Abstract
Metastasis is a key event of malignant tumor progression. The capability to metastasize depends on the ability of the cancer cell to migrate into connective tissue, adhere, and possibly transmigrate through the endothelium. Previously we reported that the endothelium does not generally act as barrier for cancer cells to migrate in three-dimensional extracellular matrices (3D-ECMs). Instead, the endothelium acts as an enhancer or a promoter for the invasiveness of certain cancer cells. How invasive cancer cells diminish the endothelial barrier function still remains elusive. Therefore, this study investigates whether invasive cancer cells can decrease the endothelial barrier function through alterations of endothelial biomechanical properties. To address this, MDA-MB-231 breast cancer cells were used that invade deeper and more numerous into 3D-ECMs when co-cultured with microvascular endothelial cells. Using magnetic tweezer measurements, MDA-MB-231 cells were found to alter the mechanical properties of endothelial cells by reducing endothelial cell stiffness. Using spontaneous bead diffusion, actin cytoskeletal remodeling dynamics were shown to be increased in endothelial cells co-cultured with MDA-MB-231 cells compared with mono-cultured endothelial cells. In addition, knockdown of the α5 integrin subunit in highly transmigrating α5β1high cells derived from breast, bladder, and kidney cancer cells abolished the endothelial invasion-enhancing effect comparable with the inhibition of myosin light chain kinase. These results indicate that the endothelial invasion-enhancing effect is α5β1 integrin-dependent. Moreover, inhibition of Rac-1, Rho kinase, MEK kinase, and PI3K reduced the endothelial invasion-enhancing effect, indicating that signaling via small GTPases may play a role in the endothelial facilitated increased invasiveness of cancer cells. In conclusion, decreased stiffness and increased cytoskeletal remodeling dynamics of endothelial cells may account for the breakdown of endothelial barrier function, suggesting that biomechanical alterations are sufficient to facilitate the transmigration and invasion of invasive cancer cells into 3D-ECMs.
Keywords: Cell Migration, Cell Surface Receptor, Endothelium, Integrin, RNA Interference (RNAi), Cancer Cell Invasion, Cellular Stiffness, Cytoskeletal Remodeling Dynamics, Microrheology, Transendothelial Migration
Introduction
The malignancy of tumors is responsible for most cancer-related deaths. A benign tumor becomes malignant when cancer cells spread from the primary tumor and form metastases (1–4). The process of metastasis can be described in several steps that involve the dissemination of cancer cells from the primary tumor into the extracellular matrix (ECM),2 the invasion of cancer cells through connective tissue, adhesion of cancer cells to the endothelium of blood or lymph vessels, possibly the transmigration of cancer cells through the endothelium (intravasation and/or extravasation), and subsequently, the formation of a secondary tumor in a distant targeted organ (5, 6).
The impact of endothelial cells on the regulation of cancer cell invasiveness into 3D-ECMs is so far unknown. The regulation of cancer cell invasiveness may be a complex scenario that is not fully characterized yet (7). In many previous studies the endothelium acts as a barrier against the invasion of cancer cells (6, 8). Furthermore, the endothelium reduces pronouncedly the invasion of cancer cells and, hence, metastasis formation (9). However, several recent reports propose a novel paradigm in which endothelial cells modulate the invasiveness of several cancer cells by increasing their dissemination through vessels (10) or by increasing the invasive capability of cancer cells to migrate into the ECM (11). Among these cancer cells is the human breast cancer cell line MDA-MB-231.
Although several adhesion molecules have been identified that play a role in tumor-endothelial cell interactions and, hence, metastasis formation, the biomechanical properties of endothelial cells co-cultured with cancer cells are still elusive. There may be altered biomechanical properties of endothelial cells that support the ability of the endothelium to act either as a barrier or as an enhancer for cancer cell invasion. Biomechanical properties have been studied so far only on cancer cells (11–14). A main biochemical pathway of the tumor-endothelial interaction has been reported to involve cell adhesion receptors and integrins such as platelet endothelial cell adhesion molecule-1 (PECAM-1) and αvβ3 integrins, respectively (15). Integrins are transmembrane adhesion receptors that cluster after activation and assembly of focal adhesions in cell-matrix as well as in cell-cell adhesive interactions (16). In addition, integrins forge a link between the ECM and the actin cytoskeleton that can be tensioned by myosin-II motors acting on actin filaments (17–19). The connection between integrins and the actomyosin cytoskeleton is facilitated through the mechano-coupling focal adhesion and cytoskeletal adaptor protein vinculin (20). Furthermore, this connection determines the amount of cellular counterforces that maintain the morphology and shape of cells and, hence, provide the cellular stiffness (21). Taken together, until now a biomechanical approach investigating the endothelial barrier breakdown in the presence of co-cultured invasive cancer cells is still elusive.
As microrheologic measurements such as magnetic tweezer rheology turn out to be adequate for the identification of cell mechanical properties such as cellular stiffness, in this study endothelial stiffness using magnetic tweezers was measured in co-culture with invasive MDA-MB-231 cells as well as in mono-culture. The results are that highly invasive breast cancer cells influenced the cellular mechanical properties of co-cultured microvascular endothelial cells by lowering the stiffness of endothelial cells. In addition, nanoscale particle tracking method diffusion measurements of actomyosin cytoskeletal-bound beads, which serve as markers, are suitable to determine the actomyosin cytoskeletal remodeling dynamics. Thus, cytoskeletal remodeling dynamics of endothelial cells using nanoscale particle tracking were measured either in mono-culture or in co-culture with highly invasive MDA-MB-231 cells. Here, the cytoskeletal remodeling dynamics of endothelial cells were increased in co-culture with highly invasive cancer cells. These findings indicate that highly invasive breast cancer cells actively altered the biomechanical properties of co-cultured endothelial cells. Hence, these results may provide an explanation for the breakdown of the endothelial barrier function of monolayers.
In conclusion, a mechanism in which a decrease in endothelial cell-cell adhesion molecule expression such as PECAM-1 and vascular endothelial-cadherin (VE-cadherin), a decrease in endothelial cell stiffness, and an increase in cytoskeletal remodeling dynamics account for the endothelial barrier breakdown by highly invasive MDA-MB-231 cells was provided in this study.
EXPERIMENTAL PROCEDURES
Cell Culture
Human MDA-MB-231 and MCF-7 breast cancer cell lines, human T-24 bladder, and 786-O kidney cancer cell lines were purchased from ATCC-LGC-Promochem (Wesel, Germany). Human umbilical vein endothelial cells (HUVECs) were isolated as described (20, 22). Human dermal microvascular endothelial cells (HDMECs) and human pulmonary microvascular endothelial cells (HPMECs) were purchased (Promocell, Heidelberg, Germany). Endothelial cell purity was determined by flow cytometric analysis using VE-cadherin (Coulter, Krefeld, Germany) and PECAM-1 (Biozol, Eching, Germany). Endothelial cell isolations contained less than 0.3% contaminating cells. HPMECs and HDMECs were used in passage 4–6 and cultured in Endothelial Cell Growth Medium MV 2 (Promocell) containing 5% FCS.
Breast, bladder, and kidney cancer cells were maintained in low glucose (1 g/liter) Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (low endotoxin), 2 mm l-glutamine, and 100 units/ml penicillin-streptomycin (DMEM complete medium, all from Biochrom, Berlin, Germany). 80% confluent cancer cells were used in passages 5–30. Accutase was used for cell harvesting, and the percentage of dead cells was less than 1%. Mycoplasma contamination was excluded using the Mycoplasma detection kit (Roche Diagnostics). All chemicals used were purchased from Sigma unless otherwise indicated.
