Background: Leu1p is an abundant cytoplasmic [4Fe-4S] enzyme thought to require Fe-S clusters (ISCs) exported from mitochondria via the Atm1p transporter.
Results: Active Leu1p is synthesized at the expense of mitochondrial aconitase independently of Atm1p overexpression or depletion.
Conclusion: Atm1p is not implicated in exporting ISCs for Leu1p activation.
Significance: Further studies should identify molecules and mechanisms controlling ISC distribution between mitochondria and cytoplasm.
Keywords: ABC Transporter, Iron Metabolism, Iron-Sulfur Protein, Mitochondria, Yeast, Leucine Synthesis
Abstract
Fe-S clusters (ISCs) are versatile cofactors utilized by many mitochondrial, cytoplasmic, and nuclear enzymes. Whereas mitochondria can independently initiate and complete ISC synthesis, other cellular compartments are believed to assemble ISCs from putative precursors exported from the mitochondria via an ATP binding cassette (ABC) transporter conserved from yeast (Atm1p) to humans (ABCB7). However, the regulatory interactions between mitochondrial and extramitochondrial ISC synthesis are largely unknown. In yeast, we found that mitochondrial ISC synthesis is regulated by the leucine biosynthetic pathway, which among other proteins involves an abundant cytoplasmic [4Fe-4S] enzyme, Leu1p. Enzymatic blocks in the pathway (i.e. leu1Δ or leu2Δ gene deletion mutations) induced post-transcriptional up-regulation of core components of mitochondrial ISC biosynthesis (i.e. the sulfur donor Nfs1p, the iron donor Yfh1p, and the ISC scaffold Isu1p). In leu2Δ cells, transcriptional mechanisms also led to dramatic up-regulation of Leu1p with concomitant down-regulation of mitochondrial aconitase (Aco1p), a [4Fe-4S] enzyme in the tricarboxylic acid cycle. Accordingly, the leu2Δ deletion mutation exacerbated Aco1p inactivation in cells with mutations in Yfh1p. These data indicate that defects in leucine biosynthesis promote the biogenesis of enzymatically active Leu1p at the expense of Aco1p activity. Surprisingly, this effect is independent of Atm1p; previous reports linking the loss of Leu1p activity to Atm1p depletion were confounded by the fact that LEU2 was used as a selectable marker to create Atm1p-depleted cells, whereas a leu2Δ allele was present in Atm1p-repleted controls. Thus, still largely unknown transcriptional and post-transcriptional mechanisms control ISC distribution between mitochondria and other cellular compartments.
Introduction
ISCs2 are highly versatile enzyme cofactors that can perform a broad range of functions including electron transfer; substrate binding and activation; enzyme regulation; and sensing of iron, oxygen, or reactive oxygen species (for a review, see Ref. 1). This versatility explains why ISC enzymes are extensively utilized by cells in different pathways localized to the mitochondria (e.g. Krebs cycle and electron transport), the cytoplasm (e.g. leucine biosynthesis and ribosome biogenesis), and the nucleus (e.g. DNA synthesis and repair mechanisms) (for reviews, see Refs. 2 and 3). The number of newly identified ISC enzymes continues to increase, and it is likely that examples of these proteins are present in most if not all cellular pathways.
In yeast and animal cells, ISC synthesis is carried out by multienzyme machineries localized to the mitochondrial matrix, the cytoplasm, and probably the nucleus (3). The components of the mitochondrial matrix machinery are highly conserved from yeast to humans, and most were inherited from prokaryotes (for a review, see Ref. 4). They include enzymes that provide elemental iron and sulfur and reducing equivalents needed for the assembly of [2Fe-2S] and [4Fe-4S] clusters on molecular scaffolds as well as other proteins that help transfer the clusters to the appropriate apoenzymes (2–4). Although this mitochondrial machinery is able to independently initiate and complete ISC synthesis in vivo and in vitro (5–8), the cytoplasmic machinery is believed to depend on ISC precursors synthesized in the mitochondrial matrix and exported via a conserved ABC transporter localized to the inner mitochondrial membrane (9–12).
In yeast, this situation is exemplified by the leucine biosynthetic pathway (for a review, see Ref. 13). Leucine synthesis is initiated with the generation of α-isopropylmalate (α-IPM), which can be produced in both the mitochondrial matrix and the cytoplasm from α-ketoisovalerate and acetyl coenzyme A. The subsequent steps take place in the cytoplasm where an isopropylmalate isomerase (Leu1p) first converts α-IPM to β-isopropylmalate, a β-isopropylmalate dehydrogenase (Leu2p) subsequently oxidizes β-isopropylmalate to α-ketoisocaproate, and lastly a branched-chain amino-acid transaminase (Bat2p) yields leucine (14). The expression of most components of the pathway is under the control of a transcription factor, Leu3p, which can function as transcriptional activator or repressor depending on the presence or absence, respectively, of α-IPM (13).
It is well known that yeast cells respond to blocks in this pathway (e.g. leu1Δ or leu2Δ deletion mutations) with a robust, although futile, up-regulation of enzymes upstream and downstream of the block. This up-regulation is triggered by the accumulation of α-IPM, which elicits the transcriptional activator function of Leu3p (15). The activity of Leu1p, a [4Fe-4S] cluster enzyme in this pathway, is thought to depend on mitochondrial biosynthesis and subsequent exportation of ISC precursors (9). Indeed, the dramatic loss of Leu1p activity associated with defects in mitochondrial ISC assembly factors (e.g. the iron donor Yfh1p) is generally regarded as evidence that mitochondria are essential for cytoplasmic ISC assembly (9, 16–19). Moreover, the dramatic loss of Leu1p activity apparently associated with depletion of the inner mitochondria membrane transporter Atm1p is regarded as evidence that assembly of the Leu1p [4Fe-4S] cluster requires ISC precursors synthesized in the mitochondrial matrix and exported by Atm1p (9, 10). The nature of the putative ISC precursors has thus far remained elusive, although a sulfur-containing compound has been hypothesized (20). In addition, the regulatory mechanisms controlling the distribution of ISCs or their precursors between the mitochondria and the cytoplasm are largely unknown.
We present evidence that defects in leucine synthesis induce changes in the levels of mitochondrial and cytoplasmic proteins, which together promote the biogenesis of enzymatically active Leu1p. We further show that the maintenance of Leu1p activity is largely independent of the Atm1p transporter, supporting the possibility that Atm1p is not directly involved in mitochondrial ISC export. Our results also reveal that leucine auxotrophic strains, which represent the vast majority of laboratory yeast strains, are not suitable to study ISC synthesis under physiological conditions.
