Background: The mental retardation p21-activated kinase (PAK3) protein regulates synaptic plasticity through the regulation of cytoskeleton dynamics of dendritic spines.
Results: PAK3 binds the Grb4/Nck2 adaptor in brain, and inhibition of this complex alters synaptic currents but not spine morphology.
Conclusion: The PAK3-Nck2 complex regulates post-synaptic transmission independently of spine dynamics.
Significance: This opens perspectives in understanding the PAK3 implication in synaptic plasticity and mental retardation.
Keywords: Adaptor Proteins, Protein-Protein Interactions, Serine/Threonine Protein Kinase, Signal Transduction, Synapses, Dendritic Spine, Mental Retardation, Nck Adaptor, Synaptic Transmission, p21-activated Kinase
Abstract
Mutations in the p21-activated kinase 3 gene (pak3) are responsible for nonsyndromic forms of mental retardation. Expression of mutated PAK3 proteins in hippocampal neurons induces abnormal dendritic spine morphology and long term potentiation anomalies, whereas pak3 gene invalidation leads to cognitive impairments. How PAK3 regulates synaptic plasticity is still largely unknown. To better understand how PAK3 affects neuronal synaptic plasticity, we focused on its interaction with the Nck adaptors that play a crucial role in PAK signaling. We report here that PAK3 interacts preferentially with Nck2/Grb4 in brain extracts and in transfected cells. This interaction is independent of PAK3 kinase activity. Selective uncoupling of the Nck2 interactions in acute cortical slices using an interfering peptide leads to a rapid increase in evoked transmission to pyramidal neurons. The P12A mutation in the PAK3 protein strongly decreases the interaction with Nck2 but only slightly with Nck1. In transfected hippocampal cultures, expression of the P12A-mutated protein has no effect on spine morphogenesis or synaptic density. The PAK3-P12A mutant does not affect synaptic transmission, whereas the expression of the wild-type PAK3 protein decreases the amplitude of spontaneous miniature excitatory currents. Altogether, these data show that PAK3 down-regulates synaptic transmission through its interaction with Nck2.
Introduction
Mental retardation, also named intellectual disability, is the most frequent human cognitive disorder and has now been associated with defects in several different genes (1, 2). Among these genes, pak3 is located on the X chromosome and encodes a p21-activated kinase (PAK)4 of group I with five mutations responsible for this disease that have been identified to date (3, 4). PAK group I also includes PAK1 and PAK2, and PAKs are known to be effectors of Rac1 and Cdc42 GTPases and to play an important role in the regulation of cytoskeleton dynamics (5). Recent data demonstrate their role in synaptic plasticity, in spinogenesis (4), and also in post-natal brain development (6). The lack of functional complementation between PAK3 and the two other PAKs in patients with mental retardation as well as in pak3 KO mice strongly suggests that PAK3 possesses specific functions in neuronal signaling (4). Indeed, pak3 knock-out leads to anomalies in plasticity (e.g. long term potentiation) and deficiencies in learning and memory (7). Expression of PAK3 genes with kinase-dead or mental retardation mutations in hippocampal neurons also alters spinogenesis (8–10). Nevertheless, how PAK3 regulates synaptic transmission and plasticity is still largely unknown (4, 11).
PAK protein activation is achieved through their recruitment to the membrane and their interaction with Rac1 and Cdc42 GTPases. One pathway for membrane recruitment is reached through the two Nck adaptors (Nck1/Nckα and Nck2/Nckβ/Grb4), each of which contains one SH2 domain and three SH3 domains (12–14). These adaptors link membrane-localized phosphotyrosine residues to proline-rich domain-containing proteins implicated in cytoskeleton regulation (15). Indeed, a mutation that abrogates Nck interaction with the Drosophila D-PAK protein generates defects of photoreceptor axon pathfinding (16). Moreover the phosphorylation-dependent regulation of the PAK1/Nck1 interaction was shown to control cell spreading, polarization, and migration (17–19). Recent data show that Nck1 and Nck2 play different roles in cell signaling, spinogenesis, and synaptic plasticity (20–23). Interestingly, Nck2 is implicated in dendritic spine morphogenesis downstream from the ephrinB reverse signaling pathway (22). The notion that interaction between PAK3 and Nck adaptors may play a role in synaptic signaling is thus particularly attractive.
