (Fig. 5, A and B) showed a clear increase in the intensity of the resonances in cells subjected to the oxidizing treatment. The lower intensity of the Fe(III) P450 resonances in the native whole cells can be explained by the presence of either low spin (S = 0) Fe(II) hemes that are EPR-silent or high spin (S = 2, D > hν) Fe(II) hemes that are also generally EPR-silent at X-band frequencies. There were no high spin (S = 5/2) Fe(III) resonances (g‖ eff = 2.0, g⊥ eff = 6.0) in the whole cell EPR spectra (results not shown), confirming the absence of endogenous ligands.
FIGURE 5.
X-band EPR spectra of purified (top panel) CYP101A1 (A; ∼300 μm in 50 mm Tris-HCl containing 50 mm KCl, pH 7.4, ν = 9.37728 GHz; top panel) and CYP176A1 (B; ∼21 μm in 100 mm potassium phosphate, pH 7.2, ν = 9.37715 GHz; top panel) and intact E. coli cells (bottom panel) expressing CYP101A1 (A) and CYP176A1 (B). Spectra of the native and oxidized samples are shown in black and gray lines, respectively, and were measured at 8.00 ± 0.02 K; g matrices are given under “Results.” The broad resonance of ∼330 mT in the purified enzymes arises from a Cu(II) impurity in the cavity. The relatively sharp resonances of ∼330 milliteslas in the whole cell spectra arise from slight differences in the concentrations of Mn(II) in the whole cell spectra and the control (no P450) sample. dχ′′/dB axes are relative.
Effect of Oxygen Availability during Measurement on P450 Whole Cell Reduction State
In the current study, Fe(II)-CO versus Fe(II) difference spectra were typically measured on resting cell resuspensions equilibrated in air for at least 30 min (prior to CO binding) and then subjected to a brief 5-s agitation immediately prior to measurement. Such resuspensions are functionally anaerobic due to the consumption of oxygen via bacterial respiration. Measured pO2 values for such stagnant resuspensions dropped to <0.1% air saturation within 3 min after aeration to 100% air saturation, measured with a Clark type polarographic electrode. No pulse in pO2 was observed by very brief agitations in air for resuspension as above. This premise was further verified by incubating CYP176A1 (previously equilibrated in air) under an argon atmosphere for up to 1 h before recording Fe(II)-CO versus Fe(II) difference spectra. The amount of endogenously reduced CYP176A1 did not change significantly when oxygen was totally excluded prior to measurement, although there was a very slight trend toward an increase in endogenously reduced P450 (55 ± 4% endogenously reduced P450 after 60 min under argon when compared with 47 ± 9% prior to argon incubation; mean ± S.D., n = 3; 72-h microaerobic cultures).
We tested whether the observation of endogenously reduced P450 was an artifact of low oxygen tension in bacterial cultures using both bacterial and representative human (CYP1A2 and CYP2C9) P450s (Table 1). When oxygen was provided by vigorous agitation in air up to the point of CO binding, all P450s tested still showed the presence of endogenously reduced Fe(II) heme (Table 1). Measured pO2 values were >90% air saturation immediately prior to CO treatment, as determined on representative resuspensions with a Clark type polarographic electrode. For CYP176A1 and CYP2C9 monocistronic cultures, the reduction percentage was similar in vigorously aerated cultures to that in cultures incubated under argon. However, CYP1A2 mono- and bicistronic, CYP2C9 bicistronic, and CYP101A1 monocistronic cultures showed less endogenously reduced P450 after vigorous aeration.
TABLE 1.
The effect of aeration and addition of substrate at measurement on the concentrations of reduced P450s measured by whole cell Fe(II)-CO versus Fe(II) difference spectroscopy
Endogenously reduced P450 concentrations were measured without external reductant. Total P450 concentrations were determined after dithionite reducing treatment. Data represent the mean ± S.D. of triplicate determinations. Asterisks indicate statistical significance between control and aerated or plus substrate samples (*, p < 0.05, **, p < 0.01, ***, p < 0.001). Data in parentheses show the percentage of total P450 measured after dithionite reducing treatment. Both control and plus substrate suspensions were equilibrated for 1 h in argon prior to collection of spectra (and after the addition of substrate where relevant). Aerobic suspensions were aerated vigorously by shaking at 800 rpm for 1 h prior to incubation with CO. Substrates were: CYP1A2, 1 mm phenacetin; CYP2C9, 1 mm warfarin; CYP176A1, 2.9 mm cineole; CYP101A1, 3.3 mm d-camphor.