EXPERIMENTAL PROCEDURES
Bacterial and Yeast Strains and Culture Conditions
Escherichia coli strain DH5-α was used for general DNA subcloning procedures. All strains used in this study were derived from Saccharomyces cerevisiae strain YPH500 (MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 +YCP50-YFH1 [Y73A]-URA3 [ρ+]) (Table 1). Wild type YFH1 was expressed from a centromeric low copy vector (YCplac22) under the control of its natural promoter. Mutant yfh1Y73A, yfh1L44A, or yfh1V150A alleles were subcloned into the TRP1-based integrating vector pRS404 under the control of the glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter and then integrated on the chromosome as described (21). High copy 2-μm vectors YEp24 (New England Biolabs) and pRS426 (22) as well as centromeric low copy vectors pRS416 (22) and p416GPD (23) were used to express LEU2, LEU1, and ACO1. For LEU2 gene integration, the DNA fragment encoding the wild type gene was amplified from a yeast genomic DNA library (24) and transformed into leu2Δ cells.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| YFH1 leu2Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] | 21 |
| yfh1Y73Aleu2Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] | 21 |
| yfh1L144Aleu2Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + pRSGPD-YFH1[L144A] [ρ+] | This study |
| yfh1L144ALEU2 (2 μm) | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + pRSGPD-YFH1[L144A] [ρ+] + Yep24-LEU2-URA3 | This study |
| yfh1V150Aleu2Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + pRSGPD-YFH1[V150A] [ρ+] | This study |
| yfh1V150A LEU2 (2 μm) | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + pRSGPD-YFH1[V150A] [ρ+] + Yep24-LEU2-URA3 | This study |
| YFH1 LEU2 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1 Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] | This study |
| yfh1Y73A LEU2 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] | This study |
| yfh1Y73A LEU2 (CEN) | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] + pRS415-LEU2 | This study |
| yfh1Y73A LEU2 (2 μm) | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] + Yep24-LEU2-URA3 | This study |
| YFH1 LEU1-FLAG LEU2-FLAG | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG LEU2-FLAG | This study |
| yfh1Y73ALEU1-FLAG LEU2-FLAG | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] LEU1-FLAG LEU2-FLAG | This study |
| YFH1 leu1Δ LEU2-FLAG | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] leu1Δ::KanMX4 LEU2-FLAG | This study |
| yfh1Y73Aleu1Δ LEU2 (CEN) | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] leu1Δ::KanMX4 + pRS415-LEU2 | This study |
| yfh1Y73Aleu1Δ LEU2 (2 μm) | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] leu1Δ::KanMX4 + Yep24-LEU2-URA3 | This study |
| yfh1Y73Aleu1Δ LEU2-FLAG | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] leu1Δ::KanMX4 LEU2-FLAG | This study |
| YFH1 LEU1-FLAG leu2Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG leu2Δ::KanMX4 | This study |
| yfh1Y73ALEU1-FLAG leu2Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] LEU1-FLAG leu2Δ::KanMX4 | This study |
| YFH1 LEU1-FLAG LEU2-FLAG leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG LEU2-FLAG leu3Δ::URA3 | This study |
| yfh1Y73ALEU1-FLAG LEU2-FLAG leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] LEU1-FLAG LEU2-FLAG leu3Δ::URA3 | This study |
| YFH1 leu1Δ-LEU2-FLAG leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] leu1Δ::KanMX4 LEU2-FLAG leu3Δ::URA3 | This study |
| yfh1Y73A leu1Δ-LEU2-FLAG leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] leu1Δ::KanMX4 LEU2-FLAG leu3Δ::URA3 | This study |
| YFH1 LEU1-FLAG leu2Δ leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG leu2Δ::KanMX4 leu3Δ::URA3 | This study |
| yfh1Y73ALEU1-FLAG leu2Δ leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] LEU1-FLAG leu2Δ::KanMX4 leu3Δ::URA3 | This study |
| YFH1 leu1Δ LEU2-FLAG leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1[Y73A]-TRP1 [ρ+] leu1Δ::KanMX4 LEU2-FLAG leu3Δ::URA3 | This study |
| yfh1Y73 Aleu1Δ LEU2-FLAG leu3Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] leu1Δ::KanMX4 LEU2-FLAG leu3Δ::URA3 | This study |
| yfh1Y73A leu2Δ aco1Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + pRSGPD-YFH1[Y73A] [ρ+] Aco1Δ::KanMX4 | This study |
| yfh1Y73LEU2 aco1Δ | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1Δ::HIS3 + YCplac22-YFH1[Y73A]-TRP1 [ρ+] Aco1Δ::KanMX4 | This study |
| LEU2 ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1 Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] ATM1-FLAG3-URA3 | This study |
| leu2Δ ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] ATM1-FLAG3-URA3 | This study |
| LEU2 Gal-ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1 Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] KanMX4-Gal-ATM1-FLAG3-URA3 | This study |
| leu2Δ Gal-ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] KanMX4-Gal-ATM1-FLAG3-URA3 | This study |
| LEU1-FLAG LEU2-FLAG ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1 Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG LEU2-FLAG ATM1-FLAG3-URA3 | This study |
| LEU1-FLAG leu2Δ ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG ATM1-FLAG3-URA3 | This study |
| LEU1-FLAG LEU2-FLAG Gal-ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 yfh1 Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG LEU2-FLAG KanMX4-Gal-ATM1-FLAG3-URA3 | This study |
| LEU1-FLAG leu2Δ Gal-ATM1 | MATα ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + YCplac22-YFH1-TRP1 [ρ+] LEU1-FLAG KanMX4-Gal-ATM1-FLAG3-URA3 | This study |
Gene deletions were carried out using a single step gene disruption procedure (25) with linear PCR products carrying kanMX (26) or Kluyveromyces lactis URA3 (27). Leu1p, Leu2p, and Atm1p were C-terminally FLAG-tagged by fusing the FLAG coding sequence to the LEU1, LEU2, or ATM1 coding sequence immediately upstream of the stop codon. A FLAG-tagged LEU2 allele was chromosomally integrated in a LEU1 leu2Δ strain, and LEU1 was deleted by replacing the coding sequence with kanMX, resulting in a leu1Δ::kanMX LEU2-FLAG strain. Next, the kanMX was replaced by a LEU1-FLAG allele, resulting in a LEU1-FLAG LEU2-FLAG strain. Finally, the LEU2 coding sequence was deleted with kanMX to construct the LEU1-FLAG leu2Δ::kanMX strain. To achieve inducible expression of ATM1, the GAL10 promoter (pGal10) of S. cerevisiae was fused with the kanMX cassette by PCR, and the fusion product was chromosomally integrated immediately upstream of the ATM1 coding sequence (Gal-ATM1). A 3× FLAG allele (ATM1-FLAG3) was fused with K. lactis URA3 by PCR, and the resultant ATM1-FLAG3 URA3 fragment was chromosomally integrated in strains containing either wild type ATM1 or Gal-ATM1, resulting in ATM1-FLAG3 URA3 and Gal-ATM1-FLAG3 URA3 strains. To obtain yeast cells in which both Leu1p and Atm1p were C-terminally tagged with FLAG, the mating type of the strains carrying LEU1-FLAG was switched; these strains were crossed with strains carrying ATM1-FLAG, diploids were sporulated, and haploid cells carrying both LEU1-FLAG and ATM1-FLAG were identified. The functionality of the FLAG-tagged proteins was verified by comparing the phenotype of strains carrying the tagged versus non-tagged versions. For Leu1p and Leu2p, the enzymatic activity and growth rate on synthetic medium omitting leucine were compared. For Atm1p, the growth rate on a non-fermentable carbon source was compared. No significant differences were observed.
To study ACO1 expression, four different constructs were subcloned into pRS416 vector: 1) ACO1 coding sequence only, 2) ACO1 coding sequence plus 1 kbp of upstream and 1 kbp of downstream flanking genomic DNA, 3) ACO1 coding sequence plus 1 kbp of upstream genomic DNA only, and 4) ACO1 coding sequence plus 1 kbp of downstream genomic DNA only. In constructs 1 and 4, ACO1 expression was driven by the GPD promoter. These vectors were transformed into strains in which the endogenous ACO1 coding sequence had been replaced by kanMX (aco1Δ::kanMX). All plasmids were verified by DNA sequencing; strains were verified by PCR analysis of genomic DNA and Western blotting as appropriate.
To test potential transcriptional regulation of ACO1 by Leu3p, 1 kbp of the ACO1 upstream flanking region was cloned into YEp356R vector. This fragment included the first seven codons of ACO1 that were fused in-frame with the E. coli lacZ gene sequence. Point mutations were introduced in the putative Leu3p binding site by PCR, and all constructs were verified by DNA sequencing. The effects of the mutations were analyzed by measuring β-galactosidase activity as described (28).
The following liquid and solid media were used: rich medium (2% peptone and 1% yeast extract) supplemented with either 2% dextrose (YPD) or 3% ethanol (YPE) and synthetic minimal medium (6.7% yeast nitrogen base without amino acids) supplemented with appropriate amino acids and either 2% dextrose (SD), 2% galactose (SGal), or 3% glycerol (SG). All cultures performed in S medium were supplemented with 20 mg/liter leucine independently of the genotype. In all experiments, yeast cells were grown aerobically on solid or liquid medium at 30 °C.
