Background: Mitochondrial single-stranded DNA-binding protein (mtSSB) coordinates the functions of the mitochondrial DNA (mtDNA) polymerase and helicase.
Results: mtSSB variants are defective in stimulating mtDNA polymerase and helicase.
Conclusion: mtSSB uses distinct structural elements to interact with mtDNA polymerase and helicase.
Significance: Novel insights are presented into the mechanism of mtDNA replication and the role of mtSSB in human diseases involving mtDNA depletion.
Keywords: DNA-binding Protein, DNA Enzymes, DNA Helicase, DNA Polymerase, DNA Replication, Mitochondria, Mitochondrial Diseases, Mitochondrial DNA
Abstract
The mitochondrial single-stranded DNA-binding protein (mtSSB) is believed to coordinate the functions of DNA polymerase γ (pol γ) and the mitochondrial DNA (mtDNA) helicase at the mtDNA replication fork. We generated five variants of the human mtSSB bearing mutations in amino acid residues specific to metazoans that map on the protein surface, removed from the single-stranded DNA (ssDNA) binding groove. Although the mtSSB variants bound ssDNA with only slightly different affinities, they exhibited distinct capacities to stimulate the DNA polymerase activity of human pol γ and the DNA unwinding activity of human mtDNA helicase in vitro. Interestingly, we observed that the variants with defects in stimulating pol γ had unaltered capacities to stimulate the mtDNA helicase; at the same time, variants showing reduced stimulation of the mtDNA helicase activity promoted DNA synthesis by pol γ similarly to the wild-type mtSSB. The overexpression of the equivalent variants of Drosophila melanogaster mtSSB in S2 cells in culture caused mtDNA depletion under conditions of mitochondrial homeostasis. Furthermore, we observed more severe reduction of mtDNA copy number upon expression of these proteins during recovery from treatment with ethidium bromide, when mtDNA replication is stimulated in vivo. Our findings suggest that mtSSB uses distinct structural elements to interact functionally with its mtDNA replisome partners and to promote proper mtDNA replication in animal cells.
Introduction
Replication of animal mitochondrial DNA (mtDNA)2 is mediated by three protein players that function directly at the replication fork: DNA polymerase γ (pol γ), which catalyzes DNA synthesis per se; mtDNA helicase (also known as Twinkle), which unwinds double-stranded DNA to provide a single-stranded DNA (ssDNA) substrate for pol γ; and mitochondrial single-stranded DNA-binding protein (mtSSB), which binds ssDNA to protect it against damage and to coordinate the functions of pol γ and mtDNA helicase (1, 2). mtSSB has been shown to stimulate the DNA polymerase activity of pol γ both in the Drosophila and human systems (3, 4) and the unwinding activity of the human mtDNA helicase (HsmtDNA helicase) (4, 5). In addition, human mtSSB (HsmtSSB) stimulates strand-displacement DNA synthesis promoted by the combined action of the human pol γ (Hspol γ) and HsmtDNA helicase (2). In vivo, absence or depletion of mtSSB protein causes reduction of mtDNA copy number in Drosophila and human cells in culture (6, 7), and lethality at the third larval stage in developing D. melanogaster (8). Although mtSSB mutations have not thus far been documented in association with human diseases, as is the case for the genes encoding pol γ and mtDNA helicase, a recent report shows a correlation between the levels of mtSSB protein, mtDNA copy number and the aggressiveness of human osteosarcoma cells, suggesting a link between mtDNA replication and cancer progression (9).
To date, mutagenesis studies of mtSSB have focused largely on biochemical and physiological properties of amino acid residues involved in tetramerization and/or ssDNA binding affinities. Li and Williams (10) demonstrated the importance of highly conserved residues among tetrameric SSBs for protomer-protomer interactions in the mouse mtSSB, including H69 (H69, HsmtSSB; H64, DmmtSSB (fly)), and for ssDNA binding, including W49, W68, and F74 (W49, W68 and F74, HsmtSSB; Y50, W63, and F69, DmmtSSB). DmmtSSB variants bearing W63A and/or F69A substitutions were defective in DNA binding in vitro as predicted, and the corresponding mutant genes promoted mtDNA depletion when expressed in Drosophila S2 cells in culture (6). Interestingly, these proteins were also defective in stimulating the DNA polymerase activity of Drosophila pol γ (Dmpol γ). The homologous residues in the prototypical tetrameric SSB from Escherichia coli (EcSSB) have been implicated in similar functions (11–13), in keeping with the high degree of conservation between bacterial and mitochondrial SSBs.
We sought to investigate properties that are specific to animal mtSSBs in the context of mtDNA replication by targeting amino acid residues that are well-conserved across animal species, but differ from those in EcSSB. Unlike mtSSBs, the catalytic subunit of pol γ and the mtDNA helicase share a common ancestry with the bacteriophage T7 DNA polymerase and primase-helicase enzymes, respectively (14). This raises the interesting question of how the bacterial-like mtSSB has evolved to function in concert with the T7-like pol γ and mtDNA helicase in the mtDNA replisome. In an earlier study, we explored biochemically the capacity of terminal deletion variants of HsmtSSB to bind ssDNA and stimulate Hspol γ and HsmtDNA helicase (4). We determined that the absence of the termini was not deleterious and in fact, the stimulation of Hspol γ was higher in the presence of the variants than with wild-type HsmtSSB. In the present report, we describe biochemical and physiological studies of additional mtSSB variants carrying amino acid substitutions or deletions in residues that map outside of the DNA binding groove, mostly in loop regions on the surface of the protein (Fig. 1). Our findings provide insight into how mtSSB coordinates the functions of pol γ and mtDNA helicase and participates in the formation of the mtDNA replisome, a multi-protein complex in which defects caused by genetic mutations can culminate in aging, and in a variety of human diseases (15–18).
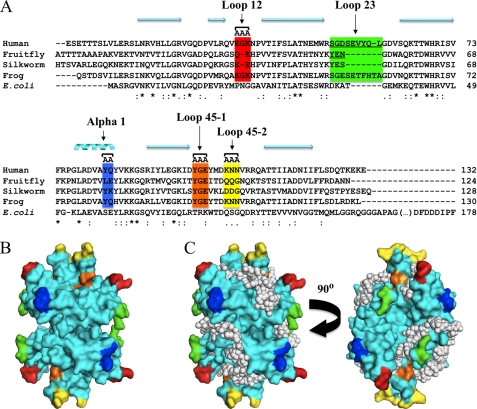
FIGURE 1.