To address this hypothesis, we investigated the interaction between PAK3 and the two Nck adaptors, using several approaches in brain lysates and in transfected cells. We identified PAK3-Nck2 as the main complex in brain and characterized the unique properties of the PAK3-Nck2 complex compared with the other PAK-Nck complexes described to date. We demonstrated that Nck2 plays an important role in evoked synaptic response. Finally, we showed that the PAK3-Nck2 complex does not influence dendritic spine and synapse formation, but does down-regulate spontaneous synaptic transmission. These results uncover new aspects of PAK3 function in neuronal signaling by linking Nck2 signaling to PAK3.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
In this report, PAK3 refers to the PAK3a splice variant (24, 25). The following plasmids were described previously (10, 24): the pcDNA3-HA-PAK3-WT, -kd, -ca, -R419X, -A365E, and -R67C plasmids encode mouse HA-tagged PAK3 wild-type, K297L kinase-defective, T421E constitutively active, mental retardation truncated mutant, missense kinase defective mutant, and the missense R67C mutated proteins, respectively. New mutants and constructs were prepared from the pcDNA3-HA-PAK3-WT plasmid with Pfu polymerase (Promega) using procedures based upon the QuikChange protocol (Stratagene) and confirmed by sequencing. The mutant of the PAK3 first proline-rich domain (P12A) was created using the oligonucleotide set (5′-GCTTGGATAACGAAGAAAAACCCGCGGCTCCCCCACTGAGGATGAAC-3′ and 5′-GTTCATCCTCAGTGGGGGAGCCGCGGGTTTTTCTTCGTTATCCAAGC-3′) by PCR from pcDNA3-HA-PAK3-WT, giving the pcDNA3-HA-PAK3-P12A plasmid. The p3×-FLAG-PAK3-WT, p3×-FLAG-PAK3-P12A and p3×-FLAG-PAK3-kd constructs were obtained by KpnI/XbaI digestion of the corresponding pcDNA3-HA-PAK3 plasmids, and the inserts were ligated into the p3×-FLAG-CMV-24 vector (Sigma). The N-terminal deleted mutant of PAK3 (ΔNter) was obtained using the oligonucleotide set (5′-CGGGATCCTGGTAACAACCGAGACTCTTCAGCACTCAACC-3′ and 5′-GGCTCTAGACTAACGGCTACTGTTCTTAATTGC-3′). After KpnI/XbaI digestion, the truncated sequence of PAK3 was introduced and ligated into the KpnI/XbaI linear p3×-FLAG-CMV-24 vector, to obtain the p3×-FLAG-PAK3-ΔNter plasmid. The pGFP-PAK3-WT plasmid was described previously (10). The BamHI/XbaI fragment of the pcDNA3-HA-PAK3-P12A plasmid was subcloned into the BamHI/XbaI linear pEGFP-C1 vector (Clontech) to obtain the pGFP-PAK3-P12A plasmid. The sequence coding for peptide covering PAK3 amino acids 2–49 fused to the glutathione S-transferase (GST) moiety were amplified with the oligonucleotide set (5′-CAGGATCCTCTGACAGCTTGGATAACG-3′ and 5′-GCCTCGAGTCAGCGAAGCCTGGCTTTCTTATTCTTCTC-3′), and the PCR products were BamHI/XhoI-subcloned into pGEX-6P1 (GE Healthcare), leading to the pGST-PAK3-(2–49) constructs (WT and S20E). pGST-PAK3-S4G and pGST-PAK3-S4G/S20E constructs were prepared from pGST-PAK3-(2–49)-WT and pGST-PAK3-(2–49)-S20E plasmids, respectively, with Pfu polymerase (Promega) using procedures based upon the QuikChange protocol (Stratagene), using the oligonucleotide set (5′-CAGGGGCCCCTGGGATCCTCTGACGGCTTGGATAACGAAGAAAAACCCCCAG-3′ and 5′-CTGGGGGTTTTTCTTCGTTATCCAAGCCGTCAGAGGATCCCAGGGGCCCCTG-3′) and confirmed by sequencing. The FLAG-Pak1-KD was obtained by subcloning an amplified fragment of the pDA-PAK1-K299R kindly provided by M. Latta-Mahieu (Sanofi-Aventis, France) into the p3×-FLAG-CMV-24 vector. The N-terminal portions of mouse PAK1-(2–56) and PAK2-(2–63) were amplified with the oligonucleotide sets 5′-CCGGATCCAGGATTCAAATAACGGCGTAGACATC-3′, 5′-GCTCTAGACTATCGATAAAACCGGTCCTTC-3′ and 5′-CGCGAATTCTCTGATAACGGGGAGCTAGAG-3′, 5′-CTCGAGCTATTTGTTCCTCGGTTTTTTTTCC-3′, respectively. The PCR products were, respectively, BamHI/XbaI subcloned into pGEX-2T* (GE Healthcare), leading to pGST-PAK1-(2–56)-WT plasmid, and EcoRI/XhoI subcloned into pGEX-6P3 (GE Healthcare), leading to pGST-PAK2-(2–63)-WT plasmid.