3D-ECM Transmigration and Invasion
Three-dimensional collagen type I matrices were prepared as a 1:1 mixture of collagen R (Serva, Heidelberg, Germany), and collagen G (Biochrom) was described (11). 100,000 cancer cells were seeded on top of the 3D-ECMs. For transendothelial migration in the presence of an endothelial cell monolayer (600,000 endothelial cells per 3.5-cm dish stained with 15 μm CyTrack orange for 10 min at room temperature) and for invasion in the absence of an endothelial cell monolayer, cancer cells were seeded on 3D-ECMs. After 3 days, cancer cells cultured on and inside the collagen matrices were fixated with 2.5% glutaraldehyde solution in PBS, the fraction of cancer cells that invaded 3D-ECMs, and their invasion depth was measured by optical sectioning. After this time, differences in the invasiveness of cell lines were clearly visible. Non-invasive cells can be readily identified by their nuclei located in one layer that coincides with the location of the topmost collagen fibers. A cell was counted as invasive when its nucleus is located below the layer formed by the non-invasive cells. Because of the depth of field of a 40 × 0.6 NA objective, the uncertainty of this method is ∼5 μm. In addition, the 3D-ECM matrices contain fibronectin (FN) sequestered from the fetal calf serum in the medium and FN secreted from the cancer cells (14). As FN is the main ligand for α5β1 integrins, the matrices are suitable for analyzing α5β1-dependent cell behavior. To analyze the function of integrin ligands in more detail, three-dimensional collagen matrices were also polymerized in the presence of 100 μg/ml FN (3D-FN-ECMs). The invasion depth was determined by focusing the microscope to the center of the nucleus; the value was read from the motorized z-drive of the microscope and was corrected for the refractive index of water (1.33). The z-focus at the gel surface served as the reference.
For inhibition of contractile forces, the myosin light chain inhibitor 15 μm ML-7 (Calbiochem) was incubated with cancer cell lines 2 h before co-culture (with HPMECs) transmigration and invasion assay start. For inhibition of small GTPase signaling, 100 μm Rho kinase inhibitor (Y27632, Calbiochem), 100 μm Rac1 inhibitor, 100 μm MEK inhibitor (PD98059, Calbiochem), or 50 μm PI3K inhibitor (LY294002, Calbiochem) were incubated with cancer cells 2 h before co-culture (with HPMECs) start.
To analyze whether endothelial cell invasion was responsible for increased cell invasion during MDA-MB-231 co-culture, endothelial cells were stimulated with 100 ng/ml rhVEGF(165) and 100 ng/ml rhbFGF (both from Promocell) in the absence (mono-cultured) and presence of MDA-MB-231 cancer cells (co-cultured).
Isolation of Tumor Cell Variants
Tumor cell variants with high (α5β1high) and low α5β1 (α5β1low) integrin expression were isolated from parental MDA-MB-231 breast cancer cells, T24 bladder cancer cells (24-α5β1high and 24-α5β1low), and 786-O kidney cancer cells (786-α5β1high and 786-α5β1low) using a cell sorter, and single cells were plated into 96-well plates. Cells were expanded to subcell lines, and the expression of α5 integrin was measured by flow cytometry. Repeated measurements of α5β1 expression on the cell surface confirmed that the α5β1 expression phenotype of the subcell lines remained stable for at least 100 generations as described (14).
siRNA Transfection
200,000 80% confluent α5β1high cells, 24-α5β1high cells, and 786-α5β1high cells were seeded in 3.5-cm dishes and cultured in 2 ml of DMEM complete medium. 5 μl of a solution containing 20 μm α5-integrin (target sequence, CCCATTGAATTTGACAGCAAA) or Allstar-control RNAi-solution (control-siRNA), 12 μl of HiPerFect-Reagent (Qiagen), and 100 μl of DMEM were mixed (14, 23). RNAi-mediated α5-integrin knockdown was confirmed by flow cytometry using anti-α5-integrin and Cy2-labeled anti-mouse-IgG antibodies (Dianova). Transfection efficiency was determined by flow cytometry to be >99% using 20 μm Alexafluor546-labeled siRNA.
Magnetic Tweezers
10,000 cancer cells live-stained with 5 μg/ml carboxyfluorescein diacetate (CFDA; Invitrogen) and 10,000 HPMECs were seeded in 3.5-cm culture dishes (Nunc) and were co-cultured for 12 h. Using magnetic tweezers, step-forces ranging from 0.5 to 10 nanonewtons were applied to superparamagnetic epoxylated 4.5-μm beads coated with 100 μg/ml FN (11, 20, 24). 2 × 105 beads were sonicated, added to 105 cells, and incubated for 30 min at 37 °C and 5%CO2. Measurements were performed at 37 °C in CO2-free medium using an inverted microscope (DMI-Leica, 40× magnification). The creep response J(t) of cells during force application followed a power-law in time, J(t) = a(t/t0)b, where the prefactor a and the power-law exponent b were force-dependent, and the reference time t0 was set to 1 s. The bead displacement in response to a staircase-like force followed a superposition of power laws (25) from which the force dependence of a and b was determined by a least-squares fit (20).
The parameter a (μm/nanonewtons) characterizes the elastic cell properties and corresponds to a compliance that is the inverse of cellular stiffness (20). The force/distance relationship in units of nanonewtons/μm is related to cell stiffness in units of Pa by a geometric factor that depends on the contact area between the bead and the cell (or the degree of bead internalization) and the cell height. If those parameters are known, for example from scanning electron micrographs, the geometric factor can be estimated from a finite element analysis (26). Without knowing the height of the cell and the degree of bead internalization, one can still estimate the typical strain ϵ as the bead displacement d divided by the bead radius r, and the typical stress σ as the applied force F divided by the bead cross-sectional area πr2 such that the cellular stiffness is G = σ/ϵ = (r/d)( F/(πr2)) (27). For 4.5-μm beads, the geometric factor is 0.14 μm−1, and a cell with an apparent stiffness of 1 nanonewton/μm would have a “proper” stiffness of 140 Pa.
The power law exponent b (creep exponent) reflects the stability of force-bearing cellular structures such as actomyosin stress fibers connected to the beads. A value for b = 1 and b = 0 indicates Newtonian viscous (such as silicone oil) and elastic behavior (such as polyacrylamide gels), respectively (28). A cell shows the behavior of a viscoelastic material when it has elastic structures and viscous elements that build its cytoskeleton. A non-zero power-law exponent b denotes that during the magnetic force application, one part of the deformation energy is not elastically stored in the cytoskeleton but is dissipated in the form of heat as the cytoskeletal structures to which the bead is connected remodel (29). Hence, dissipation is directly linked to the rate at which the elastic bonds in the cytoskeleton break up and turn over. The turnover of actomyosin bonds also contributes to the dissipative properties (30), and although this is not considered a remodeling event, it enables contractility-driven shape changes in the cytoskeleton.
Spontaneous Bead Diffusion
Before cell detachment, HPMECs (15 μm CyTrack orange stained) were incubated with FN-coated beads for 30 min at 37 °C, 5% CO2, and 95% humidity. Unbound FN beads were washed away by using 1× HEPES buffer before co-culture start. FN beads bound to CFDA (5 μg/ml)-stained cancer cells are not observed in the fields of view used for bead-tracking. The binding of FN beads during co-culture with endothelial-bound FN beads was below 1% as determined under the fluorescent microscope at 20× magnification and 24 fields of view. For mono-culture 400,000 cells of 80% confluent HPMECs and for co-culture 200,000 HPMECs and 200,000 CFDA-stained MDA-MB-231 cells seeded into 3.5-cm dishes and cultured overnight at 37 °C, 5% CO2, and 95% humidity. The spontaneous bead diffusion was analyzed using an inverted fluorescent microscope, and the position of beads was tracked over 5 min. Bead movements were computed from phase contrast images recorded with 10× magnification using a Fourier-based difference-with-interpolation algorithm (31). These beads moved spontaneously with a mean square displacement (MSD) that also followed a power law with time, MSD = D(t/t0)β + c. The evolution of the MSD over time, t, can be described by an apparent diffusion coefficient, D, and the persistence of motion can be described by the power-law exponent, β (31). Depending on the power-law exponent β, the bead motion can be classified as brownian or diffusive for β = 1, subdiffusive for β < 1, superdiffusive (=persistent motion) for β >1, and ballistic for β = 2. The term c reflects random noise from thermal and non-thermal sources such as single myosin motors, and t0 was arbitrarily set to 1 s.
The fit parameters were determined by a least-squares fit (31). The measurements of spontaneous bead movements were performed after at least 16 h of bead binding, which is sufficient to connect the FN beads to the cytoskeleton. The details of the connection matter little and do not influence the bead motion once the beads are firmly connected to the cytoskeleton (32).