| P450 form | Endogenously reduced P450 |
Total P450 | ||
|---|---|---|---|---|
| Control | Aerated | Plus substrate | ||
| nm | nm | |||
| CYP1A2a | 180 ± 30 (41%) | 49 ± 4 (11%)* | 240 ± 10 (53%) | 450 ± 80 |
| CYP1A2/hCPRa | 410 ± 20 (123%) | 190 ± 20 (57%)*** | 330 ± 30 (98%)* | 330 ± 60 |
| CYP2C9 | 140 ± 10 (17%) | 190 ± 50 (24%) | 380 ± 20 (49%)*** | 780 ± 40 |
| CYP2C9/hCPR | 270 ± 10 (60%) | 190 ± 10 (42%)** | 390 ± 20 (87%)*** | 450 ± 60 |
| CYP176A1b | 750 ± 40 (17%) | 900 ± 140 (20%) | 4500 ± 100 | |
| 170 ± 40 (8%) | 660 ± 40 (31%)*** | 2120 ± 50 | ||
| CYP101A1 | 3100 ± 400 (72%) | 1980 ± 60 (46%)* | 3490 ± 40 (82%) | 4300 ± 200 |
a Measurements on CYP1A2 and CYP1A2/hCPR were performed using cultures stored for 24h at 4 °C after harvest and resuspension. This decreased the proportion of endogenously reduced P450 making possible measurement of both increases and decreases in the degree of endogenous reduction.
b The effects of aeration and addition of substrate were measured in two separate experiments for this form.
Effect of Substrate Addition Just prior to Measurement of P450 Whole Cell Redox State
The addition of substrates to resuspensions prior to measurement significantly increased the reduction percentage for monocistronic and bicistronic CYP2C9 and monocistronic CYP176A1 (Table 1). A trend toward increased reduction percentage with substrates was also seen in CYP1A2 monocistronic and CYP101A1 monocistronic cultures, but did not reach statistical significance. CYP1A2 was already ∼100% reduced in bicistronic cultures, so although substrate appeared to significantly decrease the endogenously reduced P450 concentration, this was not significantly different from the total P450 concentration.
Investigation of the Redox State of P450 in Mammalian Hepatocytes
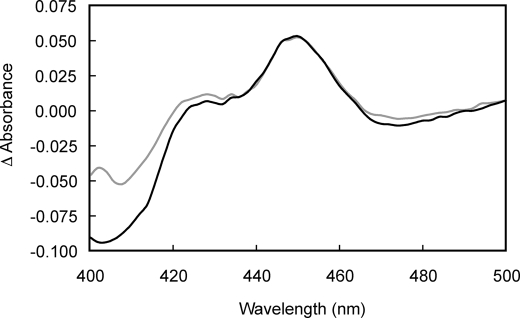
Fe(II)-CO versus Fe(II) difference spectroscopy was used to assess the redox state of P450s in rat hepatocytes. A clear peak was observed in the difference spectrum in the absence of dithionite, which was only slightly enhanced in samples treated with dithionite (reduction treatment; Fig. 6). The addition of a 100,000 × g supernatant fraction prepared from cryopreserved rat hepatocytes to diluted rat liver microsomes failed to elicit a type I binding spectrum (data not shown).
FIGURE 6.
Fe(II)-CO versus Fe(II) difference spectra recorded in intact rat hepatocytes. Rat hepatocytes were resuspended in phosphate-buffered saline to a density of 2.5 × 106 cells/ml. Fe(II)-CO versus Fe(II) difference spectra were recorded with (black) and without (gray) the addition of external reductant (10 mg/ml sodium dithionite, 13.5-min contact time).
DISCUSSION
The presence of reduced P450 in intact whole cells is unexpected for two reasons. Firstly, the conventional mechanism for P450 catalysis postulates the binding of substrate prior to the first electron reduction of ferric to ferrous heme. Secondly, the presence of ferrous P450 without bound substrate presents a potential danger to the cell in the form of uncoupled superoxide radical production when oxygen is present. Despite this, all P450 forms studied, with the exception of CYP3A4, showed a significant level of reduced P450 in the intact cell, as indicated by the observation of Fe(II)-CO versus Fe(II) difference spectra in the absence of added reductant (dithionite). Moreover, for many P450s (CYP1A2, CYP2A6, CYP2C19, and CYP101A1), this level was >80% after growth in microaerobic culture, even when cognate electron donors were not coexpressed. This was supported by the observation of nBIC ligand binding spectra of CYP101A1 and CYP176A1 that were characteristic of the Fe(II) form.