Enzymatic Activity Assays
Enzyme activities were assayed in cell lysates prepared from spheroplasts (29) obtained from 50-ml cultures grown in synthetic medium (60–200 mg of cells, wet weight). Spheroplasts were resuspended in the same buffer used to measure aconitase activity (150 mg of cells/ml of 20 mm Tris, pH 7.4, 50 mm NaCl, 1 mm phenylmethanesulfonyl fluoride) and disrupted by sonication on ice. Cell debris was removed by centrifugation (10 min at 20,000 × g) at 4 °C. Previously published methods were used to assay the activities of mitochondrial aconitase (30, 31) and isopropyl malate isomerase (Leu1p) (32). Cell lysates used for enzymatic assays were also used to perform Western blot analysis of the proteins of interest. Protein concentrations were measured by the Bio-Rad Protein Assay (Bio-Rad) or BCA (Pierce) method. Membranes after Western blotting were stained using the MemCode reversible membrane staining kit (Pierce), and total protein staining was used to assess equal protein loading.
Suppressor Screening
The yfh1Y73A leu2Δ strain was transformed with a yeast genomic library on a high copy URA3-based YEp24 vector (24), and transformants were selected at 30 °C on SD plates lacking uracil. All transformants were pooled by washing each plate with SD medium containing 100 μm FeCl3, and each pool was grown aerobically at 30 °C with shaking at 250 rpm. Cells reached stationary phase after 24 h and were harvested by centrifugation after 24, 48, or 72 h. Cell pellets were washed with fresh SD medium without iron and plated onto SG medium at a density of ∼1,000 cells/plate. Colonies were analyzed after 5 days of incubation at 30 °C. The yfh1Y73A leu2Δ strain yielded ∼90,000 initial transformants (>15 genome equivalents), 160 of which kept the ability to grow on SG. The same procedure was repeated with the yfh1V150A leu2Δ or yfh1L144A leu2Δ strain. Each yielded ∼40,000 initial transformants and 87 and 29 respiratory-competent transformants, respectively.
RNA Isolation and Quantitative RT-PCR
Yeast strains were grown in SD medium for 24 or 48 h under the conditions used for the suppressor screening but without iron supplementation. Total RNA was isolated from ∼1.5 × 107 cells using the MasterPure RNA isolation kit (Epicenter). One microgram from each RNA sample was used to synthesize cDNA with the Superscript III First Strand cDNA synthesis kit (Invitrogen). One microliter from each cDNA sample was used to determine the mRNA copy number for the ISU1, NFS1, YFH1, and ACO1 genes with ACT1 mRNA serving as an internal standard using the LightCycler FastStart DNA Master SYBR Green I kit in a LightCycler instrument (Roche Applied Science) according to the supplier's protocol. For the amplification of the specific genes, the following primers were used: ISU1: fwd, GCTCCGCCATTGCCTCCTCT; rev, GGTTGGAGTGTTTCTCTTAGATTT; NFS1: fwd, CATTGGTAGCGGGATTTGGTGA; rev, CCATCAATAAAGATTCTCCTTCCA; YFH1: fwd, GCTCATCCGGATTGTATACCTG; rev, GTTAGCTTTGTGCCATTTCTTAAC; ACO1: fwd, CTGGTATCACAACTGTCAAAGGT; rev, GGTCCTTGTGGTATAATTTAGCAA; and ACT1: fwd, CTCCTCGTGCTGTCTTCCCAT; rev, GTTGTAGAAGGTATGATGCCAGA.
RESULTS
LEU2 Gene Rescues Respiratory Function of Yeast Cells with Mutations in Yfh1p
To screen for factors that influence mitochondrial ISC biosynthesis, we used yeast cells with defects in the iron donor Yfh1p (6, 33). In earlier studies, the severe phenotype of yeast cells lacking Yfh1p completely (yfh1Δ cells) could only be suppressed by genes that blocked mitochondrial iron overload and/or limited iron toxicity (34–37). We reasoned that the milder respiratory-deficient phenotypes associated with certain Yfh1p point mutations (21, 38, 39) might enable identification of genes more directly related to mitochondrial ISC biosynthesis and its regulation. We selected point mutations in Yfh1p (Y73A, L144A, and V150A) that did not result in obvious phenotypes when yeast cells grew logarithmically in rich medium (Ref. 21 and data not shown). However, each of the three mutations caused the loss of respiratory function when yeast cells entered stationary phase upon growth in SD medium supplemented with iron (Fig. 1, A–C). This particular stationary phase is associated with a high metabolic rate and mitochondrial iron uptake as well as an increased potential for oxidative damage (40), conditions in which yeast cells are expected to rely heavily on mitochondrial ISC biosynthesis.
FIGURE 1.
LEU2 gene rescues respiratory function of yfh1 mutant yeast. The genotype of each strain is indicated in Table 1. In all cases, yeast cultures were started from freshly streaked frozen stocks, synchronized to late logarithmic phase in SD medium at 30 °C, diluted with fresh SD medium to an A600 of 0.1, and grown for 3 h at which point 100 μm FeCl3 was added to the medium and incubation continued. All cultures reached stationary phase in ∼24 h. After 24 (A, D, E, and F), 48 (B), or 72 (C) h, 10-fold serial dilutions were spotted onto YPE or YPD plates that were subsequently incubated at 30 °C and photographed after 3 (YPD) or 5 (YPE) days. + Leucine denotes cells that were cultured as described above except that 40 mg/liter leucine was added to the SD medium every 12 h throughout growth. The red pigment is due to the ade2–101ochre mutation in the genome and is indicative of mitochondrial respiratory function.
Haploid yeast cells bearing a mutant yfh1Y73A, yfh1V150A, or yfh1L144A allele were transformed with a high copy genomic DNA library (24), and transformants were analyzed for the ability to maintain respiratory function during the stressful stationary phase described above. Over 270 respiratory-competent transformants were isolated. In most cases, the vector responsible for suppression of the respiratory-deficient phenotype carried a genomic fragment containing wild type YFH1, indicating that the screening was suitable to identify genes related to Yfh1p function. Interestingly, ∼12% of the suppressing vectors contained a ∼16-kbp genomic fragment from yeast chromosome III spanning five different genes. Deletion analysis subsequently showed that one of these genes, LEU2, was solely responsible for suppression of the phenotype. The extent of the suppression depended on the yfh1 allele (yfh1L144A > yfh1V150A > yfh1Y73A) and was only partial for the yfh1V150A LEU2 and yfh1Y73A LEU2 mutants compared with YFH1 leu2Δ cells (Fig. 1, A–C).
Functional Leucine Biosynthetic Pathway Is Required for Complementation of yfh1 Mutant Yeast by LEU2
Similar to most laboratory yeast strains, our three yfh1 mutants carried an inactive leu2-Δ1 allele (henceforth designated leu2Δ), which confers leucine auxotrophy (22), whereas LEU1 and all other genes involved in leucine biosynthesis were intact (the genotypes of the strains used in this study are shown in Table 1; in the text and in the figures, strains are denoted by the status of their YFH1 and LEU2 loci only, whereas the status of the LEU1 locus and of additional chromosomal or episomal alleles is indicated as appropriate). In the stressful stationary phase described above, yfh1Y73A leu2Δ, yfh1V150 leu2Δ, and yfh1L144A leu2Δ cells remained viable (i.e. able to resume growth on YPD; Fig. 1, A–C). However, after 24, 48, and 72 h, respectively, yfh1Y73A leu2Δ, yfh1V150 leu2Δ, and yfh1L144A leu2Δ cells lost the ability to respire (i.e. resume growth on YPE; Fig. 1, A–C). At the same time points, yfh1L144A LEU2, yfh1V150A LEU2, and yfh1Y73A LEU2 cells remained at least partially capable of growing on YPE as described above (Fig. 1, A–C).