Scanning EM
The cells were fixed on and in three-dimensional collagen matrices with 2.5% glutaraldehyde-fixed, dehydrated through a graded ethanol series, washed with hexadimethylsilazane reagent (Electron Microscopy Science, Hatfield, PA), and air-dried as described (11). After sputter-coating of the samples with gold, cells were analyzed using a scanning electron microscope (ISI-SX-40, International Scientific Instruments, Milpitas, CA).
Transmission EM
The cells were fixed in 4% paraformaldehyde/0.1% glutaraldehyde, post-fixed in 2% buffered osmium tetroxide, dehydrated through a graded ethanol series, and finally embedded in epoxy resin. For orientation, 1.0-μm sections were stained with toluidine blue. The 70-nm ultrathin sections were stained with uranyl acetate and lead citrate and examined with a Transmission EM (EM906E; Zeiss, Oberkochen, Germany).
Flow Cytometry
80% confluent cells were harvested and resuspended in HEPES buffer (20 mm Hepes, 125 mm NaCl, 45 mm glucose, 5 mm KCl, 0.1% albumin, pH7.4). Cancer cells were incubated with mouse antibodies against human α5 (Biozol, Eching, Germany). Microvascular endothelial cells (HDMECs and HPMECs, passages 2–4) were mono-cultured or co-cultured with CFDA-stained (5 μg/ml carboxyfluorescein diacetate, Invitrogen) MDA-MB-231 cells or CFDA-stained MCF-7 cells for 16 h and incubated with VE-cadherin and PECAM-1. Isotype-matched antibodies were used as controls (Caltag, Burlingame, CA). After 30 min at 4 °C, cells were washed and stained with an R-phycoerythrin-labeled goat anti-mouse-IgG (F(ab)2 fragment; Dianova). Flow cytometry measurements were performed using a FACSCalibur System (BD Biosciences). For the staining of internalized receptors, endothelial cells were permeabilized using 0.1–0.3% Triton X-100 for 5 min at 37 °C.
Statistics
The data were expressed as the mean values ± S.E. if not indicated otherwise. Statistical analysis was performed using the two-tailed unpaired t test; p < 0.05; P was considered to be statistically significant.
RESULTS
Endothelial Cells Enhance MDA-MB-231 Cell Invasiveness
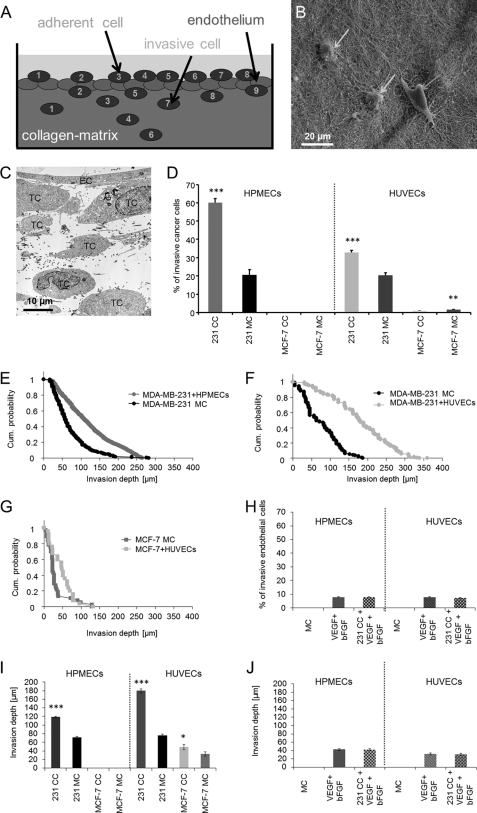
To investigate how the invasive behavior of highly invasive and weakly invasive breast cancer cells is affected by the co-culture with an endothelial cell monolayer on top of 3D-ECMs (Fig. 1A), two well established human breast cancer cell lines, the highly invasive MDA-MB-231 cells and the weakly invasive an MCF-7 cells, were cultured in the presence or absence of macrovascular (HUVECs) and microvascular (HPMECs or HDMECs) endothelial cells. In the presence of a closed endothelial cell monolayer on top of the 3D-ECMs, the invasiveness of MDA-MB-231 cells was increased (Fig. 1, D–F). The percentage of invasive MDA-MB-231 cells increased in the presence of co-cultured HPMECs (60.2% ± 2.2) as well as HUVECs (32.7% ± 1.2) (Fig. 1D). In contrast, the invasiveness of the weakly invasive MCF-7 cells was not affected (Fig. 1, D and G), and the percentage of invasive MCF-7 cells was even decreased in presence of HUVECs (Fig. 1D). These results show that the presence of a closed endothelial cell monolayer on the surface of the 3D-ECMs enhanced the invasiveness of MDA-MB-231 cells (Fig. 1, D–F), whereas the invasiveness of MCF-7 cells was not affected (Fig. 1, D and G). Taken together, these results indicate that the endothelial cell monolayer did not increase the invasiveness of all breast cancer cell lines, which was consistent with a previous report (11). The increased invasiveness of MDA-MB-231 cells into 3D-ECMs was evoked by HUVECs as well as HPMECs as indicated by increased numbers of invasive MDA-MB-231 cells (Fig. 1D) and an invasion profile which shows that MDA-MB-231 cells invaded deeper into the 3D-ECMs when co-cultured with endothelial cells (Fig. 1, E–G). For the weakly invasive MCF-7 cancer cells, HUVECs hardly evoked an increase in invasiveness (Fig. 1, D and G), and HPMECs even had no effect on enhancing the invasiveness of MCF-7 cells (Fig. 1D).
FIGURE 1.
MDA-MB-231 breast cancer cells were at least 3-fold more invasive after co-culture with an endothelial cell monolayer. A, shown is a schematic presentation of the transendothelial cancer cell invasion assay. Fluorescently labeled (CyTrack orange) endothelial cells were cultured 16 h before co-culture assay start on top of 3D-ECMs to obtain a fully closed endothelial cell monolayer. Fluorescently labeled (CFDA) cancer cells were seeded on top of the endothelial cell monolayer and were co-cultured for 3 days. Then cancer cells on top of the 3D-ECMs were counted (numbers 1–8), and cancer cells inside the 3D-ECM (collagen matrix) were counted (numbers 1–9). B, shown is an invasion assay. Single cells of MDA-MB-231 breast carcinoma cells (100,000 cells) were added on the surface 3D-ECMs of collagen type I and cultured for 3 days. Collagen matrices were fixed with 2.5% glutaraldehyde and processed for S.E. The image shows MDA-MB-231 cells adhered (right arrow) or invaded (two left arrows). C, shown is a transendothelial 3D-ECM invasion assay. The transmission scanning electron microscopic image show MDA-MB-231 breast cancer cells (TC, 100,000 cells) that had transmigrated through the microvascular endothelial cell monolayer (EC) and invaded into 3D-ECMs for 3 days. D, the endothelium enhanced MDA-MB-231 cell invasion (100,000 cells) into three-dimensional collagen matrices. Microvascular HPMECs (E) and macrovascular HUVECs (F) enhanced MDA-MB-231 breast cancer cell invasion but not MCF-7 (100,000 cells) breast cancer cell invasion (G). H, endothelial cell migration (HPMECs and HUVECs) into three-dimensional collagen matrices in the presence of the growth factors 100 ng/ml rhVEGF(165) and 100 ng/ml rhbFGF after 3 days is shown. In addition, HPMECs and HUVECs were also co-cultured with MDA-MB-231 cancer cells as well as rhVEGF(165) and rhbFGF. I, mean invasion depths of MDA-MB-231 cells and MCF-7 cells in the presence (CC) or absence of endothelial cells (MC) show that the invasion depths of MDA-MB-231 is significantly increased by co-culture with HPMECs (left) and HUVECs (right). J, mean invasion depths of HPMECs and HUVECs values show that endothelial cells cannot invade deep into the matrices. bFGF, bovine FGF. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To analyze whether the endothelial cells are able to invade the dense 3D-ECMs, endothelial cell monolayers were cultured in the presence and absence of the growth factors rhVEGF165 (100 ng/ml) and rhbFGF (100 ng/ml). Indeed, these growth factors induced the invasiveness of endothelial cells into 3D-ECMs. To investigate whether the presence of MDA-MB-231 cells could further enhance the invasiveness of endothelial cells in the presence of both growth factors, endothelial cells were co-cultured with MDA-MB-231 cells in the presence and absence of MDA-MB-231 cells. These growth factors induced the invasiveness of endothelial cells into three-dimensional collagen matrices (Fig. 1H). The growth factor-induced increased invasiveness of endothelial cells was not affected by co-culture of endothelial cells with MDA-MB-231 cells (Fig. 1H). These results indicate that the slightly induced invasiveness of endothelial cells toward 3D-ECMs could not explain the increased numbers of invasive cells observed during co-culture with invasive MDA-MB-231 cells. Furthermore, the amount of endothelial cell invasiveness is insignificant compared with the strong increase of invasive cells during the co-culture with MDA-MB-231 cells (Fig. 1, D and H). In addition, the endothelial cell invasion depth is <45 μm (Fig. 1J). Thus, the increase in invasion depths observed during MDA-MB-231 and endothelial cell co-culture could not be explained by enhanced invasiveness of endothelial cells. Thus, the increase of invasive cells during co-culture of cancer cells and endothelial cells could be explained by increased numbers of invasive, fluorescently labeled CFDA-stained MDA-MB-231 cells.
