Background: The enzymatic activities catalyzing disassembly of promoter nucleosomes in vivo are unknown.
Results: We show that nucleosomes are lost from activated PHO8 gene and promoter circles, formed in vivo, in an SWI/SNF-dependent manner.
Conclusion: SWI/SNF plays a role in nucleosome disassembly in vivo.
Significance: Our findings are the first demonstration of nucleosome disassembly in vivo dependent on a known chromatin-remodeling enzyme.
Keywords: Chromatin Remodeling, Gene Expression, Gene Regulation, Mathematical Modeling, Nucleosome, PHO5, PHO8, SWI/SNF
Abstract
Analysis of in vivo chromatin remodeling at the PHO5 promoter of yeast led to the conclusion that remodeling removes nucleosomes from the promoter by disassembly rather than sliding away from the promoter. The catalytic activities required for nucleosome disassembly remain unknown. Transcriptional activation of the yeast PHO8 gene was found to depend on the chromatin-remodeling complex SWI/SNF, whereas activation of PHO5 was not. Here, we show that PHO8 gene circles formed in vivo lose nucleosomes upon PHO8 induction, indicative of nucleosome removal by disassembly. Our quantitative analysis of expression noise and chromatin-remodeling data indicates that the dynamics of continual nucleosome removal and reformation at the activated promoters of PHO5 and PHO8 are closely similar. In contrast to PHO5, however, activator-stimulated transcription of PHO8 appears to be limited mostly to the acceleration of promoter nucleosome disassembly with little or no acceleration of promoter transitions following nucleosome disassembly, accounting for the markedly lower expression level of PHO8.
Introduction
Nucleosomes occlude transcription factor binding sites and inhibit the initiation of transcription in vitro and in vivo (1–5). Transcription therefore requires changes in the structure of nucleosomes that occupy essential promoter sequences. The isolation of enzymes that can break and reform histone-DNA interactions in an ATP-dependent manner provided a first answer to the question of how cells overcome nucleosomal inhibition (6).
In vitro experiments demonstrated the ability of chromatin remodelers to couple ATP hydrolysis to the sliding of nucleosomes along the DNA (7, 8), and evidence has been presented for nucleosome sliding in vivo (9). Although the activity of some remodelers appears to be limited to sliding, others, including SWI/SNF and its close relative RSC, were also found to catalyze the transition of mononucleosomes into a persistently altered state with increased DNA accessibility but without loss of histones (10–12), the transfer of histone octamers between DNA molecules (13, 14), and nucleosome disassembly (15, 16). Which of these activities are physiologically significant, rather than owed to the biochemical assay conditions, is unclear.
Extensive structural analysis of in vivo chromatin remodeling at the PHO5 promoter of Saccharomyces cerevisiae, which undergoes a major transition in chromatin structure upon PHO5 induction (17), suggested that remodeling results in loss of promoter nucleosomes. Loss was inferred from complementary quantitative results of limiting nuclease digestion analysis and topology measurements (18). Conforming with the loss hypothesis, regions occupied by nucleosomes under repressing conditions sedimented like naked DNA in a density gradient after release from the activated promoter by restriction endonuclease digestion, and chromatin immunoprecipitation experiments suggested that fewer histones were bound to the activated promoter than the repressed one (18, 19).
Chromatin circles formed in vivo were seen to lose nucleosomes under conditions that activate PHO5 when encompassing the PHO5 promoter, demonstrating the ability of the cell to catalyze nucleosome disassembly and suggesting that loss of promoter nucleosomes at the chromosomal PHO5 locus is due to disassembly rather than sliding of nucleosomes away from the promoter (20).
The SWI/SNF complex has been implicated in histone eviction from promoter elements in vivo (21–23). Whether SWI/SNF-dependent eviction occurred by disassembly or sliding of nucleosomes is unknown. Deletion of SNF2, which encodes the catalytic subunit of the SWI/SNF complex, was found to affect expression and promoter chromatin remodeling of PHO5 only marginally. Defects in PHO5 expression have been reported in the absence of other remodelers, but none of them alone proved to be essential for PHO5 activation (24–26). The catalytic activities that promote nucleosome disassembly in vivo remain to be determined.
In contrast, activated expression of the PHO8 gene of yeast, although induced by the same signaling pathway and transcriptional activator as PHO5, has been shown to be dependent on SNF2 (27). The structural nature of remodeled chromatin at the PHO8 promoter and the question of whether nucleosome loss occurs by disassembly rather than sliding are, therefore, of particular interest.
It has been argued previously that loss of nucleosomes from the PHO8 promoter was due to disassembly rather than sliding because expression and chromatin remodeling of PHO8 were found to require the histone chaperone Asf1 (28), which earlier had been implicated in replication-dependent nucleosome assembly (29). This argument implicitly assumed that Asf1 facilitates nucleosome disassembly. However, independent evidence to support this assumption has not been presented, and subsequent biochemical tests failed to provide such evidence (16).
Here, we addressed the question of whether nucleosomes are removed from the induced PHO8 promoter by disassembly rather than sliding. Our findings are consistent with removal of nucleosomes by disassembly, but not sliding. We observed that Snf2 was essential for disassembly of promoter nucleosomes at PHO8. Together with recent biochemical analyses, our findings support the notion that SWI/SNF catalyzes nucleosome disassembly in vivo.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
For a complete list of strains used in this study, see supplemental Table 1. The PHO8 gene, including 1500 bp upstream of the start codon and 710 bp downstream of the stop codon, was cloned by PCR and inserted into the NotI site of pM47.1 (18), generating plasmid pM75.2. Introduction of an NruI site in place of the EcoRI site most proximal to the 5′-end of the PHO8 open reading frame (ORF) yielded plasmid pM77.2. Deletion of a 2.93-kb NruI fragment from plasmid pM77.2, encompassing the PHO8 promoter and ORF, and its replacement with a 1.1-kb fragment bearing the URA3 gene generated plasmid pM78.5. The PHO8 gene circle plasmid pM79.44 was constructed in a way similar to that for the PHO5 gene circle plasmid (18). One recombination sequence (RS)4 element was inserted into the NruI site upstream of the ORF, and the other RS element and LexA cluster were inserted into the downstream NruI site on pM77.2. In pM79.44 the sequence 5′-TAGTATATAAAGAAAGAAGTGTA-3′ was replaced with 5′-TCATCGATCCCCCGGGGGACGAGT-3′, which replaced the PHO8 TATA box and downstream promoter sequences with cleavage sites for restriction endonucleases ClaI and SmaI yielding the PHO8 gene circle plasmid pM82.1. The PHO8 promoter circle plasmid pCM110.1 was derived from pM75.2 and pM82.1 by inserting the downstream RS element and LexA cluster into the SacI site 56 bp downstream of the start codon. Pho4 activation domain mutants were constructed as described previously (30).