The pRK5-HA-Nck1 and the pRK5-HA-Nck2 plasmids were a gift from W. Li (University of Southern California, Los Angeles). The pGST-Nck1-(SH3)2 was kindly provided by B. Mayer (University of Connecticut Health Center, Farmington). The pGST-Nck2-(SH3)2 plasmid was obtained by amplification of the second SH3 domain of human Nck2 from pRK5-HA-Nck2 with the oligonucleotide set 5′-CGCGGATCCGCCGACCGCATCTACGACC-3′ and 5′-CCGGAATTCAGTCCACCTCCTCCAAGACG-3′, and BamHI/EcoRI cloning into the pGEX-6P3 vector.
Plasmids expressing GFP-fused GTPases (GFP-Cdc42-V12 and GFP-Cdc42-N17) and the GST-Cdc42-V12 plasmid were a gift from P. Fort (CNRS, CRBM, Montpellier, France). The huntingtin plasmid was a gift from S. Humbert and F. Saudou. The GST-B-Raf plasmid was described previously (26).
Cell Culture and Transfection
COS-7 cells and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Plasmid DNA was transfected either using the electroporation method with an electroporator (Bio-Rad) for COS-7 cells, as described previously (24), or with Lipofectamine LTX Plus reagent (Invitrogen) for HeLa cells, as recommended by the manufacturer.
RT-Quantitative Real Time PCR
cDNAs were synthesized using the Moloney murine leukemia virus reverse transcriptase according to the manufacturer's instructions (Promega) from 2 μg of total RNA of mouse E18 and adult brains. Quantitative PCR was performed with a Lightcycler (Roche Diagnostics) using the FastStart DNA MasterPLUS SYBR Green I detection protocol. The reaction mixture was subjected to real time PCR cycling by hot start followed by multiple cycles of denaturing at 95 °C for 5 s, annealing at 60 °C for 5 s, and extension at 72 °C for 8 s. Mouse β-actin, Nck1, and Nck2 primers sets utilized for real time quantification were obtained from Qiagen (QuantiTect Primer Assay) and used following the manufacturer's instructions. The analysis of fluorescence was carried out using LC software version 3.01 (Roche Diagnostics).
Antibodies and Immunoblotting
Immunoblot analyses were performed using anti-HA (monoclonal 12CA5 from Roche Diagnostics, agarose-linked-HA antibodies from Sigma A2095, and the polyclonal sc-805 from Santa Cruz Biotechnology), anti-FLAG (F7425 from Sigma), anti-GFP (catalog no. 2555 from Cell Signaling Technology), anti-PAK3 (N19, Sc-1871 from Santa Cruz Biotechnology), anti pan-Nck directed against the SH2 domain of the Nck adaptor (catalog no. 61099 from BD Transduction Laboratories), and anti-β-actin (AC-15, sc-69879 from Santa Cruz Biotechnology). Because commercial Nck antibodies do not allow the immunoprecipitation of each Nck isoform, we generated two new antibodies against unique sequences for each Nck adaptor. The PADDSFVDPGE peptide corresponding to an internal (92–102) amino acid sequence of the human/mouse Nck1 protein was synthesized and fused to an aminohexanoic acid arm and a cysteine for linking to keyhole limpet hemocyanin (GeneCust). The DGPALHPAH peptide inside the human/mouse Nck2 protein (residues 257–265) was similarly processed. Two rabbits for each conjugated peptide were immunized with a classical procedure. For the Western blot in Fig. 1E, the Nck polyclonal antibodies were affinity-purified by Ultralink column chromatography (Perbio Science) after covalent attachment of the corresponding peptide, following the manufacturer's instructions. Immunocytochemistry experiments were performed using mouse monoclonal PSD95 (K28/43 from NeuroMab) antibodies, rabbit vGLUT1 antibody (27), and Alexa-568- and Alexa-633-conjugated secondary antibodies (Invitrogen), as described previously (10). For Western blot analysis, protein samples were separated by 10% SDS-PAGE and transferred to PVDF membranes (Millipore). Immunodetection was performed using the SuperSignal chemiluminescent reagent (Thermo Scientific Pierce). Quantification was performed after acquisition with a CCD camera (GeneSnap, SynGene) and quantification software (GeneTools, SynGene). Statistical analyses were performed using the Student's t test in GraphPad Prism (GraphPad Software, Inc.). All data shown are representative of a typical experiment from three independent experiments or more.
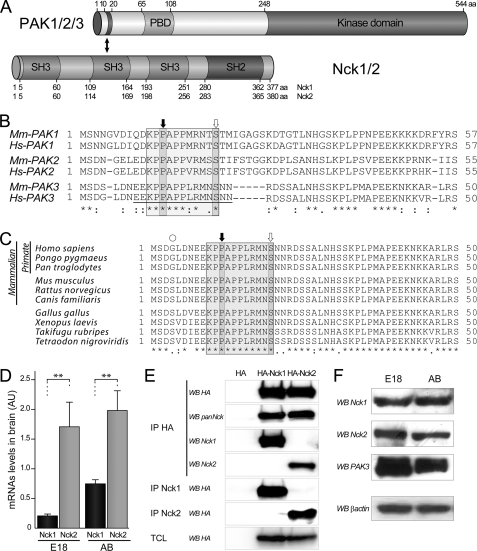
FIGURE 1.