Endothelial Cell Stiffness Is Decreased by Co-cultured Invasive MDA-MB-231 Cells
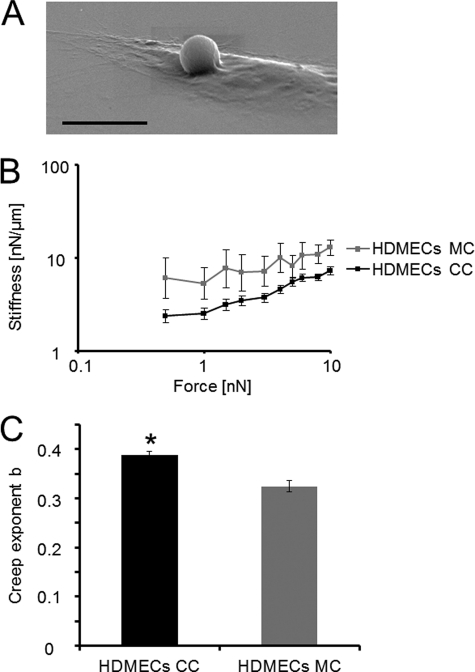
To analyze whether highly invasive cancer cells facilitate the transmigration through the endothelium by altering biomechanical properties of endothelial cells such as cell stiffness, the cell stiffness of co-cultured and mono-cultured endothelial cells was determined using magnetic tweezers. Cellular stiffness is a measure of the resistance by an elastic body such as endothelial cells to external applied forces to non-permanent deformation. The stiffness of cells is determined by their ability to generate and transmit contractile forces (14). Human microvascular endothelial cells were co-cultured with MDA-MB-231 cells. Endothelial cell stiffness was measured by applying force to FN-coated beads that were bound through cell-matrix adhesion receptors such as integrins to the actomyosin cytoskeleton of mono-cultured and co-cultured endothelial cells. FN bead binding toward endothelial cells was analyzed by staining of endothelial cells with CyTrack orange and cancer cells with CFDA. Indeed, almost all FN beads bound to endothelial cells, which had been incubated with the FN beads 2 h before co-culture start. The FN bead binding to cancer cells during the co-culture was below 1% (data not shown). Fig. 2A shows a scanning electron microscopic image of a FN bead that was attached to an endothelial cell after incubation for 30 min. The highly invasive MDA-MB-231 cells decreased cellular stiffness in co-cultured HPMECs compared with mono-cultured HPMECs (Fig. 2B). The co-culture of weakly invasive MCF-7 cells with HPMECs had no effect on endothelial cell stiffness (data not shown).
FIGURE 2.
MDA-MB-231 cells reduced the cellular stiffness of endothelial cells, which is a measure of resistance toward external deformation by forces applied to cytoskeletal-bound beads through integrins. A, shown is a scanning electron microscopic image of a FN-coated bead that was bound to endothelial cells 30 min before the co-culture assay start. The bar is 10 μm. B, the cellular stiffness of microvascular endothelial cells was decreased after co-culture with the invasive MDA-MB-231 breast cancer cells measured during increasing force application to FN-coated beads, which were, before the co-culture assay start, bound to integrins on endothelial cells. C, as the bead displacement follows a power law, the creep exponent b (=power-law exponent) can have values of b = 0 such as an elastic solid, of b = 1, such as a viscous fluid, and between 0 < b <1, such as a viscoelastic material and, thus, describes the cellular fluidity, which is determined by cytoskeletal structures. The creep exponent b of co-cultured and mono-cultured microvascular endothelial cells was not significantly altered. The values are expressed as the mean ± S.E. *, p < 0.05.
To confirm that the FN-coated beads were bound to integrins such as the FN receptors α5β1 or αvβ1, the binding was inhibited by adding 10 μg/ml concentrations of an inhibitory β1 integrin antibody to endothelial cells before FN bead incubation. The observation was a complete reduction of FN beads that were bound to endothelial cells, indicating that the FN bead binding was indeed to β1 integrins (data not shown). As bead displacement follows a power law, cellular fluidity can be measured by analyzing the power-law exponent b (creep exponent b). The creep exponent b can have two extreme values; a value of 0, such as elastic solid materials, and a value of 1, such as viscous fluids. Normally, the cells have b values between 0.1 and 0.5, which indicate viscoelastic behavior (20). Here, the creep exponent b was not significantly altered between co-cultured and mono-cultured endothelial cells (Fig. 2C).
Cytoskeletal Remodeling Dynamics of Endothelial Cells Are Increased by Co-cultured Highly Invasive MDA-MB-231 Cells
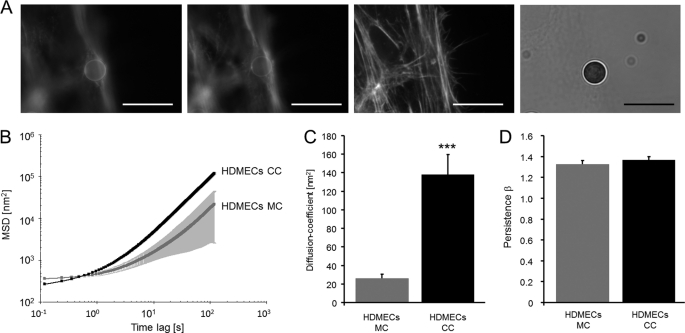
Alterations in actin cytoskeletal structures and dynamics are supposed to play a role in transendothelial migration (23). Therefore, here we investigated how the remodeling of cytoskeletal structures such as the formation or degradation of stress fibers in endothelial cells affects their barrier function in the presence and absence of the invasive MDA-MB-231 breast cancer cells. To analyze whether the cytoskeletal remodeling dynamics are altered in co-cultured endothelial cells compared with mono-cultured endothelial cells, nanoscale particle tracking was used. For this method, the displacement of FN-cell-bound beads, which were connected to the actomyosin cytoskeleton via integrin receptors expressed on the cell surface of endothelial cells, was analyzed (Fig. 3A). This method was chosen due to its high accuracy and the possibility to directly visualize the movement of particles evoked by ATP-driven cytoskeletal rearrangements. In summary, the nanoscale particle-tracking method analyzes the random walk of unforced, spontaneously diffusing beads. These beads are bound to integrins, initiate focal adhesion formation, and anchor to the actomyosin cytoskeleton of endothelial cells. The beads do not move unless the focal adhesions and/or the actin structures to which they are firmly connected reorganize (31, 32). The evolution of the MSD of beads over time (t) can be described by an diffusion coefficient (D) and the direction of bead-motion as an index of persistence (β) according to MSD = D(t/t0)β + c (31). The MSD of beads that were bound to co-cultured endothelial cells was significantly higher (p < 0.001) compared with mono-cultured endothelial cells (Fig. 3B). The diffusion coefficient D of FN beads bound to co-cultured endothelial cells (HDMECs) was markedly higher (p < 1.77 × 10−0.7) compared with mono-cultured endothelial cells, indicating that the speed (obtained by the square root of the diffusion coefficient D) of cytoskeletal remodeling is increased in MDA-MB-231 cells co-cultured endothelial cells. These results suggest that MDA-MB-231 cells induce a more dynamic remodeling of the actin cytoskeleton in co-cultured endothelial cells compared mono-cultured endothelial cells (Fig. 3C). Depending on the values of the power-law exponent β (=index of persistence), the bead motion can be classified as brownian or diffusive motion for β = 1, subdiffusive motion for β < 1, superdiffusive motion (=persistent motion such as directed motion) for β > 1, and ballistic motion for β = 2. As the FN beads were bound to the endothelial actomyosin cytoskeleton via integrins, the persistent (directed) motion of the beads reflects cytoskeletal remodeling such as reorganization of actin microfilaments. Here, the persistence of the bead movement, which is a measure of the directionality, was not altered between co-cultured or mono-cultured endothelial cells (Fig. 3D).