Additional analyses performed in cells bearing the yfh1Y73A allele showed that the levels of respiratory growth achieved with LEU2 overexpression could be achieved simply by low copy vector expression or chromosomal reintegration of LEU2 (Fig. 1D, compare yfh1Y73A LEU2 (CEN) or yfh1Y73A LEU2 with yfh1Y73A LEU2 (2 μm)). Moreover, the positive effect of LEU2 was abrogated when the LEU1 gene was deleted (Fig. 1, D and E, compare yfh1Y73A LEU2 with yfh1Y73A leu1Δ LEU2). However, addition of excess leucine to the medium during growth (double the standard amount, added fresh every 12 h) was not sufficient to maintain respiration (Fig. 1F, compare yfh1Y73A leu2Δ with yfh1Y73A leu2Δ + Leucine). Thus, complementation of yfh1 mutant cells by LEU2 required a functional leucine biosynthetic pathway but was independent of leucine per se.
Functional Leucine Biosynthetic Pathway Has Opposite Effects on Protein Levels and Enzymatic Activities of Mitochondrial Aco1p and Cytosolic Leu1p
Aco1p is a key enzyme in the mitochondrial tricarboxylic acid cycle important for providing isocitrate for 2-oxoglutarate and glutamate production, and it is also required for mitochondrial DNA integrity (41). Aco1p activity depends on a [4Fe-4S]2+ cluster highly susceptible to oxidant-induced inactivation (42) and thus provides a measure of the mitochondrial ISC biosynthetic capacity (43). In the stressful stationary phase described above, Aco1p activity was strongly reduced in yfh1Y73A leu2Δ cells but increased ∼4-fold in yfh1Y73A LEU2 cells (Fig. 2A, +Fe). LEU2 expression caused a significant increase (∼2-fold) in Aco1p activity even in the presence of wild type YFH1 (Fig. 2A, +Fe, compare YFH1 leu2Δ with YFH1 LEU2). Moreover, when iron was not included in the culture medium, LEU2 expression was sufficient to maintain basal levels of Aco1p activity even in the presence of the yfh1Y73A allele (Fig. 2A, −Fe, compare yfh1Y73A LEU2 with YFH1 LEU2). These data together indicated that Yfh1p and Leu2p independently, although synergistically, promoted Aco1p activity.
FIGURE 2.
LEU2 expression has opposite effects on protein levels and enzymatic activities of mitochondrial Aco1p and cytosolic Leu1p. Strains carrying the YFH1 or yfh1Y73A allele with intact or deleted LEU1 and/or LEU2 alleles were used in these analyses as shown. Cells were grown in SD medium as described in the legend of Fig. 1 with (+Fe) or without (−Fe) iron supplementation. Cells were harvested, and total cell lysates were used to measure enzymatic activities or protein levels as described under “Experimental Procedures.” A, three independent cell lysates were analyzed in triplicate, and the three data sets were analyzed as n = 3 experiments; data shown are mean (bar and number above each bar) ± S.D. For leu1Δ and leu2Δ strains carrying the YFH1 allele, p is 0.014 for Aco1p and 0.012 for Leu1p compared with the corresponding LEU2 strain, with (+Fe) or without (−Fe) iron supplementation as appropriate; for leu1Δ and leu2Δ strains carrying the yfh1Y73A allele, p is 0.004 for both Aco1p and Leu1p compared with the corresponding LEU2 strain, with (+Fe) or without (−Fe) iron supplementation as appropriate; p was determined with Student's t test. B, aliquots of the cell lysates described above (10 μg of total protein) were analyzed by Western blotting using anti-Aco1p polyclonal antiserum or anti-FLAG monoclonal antibody (Sigma) to detect FLAG-tagged Leu1p. Protein bands were quantified by densitometry using ImageQuant software (GE Healthcare), and values were normalized to the signal present in the YFH1 LEU2 (−Fe) (denoted by *) or yfh1Y73A LEU2 (−Fe) strain (denoted by **) as shown. For conditions with (+Fe) or without (−Fe) iron supplementation, n = 4 and n = 2 independent cell lysates, respectively, were analyzed; data shown are mean ± S.D. For Leu1p, ♦ denotes p 0.004 compared with *; ▾ denotes p 0.006 compared with **; for Aco1p, ♦ denotes p 0.003 compared with *; ▾ denotes p 0.015 compared with ** as determined by Student's t test. n.d., not detectable as expected for leu1Δ strains. The panel denoted Total protein shows the same membrane analyzed by Western blotting (upper panels) upon staining with Pierce Reversible Protein Stain to demonstrate equal total protein loading; shown is the region of the membrane corresponding to the molecular mass range of Aco1p (85.4 kDa) and Leu1p (85.8 kDa). mU, milliunits.
Leu1p is a critical enzyme in the leucine biosynthetic pathway that is structurally and functionally related to Aco1p (43). Leu1p activity depends on a [4Fe-4S]2+ cluster highly susceptible to oxidant-induced inactivation and provides a measure of the cytoplasmic ISC biosynthetic capacity (43). The activity of Leu1p was ∼3-fold lower in yfh1Y73A leu2Δ relative to YFH1 leu2Δ cells (Fig. 2A, +Fe). However, opposite to Aco1p activity, Leu1p activity was nearly undetectable upon LEU2 expression, indicating that Leu1p activity was abnormally enhanced in the leu2Δ deletion mutant. This was the case in the presence of either yfh1Y73A or YFH1 and was independent of iron supplementation (Fig. 2A, +Fe or −Fe, compare yfh1Y73A leu2Δ with yfh1Y73A LEU2 and compare YFH1 leu2Δ with YFH1 LEU2).
The upward changes in Aco1p activity associated with LEU2 expression correlated with upward changes in Aco1p protein levels, which were more evident in the yfh1Y73A background compared with the YFH1 background and were independent of iron supplementation (Fig. 2B, −Fe or +Fe). In both backgrounds, the increase in Aco1p activity was more pronounced than the increase in protein levels (Fig. 2, A versus B), suggesting that LEU2 expression influenced not only Aco1p protein levels but also the fraction of enzymatically active Aco1p. Conversely, the downward changes in Leu1p activity associated with LEU2 expression correlated with downward changes in Leu1p protein levels and were independent of iron supplementation (Fig. 2B, −Fe or +Fe), indicating that both Leu1p activity and protein expression levels were abnormally enhanced in the leu2Δ deletion mutant. The reduction in Leu1p activity was significantly more pronounced than the apparent reduction in Leu1p protein levels (Fig. 2, A versus B). For example, in yfh1Y73A LEU2 cells, Leu1p activity decreased >10-fold (Fig. 2A), whereas Leu1p protein levels decreased only ∼2-fold (Fig. 2B) as compared with yfh1Y73A leu2Δ cells. Defects in leucine biosynthesis had been shown previously to result in up-regulation of most enzymes in the pathway including Leu1p (13). Our results further indicated that LEU2 expression not only down-regulated Leu1p protein levels but also limited the fraction of enzymatically active Leu1p. It is important to emphasize that the low levels of Leu1p protein and activity present in LEU2 strains reflect the normal phenotype; in contrast, the increased expression and activity of Leu1p present in leu2Δ strains is a mutant phenotype. In agreement with the data shown in Fig. 1, D and E, the positive effect of LEU2 expression on Aco1p activity was no longer observed upon deletion of the LEU1 gene (Fig. 2, A and B, compare LEU2 strains with leu1Δ LEU2 strains).