Mutagenesis of mtSSB and model of ssDNA binding. A, multiple sequence alignment of animal mtSSBs with E. coli SSB was performed as described in Oliveira and Kaguni (4). Only the representative mtSSB sequences from humans (GenBankTM accession: NP_003134), fruit fly (Drosophila melanogaster; GenBankTM accession: AAF16936), silkworm (Bombyx mori; GenBankTM accession: ABF51293), and frog (Xenopus laevis; GenBankTM accession: NP_001095241) are shown. Targeted amino acid residues are highlighted in colored boxes: those indicated as AA or AAA were selected for double or triple alanine substitution mutagenesis, respectively; residues in loop 2,3 were targeted for deletion mutagenesis (underlined). B, mapping of targeted residues on the crystal structure of HsmtSSB (23). C, model of HsmtSSB bound to ssDNA (represented as white spheres) showing that the targeted residues do not lie on the DNA binding groove of the protein. Colored residues in B and C are the same as represented in A. The nomenclature adopted to describe the structural elements of HsmtSSB is that used by Raghunathan et al. (41) for EcSSB, in which the loop regions contain the numbers of their flanking β-sheet elements (for example, loop 1,2 lies between β-sheets 1 and 2).
EXPERIMENTAL PROCEDURES
Nucleotides and Nucleic Acids
Unlabeled deoxy- and ribonucleotides were purchased from Amersham Biosciences. [α-32P]dATP and [γ-32P]ATP were purchased from MP Biomedicals. Wild-type M13 (6,407 nt) and pBSKS+ (2,958 nt) DNAs were prepared by standard laboratory methods. Oligodeoxynucleotides complementary to these DNAs were synthesized in an Applied Biosystems oligonucleotide synthesizer. The singly primed M13 DNA used in DNA polymerase assays was prepared as described previously (19). The substrate for the DNA unwinding assays and the 48-mer oligodeoxynucleotide used in gel mobility shift assays (GMSA) were prepared as described in Oliveira and Kaguni (4). The dsRNA used for knockdown of endogenous DmmtSSB (DmmtSSBendo) in S2 cells was produced in vitro using MEGAscript® T7 kit (Ambion), according to manufacturer's specifications. The DNA used as template in the reaction to produce the dsRNA was generated via PCR with the following primers: 5′-TAATACGACTCACTATAGGGAGAAATTTAAGCCCAGATCAC-3′ and 5′-TAATACGACTCACTATAGGGAGATGGAGTACGACTACGCATG-3′, where the underlined sequences represent the minimal promoter needed for T7 RNA polymerase transcription initiation. The amplified fragment (∼120 bp) contained the entire sequence of the 3′-UTR of the mRNA of DmmtSSBendo.
Mutagenesis of HsmtSSB and DmmtSSB
HsmtSSB variants were constructed via site-directed PCR mutagenesis of the HsmtSSBwt cDNA (notwithstanding the mitochondrial presequence) cloned into pET11a vector. PCRs were performed using this vector as template, Pfu DNA polymerase (Stratagene) and standard laboratory methods. The oligonucleotides used for PCR were: 5′-TGAGACAGGTGGcAGcAgcAAATCCAGTCACAATA-3′ and 5′-TATTGTGACTGGATTTgcTgCTgCCACCTGTCTCA-3′ for HsmtSSBloop12; 5′-CTAATGAGATGTGGCGA−GGTGATGTCAGTC-3′ and 5′-GACTGACATCACC−TCGCCACATCTCATTAG-3′ for HsmtSSBloop23; 5′-CAGAGACGTGGCAgcTgcATATGTGAAAAAG-3′ and 5′-CTTTTTCACATATgcAgcTGCCACGTCTCTG-3′ for HsmtSSBα1; 5′-GGGAAAATAGACgcTGcTGcATACATGGATAAAAA-3′ and 5′-TTTTTATCCATGTATgCAgCAgcGTCTATTTTCCC-3′ for HsmtSSBloop45–1; and 5′-GTGAATACATGGATgcAgcTgcTGTGAGGCGACAAG-3′ and 5′-CTTGTCGCCTCACAgcAgcTgcATCCATGTATTCAC-3′ for HsmtSSBloop45–2. DmmtSSB variants were constructed via site-directed PCR mutagenesis of the DmmtSSBwt cDNA cloned into pMt/Hy vector. PCRs were performed using this vector as template, Pfu DNA polymerase (Stratagene) and standard laboratory methods. The oligonucleotides used for PCR were: 5′-CCGGCACGCGGG−GTTGAAAAAACTG-3′ and 5′-CAGTTTTTTCAAC−CCCGCGTGCCGG-3′ for DmmtSSBΔN; 5′-GCTGCGTGGATCCgcGGcGCATCCGGTGGTC-3′ and 5′-GACCACCGGATGCgCCgcGGATCCACGCAGC-3′ for DmmtSSBloop12; 5′-CACCAACTACAAA−GGCGACTGGGCC-3′ and 5′-GGCCCAGTCGCC−TTTGTAGTTGGTG-3′ for DmmtSSBloop23; 5′-GCGTGACACCGTGgcGGcATACTTGAAGAAGG-3′ and 5′-CCTTCTTCAAGTATgCCgcCACGGTGTCACGC-3′ for DmmtSSBα1; 5′-GGGAAAGATCACCgcTGcAGcGATCACCGACCAGC-3′ and 5′-GCTGGTCGGTGATCgCTgCAgcGGTGATCTTTCCC-3′ for DmmtSSBloop45–1; 5′-GAGAGATCACCGACgcGgcGGcCAACCAGAAGACT-3′ and 5′-AGTCTTCTGGTTGgCCgcCgcGTCGGTGATCTCTC-3′ for DmmtSSBloop45–2; and 5′-GTTGTTTTTCCGTTAAGCCAACAACTAA-3′ and 5′-TTAGTTGTTGGCTTAACGGAAAAACAAC-3′ for DmmtSSBΔC. The lowercase letters, the −, and the underlined sequences indicate the sites where mutations were introduced to create alanine substitutions, amino acid deletions, or a stop codon, respectively.
Purification of HsmtSSBs, Hspol γ, and HsmtDNA Helicase
Recombinant human mtSSB proteins were prepared from bacterial cells, as described by Oliveira and Kaguni (4). Recombinant human pol γ-α exo− and pol γ-β were prepared from Sf9 and bacterial cells, respectively, as described by Oliveira and Kaguni (20). Recombinant human mtDNA helicase was prepared from Sf9 cells, as described by Ziebarth et al. (21).
ssDNA Binding and Gel Mobility Shift Assays
Reaction mixtures (20 μl) contained 20 mm Tris-HCl, pH 7.5, 1 mm DTT, 4 mm MgCl2, 50 mm NaCl, 36 fmol 5′-end-labeled 48-mer, and the indicated amounts of the HsmtSSB proteins. Incubation was at 20 °C for 10 min. Samples were processed and electrophoresed in 6% native polyacrylamide gels. The amounts of shifted and free oligonucleotide were quantitated as follows: % ssDNA bound = (VS/(VS + VF)) x 100, where VS represents the volume of the shifted and VF the volume of unshifted oligonucleotide in the sample lane of interest.
dsDNA Unwinding Assays
Reaction mixtures (50 μl) contained 20 mm Tris-HCl, pH 7.5, 10% glycerol, 500 μg/ml bovine serum albumin, 10 mm DTT, 4 mm MgCl2, 3 mm ATP, 50 mm KCl, 0.4 nm of DNA unwinding substrate, 3.5 nm of mtDNA helicase (as hexamer), and the indicated concentrations of HsmtSSB proteins. The reactions were pre-incubated at 37 °C for 5 min prior to the addition of the helicase. Once the helicase was added, the reactions were incubated further at 37 °C for 30 min and then stopped by the addition of 5 μl of 10× stop solution (6% SDS, 100 mm EDTA, pH 8.0), followed by 5 μl of 10× loading buffer (50% glycerol, 0.25% bromphenol blue). DNA products were fractionated from substrate by electrophoresis in a 22% polyacrylamide gel (59:1 acrylamide/bisacrylamide) using 1× TBE (90 mm Tris-HCl-borate, 2 mm EDTA) at 600 V for ∼30 min. After electrophoresis, the gel was processed and the unwinding activity was quantitated as described in Oliveira and Kaguni (4).