FIGURE 3.
MDA-MB-231 cells increased the cytoskeletal remodeling dynamics of co-cultured microvascular endothelial cells. A, a FN-coated 4.5-μm bead is bound to the actin cytoskeleton of an endothelial cell that was stained with Alexa546-phalloidin after 24 h of bead-incubation. A dense network of actin fibers surrounds the bead. The images are recorded at different focal planes 2 μm apart. All bars are 10 μm. B, the MSD of spontaneous bead motion was increased in co-cultured (CC) HDMECs (n = 625) compared with mono-cultured (MC) HDMECs (n = 133). C, the diffusion coefficient (D) of cytoskeletal bound FN beads is significantly increased in co-cultured HDMECs compared with mono-cultured HDMECs. D, the persistence (=directionality) β of FN bead motion was not altered between co-cultured and mono-cultured endothelial cells. ***, p < 0.001.
Highly Invasive Cancer Cells Decrease Cell-Cell Adhesion Molecule Expression on Microvascular Endothelial Cells
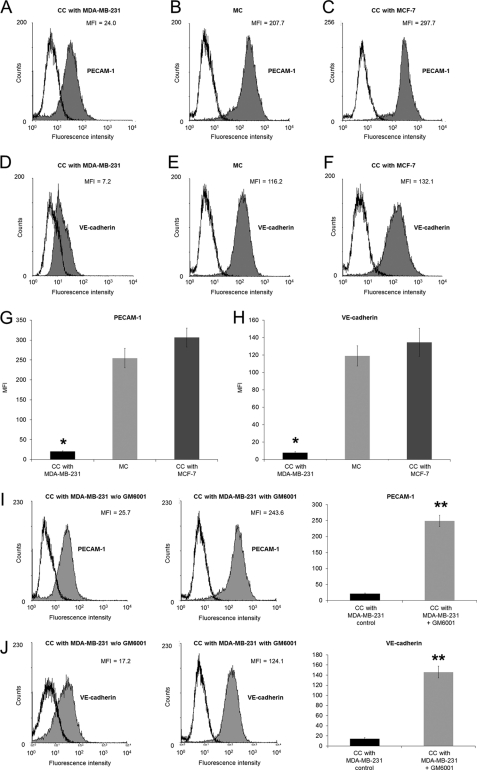
To investigate whether highly invasive cancer cells alter the cell-cell adhesion molecule expression on human microvascular endothelial cells during co-culture, HPMECs were co-cultured for 16 h with highly invasive MDA-MB-231 and weakly invasive MCF-7 cells. Using flow cytometric analysis, the co-culture of microvascular endothelial cells and highly invasive MDA-MB-231 breast cancer cells revealed that the PECAM-1 and the VE-cadherin are down-regulated during co-culture with highly invasive MDA-MB-231 cells (Fig. 4, A, D, G, and H) compared with mono-cultured endothelial cells (Fig. 4, C, F, G, and H). Moreover, the down-regulation of PECAM-1 and VE-cadherin was only observed during co-culture with highly invasive MDA-MB-231 cells compared with weakly invasive MCF-7 cells (Fig. 4, B, E, G, and H). Taken together, these results show that the co-culture of endothelial cells with highly invasive cancer cells leads to a decrease in cell-cell adhesion molecule expression, altered biomechanical properties of endothelial cells, and subsequently, breakdown in endothelial barrier function.
FIGURE 4.
MDA-MB-231 cells decreased the expression of endothelial cell-cell adhesion proteins. A–C, flow cytometric analysis of PECAM-1 expression on HPMECs co-cultured (CC) with highly invasive MDA-MB-231 cancer cells (A), on mono-cultured (MC) HPMECs (B), and on HPMECs co-cultured (CC) with weakly invasive MCF-7 cancer cells (C) - is shown. One representative experiment of three is shown. D–F, shown is a flow cytometric analysis of VE-cadherin expression on HPMECs co-cultured with invasive MDA-MB-231 cancer cells (D), on mono-cultured HPMECs (E), and on HPMECs co-cultured with weakly invasive MCF-7 cancer cells (F). One representative experiment of three is shown. G, MFI (mean ± S.E., n = 3) of PECAM-1 expression on HPMECs co-cultured with MDA-MB-231, mono-cultured HPMECs, and HPMECs co-cultured with weakly invasive MCF-7 is shown. H, MFI (mean ± S.E., n = 3) of VE-cadherin expression on HPMECs co-cultured with highly invasive MDA-MB-231, mono-cultured HPMECs, and HPMECs co-cultured with weakly invasive MCF-7 is shown. I, flow cytometric analysis of PECAM-1 expression on HPMECs co-cultured with highly invasive MDA-MB-231 cancer cells in the absence of GM6001 (left) and in the presence of GM6001 (middle) is shown. One representative experiment of three is shown. MFI (mean ± S.E., n = 3) of PECAM-1 expression on HPMECs co-cultured with MDA-MB-231 in the absence of GM6001 (black bars) and in the presence of GM6001 (gray bars) is shown. J, flow cytometric analysis of PECAM-1 expression on HPMECs co-cultured with highly invasive MDA-MB-231 cancer cells in the absence of GM6001 (left) and in the presence of GM6001 (middle) is shown. One representative experiment of three is shown. MFI (mean ± S.E., n = 3) of PECAM-1 expression on HPMECs co-cultured with MDA-MB-231 in the absence of GM6001 (black bars) and in the presence of GM6001 (gray bars) is shown. *, p < 0.05; **, p < 0.01.
To investigate whether the decreased expression of PECAM-1 and VE-cadherin receptors on endothelial cells during co-culture with MDA-MB-231 cells is due to increased membrane shedding of these receptors, the co-culture of MDA-MB-231 cells and HPMECs was performed in the presence and absence of the broad matrix-metalloproteinase inhibitor GM6001 (Fig. 4, I and J). Indeed, the reduction of PECAM-1 and VE-cadherin receptor expression on endothelial cells during co-culture with MDA-MB-231 cells was evoked by increased membrane shedding of the cell-cell adhesion receptors.
Cancer Cells with High α5β1 Expression Transmigrate More Efficiently Compared with Cancer Cells with Low α5β1 Expression
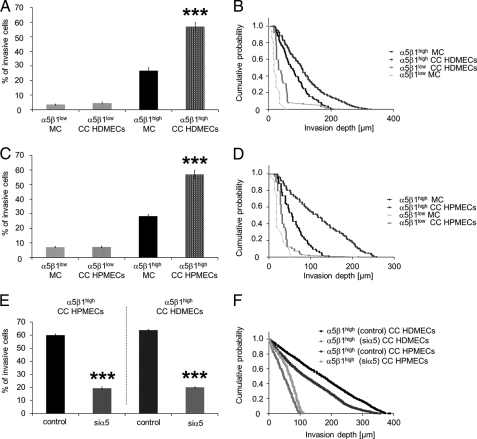
To analyze whether the increased invasiveness of MDA-MB-231 cells is facilitated by increased expression of the α5β1 integrin on their cell surface, the invasiveness of high and low α5β1 integrin-expressing subcell lines derived from parental MDA-MB-231 cells was determined using a 3D-ECM invasion assay. The results show that α5β1high cells invaded in greater quantity and deeper into 3D-ECMs during co-culture with HDMECs or HPMECs compared with mono-cultured cells (Fig. 5, A–D), whereas the invasiveness of α5β1low cells was not significantly affected (Fig. 5, A–D). Knockdown of the α5 integrin subunit in α5β1high cells decreased the endothelial-mediated increased invasiveness to the level of mono-cultured α5β1high cells as shown by the decreased numbers of invasive cells and their decreased invasion depth (Fig. 5, E and F). These results demonstrate that the knockdown (knockdown efficiency was 43.5 ± 2.1%, n = 3) of the α5 integrin subunit abolished the endothelial-facilitated increased invasiveness of α5β1high cells, suggesting an involvement of α5β1 integrin signaling in this process.