Nck adaptor expression in mouse brain. A, domain structure of PAK and Nck proteins involved in the interaction. The N-terminal part of PAK proteins contains an SH3-binding domain located between amino acids 10 and 20. This region interacts with the second SH3 domain of Nck adaptors. The amino acid (aa) numbering of PAK corresponds to those of the human PAK3 sequence. B, amino acid alignment of the N-terminal region of PAK proteins of group I encoded by mouse and human genes. The amino acid sequence of the SH3-binding domain contains a conserved proline-rich motif PPXPP and two critical residues Pro-12 and Ser-20 (numbers refer to PAK3) for Nck interaction are indicated with black and white arrows, respectively. The sequence of the P12 peptide used in the following sections, which inhibits Nck2 interaction, is underlined. C, sequence alignment of the N-terminal domains of primate, mouse, Xenopus, and fish illustrates the phylogenic conservation of the amino extremity of the PAK3 proteins. D, mRNA quantification by quantitative-PCR shows more Nck2 than Nck1 expression in brain. Total RNAs (2 μg) from mouse embryonic E18 brain (E18) and adult brain (AB) were subjected to quantitative RT-PCR. Comparison with Student's t test: **, p < 0.01, n = 4. Error bars indicate the S.E. E, characterization of the Nck sera. The two Nck isoform-specific sera were tested for their ability to specifically recognize their targets. Lysates of COS-7 cells transfected either with HA-Nck1 or HA-Nck2 plasmids were controlled for Nck expression by HA-Western blotting (WB) on total cell lysate (last panel), and HA-precipitated (IP HA) before being analyzed by Western blotting, using a polyclonal HA or the pan-Nck monoclonal antibodies (1st and 2nd panels), or immune Nck1 or Nck2 sera (3rd and 4th panels). Ability for immunoprecipitation of the two new sera was tested from similarly transfected cells, and precipitated complexes were analyzed by HA Western blotting (5th and 6th panels). F, the two Nck and PAK3 proteins were detected in mouse brain by Western blot analysis. E18 and adult brain lysates (25 μg of proteins) were probed with either the Nck1 and the Nck2 antibodies or the PAK3 antibodies and β-actin antibodies for loading control. Data shown in the D–F are representative of a typical experiment.
Pulldown Assay and Co-immunoprecipitation
The GST-fused proteins were expressed in Escherichia coli BL21 and purified on glutathione-agarose beads as described by the manufacturer (GE Healthcare). Pulldown assays were performed as described previously (24). Briefly, COS-7 cells were transfected with constructs expressing HA-PAK3-WT. Forty hours later, cells were washed in cold phosphate-buffered saline (PBS) containing 0.1 mm orthovanadate and lysed in 50 mm HEPES, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 15 mm NaF, 2 mm EDTA, and protease inhibitor mixture. After 15 min on ice, lysates were cleared by centrifugation at 20,000 × g for 15 min. Cleared cell extracts were incubated with 10 μg of GST-Nck1-(SH3)2 or GST-Nck2-(SH3)2 recombinant proteins and immobilized on glutathione beads, for 2 h at 4 °C. The beads were washed with the lysis buffer, and affinity-precipitated proteins were analyzed by Western blotting using the appropriate antibodies. For co-immunoprecipitation from COS-7 and HeLa cells expressing FLAG-PAK3 constructs and/or the HA-Nck plasmids, cells lysed in modified Robert's lysis buffer (0.1% Triton X-100, 10% glycerol, 20 mm Tris, pH 8.0, 137 mm NaCl, 50 mm NaF supplemented with protease and phosphatase inhibitors mixture) were immunoprecipitated overnight at 4 °C, with anti-HA agarose-conjugate (Sigma). For immunoprecipitation in the presence of synthetic peptides, the wild-type inhibitory peptide named P12 (EEKPPAPPLRMNSNN) or the mutant peptide named A12 (EEKPAAPPLRMNSNN), purchased from GeneCust at >90% purity, was added at 1 mm after cell lysis. Immune complexes were separated by electrophoresis and transferred to PVDF membranes, before Western blotting analysis, with appropriate antibodies. Expression of transfected proteins was checked in each experiment by Western blotting of total cell lysate (TCL) sample. The competing P12 or the control A12 peptides were added at 250 μm in cell lysate before PAK proteins were pulled down by GST-Nck2(SH3)2 beads, as described above. For brain co-immunoprecipitation, adult brain or 18-day embryonic mouse brains were dissected and washed in ice-cold PBS, homogenized with a Potter-Elvehjem homogenizer in modified Robert's lysis buffer as described above, and incubated for 20 min on ice. Lysates were cleared by centrifugation at 20,000 × g for 15 min, and 10 mg of total protein was used for each assay. A clarification step with protein G-agarose (Sigma) for 2 h at 4 °C was performed. 10 μl of anti-Nck1 or anti-Nck2 serum or the corresponding preimmune sera, in the presence of 50 μl of protein G-agarose, were added for overnight incubation at 4 °C. After extensive washing, precipitated complexes were analyzed by Western blotting, with appropriate antibodies.