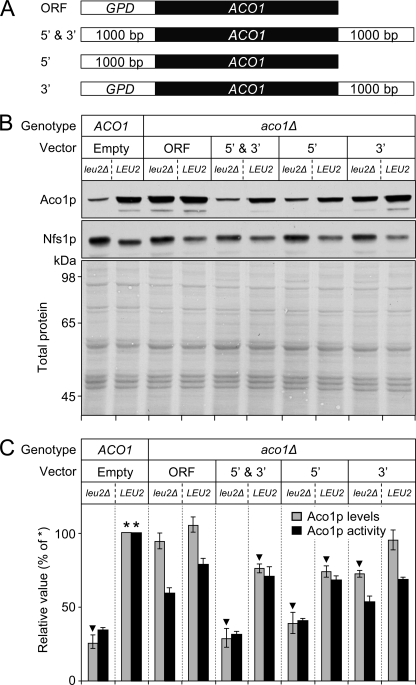
5′ DNA Flanking Region of ACO1 Mediates Aco1p Down-regulation in leu2Δ Cells
Leu1p was consistently up-regulated in leu2Δ versus LEU2 cells (Fig. 2B, −Fe or +Fe), which most likely resulted from accumulation of α-IPM in leu2Δ cells, making Leu3p function as a transcriptional activator of the LEU1 gene (15). Accordingly, this effect was no longer observed when LEU3 was deleted (data not shown). We investigated the possibility that the down-regulation of Aco1p protein levels observed in leu2Δ cells might also be transcriptionally driven. We measured ACO1 mRNA levels by quantitative RT-PCR in LEU2 and leu2Δ strains after 24 or 48 h of growth under the conditions used in Fig. 2, A and B. After 24 h, ACO1 mRNA was reduced ∼1.4-fold in the leu2Δ versus LEU2 background; this difference was no longer present at 48 h, the time point at which Aco1p protein levels were measured in Fig. 2B (data not shown). Next, we made plasmids carrying different ACO1 alleles (Fig. 3A), expressed them in the LEU2 or leu2Δ background upon deletion of the endogenous ACO1 gene, and measured Aco1p protein levels and activity (Fig. 3, B and C). Cells bearing a vector with ACO1 plus its upstream and downstream flanking regions (Fig. 3B, 5′ & 3′) expressed levels of Aco1p protein close to those present in cells with chromosomal ACO1 (Fig. 3B, Empty). In both of these strains, Aco1p protein levels increased over 2-fold in the LEU2 background compared with the leu2Δ background. The use of plasmids with ACO1 plus either its upstream or downstream flanking region demonstrated that the upstream region was largely responsible for the down-regulation of Aco1p protein levels in the leu2Δ background (Fig. 3B, 5′ versus 3′). Accordingly, with a construct carrying only the ACO1 open reading frame under the control of the constitutive glyceraldehyde-3-phophate dehydrogenase (GPD) promoter, we observed equivalent levels of Aco1p expression in the leu2Δ and LEU2 backgrounds (Fig. 3B, ORF). In all cases, Aco1p activity showed good correlation with Aco1p protein levels (Fig. 3C). As a control, Nfs1p expression was analyzed in parallel and was consistently down-regulated in the LEU2 versus the leu2Δ background, opposite to what was observed for Aco1p (Fig. 3B).
FIGURE 3.
5′-Flanking region of ACO1 mediates Aco1p down-regulation in leu2Δ cells. A, schematic representation of the constructs used in this analysis. ORF denotes a construct in which the ACO1 coding sequence was under the control of the GPD promoter; 5′ & 3′ denotes a constructs in which the ACO1 coding sequence was under the control of the natural upstream and downstream regions (1,000 bp each), 5′ denotes a construct lacking the downstream region, and 3′ denotes a construct in which the GPD promoter replaced the upstream region. B and C, strains carrying the yfh1Y73A allele with an intact LEU2 or a deleted leu2Δ allele were used in these analyses. These strains were transformed with centromeric pRS416 vectors carrying the constructs described above after which the endogenous ACO1 gene was deleted. “Empty” denotes strains carrying an intact chromosomal ACO1 gene and an empty pRS416 vector. Cells were grown in SD medium as described in the legend of Fig. 1 with 100 μm iron supplementation. B, cells were harvested, and total cell lysates were used to measure Aco1p or Nfs1p protein levels by Western blotting with specific antibodies (21). After Western blotting, the membrane was stained to verify equal protein loading; shown is the region of stained membrane encompassing the molecular mass range of Aco1p (85.4 kDa) and Nfs1p (54.5 kDa) (panel denoted Total protein). C, Aco1p protein levels were determined by densitometry and normalized to the levels present in the yfh1Y73A LEU2 strain transformed with empty pRS416 vector (denoted by *). For each of the indicated strains, Aco1p protein levels were determined from three independently prepared cell lysates and one or two separate Western blots per lysate. The three data sets were analyzed as n = 3 experiments; data shown are mean ± S.D. ▾ denotes p 0.005 compared with *. Aco1p activity was measured in triplicate in one of the three lysates to demonstrate good agreement with protein levels; values were normalized as above.
Leu3p recognizes a consensus CCG(N)4CCG sequence in genes involved in leucine synthesis (44, 45). Analysis of the ACO1 upstream flanking region revealed a putative Leu3p binding site (CCGAAATCGG; 312 bases upstream of the ATG codon). We analyzed the effects of mutations in this putative site by use of a reporter construct in which 1 kbp of the ACO1 upstream flanking region was fused to the β-galactosidase coding sequence (28). As compared with a construct carrying the wild type sequence (CCGAAATCGG), constructs carrying modified sequence (44) (TCGAAATCGG or AAGAAATCAA) yielded equally up- and down-regulated levels of β-galactosidase activity in the LEU2 and leu2Δ background, respectively (data not shown). This suggested that Leu3p was probably not involved in the transcriptional regulation of ACO1. Similarly, an earlier study did not identify ACO1 as a target of Leu3p regulation using DNA microarray analysis (46).
Defects in Leucine Biosynthesis Up-regulate Core Machinery for Mitochondrial ISC Assembly
The first step in ISC assembly in the mitochondrial matrix involves a sulfur donor (Nfs1p-Isd11p complex), an iron donor (Yfh1p), and a protein scaffold (Isu1p) upon which a [2Fe-2S] cluster is assembled (for a review, see Ref. 2). In the studies described above, we found that LEU2 acted as a suppressor of mutations in Yfh1p under conditions predicted to challenge the mitochondrial ISC biosynthetic capacity. Under the same conditions, we analyzed by Western blotting the levels of Nfs1p, Isu1p, and Yfh1p in strains carrying the YFH1 or yfh1Y73A allele together with deletions of the LEU1 and/or LEU2 gene. We found that the protein levels of Nfs1p, Isu1p, and Yfh1p were consistently higher in leucine pathway-deficient mutants (leu1Δ, leu2Δ, or combinations thereof) compared with LEU1 LEU2 cells (Fig. 4A).
FIGURE 4.
Defects in leucine biosynthesis up-regulate core machinery for mitochondrial ISC assembly. A–D, strains carrying the YFH1 or yfh1Y73A allele with intact or deleted LEU1 and/or LEU2 alleles were used in these analyses as shown. A and C, cells were grown for 48 h as described in the legend of Fig. 1 without iron supplementation, and cell lysates were analyzed by Western blotting using 10 μg of total protein for detection of Nfs1p and 40 μg for detection of Isu1p or Yfh1p. B, protein bands were quantified by densitometry, and values were normalized to the signal present in the YFH1 LEU2 (denoted by *) or yfh1Y73A LEU2 (denoted by **) strain as appropriate. For each of the indicated strains, protein levels were determined from three independently prepared cell lysates and one Western blot per lysate and analyzed as n = 3 experiments; data shown are mean ± S.D. ♦ denotes p 0.037 compared with *; ▾ denotes p 0.045 compared with **. C, after Western blotting, membranes were stained to verify equal protein loading; shown is the region of two stained membranes encompassing the molecular mass ranges of Nfs1p (54.5 kDa) (top) and Yfh1p (13.7 kDa; migrates close to ∼20 kDa) and Isu1p (14.9 kDa) (bottom). D, the indicated strains were grown as described above, and mRNA levels were determined by RT-PCR as described under “Experimental Procedures.” NFS1, ISU1, and YFH1 or yfh1Y73A mRNA levels were normalized to ACT1 mRNA. Data shown are mean ± S.D. of three independent experiments. For NFS1 mRNA levels, ♦ denotes p 0.02 compared with the YFH1 LEU2 strain. No other statistically significant differences were observed by Student's t test analysis of NFS1, ISU1, and YFH1 or yfh1Y73A mRNA levels.