DNA Polymerase γ Stimulation Assays
Reaction mixtures (50 μl) contained 50 mm Tris-HCl, pH 8.5, 4 mm MgCl2, 400 μg/ml bovine serum albumin, 10 mm DTT, 20–100 mm KCl, 20 μm each dGTP, dATP, dCTP, and dTTP, [α-32P]dATP (2 μCi), 3 μm (as nucleotide) singly primed wild-type M13 DNA (6,407 nt), 10 ng Hspol γ-α exo− Fr IV, 48 ng Hspol γ-β Fr III, and the indicated amounts of HsmtSSB proteins. Incubation was at 37 °C for 30 min. Samples were processed, and nucleotide incorporation was quantitated in a liquid scintillation counter.
Generation of Stable Cell Lines, Knockdown of DmmtSSBendo, and Induction of DmmtSSB Variants
Drosophila Schneider S2 cells were cultured at 25 °C in Drosophila Schneider medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin G and 50 μg/ml streptomycin sulfate (Invitrogen). For establishment of stable lines, ∼1.2 × 107 cells were transfected with 1 μg of pMt/Hy vector carrying DmmtSSBwt or variant genes using the Effectene kit (Qiagen), according to manufacturer's specifications. Hygromycin-resistant cells were selected with 0.2 mg/ml hygromycin for at least 5 passages, before transfer to standard growth medium. For knockdown of endogenous DmmtSSB protein, 3 μg of dsRNA (representing the 3′-UTR of DmmtSSBendo mRNA) were delivered to 3 × 106 cells using the Effectene kit. Cells were cultured for 3 days before reduction in levels of DmmtSSBendo protein was observed. In the experiments with EtBr, cells were treated with 0.2 μg of EtBr per ml of growth medium 3 days prior to treatment with dsRNA. For induction of expression of DmmtSSB variants, cells treated with dsRNA for 3 days were subjected to the indicated concentrations of CuSO4 to promote appropriate expression from the metallothionein promoter (22).
Immunoblotting
For DmmtSSB detection, total protein extracts from S2 cells were separated in 17% SDS-polyacrylamide gels following standard laboratory methods. Proteins were transferred to nitrocellulose membranes (Protran BA83, Whatman), which were preincubated for 1 h with 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBST), followed by incubation of 1 h with DmmtSSB antibody (1:8000 dilution in TBST/1% milk). The membranes were washed several times with TBST for 1.5 h, incubated with horseradish peroxidase-conjugated anti-rabbit IgG (1:16000 dilution in TBST/1% milk, Bio-Rad) for 1 h, and washed again several times with TBST for 1.5 h. DmmtSSB bands were visualized using ECL Western blotting reagents (Amersham Biosciences). For β-tubulin detection, a similar procedure was performed, except that protein extracts were run on 12% SDS-polycrylamide gels and the antibodies used were E7 supernatant (1:50 dilution in TBST/1% milk, DSHB, University of Iowa) and alkaline phosphatase-conjugated anti-mouse IgG (1:2000 dilution in TBST/1% milk, Bio-Rad). β-tubulin bands were visualized using alkaline phosphatase buffer (100 mm Tris-HCl, pH 9.5, 100 mm NaCl and 5 mm MgCl2) containing 330 μg/ml nitro blue tetrazolium and 165 μg/ml 5-bromo-4-chloro-3-indolyl phosphate.
Quantitative Real Time-PCR
Total DNA was extracted from S2 cells using phenol/chloroform protocol and standard laboratory procedures. mtDNA copy number was determined by quantification of relative amounts of mtDNA to nuclear DNA via real time amplification of a fragment of the mitochondrial 16 S gene (primers: 5′-AAAAAGATTGCGACCTCGAT-3′ and 5′-AAACCAACCTGGCTTACACC-3′) and the nuclear RpL32 gene (primers: 5′-AGGCCCAAGATCGTGAAGAA-3′ and 5′-TGTGCACCAGGAACTTCTTGAA-3′). Reactions were performed using SYBR® Green JumpStartTM Taq ReadyMixTM (Sigma-Aldrich) on a 7500 Real Time PCR System instrument (Applied Biosystems), according to manufacturer's specifications.
Molecular Modeling
To construct the model of HsmtSSB bound to ssDNA, the crystal structure of HsmtSSB (PDB code 3ULL (23)) was aligned to the structure of EcSSB bound to ssDNA (PDB code 1EYG (13)) using PyMol (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC). The atomic coordinates of EcSSB were then removed, allowing placement of the ssDNA in the DNA binding groove of HsmtSSB without major atomic collisions.
To construct the model of the Hspol γ-HsmtSSB interaction, the regions of HsmtSSB that are disordered in the crystal structure (E1-V9, N terminus; S54-L59, loop 2,3; and D126-E132, C terminus; PDB code 3ULL (23)) were first modeled using MODELLER (24) at default parameters. The tetrameric HsmtSSB structure containing the modeled termini and loop 2,3 was submitted to the software ClusPro 2.0 (25) for docking onto the crystal structure of the Hspol γ holoenzyme (PDB code 3IKM (26)) at default parameters. All of the figures were generated and the model was analyzed using the software PyMol.
RESULTS
We selected candidate regions of the mtSSB protein for mutagenesis based on the assumption that residues important for functional and/or physical interactions with pol γ and mtDNA helicase would be well conserved among animal mtSSB sequences, and also locate on the surface of the protein structure. We avoided residues conserved between mtSSBs and EcSSB, because of their essential roles in the stabilization of the tertiary/quaternary structure of the protein and/or in contacting DNA directly; disrupting such properties would be expected to cause indirect effects on the ability of the protein to stimulate its mtDNA replisome partners (6). The selected residues and an evaluation of their locations in a molecular model of HsmtSSB bound to ssDNA that indicates none of them are likely to interact directly with ssDNA are shown in Fig. 1.