The PHO8 gene was replaced with URA3 by homologous recombination in strains YS18 and yM7.8 using plasmid pM78.5, generating strains yM51.1 and yM52.2, respectively. PHO8 gene circle strains yM53.78 and yM54.9 were generated by replacing pho8::URA3 in strains yM51.1 and yM52.2 with the TATA-less pho8 gene circle construct on plasmid pM82.1. The pho4 deletion strain yM59.20 was generated by replacing PHO4 with URA3. Strain yM120.2 was generated from strain yM54.9 by replacing PHO4 with URA3 from plasmid pCM4.5. The PHO4 activation domain mutation strains were derived from strain yM120.2 as described previously (30). The PHO8 promoter circle strain yC104.34 was derived from strain yM52.2 by replacing pho8::URA3 with a pho8 promoter circle from plasmid pCM110.1. Strain yC192.1 was generated from yC104.34 by replacing PHO4 with URA3 using plasmid pCM4.5. Strain yE22.1 was generated by replacing the PHO5p:CFP cassette in strain yE2.1 with URA3. Exchanging LEU2 in the PHO80 locus of yE22.1 with HIS3 generated strain yC57.10. The PHO5 gene circle strain yC59.11 was then generated by replacing URA3 in yC57.10 with the PHO5 gene circle construct using plasmid pM70.1 (18), and replacement of PHO4 with URA3 in yC59.11 gave strain yC87.2.
PHO8 promoter and CFP ORF sequences were fused at the PHO8 start codon by overlapping PCR (31), using plasmids pM75.2 (see above) and 14846 (Addgene) as templates. The PHO8p:CFP fusion was used to replace a 2.3-kb EcoRV-SalI fragment in pM75.2 bearing the PHO8 gene, including its promoter, except for the last 3′-terminal 238 bp of the ORF, generating plasmid pJL2.2. Plasmid pJL3.4, which bears the fusion between the PHO8 promoter and YFP ORF, was constructed analogously. YFP ORF sequences were amplified by PCR from plasmid 14840 (Addgene). The wild-type PHO5 gene was restored in strains yC56.11 and yC57.10 (30) using plasmid pM50.1 (18) to generate strains yJL5.7 and yJL4.4, respectively. The PHO8 gene in these two strains was then replaced with URA3 using plasmid pM78.5 to generate strains yJL7.4 and yJL6.4. Strains yJL8.1 and yJL9.1 were then derived from yJL6.4 and yJL7.4 by homologous recombination with plasmids pJL2.2 and pJL3.4, respectively, replacing pho8::URA3 with PHO8p:CFP and PHO8p:YFP. Strain yJL12.1 was generated from strain yJL9.1 by knocking out PHO4 with URA3 using plasmid pCM4.5. The diploid strain yJL10.1 was generated by mating yJL8.1 and yJL9.1.
An asf1::KanMX knock-out cassette was amplified from a heterozygous diploid (GSA-6 Heterodiploid Knock-out Collection, ATCC) using primers 5′-AATGCTGTTTTATTCCGTTCTTACA-3′ and 5′-GATTTTTATTCCAACATGTTCGTTC-3′. The appropriate band was purified and used to transform PHO8 and PHO5 gene circle strains yM53.78, yM54.9, and yC58.8, yC59.11, yC72.1, and yC87.2, respectively, creating strains CBY11.4, CBY12.1, and yC117.1, yC118.1, yC119.1, and yC120.1, respectively. A snf2::KanMX knock-out cassette was amplified from a heterozygous diploid (GSA-6 Heterodiploid Knock-out Collection, ATCC) using primers 5′-TTAATTCCAATACATTCGCCACTAT-3′ and 5′-CAAGAACAAGTTCACTATGATGACG-3′. The appropriate band was purified and used to transform PHO8 gene circle strains yM53.78, yM54.9, and yM59.20, creating strains CBY14.5, CBY15.5, and CBY16.1, respectively. The snf2::KanMX knock-out cassette was also used to transform PHO5 gene circle strains yM63.19 and yM19.2 (30), creating strains yC45.3 and yC44-1, respectively. A SNF2 expression plasmid, pSR127 (kindly supplied by Fred Winston, Harvard University), was used to transform strains CBY14.5, CBY15.5, and CBY16.1, creating strains CBY14.5P, CBY15.5P, and CBY16.1P, respectively. All yeast transformations were performed using the lithium acetate method.
Limiting Digestion Analysis
Strains yM53.78 and yM54.9 were grown overnight in 500 ml of 2×SCR-L medium to a density of 3 × 107 cells/ml. Upon reaching this density, 50 ml of each culture was removed and placed into a new flask, to which 5 ml of 20% galactose was added to induce PHO8 gene circle formation. These smaller cultures were allowed to incubate for an extra 1.5 h in the presence of 2% galactose, after which topoisomers were isolated as described previously (18). Nuclei were prepared from the remaining 450-ml cultures as described previously (18, 32). Frozen nuclei pellets were resuspended in micrococcal nuclease (MNase; Sigma) buffer (15 mm Tris, pH 8, 50 mm NaCl, 1 mm CaCl2, 5 mm β-mercaptoethanol) and divided into 5 equal volumes to be digested with 0, 0.5, 1, 2, or 4 units of MNase for 20 min at 37 °C. Undigested (0 unit) samples were subsequently digested with either ApaLI and HaeIII (yM53.78) or EcoRV and XhoI (yM53.78 and yM54.9) to cut out a 941-bp or 930-bp fragment of the PHO5 or PHO8 promoter, respectively. MNase-digested samples and cut promoter fragments were run on a 1% agarose gel and prepared for Southern blotting. Blots were first probed with a 690-bp PHO8 probe spanning the four putative promoter nucleosome positions (27). Primers used to generate the PHO8 promoter probe were P164 5′-TCAAGAATGGCACTATAAGTGTGG-3′ and P165 5′-GCTAATGCGCGTTCAAATAATGTC-3′. Blots were stripped and rehybridized with a 690-bp PHO5 probe spanning PHO5 promoter nucleosomes N-1 to N-4 (33). Primers used to generate the PHO5 promoter probe were P232 5′-GGTCCGCTCCTTCTAATAATCG-3′ and P15 5′-TGGTAATCTCGAATTTGCTTG-3′. The signal from the EcoRV/XhoI-digested PHO8 promoter (yM53.78) probed with P164/P165 was compared with the ApaLI/HaeIII-digested PHO5 promoter (yM53.78) probed with P232/P15 to normalize the signal intensity of the two different probes. MNase profiles for yM53.78 were used to generate a limiting value to compare the absolute number of nucleosomes on the PHO8- and PHO5-repressed promoters (PHO8 probe signal/PHO5 probe signal). To determine the number of nucleosomes remaining on the activated PHO8 promoter we first normalized the PHO8 promoter signal from yM53.78 and yM54.9 (using the EcoRV/XhoI-digested promoter samples from strains yM53.78 and yM54.9) prior to taking the yM53.78/yM54.9 signal ratio.