To investigate the mechanism of Nfs1p and Isu1p regulation, we measured the levels of NFS1 and ISU1 mRNA in the LEU2 and leu2Δ backgrounds under the same conditions used to analyze Nfs1p and Isu1p protein levels. Because protein levels were measured after 48 h of growth, mRNA levels were measured after 24 and 48 h of growth. In both LEU2 and leu2Δ strains, NFS1 and ISU1 mRNA levels were progressively up-regulated over the course of the experiment; this up-regulation was especially evident in the yfh1Y73A background (Fig. 4D). At 24 or 48 h, NFS1 and ISU1 mRNA levels were not significantly different between LEU2 and leu2Δ strains except in the case of the YFH1 LEU2 strain, which contained ∼2-fold higher levels of NFS1 mRNA compared with the YFH1 leu2Δ strain (Fig. 4D). We also measured the levels of YFH1 or yfh1Y73A mRNA. This was of interest because expression of the YFH1 allele was driven by its natural promoter, whereas expression of the yfh1Y73A allele was driven by the GPD promoter (21). Similar to NFS1 and ISU1 mRNAs, YFH1 and yfhY73A mRNAs were progressively up-regulated over the course of the experiment, and at both time points analyzed, they were not significantly different between LEU2 and leu2Δ strains (Fig. 4D). Together, these data suggested that the higher levels of Nfs1p, Isu1p, and Yfh1p observed in the leu2Δ background resulted from a combination of transcriptional and post-transcriptional effects. Although transcriptional effects were present in both LEU2 and leu2Δ cells, post-transcriptional mechanisms were most active in leu2Δ cells.
Leu1p Activity Is Not Influenced by Atm1p Overexpression or Depletion
The data so far suggested that defects in leucine biosynthesis resulted in down-regulation of Aco1p expression and up-regulation of mitochondrial ISC assembly proteins and that these effects, coupled with up-regulation of Leu1p expression, enabled the dramatic increase in Leu1p activity observed in leu2Δ cells. Based on previous reports (9, 20), the mitochondrial inner membrane transporter Atm1p appeared to be a likely link between these mitochondrial and cytosolic effects. To test this, we compared LEU2 and leu2Δ cells in which ATM1 gene expression was driven by its natural promoter or by the glucose-repressible GAL10 promoter (Gal-ATM1) as described previously by others (9, 47). In these previous studies, LEU2 had been used as a selectable marker to create a Gal-ATM1 strain, and LEU2 Gal-ATM1 cells had been compared with leu2Δ ATM1 controls. We instead used a KanMX4 cassette to create both a LEU2 Gal-ATM1 and a leu2Δ Gal-ATM1 strain and compared these two strains with a LEU2 ATM1 or a leu2Δ ATM1 strain, respectively. In all cases, Atm1p was expressed with a C-terminal FLAG tag and was detected by Western blotting using a monoclonal anti-FLAG antibody.
Growing Gal-ATM1 cells for 16 h in synthetic minimal medium containing galactose resulted in >20-fold higher levels of Atm1p compared with ATM1 cells without obvious differences between the LEU2 and leu2Δ backgrounds (Fig. 5A, Galactose 16 h, lanes 3 and 4 versus lanes 1 and 2). The levels of Leu1p varied according to the LEU2 or leu2Δ background as observed above in a manner that appeared to be independent of Atm1p levels (Fig. 5A, Galactose 16 h, lanes 1 and 3 versus lanes 2 and 4). Aco1p levels were otherwise similar between the LEU2 and leu2Δ backgrounds (Fig. 5A, Galactose 16 h, lanes 1 and 3 versus lanes 2 and 4), suggesting that the down-regulation associated with leu2Δ status was not operational during growth in galactose. As observed for Leu1p, Aco1p levels appeared to be independent of changes in Atm1p levels (Fig. 5A, Galactose 16 h). We used cells analyzed as described above to measure enzyme activities. As observed in Fig. 2A, Leu1p activity decreased severalfold in the LEU2 versus the leu2Δ background (Fig. 5B, Galactose 16 h, lanes 3 and 4 versus lanes 1 and 2). Interestingly, LEU2 Gal-ATM1 and LEU2 ATM1 cells exhibited similarly low levels of Leu1p activity (Fig. 5B, Galactose 16 h, lanes 4 and 2; p > 0.3). In addition, Leu1p activity was >50% lower in leu2Δ Gal-ATM1 versus leu2Δ ATM1 cells (Fig. 5B, Galactose 16 h, lanes 1 and 3; p < 0.003). These data together indicated that Leu1p activity was not enhanced by overexpression of Atm1p. Robust Aco1p activity was present in all cases, and small differences among the four strains appeared to be largely independent of Atm1p levels (Fig. 5B, Galactose 16 h, lanes 1–4).
FIGURE 5.
Leu1p activity is independent of Atm1p overexpression or depletion. A and B, in these analyses, the LEU1-FLAG LEU2-FLAG Gal-ATM1 and LEU1-FLAG leu2Δ Gal-ATM1 strains were compared, respectively, with the LEU1-FLAG LEU2-FLAG ATM1 and LEU1-FLAG leu2Δ ATM1 strains. In all strains, the C terminus of both Atm1p and Leu1p was FLAG-tagged (see “Experimental Procedures” and Table 1 for details). The strains were grown in synthetic minimal medium containing galactose for 16 h (to induce Atm1p expression in the Gal-ATM1 strains; Galactose 16 h) or dextrose for 48 h (to deplete Atm1p in the Gal-ATM1 strains; Dextrose 48 h). Protein levels were analyzed in total cell lysates (10 μg of total protein per sample) by Western blotting with anti-FLAG antibody (for detection of Atm1p and Leu1p) or anti-Aco1p antibody. Atm1p is detected as a diffuse band as observed previously by others (12, 51). The panel denoted Total protein shows the same membrane analyzed by Western blotting upon staining with Pierce Reversible Protein Stain to demonstrate equal total protein loading; shown is the region of the membrane corresponding to the molecular mass range of Atm1p (77.5 kDa), Leu1p (85.8 kDa), and Aco1p (85.4 kDa). B, Leu1p and Aco1p activities were measured in triplicate in two independently prepared cell lysates. The two data sets were analyzed as n = 2 experiments; data shown are mean (bar and number above each bar) ± S.D. p values are provided under “Results.” mU, milliunits.