Differential Stimulation of Hspol γ and HsmtDNA Helicase by Variants of HsmtSSB
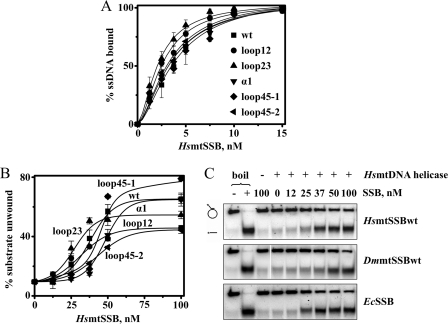
We expressed and purified to near-homogeneity recombinant wild-type HsmtSSB (HsmtSSBwt), and proteins bearing double- or triple-alanine substitutions mapping in loop 1,2 (E33A/G34A/K35A, HsmtSSBloop12), in α-helix 1 (Y83A/Q84A, HsmtSSBα1), and in loop 4,5 (Y100A/G101A/E102A, HsmtSSBloop45–1; and K106A/N107A/N108A, HsmtSSBloop45–2), or a deletion in loop 2,3 (S51-L59, HsmtSSBloop23)(supplemental Fig. S1). Velocity sedimentation indicated that all the variant HsmtSSB proteins retain their homotetrameric state (S values = 4.3; supplemental Fig. S2), whereas their ssDNA-binding affinities, as analyzed by gel mobility shift assays (GMSA), varied slightly (Fig. 2A). The variants HsmtSSBloop12 (Kd = 2.9 nm) and HsmtSSBloop23 (Kd = 2.4 nm) bound to ssDNA with affinities 1.3- and 1.6-fold higher than that of HsmtSSBwt (Kd = 3.8 nm), respectively; the affinities of the three other variants (α1: Kd = 3.8 nm; loop45–1: Kd = 4.1 nm; loop45–2: Kd = 3.7 nm) did not differ substantially from that of HsmtSSBwt, consistent with the model presented in Fig. 1C. At the same time, no significant variations were observed in the gel shift patterns among any of the variants as compared with the wild-type protein (data not shown).
FIGURE 2.
ssDNA binding affinities and stimulation of HsmtDNA helicase by HsmtSSB variants. A, ssDNA-binding affinity was evaluated by GMSA, as described under “Experimental Procedures.” The fraction of unbound and bound oligomer was quantitated, and the data were plotted as the average percent of substrate utilized from three independent experiments. B and C, DNA unwinding assays were performed as described under “Experimental Procedures,” using 3.5 nm of HsmtDNA helicase (as hexamer). “−” and “+ boil” lanes represent the intact and denatured (heated to 100 °C for 2 min prior to loading) substrate, respectively. The data in B represent the average of unwound substrate as percent from three independent experiments.
We then examined the ability of the HsmtSSB variants and non-cognate SSBs to stimulate the dsDNA unwinding activity of HsmtDNA helicase. Interestingly, helicase stimulation by HsmtSSBloop12 and HsmtSSBloop23 reflects their increased ssDNA binding, as maximal stimulation is achieved at lower concentrations of these variants (37 nm) as compared with HsmtSSBwt (50 nm). Nevertheless, maximal stimulation by HsmtSSBloop12 is reduced ∼40% as compared with that by HsmtSSBwt, whereas the reduction caused by HsmtSSBloop23 is only ∼15% (Fig. 2B). In addition, HsmtSSBloop45–2 also showed a ∼40% reduced capacity to stimulate HsmtDNA helicase in vitro, suggesting that residues in loop 1,2 and 4,5–2 are responsible for the functional interactions that we observed between HsmtSSB and HsmtDNA helicase. In comparison, we found that the maximal stimulation of the HsmtDNA helicase by HsmtSSBwt is similar to that contributed by the non-cognate DmmtSSBwt and EcSSB (Fig. 2C), in contrast to the results reported by Korhonen et al. (5).
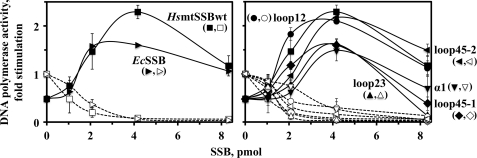
Next, we measured the stimulation of the DNA polymerase activity of Hspol γ on a singly primed ssDNA template. As we reported previously (4), we observed that the DNA polymerase activity of Hspol γ in the absence of HsmtSSB was ∼2-fold higher at 100 mm than at 20 mm KCl. In contrast, in the presence of the variant HsmtSSBs, Hspol γ was stimulated at 20 mm KCl, but inhibited completely at 100 mm KCl (Fig. 3). Furthermore, the variants HsmtSSBloop23, HsmtSSBα1, and HsmtSSBloop45–1 show a reduced capacity to stimulate DNA synthesis by Hspol γ (∼60% of that of HsmtSSBwt), which indicates that the altered amino acid residues in these variants are important for the functional interactions between HsmtSSB and Hspol γ. Notably, the stimulation of Hspol γ by these defective variants is similar to that observed with the non-cognate EcSSB, and the HsmtSSB residues identified as important for Hspol γ stimulation differ from those shown to be important for stimulation of HsmtDNA helicase (Fig. 2B).
FIGURE 3.
Variants of HsmtSSB have reduced capacity to stimulate DNA polymerase activity of Hspol γ. DNA polymerase assays were performed as described under “Experimental Procedures,” on singly primed M13 DNA using 60 fmol of Hspol γ holoenzyme and the indicated amounts of SSB. Assays were performed at 20 mm KCl (solid lines and symbols) and at 100 mm KCl (dashed lines and open symbols). The data were normalized to the amount of nucleotide incorporated by Hspol γ at 100 mm KCl in the absence of SSB (arbitrarily set to 1), and represent the average of three independent experiments.
mtDNA Depletion in Cells Overexpressing Variants of DmmtSSB under Conditions of Mitochondrial Homeostasis
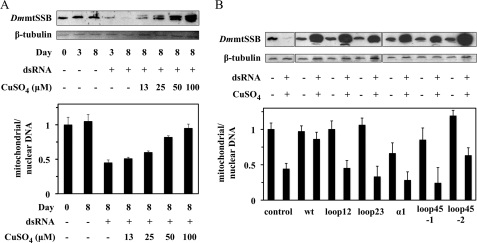
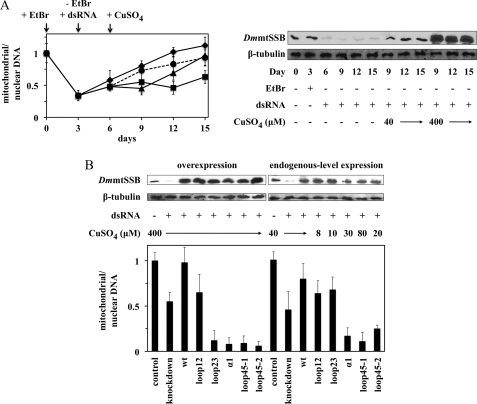
We developed a Drosophila S2 cell system in which we knocked down the DmmtSSB protein (DmmtSSBendo) by treatment with dsRNA homologous to the 3′-UTR of the endogenous gene and expressed an exogenous DmmtSSB variant, to evaluate mtDNA maintenance and replication under various physiological conditions. dsRNA treatment depleted DmmtSSBendo and mtDNA copy number to ∼10 and ∼40% of the levels in the control cells, respectively. In agreement with previous data from our laboratory (6), expression of an exogenous DmmtSSBwt gene promoted an increase in mtDNA copy number proportional to the level of protein induction (Fig. 4A).