FIGURE 5.
Microvascular endothelial cells increased the invasiveness of α5β1high cancer cells derived from parental MDA-MB-231 breast cancer cells and knockdown of α5 integrin subunit in α5β1high cells abolished the endothelial invasion-enhancing effect. A, the percentage of invasive α5β1high cells, but not of α5β1low cells, is increased in the presence of HDMECs and HPMECs (C). B and D, the invasion profile shows that the α5β1high cells invaded deeper into the 3D-ECMs when co-cultured (CC) with HDMECs (B) or HPMECs (D), whereas the invasiveness of α5β1low cells was not significantly altered. E, knockdown of the α5 integrin subunit in α5β1high cells reduced the percentage of invasive cells in co-culture with HDMECs and HPMECs below the mono-cultured level. F, the invasion profile shows that knockdown of the α5 integrin subunit in α5β1high cells reduced the invasion depths in co-culture with HDMECs and HPMECs the below mono-cultured level. ***, p < 0.001.
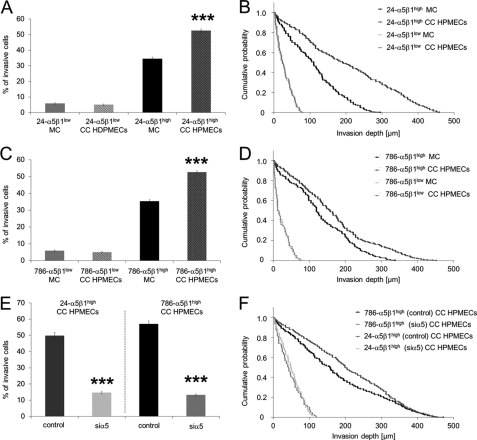
In addition, to determine whether the increased expression of α5β1 integrins on the cell surface of invasive breast cancer cells is transferable to other cancer cells such as bladder (T24) and kidney cancer cells (786-O), subcell lines with high and low α5β1 integrin expression were generated. The 24-α5β1high cells expressed mean MFI values of 274.9 ± 27.0 and the 24-α5β1low cells of 45.7 ± 7.7, which is a 6.0-fold difference. The 786-α5β1high cells expressed mean MFI values of 333.0 ± 63.1, and the 786-α5β1low cells expressed mean MFI values of 35.3 ± 6.2, which is a 9.4-fold difference. Indeed, the number of invasive cells of 24-α5β1high cells and 786-α5β1high cells was increased in the presence of HPMECs, but invasiveness of 24-α5β1low cells and 786-α5β1low cells was not affected by the presence of HPMEC (Fig. 6A, B and C, D, respectively). Knockdown of the α5 integrin subunit in 24-α5β1high cells and 786-α5β1high cells decreased the endothelial-mediated increased invasiveness to the level of mono-cultured α5β1high cells, as shown by the decreased numbers of invasive cells and their decreased invasion depth, whereas the invasiveness of control siRNA-treated co-cultured cancer cells (24-α5β1high and 786-α5β1high cells, respectively) was not affected (Fig. 6, E and F).
FIGURE 6.
Microvascular endothelial cells (HPMECs) increased the invasiveness of 24-α5β1high cells derived from T24 bladder as well as the invasiveness of 786-α5β1high cells derived from 786-O kidney cancer cells. Knockdown of α5 integrin subunit in 24-α5β1high cells and 786-α5β1high cells abolished the endothelial invasion-enhancing effect. A, the percentage of invasive 24-α5β1high cells, but not of 24-α5β1low cells, was increased in the presence of HPMECs. B, the invasion profile shows that the 24-α5β1high cells invaded deeper into the 3D-ECMs when co-cultured with HPMECs, whereas the invasiveness of 24-α5β1low cells was not significantly altered. E, knockdown of the α5 integrin subunit in 24-α5β1high cells and 786-α5β1high cells reduced the percentage of invasive cells in co-culture with HPMECs to the level of mono-cultured 24-α5β1high cells and 786-α5β1high cells, respectively. F, the invasion profile shows that knockdown of the α5 integrin subunit in 24-α5β1high and 786-α5β1high cells reduced the invasion depths in co-culture with HPMECs to the level of mono-cultured 24-α5β1high cells and 786-α5β1high cells, respectively. ***, p < 0.001.
FN Enhances the Transmigration of Highly Invasive Cancer Cells
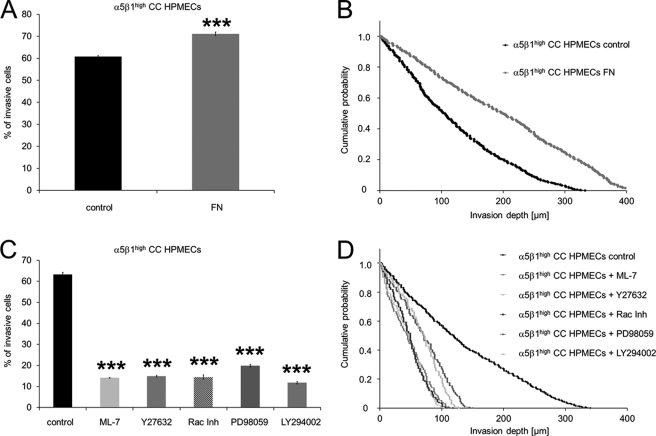
To investigate whether embedded FN in the 3D-ECMs underneath the endothelial cells can further increase the endothelial-facilitated invasiveness, the invasiveness of α5β1high cells was analyzed in the presence or absence of embedded FN. The results show that embedded FN (100 μg/ml) enhanced the endothelial-facilitated invasiveness of α5β1high cells as shown by increased numbers of invasive cells (Fig. 7A) and their increased invasion depth (Fig. 7B). These findings suggest that the stimulation of FN binding receptors with embedded FN may facilitate the increased invasiveness of highly invasive (and highly α5β1 integrin expressing) cancer cells through enhanced contractile force transmission and generation.
FIGURE 7.
The endothelial-facilitated invasiveness of α5β1high cells was enhanced in the presence of embedded FN and reduced after the addition of the myosin light chain kinase inhibitor ML-7 as well as of inhibitors of small GTPase signaling. A, the percentage of invasive α5β1high cells derived from MDA-MB-231 cells was increased in the presence of HPMECs cultured on 3D-FN-ECMs. CO, co-cultured. B, the invasion profile shows that the α5β1high cells invaded deeper into the 3D-ECMs when co-cultured with HPMECs on 3D-FN-ECMs. C, incubation of the α5β1high cells with 15 μm ML-7 (myosin light chain kinase inhibitor), 15 μm Y27632 (Rho kinase inhibitor (Inh)), 100 μm Rac Inh (Rac-1 inhibitor), 100 μm PD98059 (MEK inhibitor), and 50 μm LY294002 (PI3K inhibitor) 2 h before the co-culture invasion assay start reduced the percentage of invasive cells in the presence of HPMECs. D, the invasion profile shows that the invasion depths were reduced in α5β1high cells in the presence of HPMECs pretreated with ML-7, Y27632, Rac inhibitor, PD98059, and LY294002 compared with untreated cells. ***, p < 0.001.
The Inhibition of Contractile Forces Reduces the Endothelial-facilitated Invasiveness of α5β1high Cells
To investigate whether the inhibition of contractile forces affects the endothelial-facilitated increased invasiveness of α5β1high cells, the myosin light chain kinase was inhibited by adding the inhibitor ML-7 to α5β1high cells 2 h before co-culture and invasion assay start. The inhibition of contractile forces led to decreased invasiveness of HPMEC-co-cultured α5β1high cells (Fig. 7, C and D) as shown by the decreased percentage of invasive cells (Fig. 7C) as well as their decreased invasion depth (Fig. 7D). These results indicate that the endothelial-mediated increased invasiveness of α5β1high cells depended on the generation or transmission of contractile forces of cancer cells.