Growing Gal-ATM1 cells for 48 h on synthetic minimal medium containing dextrose, thereby repressing the pGAL10 promoter (9), caused depletion of Atm1p to the point that it became undetectable by Western blotting (Fig. 5A, Dextrose 48 h, lanes 7 and 8). Under the same conditions, discrete levels of Atm1p were otherwise present in ATM1 cells (Fig. 5A, Dextrose 48 h, lanes 5 and 6). The levels of Leu1p and Aco1p varied in opposite directions according to LEU2 or leu2Δ status (Fig. 5A, Dextrose 48 h) as observed in Fig. 2B. Accordingly, Leu1p activity decreased severalfold in the LEU2 versus the leu2Δ background, whereas Aco1p activity behaved in the opposite manner (Fig. 5B, Dextrose 48 h; p 0.023 for Leu1p and p 0.014 for Aco1p). All of these changes appeared to be largely independent of changes in Atm1p levels. There was a ∼30% reduction in Leu1p activity in leu2Δ Gal-ATM1 (i.e. Atm1p-depleted) versus leu2Δ ATM1 (i.e. Atm1p-repleted) cells (Fig. 5B, Dextrose 48 h, lane 7 versus lane 5). However, Leu1p activity in leu2Δ Gal-ATM1 cells was ∼1.4-fold higher during growth in dextrose than during growth in galactose (Fig. 5B, lane 7 versus lane 3; p < 0.014), whereas Atm1p levels were undetectable during growth in dextrose and highly induced during growth in galactose (Fig. 5A, lanes 7 and 8 versus lanes 3 and 4). Therefore, the ∼30% reduction in Leu1p activity noted above could not be attributed to Atm1p depletion. These data were in contrast with the general belief that Atm1p is required to maintain Leu1p activity via export of an ISC precursor (9, 10). However, we noted that in these previous studies LEU2 had been used as selectable marker to create Atm1p-depleted cells, whereas a leu2Δ allele was present in Atm1p-repleted cells. Thus, the drastic reduction in Leu1p activity purportedly associated with Atm1p depletion (9, 10) was the normal phenotype associated with LEU2 expression as seen in Fig. 5B.
DISCUSSION
We conducted a screening for genes that could suppress the respiratory-deficient phenotype associated with point mutations in Yfh1p, the iron donor for mitochondrial ISC assembly (6, 33). The phenotype was elicited by stressful conditions expected to increase cellular demand for ISC synthesis. Under such conditions, the LEU2 gene was isolated as a suppressor at high frequency in three different yfh1 mutants. This led to the realization that the leu2Δ allele (present in our strain as in most laboratory yeast strains) caused cytoplasmic and mitochondrial effects that acted synergistically with the yfh1 mutant alleles used in the screening. In particular, we found that an incomplete leucine biosynthetic pathway profoundly altered the biogenesis and function of at least two [4Fe-4S] enzymes, Aco1p and Leu1p, localized to the mitochondrial matrix and the cytoplasm, respectively. Aco1p is a mitochondrial matrix enzyme structurally and functionally related to Leu1p. Both Aco1p and Leu1p depend on a [4Fe-4S]2+ cluster highly susceptible to oxidant-induced inactivation (9, 42). These two abundant enzymes are often used as representative markers of ISC enzyme biogenesis in their respective cellular compartments (9, 43). Leu1p activity is also used as a marker of ISC (or ISC precursor) transport from the mitochondria to the cytoplasm by Atm1p. Therefore, the focus on these two enzymes seemed a reasonable starting point to address how the leucine biosynthetic pathway might regulate the distribution of ISCs between the mitochondria and the cytoplasm. The observations that upward changes in Leu1p protein levels and enzymatic activity occurred in leu2Δ cells and that they were associated with opposite changes in Aco1p levels and activity led us to hypothesize mechanisms that promote leucine biosynthesis at the expense of Aco1p activity.
Activation of Leu3p by α-IPM followed by Leu3p-induced expression of LEU1 (as well as other genes in the leucine biosynthetic pathway) is a well characterized mechanism triggered by blocks in the pathway (e.g. leu2Δ deletion mutation) (15). Our data further indicate that the down-regulation of Aco1p protein levels in leu2Δ versus LEU2 cells was mediated by transcriptional mechanisms involving the 5′-flanking region of ACO1. Mutational analysis of a putative Leu3p consensus sequence (44, 45) within this region excluded a possible direct role of Leu3p; and thus, the mechanism that couples Aco1p down-regulation with Leu1p up-regulation remains to be established.
The observation that Nfs1p protein levels were consistently higher in leu2Δ versus LEU2 cells prompted us to extend our analyses to the three core components of the mitochondrial ISC biosynthetic machinery: Nfs1p, Isu1p, and Yfh1p. The levels of all three proteins were consistently higher in YFH1 or yfh1 cells when LEU1, LEU2, or both were deleted. At least two enzymes in the leucine biosynthetic pathway, mitochondrial Ilv3p and cytoplasmic Leu1p, are ISC-dependent enzymes. Thus, up-regulation of Nfs1p, Isu1p, and Yfh1p may be part of the cellular response to blocks in this pathway. Together, our analysis of mRNA and protein levels suggests that the up-regulation of Nfs1p, Isu1p, and Yfh1p observed in the leu2Δ background resulted from a combination of transcriptional and post-transcriptional effects. Although transcriptional effects (i.e. transcriptional activation of NFS1 and ISU1) were present in both LEU2 and leu2Δ cells, post-transcriptional mechanisms were most active in leu2Δ cells. The latter mechanisms may include mRNA stabilization, protein stabilization, or both. Up-regulation of Isu1p through a reduction in protein turnover rate has been reported previously by others (48). The up-regulation of Yfh1p observed in leu2Δ cells was probably the result of both mRNA up-regulation (likely through mRNA stabilization as the YFH1 and yfh1Y73A alleles were under the control of different promoters (21)) and protein stabilization (suggested by the fact that mRNA levels were equally up-regulated in LEU2 and leu2Δ cells but protein levels were up-regulated only in the latter). Because Nfs1p, Isu1p, and Yfh1p up-regulation was observed in both leu1Δ and leu2Δ cells, Leu1p and Leu2p can be excluded from being directly involved. Factor(s) present in both leu1Δ and leu2Δ cells, possibly factors that modulate mRNA stability and/or protein turnover, might be responsible. We could not find a Leu3p consensus sequence in the upstream regions of NFS1, ISU1, or YFH1, and accordingly, the levels of their corresponding proteins were not significantly affected by the presence or absence of LEU3 (data not shown).
Previous studies that implicated Atm1p in the maintenance of Leu1p activity had compared LEU2 Gal-ATM1 cells with leu2Δ ATM1 controls (9, 47). We instead used a KanMX4 cassette to create both a LEU2 Gal-ATM1 strain and a leu2Δ Gal-ATM1 strain and compared these two strains with a LEU2 ATM1 or leu2Δ ATM1 strain, respectively. We found that Aco1p activity was not significantly influenced by Atm1p overexpression or depletion. More surprisingly, we found that Leu1p activity was also largely independent of the levels of Atm1p. This leads us to conclude that previous studies that linked Atm1p function to the maintenance of Leu1p activity were confounded by the fact that LEU2 was used as selectable marker to create Atm1p-depleted cells, whereas a leu2Δ allele was present in Atm1p-repleted controls. We further suggest that Atm1p may not be involved in ISC export and is perhaps involved in the export of heme or intermediates in heme biosynthesis as suggested previously by others (49, 50).
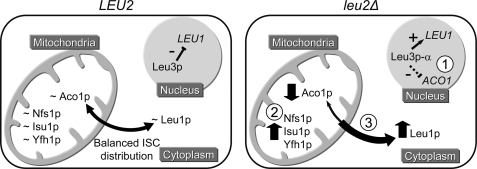
A unifying model for the findings discussed above is proposed in Fig. 6. The model highlights the complexity of the mechanisms that appear to regulate the distribution of ISCs between the mitochondria and other cellular compartments. Among a number of open questions, perhaps the most important will be to define whether mitochondria provide ISCs for cytoplasmic ISC synthesis via an as yet undefined mitochondrial transport mechanism or whether each compartment synthesizes its own ISC pool and this is regulated according to the metabolic needs of the cell.
FIGURE 6.