FIGURE 4.
Knockdown of DmmtSSBendo and overexpression of DmmtSSB variants in S2 cells causes mtDNA depletion under conditions of mitochondrial homeostasis. A, cells carrying pMt/DmmtSSBwt/Hy were cultured in the presence of dsRNA homologous to the 3′-UTR of DmmtSSBendo mRNA (1 μg per 106 cells) and the indicated concentration of CuSO4, and harvested at the indicated time points. Immunoblot analysis of DmmtSSB and β-tubulin proteins (upper panel), and qPCR to measure relative mtDNA copy number (lower panel), were performed as described under “Experimental Procedures.” The band above the exogenous DmmtSSBwt (upper panel) represents the accumulation of the pre-processed form of the protein that contains the mitochondrial targeting signal. B, cells carrying pMt/Hy plasmids with the indicated DmmtSSB variants were cultured for 14 days in the presence or absence of dsRNA (1 μg per 106 cells) and 0.4 mm CuSO4. Effects of overexpression on mtDNA levels were quantitated and are shown in the lower panel. The data were normalized to the ratio of mitochondrial/nuclear DNA in S2 cells without treatment (arbitrarily set to 1), and represent the average of experiments from at least two independent cell lines. Error bars represent the standard deviation among the lines. Bands below the exogenous DmmtSSBwt, loop12 and loop45–1 protein bands (upper panel) represent proteolytic products observed upon their overexpression, and/or in sample processing.
We also established stable cell lines carrying the genes for five DmmtSSB variants equivalent to the HsmtSSB variants described above (Q35A/E36A, DmmtSSBloop12; ΔY52-N54, DmmtSSBloop23; L68A/E69A, DmmtSSBα1; Y85A/G86A/E87A, DmmtSSBloop45–1; and Q91A/Q92A/G93A, DmmtSSBloop45–2). Additionally, we established cell lines to express DmmtSSBΔN (Δ1–11) and DmmtSSBΔC (Δ121–124), which we have recently shown to modulate pol γ stimulation in vitro in the human system (4). However, we observed that their induction is consistently poor (data not shown), indicating that the deletion variants are unstable in the cellular environment. Immunoblot analysis of cell extracts showed that expression of the other five variants occurs at high levels after 14 days of induction with 0.4 mm CuSO4 (determined quantitatively to be 5–10-fold higher than DmmtSSBend, data not shown and Fig. 4B, upper panel). Relative mtDNA copy number in cells overexpressing DmmtSSBwt was unchanged, but was reduced significantly (25–60% of that of control cells) in cells overexpressing DmmtSSBloop12, loop23, α1, loop45–1, and loop45–2, and in DmmtSSBendo-knockdown cells (Fig. 4B, lower panel).
Defects in mtDNA Repletion upon Expression of DmmtSSB Variants
We then investigated mtDNA repletion in S2 cells expressing the DmmtSSB variants after mtDNA depletion with ethidium bromide (EtBr). Application of low concentrations of EtBr in the cell culture (0.2 μg/ml growth media) depleted mtDNA to ∼30% of that of control cells after 3 days of treatment, without affecting DmmtSSBendo levels (Fig. 5A). After removal of EtBr from the culture, control cells recovered completely from mtDNA depletion in ∼9 days, whereas mtDNA levels in DmmtSSBendo-knockdown cells remained substantially lower (55% of that of control cells) for as long as 15 days. Cells carrying the exogenous DmmtSSBwt gene were treated with dsRNA to knock down DmmtSSBendo, and with CuSO4 to induce the expression of DmmtSSBwt after EtBr treatment. When DmmtSSBwt was expressed at a level comparable to DmmtSSBendo in control cells (in the presence of 0.04 mm CuSO4), recovery from mtDNA depletion also occurred in ∼9 days, after a 3-day lag time. This delay may result from our experimental design, which requires CuSO4 addition to the cells 3 days after the dsRNA treatment (see “Experimental Procedures” for details). In contrast to endogenous-level DmmtSSBwt expression, the 3-day lag time was not observed upon DmmtSSBwt overexpression, and mtDNA copy number was restored to normal levels at a faster rate (Fig. 5A, right panel).
FIGURE 5.
Expression of DmmtSSB variants impedes recovery from mtDNA depletion in Drosophila S2 cells. A, left panel, 0.2 μg/ml of EtBr was applied for 3 days to the growth media of cells carrying the DmmtSSBwt gene, followed by a recovery time of 12 days upon removal of EtBr. Control cells had neither dsRNA or CuSO4 treatment (diamonds and solid line), whereas experimental lines were treated with dsRNA only (squares and solid line), dsRNA and 40 μm CuSO4 (triangles and solid line), or dsRNA and 400 μm CuSO4 (circles and dashed line). The time point in which each treatment was initiated is indicated with arrows. Relative mtDNA copy number was analyzed by qPCR, as described under “Experimental Procedures.” Right panel, DmmtSSB and β-tubulin protein levels were analyzed by immunoblot, as described under “Experimental Procedures.” B, the experimental design was as in A using cells expressing different DmmtSSB variant genes. The concentrations of CuSO4 used to induce the overexpression and endogenous-level expression of DmmtSSB proteins are indicated. Cells were harvested at day 12 (9 days of recovery from EtBr treatment). DmmtSSB and β-tubulin protein levels were analyzed by immunoblot (upper panel), and relative mtDNA copy number was analyzed by qPCR (lower panel). The data were normalized to the ratio of mitochondrial/nuclear DNA in control S2 cells (arbitrarily set to 1), and represent the average of experiments with two independent cell lines. Error bars represent the standard deviation among the lines.
Next, we induced the five DmmtSSB variant proteins at two expression levels after the combined EtBr and dsRNA treatments: expression levels equivalent to DmmtSSBendo under normal growth conditions (endogenous-level) and overexpression (Fig. 5B). We observed distinct phenotypes of mtDNA depletion, allowing us to categorize the variants in three groups. The first comprises DmmtSSBloop12, which is the only protein that caused strong reduction of mtDNA copy number under conditions of mitochondrial homeostasis (40% of that of control cells), but showed only a small-to-moderate effect under recovery from depletion, either upon endogenous-level expression or overexpression of the protein (60–80% of that of cells expressing DmmtSSBwt). The second group comprises DmmtSSBloop23, which caused a clear mtDNA depletion (15–30% of that of control cells) only when high levels of the protein were achieved (overexpression conditions). The third group comprises DmmtSSBα1, loop45–1 and loop45–2, which caused reduction in mtDNA copy number under all conditions tested (10–50% of that of control cells), with more severe phenotypes revealed upon overexpression of the protein during recovery from the EtBr treatment. Interestingly, the knockdown of DmmtSSBendo (dsRNA treatment only) caused about the same reduction in mtDNA copy number under both homeostasis and EtBr recovery conditions (Figs. 4B and 5B).