Topoisomer Analysis
Analysis of topoisomer distributions was performed as described previously (18, 34). PHO8 and PHO5 gene and promoter circle strains all bore a mutated TATA box.
Phosphatase Assay
Phosphatase assays were performed as described previously (5).
Northern Blot Analysis
Northern blot analysis was performed as described previously (30).
Expression Analysis by FACS
PHO5 and PHO8 promoter-controlled YFP expression was measured in live haploid cells by flow cytometry (BD Influx, BD Biosciences). The YFP proteins were excited by an air-cooled Solid State “Sapphire” (Coherent), 200-mW, 488-nm laser. The level of expression for each strain was referenced to strain PHO5A (yE1.1) (PHO4 pho80Δ), which was always analyzed in parallel. The level of yE1.1 expression was set to 100%. Strain yE3.1 (pho4Δ pho80Δ) was employed as a negative control to measure the level of autofluorescence and set to 0% level of expression. The YFP fluorescence was collected in the 531/40-nm channel.
For the expression analysis, cells were grown in SCD medium. Data from 100,000 cells were collected for each measurement; each sample was remeasured at least four times. No fewer than two independent cell cultures at two different cell density levels were evaluated for each strain.
Data from flow cytometry measurements were exported into FSC3.0 files, and data analysis was carried out by FlowJo software (Tree Star). To ensure sufficient data quality, data were analyzed in FSC and SSC channels, and scatters were rejected. Outliers were identified visually in the YFP channel. These procedures reduced the number of cells by 20–25%. Further reduction in cell number by imposing more stringent selection criteria did not significantly change relative expression levels.
Fluorescence Microscopy and Expression Noise Measurements
CFP and YFP expression under control of the PHO8 promoter were measured in single diploid cells by quantitative fluorescence microscopy (30). CFP and YFP fluorescence signals for individual cells were divided by the mean CFP and YFP signal, respectively, and plotted in a scatter plot. The intrinsic noise is given by the mean squared distance of points from the diagonal of the plot (coefficient of variation squared). The intrinsic noise was determined from four independent measurements of at least 250 cells each. The variability in intrinsic noise between samples was larger than observed previously for PHO5 (30), consistent with larger variations in expression and topology measurements, too (see below).
Computation and Modeling
Predictions of PHO8 expression noise were made using the previously described stochastic model of PHO5 gene expression (30). Adjustments are outlined under “Results.” Calculations were performed using Mathematica 7 (Wolfram Research).
RESULTS
Transcription of PHO8 and PHO5 is activated upon sequence-specific binding of the transcriptional activator Pho4 at specific promoter elements (35). Entry of Pho4 into the nucleus is inhibited by the cyclin-cyclin-dependent kinase complex Pho80/Pho85. Lack of inorganic phosphate in the medium results in inhibition of the Pho80/Pho85 kinase and translocation of Pho4 into the nucleus. Thus, cells that lack Pho80 activate PHO8 constitutively, whereas cells that lack Pho4 fail to activate PHO8 under inducing conditions. To approach steady-state conditions as closely as possible, induced PHO8 promoter chromatin was analyzed, unless indicated otherwise, in pho80Δ cells cultured in phosphate-containing media.
The Noninduced PHO8 Promoter Is Occupied by Three Nucleosomes Rather Than Four
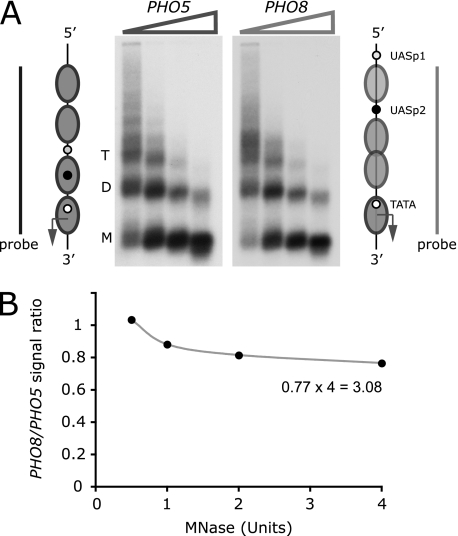
The noninduced PHO8 promoter is believed to be occupied by four nucleosomes, three of which were proposed to be partially unfolded to account for elevated restriction enzyme accessibilities (36). To test this hypothesis, we analyzed PHO8 and PHO5 promoter chromatin by limiting digestion analysis (18). To this end, we partially digested nuclei from noninduced cells with increasing concentrations of MNase, which preferentially digests linker DNA. The amount of undigested promoter DNA was measured by hybridization with 32P-labeled PHO5 and PHO8 promoter DNA probes after gel electrophoresis and Southern blotting of isolated DNA (Fig. 1A). Both probes spanned a four-nucleosome-wide domain (Fig. 1A). The same domains were released from genomic DNA by restriction endonuclease digestion and run on the same gel as the MNase-digested DNA (data not shown). This allowed us to account for differences in probe activity.
FIGURE 1.
The repressed PHO8 promoter is occupied by three nucleosomes. A, schemes of the repressed PHO5 and PHO8 chromatin structures (32, 36). Chromatin of noninduced cells was digested with increasing concentrations of MNase. The isolated DNA was fractionated by agarose gel electrophoresis, blotted, and sequentially hybridized with 32P-labeled DNA probes spanning the PHO5 or the PHO8 promoter (bars). Positions of bands corresponding to nucleosome trimers (T), dimers (D), and monomers (M) are indicated. B, radioactivity signals integrated over the entire lane for each enzyme concentration. The ratio of PHO8 and PHO5 promoter probe signals corrected for differences in probe activity was plotted as a function of enzyme concentration.
The ratio of undigested PHO8 and PHO5 promoter DNA reached a limiting value of 0.77 ± 0.1 (two independent experiments) at high enzyme concentrations (Fig. 1B). The low restriction enzyme accessibilities of sequences occupied by nucleosomes suggest that nucleosome positions are close to fully occupied at the repressed PHO5 promoter (17). Thus, the limiting value indicated that the noninduced PHO8 promoter is occupied, on average, by 0.77 × 4 = 3.1 nucleosomes that are equally resistant to MNase attack as nucleosomes at the PHO5 promoter. Notably, the length of DNA protected against MNase attack was identical for the nucleosomes of both promoters (Fig. 1A). The paucity of distinct digestion products shorter than the expected length of the nucleosome core particle argued against the existence of stable, partially unfolded nucleosomes. Differences in the cleavage frequencies of restriction endonucleases are likely to reflect differences in nucleosome occupancy rather than structure. The same conclusion was reached for activated PHO5 promoter chromatin (18).