The measurement of a CO difference spectrum is itself capable of thermodynamically perturbing the P450 oxidation state by removing unbound ferrous P450 heme as the CO-bound form. The same is true of any ligand (e.g. nBIC) that binds to the heme unless it has an identical affinity for the Fe(II) and Fe(III) forms. X-band EPR spectroscopy provides a means by which to assess the natural redox state of the heme without perturbing the relative proportions of Fe(III) P450 and Fe(II) P450 by the addition of a ligand. The ferric form of the heme is paramagnetic, whereas the ferrous form is generally silent (low spin, S = 0; high spin, S = 2 with D > hν) at X-band microwave frequencies. To test whether any P450 was present in the reduced, EPR-silent form, we sought a reagent capable of oxidizing the P450 within intact cells. The prototypical oxidant potassium ferricyanide fails to enter cells. However, vigorous agitation in air of cultures containing the cell-permeable reducing agent, sodium dithionite (typically used to reduce P450 in intact E. coli (16, 17, 38)), significantly decreased the fraction of endogenously reduced P450 in cells. This counterintuitive effect is most likely to be mediated by hydrogen peroxide produced within cells upon aerobic decomposition of dithionite; S2O42− breaks down to SO2−, which has been shown to react rapidly with dioxygen in solution generating superoxide and ultimately hydrogen peroxide by dismutation or further reaction with SO2− (28, 29, 39).
Identical bacterial suspensions were examined by EPR before and after the oxidizing treatment. A clear increase was seen in the intensity of the characteristic resonances from the Fe(III) forms of CYP101A1 and CYP176A1 in cells subjected to the oxidizing treatment, consistent with conversion of prereduced (EPR-silent) P450 to the Fe(III) form upon oxidation.
This oxidizing treatment could cause damage to the P450. However, no resonances were observed over a wider scan range, which could be attributed to the breakdown of the P450 prosthetic group to either hemin (g⊥ eff = 6.0, g‖ eff = 2.0) or non-heme (geff = 4.3) Fe(III). In addition, the augmentation of the type I response to camphor in oxidized cell suspensions (supplemental Fig. S1) argues against the reduction of the Fe(II)-CO being due to any obvious P450 destruction. Indeed the type I spectra may underestimate the amount of Fe(III) produced by the oxidizing treatment because the addition of camphor for the type I measurement will potentiate reduction of the P450 and turnover of the camphor in whole cells.
The transfer of the first electron to the P450 forms tested here can be attributed to endogenous proteins naturally present in the whole cells. Cellular redox conditions are unlikely to enable straightforward reduction by freely diffusing intermediates without the mediation of a redox partner, especially in aerobic conditions (Table 1). Previous studies have shown that flavodoxin (Fd), coupled to FdR, can reconstitute a functional electron transfer pathway to P450s in E. coli (40). Like the cognate redox partners, these endogenous electron donors couple to NAD(P)H and can be considered electron conduits to the same cellular redox state. The smaller proportion of endogenously reduced P450 seen in CYP2C9, CYP3A4, and CYP176A1 monocistronic cultures may reflect a reduced ability to couple to the endogenous redox partners relative to the other forms examined.
It was possible to augment the endogenous redox partners with the cognate electron donors for each P450 form: hCPR, for the human forms, cindoxin plus the surrogate endogenously expressed E. coli FdR for CYP176A1 (41), and putidaredoxin plus putidaredoxin reductase for CYP101A1. The effect of coexpressed electron donors on the P450 reduction state was most apparent for forms that showed less reduction by the endogenous electron donors, i.e. CYP2C9, CYP3A4, and CYP176A1 (Fig. 3). This indicated that although stable, the balance of Fe(II) to Fe(III) P450 in whole cells is likely to be in steady state and not in a thermodynamically controlled equilibrium because the presence of cognate electron donors represents an additional electron transfer pathway to the P450, and thus, a kinetic rather than a thermodynamic intervention to the P450 reduction state. All P450s were significantly reduced (>80%) upon coexpression of cognate electron donors, showing that the redox state of intact cells was sufficiently reduced under the experimental conditions, once electron transfer pathways were in place, to maintain the P450s in a largely reduced state.