DISCUSSION
DNA replisomes are well-coordinated machines in which the activity of one protein component is regulated by that of the others. For example, replicative helicases from bacteria and viruses that unwind dsDNA at low catalytic rates become highly efficient when coupled to DNA polymerases and/or SSBs (reviewed in Refs. 27, 28). At the animal mtDNA replication fork, little is known about how the three key components of the replisome function coordinately to copy the mitochondrial genome. Studies to date have focused largely on structure-function relationships in pol γ and mtDNA helicase, and their association with human diseases, such as progressive external ophthalmoplegia (PEO) and Alpers syndrome (1, 17, 18, 29, 30). Furthermore, deregulation of replisome function in vivo has been shown in human cell lines expressing active site mutants of pol γ and mtDNA helicase: dysfunctional pol γ mutants induced delayed lagging-DNA strand synthesis causing replication stalling, whereas defective helicases caused an increased rate of initiation of lagging-DNA strand synthesis relative to the rate of mtDNA fork progression (31).
Here, we present a biochemical and physiological analysis of the role of mtSSB at the mtDNA replication fork. We generated five recombinant HsmtSSB variants designed not to exhibit defects in ssDNA binding, and found that two of them (HsmtSSBloop12 and loop23) actually bound ssDNA with slightly higher affinity than HsmtSSBwt, indicating that loops 1,2 and 2,3 in HsmtSSB are either closer to the DNA binding channel and/or participate negatively in an alternative binding mode. We speculate that loop 2,3 of vertebrate mtSSBs, which contain a 6–7 amino acid insertion relative to this region in invertebrate mtSSBs and bacterial SSBs, can form a flexible domain that may disturb ssDNA binding in the absence of pol γ. Most of the amino acid residues in loop 2,3 are disordered in the crystal structure of HsmtSSB (23), the loop carries a slight negative charge as analyzed by electrostatic surface potential, and it appears to be important for pol γ stimulation in vitro and in vivo, as discussed below.
Our group and others have shown that HsmtSSBwt interacts functionally with HsmtDNA helicase using a preformed replication fork-like substrate to stimulate its dsDNA unwinding activity (4, 5). On such a simple substrate one might predict that the helicase would be stimulated in vitro by any SSB capable of preventing reannealing of the unwound DNA. Indeed, we show here that both EcSSB and DmmtSSB can stimulate the activity of HsmtDNA helicase similarly to HsmtSSBwt. In contrast, Korhonen et al. (5) failed to show a stimulation by EcSSB and interpreted their findings as a specific interaction between HsmDNA helicase and HsmtSSBwt. Though we cannot document conclusively an explanation for the different observations discussed here, we note that the stimulation of HsmtDNA helicase by HsmtSSBwt reported by Korhonen et al. (5) was only moderate (∼2.5-fold), whereas our data show a ∼7-fold effect ((4), Fig. 2B) that may allow a discrimination of specificity.
Our studies of the fly and human mtSSB variants suggest that residues in loop 1,2 and loop 4,5 serve important but distinct roles in interacting with the mtDNA helicase. Interestingly, loop 1,2 of DmmtSSB appears to be involved only in mtDNA homeostasis in S2 cells, whereas DmmtSSBloop45–2 induces moderate and severe mtDNA depletion under homeostasis and EtBr recovery conditions, respectively. During recovery from EtBr treatment, mtDNA replication is apparently stimulated and may occur by a different mechanism and/or more independently from the cell cycle than under homeostasis conditions. We speculate that loop 4,5–2 provides the primary point of interaction between mtSSB and mtDNA helicase that promotes efficient dsDNA unwinding during mtDNA replication. Loop 1,2 may also contact the helicase, perhaps facilitating enzyme function only to maintain mtDNA at normal levels under homeostasis conditions. This hypothesis correlates with our findings that HsmtSSBloop45–2 shows the lowest stimulation of HsmtDNA helicase in vitro, even at protein levels 3-fold higher than that required to saturate the ssDNA substrate (data not shown). Alternatively, disruption of loop 4,5 may cause a primary biochemical defect that we have not yet tested but that results in reduced DNA polymerase and/or helicase stimulation in vitro, and depletion of mtDNA in cultured cells. Though cooperativity in ssDNA binding is debated in mtSSBs (32, 33), loss of this property would represent a plausible explanation, because loop 4,5 of EcSSB has been implicated in cooperative DNA binding in the bacterial protein (13).
Functional interactions between pol γ and mtSSB have been shown clearly with native and recombinant forms of the Drosophila proteins (3, 6, 8, 33). Both the DNA polymerase and exonuclease activities of the Dmpol γ holoenzyme are stimulated 15- to 20-fold on singly primed ssDNA templates by DmmtSSB over a broad range of KCl concentrations. DmmtSSB increases primer recognition, DNA synthesis, processivity, and mispair hydrolysis during proofreading DNA synthesis by Dmpol γ. In the human system, Hspol γ holoenzyme is stimulated by HsmtSSB only at very low ionic strength, and the overall stimulation is only ∼2.5-fold (Ref. 4 and Fig. 3). We showed here that EcSSB can also interact functionally with Hspol γ and stimulate its DNA polymerase activity ∼1.5-fold at 20 mm KCl, establishing a baseline level of Hspol γ stimulation in vitro. The HsmtSSB variants in loop23, α1 and loop45–1 were able to stimulate the mitochondrial replicase similarly to EcSSB, suggesting possible defects in pol γ-mtSSB interactions. We found that DmmtSSBα1 and loop45–1 caused mtDNA depletion in S2 cells under all conditions tested, whereas DmmtSSBloop23 only reduced mtDNA copy number when expressed >50-fold higher than DmmtSSBendo. Residual DmmtSSBendo, and mixed DmmtSSBendo-DmmtSSBloop23 tetramers in cells expressing DmmtSSBloop23 at levels equivalent to the endogenous level (resulting in a ratio of ∼10 DmmtSSBloop23 to 1 DmmtSSBendo) appear to be sufficient to maintain mtDNA at the same level as in DmmtSSBendo-knockdown cells (40–50% of control S2 cells). Thus, at endogenous-level expression, the increased ssDNA-binding ability of DmmtSSBloop23 may compensate for the loss of native pol γ-mtSSB interactions. However, as indicated above, loop 2,3 is one of the most variable regions among animal mtSSBs, suggesting that the biochemical results observed with HsmtSSBloop23 and the physiological response observed with DmmtSSBloop23 are likely species-specific effects that represent distinct mechanisms of pol γ regulation. In fact, as we have speculated previously (4), the presence of a dimeric accessory subunit exclusive to vertebrate pol γs might increase DNA synthesis by the mitochondrial replicase to a level that makes the contribution of mtSSB to the overall rate of DNA synthesis only moderate. By comparison, the substantial stimulation of DmmtSSB on the DNA polymerase activity of Dmpol γ holoenzyme appears to correlate with the presence of a monomeric accessory subunit in insect pol γs, and to a short loop 2,3 in mtSSBs.