Chromatin Remodeling Removes Nucleosomes from the Induced PHO8 Promoter
Loss of nucleosomes from the induced PHO8 promoter was previously inferred from nuclease accessibility and chromatin immunoprecipitation assays (28, 36). However, the existence of stable partially unfolded nucleosomes may be, and has been (see above), invoked to explain elevated nuclease accessibility, and nucleosome-bound remodelers that encapsulate the nucleosome upon binding, such as RSC (37–39), may interfere with cross-linking of histones to DNA or the recognition of histone epitopes by antibodies.
To clarify the structural nature of induced PHO8 promoter chromatin, we compared activated and repressed promoter chromatin at steady state by limiting digestion and topology analyses. For these experiments, we used strains in which the PHO8 gene or promoter was flanked by the recognition sequences for the site-specific R recombinase of Zygosaccharomyces rouxii, which was expressed under control of the inducible GAL1 promoter (40). The position of the upstream RS is indicated in Fig. 2A. The second RS element was inserted 250 bp downstream of the PHO8 stop codon for gene circles or 56 bp downstream of the PHO8 start codon for promoter circles. Recombination results in removal of the PHO8 gene in circular form from its chromosomal locus. To exclude topological effects due to transcription (18), strains furthermore bore a mutation in the PHO8 TATA box.
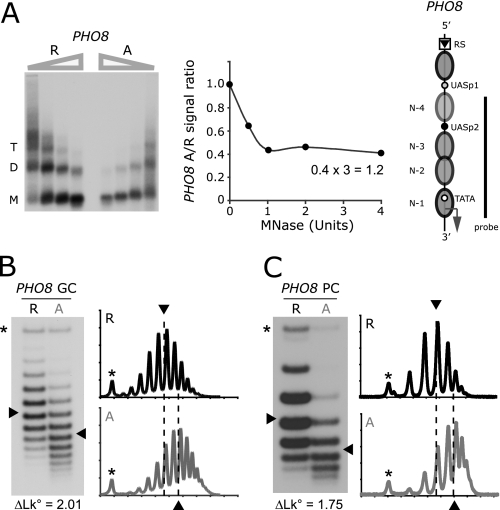
FIGURE 2.
Nucleosomes are removed from the induced PHO8 promoter. A, repressed (R) and activated (A) PHO8 promoter chromatin digested with increasing concentrations of MNase. The isolated DNA was fractionated by agarose gel electrophoresis, blotted, and hybridized with a 32P-labeled DNA probe spanning the PHO8 promoter. The signal:ratio for DNA from activated and repressed cells on corresponding lanes was plotted as a function of enzyme concentration. The position of the upstream recombination sequence, RS, is indicated on the PHO8 promoter diagram to the right. B, topoisomer analysis of repressed and activated PHO8 gene circles (GC). DNA topoisomers were resolved by gel electrophoresis in the presence of chloroquine (18), blotted, and hybridized with a 32P-labeled DNA probe spanning the PHO8 gene. The linking number of topoisomers increases from top to bottom. An asterisk indicates the position of nicked circles. Line profiles of the topoisomer distributions are shown to the right of each blot. ΔLk refers to the linking difference measured between the centers of the topoisomer distributions of activated and repressed PHO8 gene circles. C, topoisomer analysis of repressed and activated PHO8 promoter circles (PC) spanning five nucleosome positions.
The digestion of equal amounts of activated and repressed promoter chromatin with increasing concentrations of MNase showed that the ratio between activated and repressed promoter DNA reached a limiting value of 0.41 ± 0.1 (Fig. 2A, two independent experiments). Assuming that the repressed promoter bears three nucleosomes, on average, this result suggests that about 0.4 × 3 = 1.2 nucleosomes are retained at the activated promoter, equally resistant to MNase attack and protecting the same length of DNA as repressed promoter nucleosomes (Fig. 2A).
What was the fate of the 1.8 altered nucleosomes? To address this question, we induced formation of PHO8 gene circles in the same cultures that were used for limiting digestion analysis and isolated gene circle DNA topoisomers. If the altered nucleosomes unfolded completely, the linking difference between activated and repressed circles should be similar to the number of altered nucleosomes, given that nucleosome formation on a closed DNA circle reduces the linking number of the DNA circle by about −1/nucleosome in relaxation equilibrium (41–43). Topoisomer distributions were resolved by chloroquine-agarose gel electrophoresis, blotted, and hybridized with a 32P-labeled DNA probe encompassing the PHO8 gene. The average linking difference between the topoisomers of the activated and repressed gene circles was indeed closely similar to the number of altered nucleosomes determined by limiting digestion analysis (Fig. 2B). For unknown reasons, linking differences between PHO8 gene circles varied more strongly than observed previously for PHO5 gene circles (18). However, the average linking difference of 1.82 (S.D. ± 0.33) from 20 measurements of 10 independent preparations was in excellent agreement with our expectation for complete unwrapping of DNA from the altered nucleosomes.
Topology analysis of promoter circles that encompass promoter nucleosome positions N-1 to N-4 (Fig. 2A) plus sequences for one additional upstream nucleosome led to a similar result (Fig. 2C). We found an average linking difference between activated and repressed circles of 1.61 (S.D. ± 0.28), suggesting that most if not the entire linking difference between activated and repressed gene circles was due to changes in promoter chromatin.
Removal of PHO8 Promoter Nucleosomes Occurs by Disassembly Rather Than Sliding
Promoter nucleosomes may be removed either by sliding away from the promoter or by disassembly. To distinguish between both possibilities, we formed PHO8 gene circles in pho4Δ and PHO4 wild-type cells grown in high phosphate medium and isolated topoisomers before and after transfer of cells into phosphate-free medium. As expected, topoisomer distributions from both cell types were virtually identical prior to transfer of cells into phosphate-free medium (Fig. 3A). However, after incubation of cells in phosphate-free medium, the average linking number of PHO8 circles in PHO4 wild-type cells increased relative to circles from pho4Δ cells by 1.9 (Fig. 3A), suggesting the loss of about 2 nucleosomes from induced PHO8 circles. Removal of promoter nucleosomes by sliding away from the promoter would have preserved the number of circle nucleosomes and the circle linking number.
FIGURE 3.
Nucleosomes continually disassemble and reassemble at the activated PHO8 promoter. A, PHO8 gene circles (GC) formed in pho4Δ (Δ) or PHO4 (wt) cells cultured in high phosphate media. Topoisomers were obtained from cells either before (+Pi) or after (−Pi) transfer of cells into phosphate-free medium, separated by gel electrophoresis, blotted, and hybridized as described above (Fig. 2). The linking difference of 0.05 for repressed circles is within the margin of measurement error and sample variability, but may also be explained by the finding that Pho4 briefly enters the nucleus during S phase under repressing conditions (50). ΔLk refers to the linking difference measured between the centers of the topoisomer distributions of activated and repressed PHO8 gene circles. B, topoisomer distributions of PHO8 gene circles formed in strains bearing mutations in the activation domain of the Pho4 activator. Mutants are arranged, from left to right, in order of decreasing activator strength (PHO8 expression level, see C). Linking differences relative to repressed PHO8 (pho4Δ) gene circles are indicated at the bottom of the image. C, Northern blot analysis of PHO8 expression in pho4 mutants. ACT1 mRNA was used as a loading control.