Complex mechanisms regulate ISC distribution between mitochondria and cytoplasm in leu2Δ background. The sign ∼ denotes normal protein levels; upward and downward arrows denote increased or decreased protein levels; + and − denote transcriptional activation and repression; Leu3p-α denotes α-IPM-activated Leu3p. In wild type (LEU2) cells, there is a balanced distribution of ISCs between the mitochondria and the cytoplasm. In leu2Δ cells, accumulation of α-IPM makes Leu3p function as a transcriptional activator of the LEU1 gene (15), leading to up-regulation of Leu1p protein levels (as observed in Fig. 2). Leu3p may also be indirectly involved in the transcriptional repression of ACO1, leading to down-regulation of Aco1p protein levels (as observed in Figs. 2 and 3). Leu1p up-regulation together with Aco1p down-regulation shifts the mitochondrial-cytoplasmic ISC balance in a manner that promotes synthesis of enzymatically active Leu1p at the expense of enzymatically active Aco1p. Mostly post-transcriptional mechanisms lead to up-regulation of Nfs1p, Isu1p, and Yfh1p in leu2Δ cells (as observed in Fig. 4). This may reflect a direct role of mitochondria in providing ISCs (or ISC precursors) for Leu1p biogenesis as proposed previously (9) although via an as yet unknown transporter different from Atm1p; alternatively, it could simply represent a secondary response to the loss of Aco1p activity. Open questions that remain to be addressed include the following. 1) What are the factors that mediate ACO1 down-regulation in leu2Δ cells? 2) What are the mechanisms involved in post-transcriptional up-regulation of Nfs1p, Isu1p, and Yfh1p in leu2Δ cells? 3) Are mitochondria directly or indirectly involved in providing ISCs or ISC precursors to Leu1p and other cellular Fe-S enzymes?
Acknowledgments
We thank Dr. David Katzmann and Mrs. Johanna Payne, Mayo Clinic Department of Biochemistry and Molecular Biology, for their generous help with generation of some yeast strains, and Dr. Nicholas F. LaRusso, Mayo Clinic Center for Basic Research in Digestive Diseases, for sharing laboratory equipment in the quantitative RT-PCR experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant AG15709 (to G. I.). This work was also supported by grants from the Friedreich Ataxia Research Alliance and Muscular Dystrophy Association (to G. I.) and the Mayo Clinic Department of Pediatric and Adolescent Medicine (to T. B. and H. L.).
- ISC
- Fe-S cluster
- α-IPM
- α-isopropylmalate
- fwd
- forward
- rev
- reverse
- ABC
- ATP binding cassette.
REFERENCES
- 1. Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 2. Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 3. Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fontecave M., Ollagnier-de-Choudens S. (2008) Arch. Biochem. Biophys. 474, 226–237 [DOI] [PubMed] [Google Scholar]
- 5. Gakh O., Bedekovics T., Duncan S. F., Smith D. Y., 4th, Berkholz D. S., Isaya G. (2010) J. Biol. Chem. 285, 38486–38501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li H., Gakh O., Smith D. Y., 4th, Isaya G. (2009) J. Biol. Chem. 284, 21971–21980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mühlenhoff U., Gerber J., Richhardt N., Lill R. (2003) EMBO J. 22, 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schilke B., Voisine C., Beinert H., Craig E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lill R. (2009) Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 11. Cavadini P., Biasiotto G., Poli M., Levi S., Verardi R., Zanella I., Derosas M., Ingrassia R., Corrado M., Arosio P. (2007) Blood 109, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 12. Chloupková M., Reaves S. K., LeBard L. M., Koeller D. M. (2004) FEBS Lett. 569, 65–69 [DOI] [PubMed] [Google Scholar]
- 13. Kohlhaw G. B. (2003) Microbiol. Mol. Biol. Rev. 67, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prohl C., Kispal G., Lill R. (2000) Methods Enzymol. 324, 365–375 [DOI] [PubMed] [Google Scholar]
- 15. Sze J. Y., Woontner M., Jaehning J. A., Kohlhaw G. B. (1992) Science 258, 1143–1145 [DOI] [PubMed] [Google Scholar]
- 16. Gerber J., Neumann K., Prohl C., Mühlenhoff U., Lill R. (2004) Mol. Cell. Biol. 24, 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lange H., Kaut A., Kispal G., Lill R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mühlenhoff U., Richhardt N., Ristow M., Kispal G., Lill R. (2002) Hum. Mol. Genet. 11, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 19. Sipos K., Lange H., Fekete Z., Ullmann P., Lill R., Kispal G. (2002) J. Biol. Chem. 277, 26944–26949 [DOI] [PubMed] [Google Scholar]
- 20. Balk J., Aguilar Netz D. J., Tepper K., Pierik A. J., Lill R. (2005) Mol. Cell. Biol. 25, 10833–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gakh O., Smith D. Y., 4th, Isaya G. (2008) J. Biol. Chem. 283, 31500–31510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mumberg D., Müller R., Funk M. (1994) Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlson M., Botstein D. (1982) Cell 28, 145–154 [DOI] [PubMed] [Google Scholar]
- 25. Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. (1993) Nucleic Acids Res. 21, 3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 27. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) Nucleic Acids Res. 30, e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guarente L. (1983) Methods Enzymol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 29. Daum G., Böhni P. C., Schatz G. (1982) J. Biol. Chem. 257, 13028–13033 [PubMed] [Google Scholar]
- 30. Gardner P. R., Nguyen D. D., White C. W. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12248–12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fansler B., Lowenstein J. M. (1969) Methods Enzymol. 13, 26–30 [Google Scholar]
- 32. Kohlhaw G. B. (1988) Methods Enzymol. 166, 423–429 [DOI] [PubMed] [Google Scholar]
- 33. Gerber J., Mühlenhoff U., Lill R. (2003) EMBO Rep. 4, 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Campanella A., Isaya G., O'Neill H. A., Santambrogio P., Cozzi A., Arosio P., Levi S. (2004) Hum. Mol. Genet. 13, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 35. Chen O. S., Kaplan J. (2000) J. Biol. Chem. 275, 7626–7632 [DOI] [PubMed] [Google Scholar]
- 36. Desmyter L., Dewaele S., Reekmans R., Nystrom T., Contreras R., Chen C. (2004) Exp. Gerontol. 39, 707–715 [DOI] [PubMed] [Google Scholar]
- 37. Foury F., Roganti T. (2002) J. Biol. Chem. 277, 24475–24483 [DOI] [PubMed] [Google Scholar]
- 38. Foury F., Pastore A., Trincal M. (2007) EMBO Rep. 8, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gakh O., Park S., Liu G., Macomber L., Imlay J. A., Ferreira G. C., Isaya G. (2006) Hum. Mol. Genet. 15, 467–479 [DOI] [PubMed] [Google Scholar]
- 40. Fabrizio P., Longo V. D. (2007) Methods Mol. Biol. 371, 89–95 [DOI] [PubMed] [Google Scholar]
- 41. Chen X. J., Wang X., Kaufman B. A., Butow R. A. (2005) Science 307, 714–717 [DOI] [PubMed] [Google Scholar]
- 42. Gardner P. R. (2002) Methods Enzymol. 349, 9–23 [DOI] [PubMed] [Google Scholar]
- 43. Kispal G., Csere P., Guiard B., Lill R. (1997) FEBS Lett. 418, 346–350 [DOI] [PubMed] [Google Scholar]
- 44. Hellauer K., Rochon M. H., Turcotte B. (1996) Mol. Cell. Biol. 16, 6096–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu X., Lee C. K., Granek J. A., Clarke N. D., Lieb J. D. (2006) Genome Res. 16, 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boer V. M., Daran J. M., Almering M. J., de Winde J. H., Pronk J. T. (2005) FEMS Yeast Res. 5, 885–897 [DOI] [PubMed] [Google Scholar]
- 47. Miao R., Kim H., Koppolu U. M., Ellis E. A., Scott R. A., Lindahl P. A. (2009) Biochemistry 48, 9556–9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andrew A. J., Song J. Y., Schilke B., Craig E. A. (2008) Mol. Biol. Cell 19, 5259–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leighton J., Schatz G. (1995) EMBO J. 14, 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ye H., Rouault T. A. (2010) Biochemistry 49, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuhnke G., Neumann K., Mühlenhoff U., Lill R. (2006) Mol. Membr. Biol. 23, 173–184 [DOI] [PubMed] [Google Scholar]