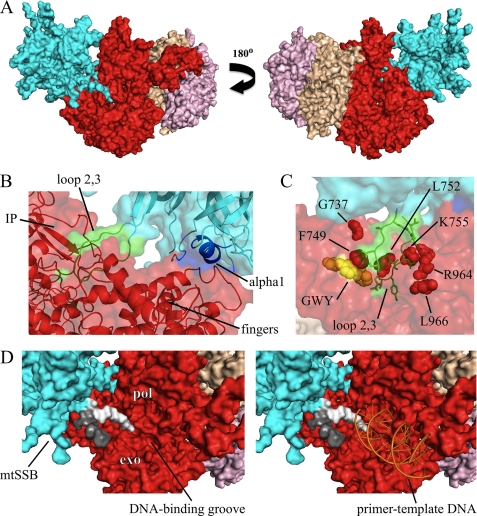
To provide structural support for our current results and hypotheses, we developed a computational model in which we docked the crystal structure of HsmtSSB onto the structure of Hspol γ (Fig. 6). The model offers insight into how loop 2,3 and α-helix 1 of HsmtSSB may interact with the intrinsic processivity (IP) and fingers subdomains of the catalytic subunit (Hspol γ-α) of the mitochondrial replicase (Fig. 6, A and B). We did not observe any evidence of protein-protein interactions involving loop 4,5–1 of HsmtSSB, presumably because the amino acid residues of additional sites of interaction with mtSSB are disordered in the crystal structure of Hspol γ-α, such as residues 319–344 in the exonuclease domain and 674–709 in the IP subdomain of the spacer domain (26). Notably, amino acid substitutions in Hspol γ-α alleles that are associated with Alpers, Parkinson, Charcot-Marie Tooth, PEO, and ataxia-neuropathy diseases in human patients (17, 34–38) locate in the vicinity of the region of potential interaction between loop 2,3 of HsmtSSB and Hspol γ-α (Fig. 6C), suggesting that defects in Hspol γ stimulation by HsmtSSB can explain the phenotype of mtDNA depletion and/or deletions associated with such diseases. Interestingly, amino acid residues G619, W620, and Y622 of Hspol γ-α are found in the same region (Fig. 6C), supporting our previous results that Dmpol γ variants bearing G575A/W576A/F578A substitutions had reduced stimulation by DmmtSSB in vitro (39). Additionally, the model suggests that either of the negatively charged termini of HsmtSSB could occupy partially the positively charged DNA-binding pocket of the mitochondrial replicase, disrupting the progression of DNA synthesis by dislodging transiently the primer-template DNA from the DNA polymerase active site of pol γ (Fig. 6D). HsmtSSB proteins lacking either or both termini would then render the holoenzyme readily available for DNA binding, thus promoting increased DNA synthesis, as shown in our previous report (4). Unfortunately, DmmtSSB variants lacking the N or C terminus are not stable when expressed in Drosophila S2 cells, which prevents us from exploring this phenomenon further at present.
FIGURE 6.
Structural model of pol γ-mtSSB interaction. The model was generated by docking the crystal structure of HsmtSSB (PDB code 3ULL (23)) onto that of the Hspol γ holoenzyme (PDB code 3KIM (26)), using the software ClusPro 2.0 server (25). Because the termini and loop 2,3 are disordered in the crystal structure of HsmtSSB, we modeled their residues using the software MODELLER (24) prior to docking HsmtSSB onto the Hspol γ structure. A, general view of the model. HsmtSSB tetramer is depicted in cyan; the catalytic subunit, and the proximal and the distal protomers of the accessory subunit of Hspol γ are shown in red, wheat, and pink, respectively. B, detailed view of the interactions of loop 2,3 and α-helix 1 of HsmtSSB with the intrinsic processivity (IP) and fingers subdomains of Hspol γ-α. Residues in loop 2,3 and α-helix 1 of HsmtSSB are represented in green and blue, respectively. C, location of residues of Hspol γ-α that have been found associated with various human diseases (red spheres), and residues G619, W620, and Y622 (yellow spheres) that have been implicated in functional interactions with mtSSB (39). The disease allele G737R is associated with PEO, Alpers, Parkinson, and Charcot Marie Tooth diseases; F749S, L752P, K755E, and L966R are found in patients with Alpers disease; and R964C is found in ataxia-neuropathy (reviewed in Ref. 17). D, left panel, positioning of the N- (light gray) and C termini (dark gray) of HsmtSSB in relation to the DNA binding groove of Hspol γ-α, based upon the docking model. pol, DNA polymerase domain; exo, 3′-5′ exonuclease domain. Right panel, model of primer-template DNA binding by Hspol γ (18) and the possible interference by the HsmtSSB termini.
Although physical interaction data are not presented here and similar structural analyses and modeling predictions cannot currently be performed reliably for mtSSB interactions with mtDNA helicase, our findings suggest that mtSSB uses a repertoire of structural elements in stimulating pol γ and mtDNA helicase to ensure proper mtDNA maintenance in animal cells. Its functional properties may result from multiple sites of physical interaction that may also be associated with different mtDNA replication modes in vivo. In any case, it is evident that the mechanisms by which mtSSBs function at the mtDNA replication fork are distinct from those that bacterial, viral and nuclear eukaryotic SSBs employ in their respective replisomes, despite their shared features in ssDNA binding (23, 40–43). Further biochemical, structural and physiological studies are clearly needed to test the relevant hypotheses and to help understand the mechanisms of mtDNA replication in healthy cells and in myriad disease states.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM45295.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- mtDNA
- mitochondrial DNA
- SSB
- single-stranded DNA-binding protein
- mtSSB
- mitochondrial SSB
- ssDNA
- single-stranded DNA
- EcSSB
- E. coli SSB
- pol γ
- DNA polymerase γ
- DmmtSSB
- D. melanogaster mtSSB
- DmmtSSBendo
- endogenous DmmtSSB from S2 cells
- Dmpol γ
- D. melanogaster pol γ
- HsmtSSB
- human mtSSB
- HsmtDNA helicase
- human mtDNA helicase
- Hspol γ
- human pol γ
- GMSA
- gel mobility shift assay
- qPCR
- quantitative real-time PCR.