Nucleosome Disassembly Is Counterbalanced by Reassembly
On average, the activated PHO8 promoter is not nucleosome-free (Fig. 2A). What is the mechanism that retains promoter nucleosomes? The simplest possibility, perhaps, is that disassembly is counterbalanced by reformation of nucleosomes (18). If nucleosomes reformed under activating conditions, mutations in the Pho4 activation domain, which is essential for promoter chromatin remodeling (44), were expected to shift the equilibrium gradually between nucleosome removal and reformation. If reassembly did not occur, the maximal number of nucleosomes would always be removed, even in cells expressing weak activators, albeit at a slower rate. We determined the extent of nucleosome loss from the induced PHO8 promoter by topology analysis in eight pho80Δ strains that bore different mutations in the Pho4 activation domain (30). Linking differences between activated and repressed gene circles gradually decreased with decreasing activator strength (Figs. 3, B and C; see also Fig. 7D), as predicted by the reformation hypothesis. This result conformed to previous topology measurements of PHO5 promoter chromatin remodeling (30) and was consistent with the observation of genome-wide histone exchange at promoters (45).
FIGURE 7.
Predicting the profile of PHO8 promoter nucleosome loss and the intrinsic noise of PHO8 expression. A, expression noise (variance divided by the squared mean, coefficient of variation) and the mean of expression calculated using a stochastic model of PHO5 promoter chromatin remodeling and expression with kinetic parameter values derived from PHO5 noise and chromatin structure data (30). The model assumes three functional promoter states (see scheme): OFF (core promoter occupied by a nucleosome, transcriptionally inactive), OFF* (no nucleosome at the core promoter, transcriptionally inactive), and ON (no nucleosome at the core promoter, transcriptionally active). For details, see Ref. 30. For predicting PHO8 expression noise, we modified the PHO5 model to account for the observation that exchange of the PHO8 core promoter for the PHO5 core promoter increased PHO8 expression 1.7-fold (47). The reverse exchange should therefore decrease PHO5 expression by a factor of 1.7. We interpreted the difference in core promoter strength in terms of the stability of general transcription factor complexes and thus decreased the kinetic parameter for transition out of the ON state (i) to reduce the level of expression by a factor of 1.7. Noise and expression were then calculated for values of the kinetic parameter for promoter activation (a) between zero and the PHO5 wild-type value of a (30). The calculation predicted a noise value of 0.053 at 7.5% of PHO5 wild-type expression, the relative expression level of PHO8 (Fig. 6). B, the intrinsic noise of PHO8 expression determined by measuring abundances of YFP and CFP in single diploid cells, expressing both chromophores under control of the PHO8 promoters. Single dots indicate YFP and CFP abundances in individual cells. Abundances were determined by quantitative fluorescence microscopy and normalized to the mean abundance of each chromophore within the analyzed cell population. The average measured noise value was 0.054. C, the adjusted PHO5 model (A) was used to calculate the quantitative relationship between promoter nucleosome loss and expression (relative to wild type) by either tuning the kinetic parameter for nucleosome disassembly alone (curve d) or equally tuning the parameters for nucleosome disassembly and promoter activation (curve d∧a). D, plot of expression (mRNA) and nucleosome loss data (linking differences) shown in Fig. 3.
Disassembly of PHO8 Promoter Nucleosomes Requires Swi2 but Not Asf1
Changes in nuclease accessibility of PHO8 promoter chromatin have previously been shown to require Snf2 (27). Thus, linking changes in gene circles due to PHO8 induction should also depend on Snf2. To test this prediction, we repeated our topology analysis of activated and repressed PHO8 gene circles in snf2Δ cells. As predicted, deletion of SNF2 markedly interfered with nucleosome removal in pho80Δ cells (Fig. 4A). The linking difference between activated and repressed PHO8 gene circles decreased by 95% to 0.087 (with a S.D. ± 0.25 in four independent experiments). In contrast, deletion of ASF1 had little effect on nucleosome disassembly in pho80Δ cells (linking difference 1.68 ± 0.11 in two independent experiments, Fig. 4B), suggesting that the rate of nucleosome disassembly at steady state was either hardly affected or that the rates of disassembly and reassembly were equally altered.
FIGURE 4.
Nucleosome removal from the PHO8 promoter requires Snf2 but not Asf1. A, topoisomer analysis of repressed (R) and activated (A) PHO8 gene circles (GC) in snf2Δ cells. B, topoisomer analysis of repressed and activated PHO8 gene circles isolated from asf1Δ cells. ΔLk refers to the linking difference measured between the centers of the topoisomer distributions of activated and repressed PHO8 gene circles.
Similarly, deletion of ASF1 in PHO5 gene circle strains had little effect on the linking difference between activated and repressed circles isolated from pho80Δ and PHO80 cells, respectively (linking difference 1.81 ± 0.25 in four independent measurements and two different strains; see supplemental Fig. 1).
Disassembly of PHO5 Promoter Nucleosomes Can Be Dependent upon SNF2
In pursuit of the possible cause for the different coactivator dependences of PHO5 and PHO8, we unexpectedly found, in the same strains as those used for PHO8 gene circle formation, that constitutive PHO5 expression in pho80Δ cells was abolished upon deletion of SNF2 (Fig. 5A). The activation defect could be fully suppressed by transforming snf2Δ cells with a SNF2 expression plasmid, ruling out the possibility that other genes critical for PHO5 expression had accidentally been damaged upon SNF2 deletion (Fig. 5A). A similar result was obtained in PHO80 wild-type cells, induced by transfer into phosphate-free medium (Fig. 5A). Only after long periods of phosphate starvation, and even then in only half of our experiments (two of four replicates), did we observe a transient Pho4-dependent increase in PHO5 expression in snf2Δ cells (Fig. 5A). In contrast, PHO5 expression in SNF2 wild-type cells decreased when cultured for long times in phosphate-free medium, raising the possibility that Snf2, depending on the physiological context, may function as a repressor as well as a coactivator of PHO5 expression.
FIGURE 5.