Kinase Assay
For GST-PAK3-(2–49)-WT or GST-PAK3-(2–49)-S20E phosphorylation by CaMKII, 10 μg of GST recombinant PAK3 proteins were incubated with 50 units of recombinant αCaMKII-(1–325) (New England Biolabs) for 45 min at 30 °C in the presence of 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 2 mm DTT, 0,1 mm EDTA, 1.2 mm calmodulin, 2 mm CaCl2, 5 μCi/tube of [γ-32P]ATP (4500 Ci/mmol, PerkinElmer Life Sciences), 10 μm ATP. For Cdc42-dependent activation, COS-7 cells were transfected with 10 μg of HA-PAK3-WT plasmid. To activate the kinase, cleared cell extracts were incubated with 5 μg of recombinant GST-Cdc42-V12 in the presence of 25 μm ATP during 30 min at room temperature. Extracts were then immunoprecipitated by incubating them with 4 μl of 12CA5 anti-HA antibody and 50 μl of protein G-agarose (Sigma) overnight at 4 °C. For kinase reactions, immunoprecipitates were washed in the kinase buffer (25 mm HEPES, pH 7.4, 25 mm MgCl2, 25 mm β-glycerophosphate, 2 mm dithiothreitol, 0.1 mm orthovanadate) and then incubated in the presence of 20 μm ATP and 5 μCi of [γ-32P]ATP for 20 min at 30 °C. Akt and PKA kinases were purchased from Cell Signaling and Sigma, respectively, and kinase assays were performed as recommended by the providers.
Tryptic Digestion and Phosphopeptide Mapping
After autophosphorylation or CaMKII kinase assays, full-length or the amino acid residues 2–49 of PAK3 protein was separated on 10 and 15% acrylamide gels, respectively. Gels were dried between two cellophane sheets, and radioactive bands corresponding to PAK3 were detected by autoradiography. Each band was excised and washed three times in 45% methanol, 10% acetic acid and three times in 50% methanol. Bands were then resuspended in 25 mm ammonium bicarbonate, pH 7.5, containing 25 units of trypsin (Promega) for 14 h at 37 °C. Two-dimensional migrations of tryptic phosphopeptides were performed as described previously (26). Briefly, the lyophilized samples were resuspended in 10% acetic acid (5000–10,000 cpm/μl), spotted onto 100-μm cellulose-coated plates (Kodak chromatogram sheet) in 1-μl aliquots, and electrophoresed for 3 h at 400 V in a horizontal electrophoresis unit in pH 3.5 buffer (acetic acid 10% and pyridine 1%). Plates were air-dried and then chromatographed in buffer containing 30% pyridine, 20% butanol, and 6% glacial acetic acid until the mobile phase reached the top of the plate. 32P-Labeled tryptic peptides were detected by autoradiography on Kodak Biomax film for up to 3 days.