Next we sought to determine whether the observation of prereduced P450 was solely due to the highly reducing environment present in cells that were functionally anaerobic during measurement. Supplying oxygen during measurement represents a complex intervention to the P450 reduction state in whole cells, with three separate interactions possible (Fig. 1). Firstly, the redox state (i.e. NAD(P)H/NAD(P)+ ratio) of the cells will alter in response to the oxygenated environment, thus changing the overall thermodynamic driving force for reduction. Secondly, oxygen binds to the ferrous heme, yielding an oxyferrous complex and potentially displacing the heme toward the ferrous form. Finally, the oxyferrous complex can enter a futile cycle by losing the electron and oxygen as the superoxide radical, regenerating the ferric state.
Although aeration of cells immediately before P450 measurement reduced the proportion of prereduced P450, a significant amount of ferrous P450 was still evident for all forms tested. The observation of endogenously reduced P450 in whole cells is therefore not an artifact of measurement under functionally anaerobic (reducing) conditions. Because of the potential for futile cycling with both ferrous P450 and oxygen present, it was surprising that the ferrous P450 pool was not totally depleted. This suggests that although futile cycling and consequent superoxide radical formation may occur in whole cells, if they do occur, these processes must be slow enough not to overrun regeneration of ferrous P450 and entirely deplete the pool of reduced P450.3
Substrate has been shown to increase the rate of reduction of many (but not all (10)) P450s in vitro, and so substrate might be expected to alter the degree of reduction in vivo. Consistent with the in vitro results, the addition of substrates immediately prior to measurement increased the proportion of endogenously reduced P450 measured by Fe(II)-CO versus Fe(II) difference spectroscopy, particularly for those forms showing less endogenous reduction of the P450 (monocistronic CYP2C9 plus warfarin and monocistronic CYP176A1 plus cineole). The observed increase can be attributed to the kinetic effect of increasing the rate of reduction of ferric P450 by introducing an alternative pathway to the ferrous form. (Interestingly, the rate of CYP2C9 reduction in vitro has been seen to be increased by substrate, whereas that of CYP1A2 was little affected (10), in parallel with the relative effects seen here.) Introduction of a more rapid pathway for heme reduction, combined with the steady state nature of the Fe(II)/Fe(III) heme cellular balance postulated above, provides a plausible explanation for the increase in ferrous P450.
The conventional representation of the P450 catalytic cycle shows ferric P450 as the resting state and substrate binding as the first step followed by transfer of the first electron (4–7). The first electron transfer to CYP101A1 has been reported to be insignificant in the absence of substrate (42), but few data exist on this key process (43). The facilitation of the first reduction step caused by substrate binding has been attributed to a positive shift in the reduction potential. However, coupling the reduction step with the binding of a (dioxygen) ligand to the ferrous enzyme should make the reduction thermodynamically favorable for both substrate-free and substrate-bound forms. Such an effect is well recognized in electron transfer processes coupled with chemical reactions (44). Rather, the smaller difference in reorganization energy associated with displacement of the aqua ligand and the consequent spin shift is more likely to explain the faster reduction of the P450 after binding of substrates, a kinetic rather than a thermodynamic effect (44). Whether controlled thermodynamically or kinetically, this perspective provides a simple mechanism by which uncoupling side reactions could be limited in the substrate-free enzyme.
The present study provides evidence that that the resting form is a mixture of ferric and ferrous forms in different ratios characteristic of the P450 in question and the nature of its exact cellular environment. This is consistent with the hypothesis that significant reduction of the P450 to the ferrous form occurs in the absence of substrate for all forms studied. Interestingly, CYP101A1 showed one of the highest proportions of endogenously reduced P450, an observation that was particularly unexpected given the prevailing view that its catalytic cycle is tightly controlled in the absence of substrate. No evidence could be found for any endogenous substrate in E. coli cell lysate that was capable of inducing a spin state shift and therefore accelerating delivery of the first electron to CYP101A1. This suggests that reduction is not gated at the first step in vivo and that some degree of futile cycling may be possible for most P450s including CYP101A1. Alternatively, another mechanism may prevent uncoupling. Notably, studies have proposed that the oxyferrous form of various P450s is highly labile, but stabilized by substrates (45). Although this would not prevent release of reactive oxygen species (i.e. superoxide), such a gating mechanism would limit further progress through the cycle in the absence of substrate. Our data do not inform us as to the pathway by which the Fe(III) P450 is reduced, i.e. reduction of low spin Fe(III) P450 versus reduction of the small amount of high spin Fe(III) P450 present at equilibrium.