SNF2 is required for PHO5 expression and promoter nucleosome removal. A, PHO5 encodes a secreted acidic phosphatase and accounts for 90% or more of the cellular acidic phosphatase activity upon induction (59). Phosphatase activity was measured in pho80Δ cells grown in high phosphate medium and in PHO80 wild-type cells after incubation for different times in phosphate-free medium. B, topoisomer distributions of repressed (R) and activated (A) PHO5 gene circles (GC) isolated from pho80Δ pho4Δ SNF2 and pho80Δ SNF2 cells, respectively. C, topoisomer distributions of repressed and activated PHO5 gene circles isolated from pho80Δ pho4Δ snf2Δ and pho80Δ snf2Δ cells, respectively. ΔLk refers to the linking difference measured between the centers of the topoisomer distributions of activated and repressed PHO5 gene circles.
The linking difference between activated and repressed PHO5 gene circles, isolated from pho80Δ and pho80Δ pho4Δ cells, respectively, was 1.85 in the presence of Snf2, conforming with earlier measurements, but only 0.18 in its absence (Fig. 5, B and C), consistent with the observed expression defect of PHO5 in snf2Δ cells (Fig. 5A).
By deleting SNF2 in other strain backgrounds, we confirmed earlier results of others that PHO5 can be activated in the absence of SNF2 (46), suggesting that the effect observed here was strain-specific (data not shown).
Activated PHO8 Expression Is Caused Primarily by Accelerated Promoter Nucleosome Disassembly
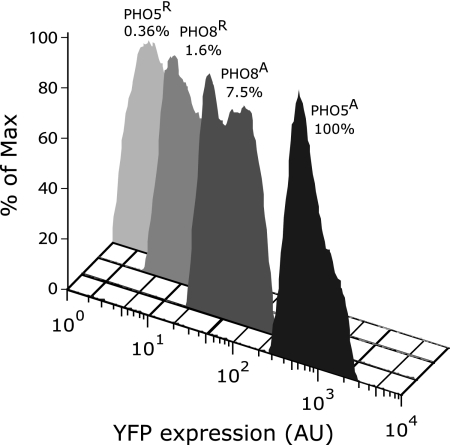
To compare promoter strengths, we measured expression of yellow fluorescent protein (YFP) from either the PHO5 or the PHO8 promoter using flow cytometry. Under repressing conditions, PHO8p:YFP expression exceeded PHO5p:YFP expression 4-fold, consistent with the assumption of partial derepression due to the lower nucleosome occupancy of the noninduced PHO8 promoter (see above). In contrast, expression of PHO8p:YFP in pho80Δ cells amounted to only 7.5% of PHO5 promoter-controlled expression (Fig. 6), although both promoters lose about two nucleosomes upon induction. How can the difference between expression levels be explained?
FIGURE 6.
Flow cytometry analysis of relative YFP expression under control of the PHO8 or PHO5 promoter in pho80Δ pho4Δ (R) or pho80Δ (A) cells. Mean values of expression distributions are indicated relative to PHO5 promoter-controlled expression in pho80Δ cells (PHO5A, 100%). AU, arbitrary units.
Because nucleosomes interfere with transcription factor binding at promoter elements and transcriptional initiation, fluctuations in promoter nucleosome occupancy contribute to the fluctuation in gene expression (expression noise). The quantitative relationship among PHO5 promoter nucleosome loss, expression level, and noise was explained on the basis of a stochastic model of chromatin remodeling and gene expression, assuming that promoter nucleosome disassembly and preinitiation complex formation (“promoter activation”) are the rate-limiting, activator-controlled, steps of PHO5 expression (30).
The PHO5 and the PHO8 promoters lose the same numbers of nucleosomes upon induction, consistent with similar rates of nucleosome disassembly. Given the markedly lower level of PHO8 expression, this suggested that promoter activation is much less efficient at PHO8 than PHO5. We therefore speculated that activated PHO8 expression is due only to the acceleration of promoter nucleosome disassembly but not promoter activation, in contrast to PHO5 (30).
To test this hypothesis, we employed the PHO5 model, with kinetic parameter values previously derived from PHO5 noise and chromatin structure analyses (30), to predict the noise of PHO8 expression, and then we compared predicted and measured noise. We took into account that replacement of the PHO8 core promoter with the PHO5 core promoter increased PHO8 expression 1.7-fold (47), suggesting that general transcription factors bind more tightly to the PHO5 promoter. In the PHO5 model, we therefore increased the rate of transitions out of the transcriptionally active state accordingly (Fig. 7A, parameter i) before calculating noise values for PHO8 expression. The tuning of the kinetic parameter for promoter activation in the PHO5 model (Fig. 7A, parameter a) generated a noise profile predicting a noise value of 0.053 at the PHO8 expression level, 7.5% of PHO5 expression (Fig. 7A, 6).
To measure the intrinsic noise of PHO8 expression (48), we constructed a diploid pho80Δ strain that expresses YFP and CFP under control of the PHO8 promoter. CFP and YFP expression in single cells was analyzed by quantitative fluorescence microscopy. Plotting YFP against CFP expression generates a scatter plot for a population of cells (Fig. 7B). With fluorescent signals normalized to a mean of 1, the intrinsic noise equals the mean squared distance of points from the plot diagonal (49). We found a noise value of 0.054 (±0.018) for PHO8 expression (Fig. 7B), in excellent agreement with the theoretical prediction of 0.053 (Fig. 7A).
If indeed Pho4 controlled the rate of nucleosome disassembly, but not promoter activation, a linear relationship between promoter nucleosome loss and expression was expected (Fig. 7C, curve d). In contrast, if Pho4 controlled the rates of nucleosome disassembly and promoter activation, a nonlinear function was predicted (Fig. 7C, curve d∧a). Consistent with the first assumption and our noise analysis, the data shown in Fig. 3, B and C, were best approximated by a linear graph (Fig. 7D). Only the data point for the gain-of-function mutation D78A was poorly explained by this linear approximation.
DISCUSSION
By limiting digestion, topology, and expression noise analyses, we provided evidence corroborating the following conclusions. The noninduced PHO8 promoter is occupied by three normally folded nucleosomes rather than four nucleosomes of which some are partially unfolded. Chromatin remodeling upon PHO8 induction removes nucleosomes from the activated promoter. Removal occurs by disassembly in a Snf2-dependent manner and is counterbalanced by continuous reformation of nucleosomes. Disassembly does not require the histone chaperone Asf1. Activated expression of PHO8, in contrast to PHO5, is due primarily to accelerated promoter nucleosome disassembly.
The observed defect in nucleosome removal from the activated PHO5 promoter in snf2Δ cells was surprising, given previous reports that PHO5 expression can be induced in the absence of Snf2 (46). However, SWI/SNF has been implicated earlier in PHO5 expression. PHO5 was found to be weakly expressed during S phase, in high phosphate media (50). This expression required Pho4 as well as Snf2. The structural characteristics of promoter chromatin during S phase expression are unknown. Furthermore, deletion of SNF6, which encodes a subunit of the SWI/SNF complex, reduced PHO5 expression to 60% of wild-type expression (51).