Dissociated Hippocampal Neuron Culture, Immunocytochemistry, Imaging, and Synaptogenesis Analysis
Primary cultures of dissociated hippocampal neurons were prepared from embryonic day 17 (E17) mice. Briefly, 300–400 cells/mm2 were plated on 18-mm coverslips previously coated with 0.5 mg/ml poly-l-ornithine and grown in B27-supplemented Neurobasal medium (Invitrogen). Neurons were transfected with GFP-tagged constructs using Lipofectamine 2000 (Invitrogen) at 18 days in vitro (DIV) for synapse analysis, before paraformaldehyde fixation at DIV21. After fixation, neurons were imaged with a Zeiss LSM 510 confocal microscope using the 488-nm band of an argon laser, the 561-nm band of a diode-pumped solid-state laser, and the 633-nm band of a He:Ne laser. Healthy and differentiated transfected neurons were selected, and cells with a high expression of the transgene were excluded. Images were acquired using a ×63 objective (1.4 numerical aperture), using a frame size of 1024 × 1024 pixels, with an optic zoom to generate 50 × 50 μm cropped images. Because protrusion density could be variable depending on the distance from the soma, dendritic segments were acquired at a distance restricted between 50 and 150 μm from the soma. Furthermore, z series stacks of 1.8–3 μm (typically five to eight frames) were taken from the bottom to the top of dendrites and used ImageJ software (National Institutes of Health) to generate image projections. All images were acquired and analyzed while blinded to the experimental condition. Each experiment was repeated two to four times from different cultures. For the morphometric study, the parameters analyzed were protrusion density and length, measured from the limit of the dendrite to the tip of the protrusion using SimplePCI image analysis software (Hamamatsu Inc.). The classification of dendritic protrusions, i.e. filopodia versus spines, was verified at the same time using the three-dimensional image stack. All data are expressed as means ± S.E. For synapse analysis, after paraformaldehyde fixation at DIV21, GFP-transfected neurons were double immunolabeled for native proteins using anti-PSD-95 (1:200) and anti-vGLUT1 (1:2000) followed by Alexa-568-conjugated anti-mouse IgG and Alexa-633-conjugated anti-rabbit IgG. PSD-95-positive protrusions were counted from GFP and PSD-95 images. Synapses were identified as PSD95-positive spines with adjacent staining of vGLUT1 from the three-color images. Immunofluorescent puncta were counted for 100 μm lengths of dendrites. All images were acquired and analyzed as described above while blinded to experimental conditions using identical acquisition parameters. Statistical comparisons of PSD-95 clusters and densities were done using the Student's t test in GraphPad Prism (GraphPad Software, Inc). All data are expressed as means ± S.E.
Peptide Perfusion and Patch Clamp Recording in Cortical Slices and mEPSCs Recording
In accordance with the guidelines of the American Physiological Society, 2 weak-old Wistar rats were decapitated and their brains quickly removed. A hemisection of the left hemisphere was removed, attached to the stage of a Vibratome tissue slicer (Campden Instrument), and immersed in ice-cold artificial cerebrospinal fluid (aCSF) containing (in mm) the following: 126 NaCl, 26 NaHCO3, 10 glucose, 2 CaCl2, 1.5 KCl, 1.5 MgSO4, and 1.25 KH2PO4 (pH 7.5, 310–330 mosm). 250-μm parasagittal slices containing primary visual cortex were cut and transferred, after at least 1 h of recovery, to the recording chamber mounted on the X-Y translation stage of an upright microscope (Zeiss Axioscop 2 FS+). Slices were perfused with aCSF, continuously bubbled with 95% O2, 5% CO2, and maintained at 32 °C using an in-line solution heater (Warner Instruments). Cells were visualized using video-enhanced differential interference contrast optics and ×40 long-working distance water immersion lens. Stable whole-cell voltage clamp recordings of evoked excitatory postsynaptic currents (eEPSCs) were performed in layer 5 pyramidal neurons using a Multiclamp 700A amplifier (Axon Instruments). As described previously (28), eEPSCs were recorded, 10 min after patch stabilization, following electrical stimulations (1–10 μA, 0.2 ms, every minute) of the layer 2/3 with 1 megohm impedance bipolar tungsten electrodes (TST33A10KT, WPI, Hertfordshire, UK). Stimulation intensity was set to obtain subliminal eEPSCs, and cells were held at −80 mV. This value is the reversal potential for inhibition in our experimental conditions so that we record only excitatory responses (29, 30). To monitor the stability of patch seals, access resistance was also measured over the whole time of the experiment using a 50-ms hyperpolarizing step injected 100 ms before the onset of stimulation (as is illustrated in Fig. 3H). In addition, to monitor eEPSCs stability during the experiments, a control recording was systematically performed at the same time in a surrounding layer 5 pyramidal neuron using a patch pipette that did not contain the peptide. Data were filtered at 2 kHz by a low pass Bessel filter and sampled at 4 kHz using a Digidata 1322A acquisition board (Axon Instruments). Peptide perfusion was achieved through patch clamp pipettes (3–5 megohms) that contained 100 μm of either the WT uncoupling peptide P12 (EEKPPAPPLRMNSNN) or the A12 mutant (i.e. noninteracting) peptide (EEKPAAPPLRMNSNN) and diluted in the following internal solution (in mm): 140 potassium gluconate, 10 HEPES, 4 ATP, 2 MgCl2, 0.4 GTP, and 0.5 EGTA (pH 7.3 adjusted with KOH, 270–290 milliosmolar). The synthetic peptides used in this study were purchased from GeneCust at >90% purity and dialyzed against a patch pipette solution. Statistical analyses were performed blind to experimental conditions using the Mann-Whitney test in GraphPad Prism (GraphPad Software, Inc). As described in Seeburg et al. (31), electrophysiological recordings of mEPSCs were performed in dissociated mouse hippocampal cultures placed in a recording chamber mounted on the X-Y translation stage of an upright microscope (Zeiss Axioscop 2 FS+) and perfused (∼2 ml/min) at room temperature (∼25 °C) with the following extracellular solution (in mm): 119 NaCl, 2.5 KCl, 2.8 CaCl2, 2 MgCl2, 26 NaHCO3, 1 NaH2PO4, 11 glucose, 0.01 bicuculline, gassed with 5% CO2, 95% O2 at pH 7.4. Tetrodotoxin (1 μm) was added to block action potentials. Whole-cell voltage clamp recordings were performed as described above, and mEPSCs were recorded after 5 min of patch stabilization and during roughly 5 min to obtain at least 100 events per cell. Patch pipettes were filled with an internal solution containing (in mm) the following: 115 cesium methanesulfonate, 20 CsCl, 10 HEPES, 2.5 MgCl2, 4 ATP disodium salt, 0.4 GTP trisodium salt, 10 sodium phosphocreatine, and 0.6 EGTA, at pH 7.25. Analysis and quantifications were performed off-line with the specialized ElphyTM software (Gérard Sadoc, Biologic UNIC-CNRS, France), blinded to experimental conditions. Statistical analyses were performed using the Mann-Whitney test in GraphPad Prism (GraphPad Software, Inc.).