The Inhibition of Small GTPase Signaling Reduces the Endothelial-facilitated Invasiveness of α5β1high Cells
To analyze whether small GTPase signaling pathways play a role in endothelial cell-facilitated increased invasion of MDA-MB-231 cells, inhibitors against Rho kinase (Y27632), Rac-1 (Rac Inh), MEK (PD98059), and PI3K (LY294002) were added 2 h before co-culture invasion assay start. The results show reduced numbers of invasive cells in the presence of HPMECs as well as reduced invasion depths after the addition of Rho kinase inhibitor (Y27632), Rac-1 inhibitor (Rac Inh), MEK inhibitor (PD98059), and PI3K inhibitor (LY294002) (Fig. 7, C and D), indicating that small GTPases are involved in the endothelial-facilitated MDA-MB-231 cancer cell invasion.
DISCUSSION
Cancer cell transendothelial migration and invasion are complex events and depend on biomechanical and biochemical properties of the endothelium and the connective tissue. This study demonstrates that the invasiveness of MDA-MB-231 breast cancer cells is increased in co-culture with human microvascular or macrovascular endothelial cells as indicated by increased numbers of invasive cancer cells and their increased invasion depth. In addition, this article shows that invasive cancer cells altered the biomechanics of endothelial cells.
To reveal the function of the endothelium in cancer cell invasion, highly invasive MDA-MB-231 cells and weakly invasive MCF-7 cells were co-cultured with two different types of microvascular endothelial cells. In agreement with a previous report (11), endothelial cells increased the numbers of invasive MDA-MB-231 breast cells as well as their invasion depth in 3D-ECMs. In contrast to MDA-MB-231 cells, MCF-7 breast cancer cells did not transmigrate through an HPMEC monolayer, and only less than 2% of MCF-7 cells were able to migrate through an HUVEC monolayer. These results indicate that the invasiveness of MCF-7 was not enhanced by endothelial cells.
Because of these findings, the suggestion was that certain cancer cells such as MDA-MB-231 cells may alter the biomechanical properties of endothelial cells in order to transmigrate and invade 3D-ECMs. Hence, this study focuses on the biomechanical mechanism that leads to higher invasiveness of MDA-MB-231 cells after transendothelial migration. To identify this mechanism, stiffness and cytoskeletal remodeling dynamics of endothelial cells were investigated in co-culture with MDA-MB-231 cancer cells and compared with mono-cultured endothelial cells.
Recent results have been shown that single or loosely clustered endothelial as well as a closed endothelial cell monolayer facilitate MDA-MB-231 cell invasion (11). Therefore, a co-culture of mixed MDA-MB-231 cells and endothelial cells was used to analyze the mechanical properties of endothelial cells. Here, the cellular stiffness of with MDA-MB-231 cells co-cultured endothelial cells was decreased compared with mono-cultured endothelial cells. This result suggests that MDA-MB-231 cells broke down the endothelial barrier by lowering the cell stiffness. In contrast, the weakly invasive MCF-7 cancer cells were not able to regulate the biomechanical properties of co-cultured endothelial cells (data not shown), suggesting that only highly invasive cancer cells are able to decrease the mechanical stiffness and, hence, increase the deformability of endothelial cells to transmigrate through the endothelial monolayer. Furthermore, this behavior may be a prerequisite to overcome and break down the endothelial barrier.
In a previous study, our group found that force fluctuations in an endothelial cell monolayer are widely spread to far distant endothelial cells, suggesting that these force fluctuations are not locally restricted (31). Thus, this study analyzed whether the cytoskeletal remodeling dynamics of endothelial cells are altered during co-culture with MDA-MB-231 cells compared with mono-cultured endothelial cells. Indeed, the MDA-MB-231 cells increased the cytoskeletal remodeling dynamics in co-cultured endothelial cells by 5-fold. In contrast to MDA-MB-231 cancer cells, the non-invasive MCF-7 cancer cells did not alter the cytoskeletal remodeling dynamics of endothelial cells when co-cultured. These results indicate that the ability of highly invasive cancer cells to regulate biomechanical properties of co-cultured endothelial cells may help these cancer cells to transmigrate and invade connective tissue.
Previous studies have reported that endothelial cells are exposed to high forces and, hence, respond to them with disruption of their cell-cell adhesions and, subsequently, of the whole endothelial monolayer when neutrophiles or lymphocytes adhere to the endothelium (33, 34). Hence, neutrophiles and lymphocytes transmit (shear) forces to transmigrate through the endothelium and, finally, into the connective tissue (33, 34). Therefore, the cell surface expression of cell-cell adhesion molecules such as VE-cadherin and PECAM-1 was analyzed during co-culture with highly invasive MDA-MB-231 cells and weakly invasive MCF-7 cells. Indeed, the expression of VE-cadherin and PECAM-1 on the cell surface of microvascular endothelial cells was reduced after co-culture with highly invasive MDA-MB-231 cells but not with weakly invasive MCF-7 cells, indicating that only the highly invasive cells are able to reduce the cell-cell interactions between neighboring endothelial cells to mediate cancer cell transmigration. The reduction of cell-cell adhesion molecules on endothelial cells and endothelial cell stiffness led to a reduction in endothelial cell barrier function. Whether the reduced cellular stiffness of endothelial cells is mediated by a decrease in cell-cell adhesion molecule expression or vice versa has to be further investigated.
For cancer cell transmigration, a dual function of the endothelium was suggested acting either as an enhancer or inhibitor of cancer cell invasion depending on the cancer cell type co-cultured (35). Here, it was confirmed that highly invasive MDA-MB-231 breast cancer cells transmigrate through HUVEC and HPMEC monolayers, indicating that their invasive capability was increased in the presence of macrovascular and microvascular endothelial cells. In contrast to previous reports presenting the endothelium as a “passive” barrier for cell invasion (36, 37), this study shows that the co-culture with highly invasive MDA-MB-231 cells caused biomechanical alterations in endothelial cells that reduced the expression of cell-cell adhesion molecules and, subsequently, the stiffness of endothelial cells. These mechanical alterations may enable cancer cells to transmigrate through an endothelial monolayer and migrate into connective tissue. Thus, the endothelial barrier function seems to be lowered by alterations in endothelial biomechanical properties. These biomechanical results are consistent with an up-regulation of the Erk phosphorylation reported in HUVECs during co-culture with colon cancer cells (38). Moreover, this study shows that the inhibition of the upstream regulatory protein of Erk MEK abolished the endothelial-facilitated increased invasiveness of α5β1high breast cancer cells. These results suggest that cancer cells may apply forces toward the endothelial cell monolayer to alter endothelial mechanical properties and, hence, facilitate cancer cell transendothelial migration. Indeed, for the transmigration and invasion of highly invasive MDA-MB231 cells, contractile forces are necessary to overcome the endothelial barrier because inhibition of the myosin light chain kinase and subsequently the transmission or generation of contractile forces reduces the number of transmigrating and invading cells, indicating that the endothelial-facilitated increased invasiveness depends on the α5β1 integrin expression. Another supporting point was that α5β1high cells are able to transmit and generate 7-fold higher contractile forces compared with α5β1low cells (14).
To reveal by which mechanism cancer cell invasiveness is enhanced by endothelial cells, the transendothelial migration and invasion assay was performed in the presence of embedded FN in 3D-FN-ECMs. As expected, the number of transmigrating and invasive α5β1high cells as well as their invasion depths was increased in the presence of an HPMEC cell monolayer, whereas the invasiveness of α5β1low cells was not affected. These results suggest that FN activates the α5β1 integrins and, subsequently, increases the transmission and generation of contractile forces in α5β1high cells, which further increased their ability to transmigrate and finally to invade. Furthermore, the inhibition of the small GTPases Rho kinase and Rac-1 and of its signaling pathway by PI3K inhibitor revealed that small GTPase signaling may play a role in endothelial-facilitated increased invasiveness of α5β1high cells. In addition, the endothelial-facilitated increased invasiveness of α5β1high cells is not restricted to breast cancer cells, as the invasiveness of α5β1high cells derived from bladder and kidney cancer cells was similarly increased by endothelial cells.
Taken together, biophysical measurements may shed light on the biomechanical mechanism that facilitates transendothelial migration of cancer cells. Furthermore, this study indicates that MDA-MB-231 cells break down the endothelial barrier function by lowering cellular stiffness through remodeling of the actin cytoskeleton. In addition, endothelial cells may secrete FN, which then further activates α5β1 integrins on cancer cells by increased transmission or generation of contractile forces and, subsequently, enhances their invasiveness.