The discrepancy between our results and those of others may be due to differences in genetic background, implying that the Pho4 activator recruits multiple factors that are each capable of catalyzing nucleosome disassembly. The inefficiency of competing activities would then have revealed SWI/SNF catalysis of nucleosome disassembly at PHO5 in our strain background. This interpretation is consistent with a recent report that PHO5 expression was marginally compromised in the absence of either Chd1 or Isw1, but abolished in the absence of both chromatin remodelers (25).
Delayed induction kinetics in mutant strains has frequently been construed as evidence of the importance of the missing factor for PHO5 chromatin remodeling (24). However, such kinetic effects may also be due to defects in the PHO signaling pathway rather than nucleosome disassembly. This latter possibility was ruled out by the persistence of the disassembly defect in pho80Δ snf2Δ cells, but the possibility of an indirect effect of SNF2 deletion on nucleosome disassembly cannot, by any means, be ruled out.
In vitro, SWI/SNF catalyzes the sliding of nucleosomes (8). The situation is less clear in the case of nucleosome disassembly. SWI/SNF has been seen to release histone H3 from chromatin templates that bore at least two nucleosomes (15), and RSC was found to disassemble mononucleosomes in the presence of the histone chaperone Nap1 (16). The ability of SWI/SNF and RSC to slide the histone octamer off the DNA end by up to 50 bp (52), however, raises the possibility that DNA ends may have played an important part in the mechanism of disassembly. In vivo, disassembly must occur in the absence of DNA ends. Optical tweezer experiments where DNA ends were blocked by attachment to polystyrene beads did not provide evidence for nucleosome disassembly by RSC or SWI/SNF (53). However, nucleosome disassembly may require specific histone modifications that are absent from in vitro reconstituted nucleosomes. A recent study of isolated PHO5 chromatin gene circles showed that relaxation of repressed circles with topoisomerase after RSC treatment led to a linking change that matched complementary results for limiting digestion analysis of the circles, demonstrating that RSC removed nucleosomes from an end-less substrate (54). Similar results were obtained with promoter circles but not ORF circles, suggesting that disassembly was limited to promoter nucleosomes, which had to be marked specifically to allow for their removal. Indeed, treatment of circles with histone deacetylase diminished the removal of promoter nucleosomes by RSC (54). Our in vivo findings, together with the demonstration of promoter nucleosome disassembly by RSC in the absence of DNA ends, support the notion that SWI/SNF catalyzes the disassembly of promoter nucleosomes upon recruitment by activators in vivo.
How can this conclusion be reconciled with the conclusion that SWI/SNF catalyzes nucleosome sliding? An answer to this question is suggested by the notion that sliding of a nucleosome within the chromatin fiber unspools the DNA from its neighbor (55). Some evidence has been presented in support of this notion (15, 56, 57). Only nucleosomes marked by specific histone modifications may be conducive to this sliding-mediated disassembly (54). Loss of the crucial modifications may then limit the remodeler activity to sliding, which at PHO5 is likely to result in repression of transcription because nucleosomes preferentially form over the transcription start sites rather than other sequences of the promoter (18). This might explain how SWI/SNF could function as a corepressor as well as coactivator of transcription (58), depending on the molecular context (see above).
The chromatin transitions at the PHO8 and PHO5 promoter are remarkably similar, pointing at the generality of our findings (18, 30). In both instances, activated promoter chromatin is generated by continual disassembly and reformation of nucleosomes. Of three promoter nucleosomes about two, on average, are removed upon Pho4 binding in each case, indicating that the ratio of the kinetic parameters for nucleosome disassembly and reassembly are about the same. The parameter values must be similar too, as we could correctly predict the magnitude of intrinsic PHO8 expression noise on the basis of a stochastic model of chromatin remodeling and gene expression with kinetic parameters derived from PHO5 expression noise and chromatin structure data (30). This only required adjustments in the kinetic parameters for transitions into and out of the transcriptionally active state to account for differences in steady-state expression between the two promoters.
Thus, we found that a lower frequency of promoter activation accounts mostly for the much reduced expression level of PHO8 relative to PHO5 (Fig. 6), suggesting that activated PHO8 expression is due primarily to the acceleration of promoter nucleosome disassembly in contrast to PHO5 where activator binding also accelerates the rate of promoter activation (30). As predicted by this assumption, we found a linear relationship between promoter nucleosome loss and PHO8 expression level (Fig. 7D), whereas an exponential relationship was observed for PHO5 (30).
How can the failure of Pho4 to stimulate the rate of promoter activation effectively at PHO8 be explained? We note that the distance between the TATA box and the proximal Pho4 binding site (UASp2) of the PHO8 promoter measures 400 bp, whereas Pho4 binding sites at the PHO5 promoter are located 150 and 250 bp upstream of the TATA box. The distance between activator binding sites and the core promoter may be too large at PHO8, such that recruitment by activators would not increase the local concentration of recruited factors at the core promoter. In contrast, the efficiency of nucleosome disassembly at core promoter sequences may be relatively insensitive to the distance between activator binding site and core promoter; disassembly of nucleosomes upon recruitment of SWI/SNF may reduce the nucleosome occupancy at a distant core promoter by subsequent sliding of nucleosomes toward the site of disassembly.
Shortening of the distance between UASp2 and the PHO8 TATA box increased the level of PHO8 expression (47), consistent with our expectation that the distance between UASp2 and the TATA box of PHO8 is suboptimal for promoter activation. A suboptimal distance may be partially compensated for by a stronger affinity between the activator and the recruited factor, which would increase the dwell time of the factor at the promoter and thus its chance to interact with core promoter sequences. This might explain the deviation of the activator gain-of-function mutant D78A from our theoretical expectations (Fig. 7D).
Supplementary Material
Acknowledgments
We thank Drs. Craig Kaplan and Joachim Griesenbeck for critical comments on the manuscript and Dr. Fred Winston for plasmid pSR127. We also thank the UCSC MEGAMER facility and its manager Dr. Brandon Carter for assistance with flow cytometry.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
- RS
- recombination sequence
- CFP
- cyan fluorescent protein
- MNase
- micrococcal nuclease
- RSC
- remodel the structure of chromatin.