REFERENCES
- 1. Kaguni L. S. (2004) Annu. Rev. Biochem. 73, 293–320 [DOI] [PubMed] [Google Scholar]
- 2. Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M. (2004) EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farr C. L., Wang Y., Kaguni L. S. (1999) J. Biol. Chem. 274, 14779–14785 [DOI] [PubMed] [Google Scholar]
- 4. Oliveira M. T., Kaguni L. S. (2010) PLoS One 5, e15379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korhonen J. A., Gaspari M., Falkenberg M. (2003) J. Biol. Chem. 278, 48627–48632 [DOI] [PubMed] [Google Scholar]
- 6. Farr C. L., Matsushima Y., Lagina A. T., 3rd, Luo N., Kaguni L. S. (2004) J. Biol. Chem. 279, 17047–17053 [DOI] [PubMed] [Google Scholar]
- 7. Ruhanen H., Borrie S., Szabadkai G., Tyynismaa H., Jones A. W., Kang D., Taanman J. W., Yasukawa T. (2010) Biochim. Biophys. Acta 1803, 931–939 [DOI] [PubMed] [Google Scholar]
- 8. Maier D., Farr C. L., Poeck B., Alahari A., Vogel M., Fischer S., Kaguni L. S., Schneuwly S. (2001) Mol. Biol. Cell 12, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shapovalov Y., Hoffman D., Zuch D., de Mesy Bentley K. L., Eliseev R. A. (2011) J. Biol. Chem. 286, 22331–22338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li K., Williams R. S. (1997) J. Biol. Chem. 272, 8686–8694 [DOI] [PubMed] [Google Scholar]
- 11. Bujalowski W., Lohman T. M. (1991) J. Biol. Chem. 266, 1616–1626 [PubMed] [Google Scholar]
- 12. Lohman T. M., Ferrari M. E. (1994) Annu. Rev. Biochem. 63, 527–570 [DOI] [PubMed] [Google Scholar]
- 13. Raghunathan S., Kozlov A. G., Lohman T. M., Waksman G. (2000) Nat. Struct. Biol. 7, 648–652 [DOI] [PubMed] [Google Scholar]
- 14. Shutt T. E., Gray M. W. (2006) Trends Genet. 22, 90–95 [DOI] [PubMed] [Google Scholar]
- 15. Spelbrink J. N., Li F. Y., Tiranti V., Nikali K., Yuan Q. P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., Santoro L., Toscano A., Fabrizi G. M., Somer H., Croxen R., Beeson D., Poulton J., Suomalainen A., Jacobs H. T., Zeviani M., Larsson C. (2001) Nat. Genet. 28, 223–231 [DOI] [PubMed] [Google Scholar]
- 16. Edgar D., Trifunovic A. (2009) Aging 1, 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stumpf J. D., Copeland W. C. (2011) Cell Mol. Life Sci 68, 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Euro L., Farnum G. A., Palin E., Suomalainen A., Kaguni L. S. (August 8, 2011) Nucleic Acids Res., doi: 10.1093/nar/gkr618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams A. J., Wernette C. M., Kaguni L. S. (1993) J. Biol. Chem. 268, 24855–24862 [PubMed] [Google Scholar]
- 20. Oliveira M. T., Kaguni L. S. (2009) Methods Mol. Biol. 554, 37–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ziebarth T. D., Farr C. L., Kaguni L. S. (2007) J. Mol. Biol. 367, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bunch T. A., Grinblat Y., Goldstein L. S. (1988) Nucleic Acids Res. 16, 1043–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang C., Curth U., Urbanke C., Kang C. (1997) Nat. Struct. Biol. 4, 153–157 [DOI] [PubMed] [Google Scholar]
- 24. Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995) Proteins 23, 318–326 [DOI] [PubMed] [Google Scholar]
- 25. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis I., Vajda S. (2010) Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee Y. S., Kennedy W. D., Yin Y. W. (2009) Cell 139, 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamdan S. M., Richardson C. C. (2009) Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 28. Langston L. D., Indiani C., O'Donnell M. (2009) Cell Cycle 8, 2686–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korhonen J. A., Pande V., Holmlund T., Farge G., Pham X. H., Nilsson L., Falkenberg M. (2008) J. Mol. Biol. 377, 691–705 [DOI] [PubMed] [Google Scholar]
- 30. Holmlund T., Farge G., Pande V., Korhonen J., Nilsson L., Falkenberg M. (2009) Biochim. Biophys. Acta 1792, 132–139 [DOI] [PubMed] [Google Scholar]
- 31. Wanrooij S., Goffart S., Pohjoismäki J. L., Yasukawa T., Spelbrink J. N. (2007) Nucleic Acids Res. 35, 3238–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Curth U., Urbanke C., Greipel J., Gerberding H., Tiranti V., Zeviani M. (1994) Eur. J. Biochem. 221, 435–443 [DOI] [PubMed] [Google Scholar]
- 33. Thömmes P., Farr C. L., Marton R. F., Kaguni L. S., Cotterill S. (1995) J. Biol. Chem. 270, 21137–21143 [DOI] [PubMed] [Google Scholar]
- 34. Davidzon G., Greene P., Mancuso M., Klos K. J., Ahlskog J. E., Hirano M., DiMauro S. (2006) Ann. Neurol. 59, 859–862 [DOI] [PubMed] [Google Scholar]
- 35. Nguyen K. V., Sharief F. S., Chan S. S., Copeland W. C., Naviaux R. K. (2006) J. Hepatol. 45, 108–116 [DOI] [PubMed] [Google Scholar]
- 36. Yamanaka H., Gatanaga H., Kosalaraksa P., Matsuoka-Aizawa S., Takahashi T., Kimura S., Oka S. (2007) J. Infect Dis. 195, 1419–1425 [DOI] [PubMed] [Google Scholar]
- 37. Zsurka G., Baron M., Stewart J. D., Kornblum C., Bös M., Sassen R., Taylor R. W., Elger C. E., Chinnery P. F., Kunz W. S. (2008) J. Neuropathol. Exp. Neurol. 67, 857–866 [DOI] [PubMed] [Google Scholar]
- 38. Bortot B., Barbi E., Biffi S., Lunazzi G., Bussani R., Burlina A., Norbedo S., Ventura A., Carrozzi M., Severini G. M. (2009) Dig. Liver Dis. 41, 494–499 [DOI] [PubMed] [Google Scholar]
- 39. Luo N., Kaguni L. S. (2005) J. Biol. Chem. 280, 2491–2497 [DOI] [PubMed] [Google Scholar]
- 40. Shamoo Y., Friedman A. M., Parsons M. R., Konigsberg W. H., Steitz T. A. (1995) Nature 376, 362–366 [DOI] [PubMed] [Google Scholar]
- 41. Raghunathan S., Ricard C. S., Lohman T. M., Waksman G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bochkarev A., Pfuetzner R. A., Edwards A. M., Frappier L. (1997) Nature 385, 176–181 [DOI] [PubMed] [Google Scholar]
- 43. Hollis T., Stattel J. M., Walther D. S., Richardson C. C., Ellenberger T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9557–9562 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.