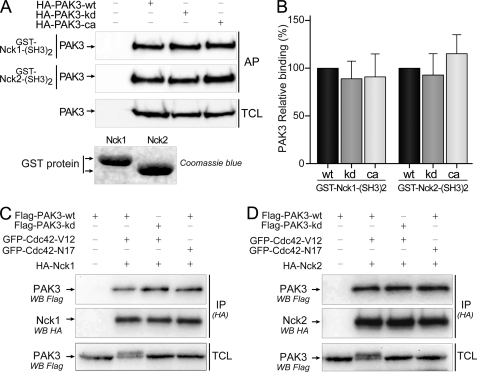
FIGURE 3.
PAK3 binds Nck2 independently of its kinase activity. A, in vitro interaction of different PAK3 kinase mutants with the second SH3 domains of Nck1 and Nck2. WT, kinase-dead (kd), and constitutively active (ca) forms of HA-PAK3 were expressed in COS-7 cells, and lysates were incubated with recombinant GST proteins containing the second SH3 domain of Nck1 (1st panel) or Nck2 (2nd panel). Affinity-purified (AP) PAK3 proteins and COS-7 expressed PAK3 proteins (TCL) were revealed by Western blotting (WB) with HA antibodies. Data shown are representative of a typical experiment from three independent experiments. Amount of GST proteins was controlled by Coomassie Blue staining. Note that the lower electrophoretic migration of the GST-Nck1-(SH3)2 protein is due to the presence of an HA tag. B, quantification of the amount of different PAK3 proteins precipitated with the second SH3 domain of Nck adaptors from the experiments illustrated in A, n = 3. Error bars indicate the S.E. C and D, Cdc42 activation of PAK3 does not suppress its interaction with Nck1 (C) or Nck2 (D). COS-7 cells were co-transfected with plasmids coding for FLAG-PAK3-WT or FLAG-PAK3-kd (kinase-dead), with either active Val-12 or inactive Asn-17 mutants of GFP-Cdc42 vectors, together with HA-tagged Nck1 (C) or Nck2 (D) plasmids. Nck1 or Nck2 proteins were HA-immunoprecipitated (IP (HA)) and visualized after HA immunoblotting (middle panels). PAK3 proteins in immune complexes (upper panels) and in TCLs were revealed by anti-FLAG immunoblotting. GTPase expression was verified using anti-GFP antibody (data not shown). Note that activation of PAK3 by Cdc42-Val-12 induced an electrophoretic shift of PAK3-WT protein as previously reported (25).