REFERENCES
- 1. Griesenbeck J., Boeger H., Strattan J. S., Kornberg R. D. (2003) Mol. Cell. Biol. 23, 9275–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Han M., Grunstein M. (1988) Cell 55, 1137–1145 [DOI] [PubMed] [Google Scholar]
- 3. Kaplan C. D., Laprade L., Winston F. (2003) Science 301, 1096–1099 [DOI] [PubMed] [Google Scholar]
- 4. Lorch Y., LaPointe J. W., Kornberg R. D. (1987) Cell 49, 203–210 [DOI] [PubMed] [Google Scholar]
- 5. Mao C., Brown C. R., Griesenbeck J., Boeger H. (2011) PLoS One 6, e17521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kornberg R. D., Lorch Y. (1999) Curr. Opin. Genet. Dev. 9, 148–151 [DOI] [PubMed] [Google Scholar]
- 7. Längst G., Bonte E. J., Corona D. F., Becker P. B. (1999) Cell 97, 843–852 [DOI] [PubMed] [Google Scholar]
- 8. Whitehouse I., Flaus A., Cairns B. R., White M. F., Workman J. L., Owen-Hughes T. (1999) Nature 400, 784–787 [DOI] [PubMed] [Google Scholar]
- 9. Fazzio T. G., Tsukiyama T. (2003) Mol. Cell 12, 1333–1340 [DOI] [PubMed] [Google Scholar]
- 10. Côté J., Peterson C. L., Workman J. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4947–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorch Y., Cairns B. R., Zhang M., Kornberg R. D. (1998) Cell 94, 29–34 [DOI] [PubMed] [Google Scholar]
- 12. Schnitzler G., Sif S., Kingston R. E. (1998) Cell 94, 17–27 [DOI] [PubMed] [Google Scholar]
- 13. Lorch Y., Zhang M., Kornberg R. D. (1999) Cell 96, 389–392 [DOI] [PubMed] [Google Scholar]
- 14. Phelan M. L., Schnitzler G. R., Kingston R. E. (2000) Mol. Cell. Biol. 20, 6380–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dechassa M. L., Sabri A., Pondugula S., Kassabov S. R., Chatterjee N., Kladde M. P., Bartholomew B. (2010) Mol. Cell 38, 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lorch Y., Maier-Davis B., Kornberg R. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3090–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Almer A., Rudolph H., Hinnen A., Hörz W. (1986) EMBO J. 5, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D. (2003) Mol. Cell 11, 1587–1598 [DOI] [PubMed] [Google Scholar]
- 19. Reinke H., Hörz W. (2003) Mol. Cell 11, 1599–1607 [DOI] [PubMed] [Google Scholar]
- 20. Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D. (2004) Mol. Cell 14, 667–673 [DOI] [PubMed] [Google Scholar]
- 21. Gkikopoulos T., Havas K. M., Dewar H., Owen-Hughes T. (2009) Mol. Cell. Biol. 29, 4057–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwabish M. A., Struhl K. (2007) Mol. Cell. Biol. 27, 6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takahata S., Yu Y., Stillman D. J. (2009) Mol. Cell 34, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barbaric S., Luckenbach T., Schmid A., Blaschke D., Hörz W., Korber P. (2007) J. Biol. Chem. 282, 27610–27621 [DOI] [PubMed] [Google Scholar]
- 25. Ehrensberger A. H., Kornberg R. D. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 10115–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O'Shea E. K. (2003) Science 299, 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gregory P. D., Schmid A., Zavari M., Münsterkötter M., Hörz W. (1999) EMBO J. 18, 6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adkins M. W., Howar S. R., Tyler J. K. (2004) Mol. Cell 14, 657–666 [DOI] [PubMed] [Google Scholar]
- 29. Tyler J. K., Adams C. R., Chen S. R., Kobayashi R., Kamakaka R. T., Kadonaga J. T. (1999) Nature 402, 555–560 [DOI] [PubMed] [Google Scholar]
- 30. Mao C., Brown C. R., Falkovskaia E., Dong S., Hrabeta-Robinson E., Wenger L., Boeger H. (2010) Mol. Syst. Biol. 6, 431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ge L., Rudolph P. (1997) BioTechniques 22, 28–30 [DOI] [PubMed] [Google Scholar]
- 32. Almer A., Hörz W. (1986) EMBO J. 5, 2681–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fascher K. D., Schmitz J., Hörz W. (1993) J. Mol. Biol. 231, 658–667 [DOI] [PubMed] [Google Scholar]
- 34. Depew D. E., Wang J. C. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lenburg M. E., O'Shea E. K. (1996) Trends Biochem. Sci. 21, 383–387 [PubMed] [Google Scholar]
- 36. Barbari S., Fascher K. D., Hörz W. (1992) Nucleic Acids Res. 20, 1031–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaban Y., Ezeokonkwo C., Chung W. H., Zhang F., Kornberg R. D., Maier-Davis B., Lorch Y., Asturias F. J. (2008) Nat. Struct. Mol. Biol. 15, 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., Spagna D., Alvarez D., Kendall J., Krasnitz A., Stepansky A., Hicks J., Bryant G. O., Ptashne M. (2010) Cell 141, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leschziner A. E., Saha A., Wittmeyer J., Zhang Y., Bustamante C., Cairns B. R., Nogales E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4913–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ansari A., Gartenberg M. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keller W. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 4876–4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prunell A. (1998) Biophys. J. 74, 2531–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simpson R. T., Thoma F., Brubaker J. M. (1985) Cell 42, 799–808 [DOI] [PubMed] [Google Scholar]
- 44. Svaren J., Schmitz J., Hörz W. (1994) EMBO J. 13, 4856–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dion M. F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O. J. (2007) Science 315, 1405–1408 [DOI] [PubMed] [Google Scholar]
- 46. Gaudreau L., Schmid A., Blaschke D., Ptashne M., Hörz W. (1997) Cell 89, 55–62 [DOI] [PubMed] [Google Scholar]
- 47. Munsterkötter M., Barbaric S., Hörz W. (2000) J. Biol. Chem. 275, 22678–22685 [DOI] [PubMed] [Google Scholar]
- 48. Swain P. S., Elowitz M. B., Siggia E. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. (2002) Science 297, 1183–1186 [DOI] [PubMed] [Google Scholar]
- 50. Neef D. W., Kladde M. P. (2003) Mol. Cell. Biol. 23, 3788–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raser J. M., O'Shea E. K. (2004) Science 304, 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Flaus A., Owen-Hughes T. (2003) Mol. Cell. Biol. 23, 7767–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y., Smith C. L., Saha A., Grill S. W., Mihardja S., Smith S. B., Cairns B. R., Peterson C. L., Bustamante C. (2006) Mol. Cell 24, 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lorch Y., Griesenbeck J., Boeger H., Maier-Davis B., Kornberg R. D. (2011) Nat. Struct. Mol. Biol. 18, 881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cairns B. R. (2007) Nat. Struct. Mol. Biol. 14, 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boeger H., Griesenbeck J., Kornberg R. D. (2008) Cell 133, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Engeholm M., de Jager M., Flaus A., Brenk R., van Noort J., Owen-Hughes T. (2009) Nat. Struct. Mol. Biol. 16, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. (1998) Cell 95, 717–728 [DOI] [PubMed] [Google Scholar]
- 59. Svaren J., Hörz W. (1997) Trends Biochem. Sci 22, 93–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.