In conclusion, this study shows that the endothelium was no passive barrier for cancer cell invasion. Instead, the endothelium seems to act cancer cell-specific, as it increased or decreased cancer cell transmigration and invasion of certain cancer cell lines. In addition, biomechanical properties of endothelial cells were altered by a certain type of cancer cells with distinct biomechanical properties, indicating that these biomechanical alterations may play a role in the transendothelial migration process of cancer cells. Finally, this article suggests that biomechanical alterations in endothelial cells evoked by certain cancer cells might provide a biomechanical selection process toward higher invasiveness of cancer cells. In conclusion, biomechanical interactions between highly invasive cancer cells and endothelial cells facilitate the transmigration of cancer cells, further enhance their invasion into connective tissue, and subsequently, may determine the malignancy of tumors.
Acknowledgments
I thank Robert Schmiedl for excellent help with the scanning EM, Ursula Schlötzer-Schrehardt for excellent help with the transmission EM, Philip Kollmannsberger for excellent help with the magnetic tweezers, Barbara Reischl and Christine Albert for excellent technical assistance, Ulrike Scholz for excellent secretarial assistance, and Ben Fabry and Wolfgang H. Goldmann for helpful discussions. I thank Thomas Mierke for excellent proofreading and editing of the manuscript.
This work was supported by the Deutsche Krebshilfe (109432).
- ECM
- extracellular matrix
- 3D-ECM
- three-dimensional ECM
- HPMEC
- human pulmonary microvascular endothelial cell
- HDMEC
- human dermal microvascular endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- FN
- fibronectin
- PECAM-1
- platelet endothelial cell adhesion molecule-1
- VE-cadherin
- vascular endothelial-cadherin
- MSD
- mean square displacement
- CFDA
- carboxyfluorescein diacetate
- MFI
- mean fluorescence intensity
- MC
- mono-cultured
- CC
- co-cultured.
REFERENCES
- 1. Frixen U. H., Behrens J., Sachs M., Eberle G., Voss B., Warda A., Löchner D., Birchmeier W. (1991) J. Cell Biol. 113, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 3. Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 4. De Craene B., Gilbert B., Stove C., Bruyneel E., van Roy F., Berx G. (2005) Cancer Res. 65, 6237–6244 [DOI] [PubMed] [Google Scholar]
- 5. Steeg P. S. (2006) Nat. Med. 12, 895–904 [DOI] [PubMed] [Google Scholar]
- 6. Al-Mehdi A. B., Tozawa K., Fisher A. B., Shientag L., Lee A., Muschel R. J. (2000) Nat. Med. 6, 100–102 [DOI] [PubMed] [Google Scholar]
- 7. Discher D. E., Janmey P., Wang Y. L. (2005) Science 310, 1139–1143 [DOI] [PubMed] [Google Scholar]
- 8. Zijlstra A., Lewis J., Degryse B., Stuhlmann H., Quigley J. P. (2008) Cancer Cell 13, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Sluis G. L., Niers T. M., Esmon C. T., Tigchelaar W., Richel D. J., Buller H. R., Van Noorden C. J., Spek C. A. (2009) Blood 114, 1968–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kedrin D., Gligorijevic B., Wyckoff J., Verkhusha V. V., Condeelis J., Segall J. E., van Rheenen J. (2008) Nat. Methods 5, 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mierke C. T., Zitterbart D. P., Kollmannsberger P., Raupach C., Schlötzer-Schrehardt U., Goecke T. W., Behrens J., Fabry B. (2008) Biophys. J. 94, 2832–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guck J., Schinkinger S., Lincoln B., Wottawah F., Ebert S., Romeyke M., Lenz D., Erickson H. M., Ananthakrishnan R., Mitchell D., Käs J., Ulvick S., Bilby C. (2005) Biophys J 88, 3689–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fritsch A., Höckel M., Kiessling T., Nnetu K. D., Franziska Wetzel F., Zink M., Käs J. A. (2010) Nature Physics 6, 730–732 [Google Scholar]
- 14. Mierke C. T., Frey B., Fellner M., Herrmann M., Fabry B. (2011) J. Cell Science 124, 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voura E. B., Chen N., Siu C. H. (2000) Clin. Exp. Metastasis 18, 527–532 [DOI] [PubMed] [Google Scholar]
- 16. Geiger B., Bershadsky A., Pankov R., Yamada K. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 793–805 [DOI] [PubMed] [Google Scholar]
- 17. Neff N. T., Lowrey C., Decker C., Tovar A., Damsky C., Buck C., Horwitz A. F. (1982) J. Cell Biol. 95, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Damsky C. H., Knudsen K. A., Bradley D., Buck C. A., Horwitz A. F. (1985) J. Cell Biol. 100, 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riveline D., Zamir E., Balaban N. Q., Schwarz U. S., Ishizaki T., Narumiya S., Kam Z., Geiger B., Bershadsky A. D. (2001) J. Cell Biol. 153, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mierke C. T., Kollmannsberger P., Zitterbart D. P., Smith J., Fabry B., Goldmann W. H. (2008) Biophys. J. 94, 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rape A. D., Guo W. H., Wang Y. L. (2011) Biomaterials 32, 2043–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mierke C. T., Ballmaier M., Werner U., Manns M. P., Welte K., Bischoff S. C. (2000) J. Exp. Med. 192, 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mierke C. T. (2008) J. Biophys. 2008 183516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kollmannsberger P., Mierke C. T., Fabry B. (2011) Soft Matter 7, 3127–3132 [Google Scholar]
- 25. Hildebrandt J. (1969) Bull. Math Biophys. 31, 651–667 [DOI] [PubMed] [Google Scholar]
- 26. Mijailovich S. M., Kojic M., Zivkovic M., Fabry B., Fredberg J. J. (2002) J. Appl. Physiol. 93, 1429–1436 [DOI] [PubMed] [Google Scholar]
- 27. Kasza K. E., Nakamura F., Hu S., Kollmannsberger P., Bonakdar N., Fabry B., Stossel T. P., Wang N., Weitz D. A. (2009) Biophys. J. 96, 4326–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fabry B., Maksym G. N., Butler J. P., Glogauer M., Navajas D., Fredberg J. J. (2001) Phys. Rev. Lett. 87, 148102. [DOI] [PubMed] [Google Scholar]
- 29. Kollmannsberger P., Fabry B. (2009) Soft Matter RSC 5, 1771–1774 [Google Scholar]
- 30. Fredberg J. J., Jones K. A., Nathan M., Raboudi S., Prakash Y. S., Shore S. A., Butler J. P., Sieck G. C. (1996) J. Appl. Physiol. 81, 2703–2712 [DOI] [PubMed] [Google Scholar]
- 31. Raupach C., Zitterbart D. P., Mierke C. T., Metzner C., Müller F. A., Fabry B. (2007) Phys. Rev. E Stat. Nonlin Soft Matter Phys. 76, 011918 [DOI] [PubMed] [Google Scholar]
- 32. Metzner C., Raupach C., Mierke C. T., Fabry B. (2010) J. Phys. Condens. Matter 22, 194105. [DOI] [PubMed] [Google Scholar]
- 33. Cinamon G., Shinder V., Alon R. (2001) Nat. Immunol. 2, 515–522 [DOI] [PubMed] [Google Scholar]
- 34. Rabodzey A., Alcaide P., Luscinskas F. W., Ladoux B. (2008) Biophys. J. 95, 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mierke C. T. (2011) Cell Biochem. Biophys., in press [Google Scholar]
- 36. Verin A. D., Patterson C. E., Day M. A., Garcia J. G. (1995) Am. J. Physiol. 269, L99–L108 [DOI] [PubMed] [Google Scholar]
- 37. Nottebaum A. F., Cagna G., Winderlich M., Gamp A. C., Linnepe R., Polaschegg C., Filippova K., Lyck R., Engelhardt B., Kamenyeva O., Bixel M. G., Butz S., Vestweber D. (2008) J. Exp. Med. 205, 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tremblay P. L., Auger F. A., Huot J. (2006) Oncogene 25, 6563–6573 [DOI] [PubMed] [Google Scholar]