E. coli provides a technically simple, semiphysiological model in which the P450 redox state can be examined in the presence of a complex cellular environment, including functional redox systems maintaining NAD(P)H levels. The regulation of the redox state is similar in eukaryotic and prokaryotic cells, i.e. NAD is found predominantly in the oxidized form, whereas NADP is found predominantly in the reduced form (46), so this model yields useful insights into the probable behavior of P450s in native environments.4 The results of this study suggest that ferrous P450 is present during aerobic growth. Detrimental effects on the cell might be expected from futile cycling of P450s in the absence of substrates. However, no evidence of deleterious effects was seen on bulk culture parameters (see supplemental Figs. S5 and S6). Thus, the possible damage due to futile cycling may be limited effectively by bacterial systems for scavenging reactive oxygen species, without apparent cost to the cell. In the case of CYP101A1, P450 expression in the natural host, Pseudomonas putida, is known to be tightly controlled, so repression of transcription in the absence of substrate (47) may be sufficient to restrict futile cycling.
To assess the relevance of these results to mammalian cells, we undertook limited experiments with rat hepatocytes. Consistent with the results obtained in the model system, effectively all the P450 detected in hepatocytes was endogenously reduced; the addition of exogenous dithionite failed to significantly increase the amount of P450 detected. Importantly, the oxygen tension to which the hepatocytes were exposed is likely to be greater under the conditions of our experiments than in vivo. Although the cost and relative fragility of hepatocytes limited more detailed experimentation on these cells, this suggests that P450s are predominantly present in the reduced state in hepatocytes. However, the situation within intact liver, especially within the relatively low pO2 regions where the P450s studied here are concentrated, remains to be clarified. It is worth noting that the experiments in E. coli in the absence of cognate redox partners represent the most stringent test of our hypothesis that the ferrous form of P450 exists in vivo. In the native cell in the presence of both the normal redox partners and likely endogenous substrates (fatty acids, etc.), we would expect to see even more facile reduction of the P450 heme iron.
The current work is the first to our knowledge in which the redox state of P450s has been monitored within intact, viable cells. This study points to the need for further investigations using systems that more effectively model the physiological state of P450s in intact cells.
Supplementary Material
Acknowledgment
We thank Prof. Maree Smith of the Centre for Integrated Preclinical Drug Development, University of Queensland, for the gift of rat hepatocytes.
This work was supported in part by Australian Research Council (ARC) Discovery Project Grant DP0210635 (to E. M. J. G. and J. J. D. V.) and ARC Linkage Project Grant LP0560595 funded in partnership with AstraZeneca R&D (to E. M. J. G., J. J. D. V., and M. A. H.).
This paper is dedicated to the memory of Lisa M. Notley (1972–2011).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S6.
The Fe(II)-CO versus Fe(II) difference peak measured after aeration represents the minimum proportion of ferrous P450 because some masking of Fe(II) heme as the oxyferrous form may occur due to the failure of CO to immediately displace all oxygen bound to heme.
A caveat applies in the case of CYP101A1. For all of the P450s examined here except CYP101A1, NADPH, present at high levels in the cell relative to NADP+, is the natural coenzyme. However, CYP101A1 is usually coupled to NADH, which is maintained at low levels relative to NAD+. Therefore if the Fd/FdR or another NADPH-dependent system enables reduction of CYP101A1 by NADPH rather than NADH, the higher levels of NADPH when compared with the natural coenzyme NADH in the original host provide a greater thermodynamic driving force for reduction of CYP101A1 in the model system. Unfortunately, the tight transcriptional regulation of CYP101A1 in P. putida confounded an attempt to assess its redox state in the host organism in the absence of substrate.
- nBIC
- n-butylisocyanide
- Fd
- flavodoxin/ferredoxin
- FdR
- flavodoxin reductase
- hCPR
- human NADPH cytochrome P450 reductase.
REFERENCES
- 1. Guengerich F. P. (2001) Chem. Res. Toxicol. 14, 611–650 [DOI] [PubMed] [Google Scholar]
- 2. Isin E. M., Guengerich F. P. (2007) Biochim. Biophys. Acta 1770, 314–329 [DOI] [PubMed] [Google Scholar]
- 3. De Voss J. J., Cryle M. J. (2007) in The Ubiquitous Roles of Cytochrome P450 Proteins (Sigel A., Sigel H., Sigel R. K. eds) Vol. 3, pp. 397–435, John Wiley and Sons, Ltd., New York [Google Scholar]
- 4. Denisov I. G., Makris T. M., Sligar S. G., Schlichting I. (2005) Chem. Rev. 105, 2253–2277 [DOI] [PubMed] [Google Scholar]
- 5. Luthra A., Denisov I. G., Sligar S. G. (2011) Arch. Biochem. Biophys. 507, 26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumar D., Altun A., Shaikh S., Thiel W. (2011) Faraday Discuss. 148, 373–383 [DOI] [PubMed] [Google Scholar]
- 7. Nouri-Nigjeh E., Bischoff R., Bruins A. P., Permentier H. P. (2011) Curr. Drug Metab. 12, 359–371 [DOI] [PubMed] [Google Scholar]
- 8. Backes W. L., Sligar S. G., Schenkman J. B. (1982) Biochemistry 21, 1324–1330 [DOI] [PubMed] [Google Scholar]
- 9. Das A., Grinkova Y. V., Sligar S. G. (2007) J. Am. Chem. Soc. 129, 13778–13779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guengerich F. P., Johnson W. W. (1997) Biochemistry 36, 14741–14750 [DOI] [PubMed] [Google Scholar]
- 11. Meunier B., de Visser S. P., Shaik S. (2004) Chem. Rev. 104, 3947–3980 [DOI] [PubMed] [Google Scholar]
- 12. Hlavica P. (2007) Curr. Drug Metab. 8, 594–611 [DOI] [PubMed] [Google Scholar]
- 13. Mak P. J., Im S. C., Zhang H., Waskell L. A., Kincaid J. R. (2008) Biochemistry 47, 3950–3963 [DOI] [PubMed] [Google Scholar]
- 14. Shaik S., Cohen S., Wang Y., Chen H., Kumar D., Thiel W. (2010) Chem. Rev. 110, 949–1017 [DOI] [PubMed] [Google Scholar]
- 15. Omura T., Sato R. (1962) J. Biol. Chem. 237, 1375–1376 [PubMed] [Google Scholar]
- 16. Otey C. R. (2003) in Methods in Molecular Biology (Arnold F. H., Georgiou G. eds) Vol. 230, pp. 137–139, Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 17. Johnston W. A., Huang W., De Voss J. J., Hayes M. A., Gillam E. M. (2008) J. Biomol. Screen. 13, 135–141 [DOI] [PubMed] [Google Scholar]
- 18. Guengerich F. P. (1994) in Principles and Methods of Toxicology (Hayes A. W. ed), Third Ed., pp. 1259–1313, Raven Press, Ltd., New York [Google Scholar]
- 19. Fisher C. W., Caudle D. L., Martin-Wixtrom C., Quattrochi L. C., Tukey R. H., Waterman M. R., Estabrook R. W. (1992) FASEB J. 6, 759–764 [DOI] [PubMed] [Google Scholar]
- 20. Soucek P. (1999) Arch. Biochem. Biophys. 370, 190–200 [DOI] [PubMed] [Google Scholar]
- 21. Cuttle L., Munns A. J., Hogg N. A., Scott J. R., Hooper W. D., Dickinson R. G., Gillam E. M. (2000) Drug Metab. Dispos. 28, 945–950 [PubMed] [Google Scholar]
- 22. Gillam E. M., Baba T., Kim B. R., Ohmori S., Guengerich F. P. (1993) Arch. Biochem. Biophys. 305, 123–131 [DOI] [PubMed] [Google Scholar]
- 23. Parikh A., Gillam E. M., Guengerich F. P. (1997) Nat. Biotechnol. 15, 784–788 [DOI] [PubMed] [Google Scholar]
- 24. Hawkes D. B., Adams G. W., Burlingame A. L., Ortiz de Montellano P. R., De Voss J. J. (2002) J. Biol. Chem. 277, 27725–27732 [DOI] [PubMed] [Google Scholar]
- 25. Huang W., Johnston W. A., Hayes M. A., De Voss J. J., Gillam E. M. (2007) Arch. Biochem. Biophys. 467, 193–205 [DOI] [PubMed] [Google Scholar]
- 26. Nishihara K., Kanemori M., Kitagawa M., Yanagi H., Yura T. (1998) Appl. Environ. Microbiol. 64, 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanson G. R., Gates K. E., Noble C. J., Griffin M., Mitchell A., Benson S. (2004) J. Inorg. Biochem. 98, 903–916 [DOI] [PubMed] [Google Scholar]
- 28. Lambeth D. O., Palmer G. (1973) J. Biol. Chem. 248, 6095–6103 [PubMed] [Google Scholar]
- 29. Dixon M. (1971) Biochim. Biophys. Acta 226, 241–258 [DOI] [PubMed] [Google Scholar]
- 30. Tomita T., Ogo S., Egawa T., Shimada H., Okamoto N., Imai Y., Watanabe Y., Ishimura Y., Kitagawa T. (2001) J. Biol. Chem. 276, 36261–36267 [DOI] [PubMed] [Google Scholar]
- 31. Holman D. A., Bennett D. W. (1994) J. Phys. Chem. 98, 13300–13307 [Google Scholar]
- 32. Ebel R. E., O'keeffe D. H., Peterson J. A. (1977) Arch. Biochem. Biophys. 183, 317–327 [DOI] [PubMed] [Google Scholar]
- 33. Schlichting I., Berendzen J., Chu K., Stock A. M., Maves S. A., Benson D. E., Sweet R. M., Ringe D., Petsko G. A., Sligar S. G. (2000) Science 287, 1615–1622 [DOI] [PubMed] [Google Scholar]
- 34. Goldfarb D., Bernardo M., Thomann H., Kroneck P. M., Ullrich V. (1996) J. Am. Chem. Soc. 118, 2686–2693 [Google Scholar]
- 35. Thomann H., Bernardo M., Goldfarb D., Kroneck P. M., Ullrich V. (1995) J. Am. Chem. Soc. 117, 8243–8251 [Google Scholar]
- 36. Lochner M., Mu L. J., Woggon W. D. (2003) Adv. Synth. Catal. 345, 743–765 [Google Scholar]
- 37. Sono M., Dawson J. H. (1982) J. Biol. Chem. 257, 5496–5502 [PubMed] [Google Scholar]
- 38. Yim S. K., Kim D. H., Jung H. C., Pan J. G., Kang H. S., Ahn T., Yun C. H. (2010) J. Microbiol. Biotechnol. 20, 712–717 [DOI] [PubMed] [Google Scholar]
- 39. Rinker R. G., Gordon T. P., Mason D. M., Sakaida R. R., Corcoran W. H. (1960) J. Phys. Chem. 64, 573–581 [Google Scholar]
- 40. Jenkins C. M., Waterman M. R. (1994) J. Biol. Chem. 269, 27401–27408 [PubMed] [Google Scholar]
- 41. Hawkes D. B., Slessor K. E., Bernhardt P. V., De Voss J. J. (2010) ChemBiochem 11, 1107–1114 [DOI] [PubMed] [Google Scholar]
- 42. Fisher M. T., Sligar S. G. (1985) J. Am. Chem. Soc. 107, 5018–5019 [Google Scholar]
- 43. Peterson J. A., Prough R. A. (1986) in Cytochrome P450 (Ortiz de Montellano P. R. ed), First Ed., pp. 89–117, Plenum Press, New York [Google Scholar]
- 44. Honeychurch M. J., Hill A. O., Wong L. L. (1999) FEBS Lett. 451, 351–353 [DOI] [PubMed] [Google Scholar]
- 45. Denisov I. G., Grinkova Y. V., Baas B. J., Sligar S. G. (2006) J. Biol. Chem. 281, 23313–23318 [DOI] [PubMed] [Google Scholar]
- 46. Ying W. (2008) Antioxid. Redox Signal. 10, 179–206 [DOI] [PubMed] [Google Scholar]
- 47. Koga H., Aramaki H., Yamaguchi E., Takeuchi K., Horiuchi T., Gunsalus I. C. (1986) J. Bacteriol. 166, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nelson D. R. (2011) Biochim. Biophys. Acta 1814, 14–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.