RESULTS
Nck1 and Nck2 Are Both Expressed in Brain
Members of the group I p21-activated kinases (PAK1–3) interact with the Nck adaptor in both invertebrates and vertebrates through the first highly conserved proline-rich domain of PAK and the second SH3 domain of Nck (Fig. 1, A and B) (12, 13, 16, 32). However, the amino acid sequence of the first SH3-binding domain of PAK3 expected to interact with Nck differs from the conserved sequence in the other PAKs. Moreover, the residues that are unique to PAK3 such as the leucine 16, the methionine 18, and the asparagine 19 are conserved among all PAK3 orthologs (Fig. 1, B and C). In mammals, the interaction between PAK1 and Nck1 has been extensively studied (12, 13, 17–20, 32). Despite much recent data describing specific cellular functions for Nck2/Grb4 (22, 23, 33), the interaction of PAK proteins with Nck2 is poorly documented. Given the salient expression of PAK3 in neurons, we first determined the relative amount of each Nck mRNA in mouse embryonic and adult brain by quantitative RT-PCR (Fig. 1D). After RNA normalization based on β-actin quantitative-PCR, amplification of Nck1 and Nck2 with specific primer sets showed equivalent efficiency. We found that Nck2 transcripts are more abundant than those of Nck1 in embryonic E18 and adult brain. In embryos, Nck2 mRNAs were found to be over eight times more abundant than those of Nck1 (Nck1, 0.20 ± 0.02; Nck2, 1.63 ± 0.27), and in adult brain the ratio is slightly higher than 2.5 (Nck1, 0.75 ± 0.06; Nck2, 1.99 ± 0.33). These results are in accordance with independent analyses such as the Allen Brain Atlas and the neural transcriptome (34, 35). To estimate Nck1 and Nck2 protein levels in brain, we generated two polyclonal sera directed against each adaptor by choosing short unique sequences from these proteins as antigens. To characterize these new antibodies, HA-tagged Nck proteins were HA-immunoprecipitated from transfected COS-7 cells and then probed with the different antibodies by Western blotting (Fig. 1E, 1st to 4th panel). Equal amounts of immunoprecipitated Nck proteins were verified by HA antibody immunoblot (Fig. 1E, 1st panel), and the pan-Nck monoclonal antibody directed against the SH2 domain of Nck adaptors recognized both Nck1 and Nck2 proteins with the same efficiency (Fig. 1E, 2nd panel). Lack of cross-reactivity of the isoform-specific Nck antibodies in Western blotting assay was confirmed (Fig. 1E, 3rd and 4th panels). We also checked the ability of each antibody to immunoprecipitate its respective target protein (Fig. 1E, 5th and 6th panels). It is to be noted that we report here the first characterization of Nck isoform-specific antibodies with immunoprecipitation ability. Furthermore, we observed that the anti-Nck2 is less effective than the anti-Nck1 antibody in recognizing its appropriate target by direct Western blot or immunoprecipitation (Fig. 1E). Using these strictly specific Nck1 and Nck2 antibodies, we analyzed the protein expression of each adaptor in brain extracts from E18 and adult mice along with PAK3 (Fig. 1F). Nck1 protein is equally abundant in embryonic and in adult brain extracts. In contrast, we observed a slightly higher expression of Nck2 and PAK3 proteins in embryo compared with adult brains.
PAK3 Interacts Preferentially with Nck2 Adaptor in Vitro and in Mouse Brain
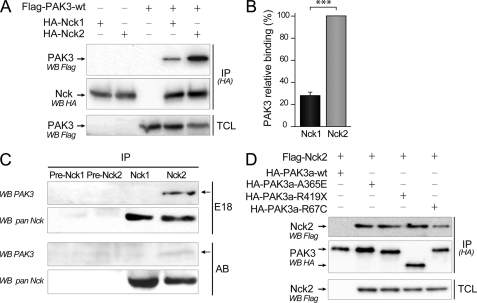
To compare the capacities of the two adaptors to interact with PAK3, COS-7 cells were co-transfected with either HA-Nck1- or HA-Nck2-expressing vectors in the presence of the FLAG-PAK3-WT plasmid. Nck proteins were immunoprecipitated with HA antibodies, and isolated complexes were analyzed for the presence of the associated FLAG-PAK3 proteins (Fig. 2A). Western blot analysis showed that specific co-immunoprecipitation of PAK3 with Nck adaptors was four times greater with Nck2 compared with PAK3 with Nck1 (27.6 ± 2.8% for the Nck1/Nck2 ratio) (Fig. 2B). The anti-Nck1 and anti-Nck2 sera were then used to co-immunoprecipitate PAK3 in mouse E18 and adult brain extracts (Fig. 2C). Preimmune sera were also used as controls. Immunoprecipitated Nck proteins were revealed using the pan-Nck antibody, indicating a similar amount of Nck proteins. Surprisingly, PAK3 protein immunoprecipitates only with Nck2 complexes, both in embryonic brain as well as in adult brain extracts, although less complex is found in adult brain. PAK3 was below the detection threshold in the Nck1 complexes, despite the fact that the anti-Nck1 serum is more efficient than the anti-Nck2 serum at immunoprecipitating its target protein. Hence, these results show that PAK3 preferentially forms complexes with the Nck2 adaptor in mouse brains and that this interaction may be particularly important during brain development. Note also that these results are the first report of co-immunoprecipitation of a PAK-Nck complex from brain tissue. We also tested the capacity of the mental retardation mutated A365E and R419X PAK3 proteins to interact with Nck2 in co-immunoprecipitation assays, and no difference was observed between these mutated proteins compared with the wild type (Fig. 2D). This result could be explained by the localization of the mutations outside of the interacting region between PAK3 and Nck adaptors (10). However, we observed that the R67C mutated PAK3 also associated with mental retardation (36) binds less Nck2, in agreement with the localization of the mutation proximal to the first proline-rich domain.
FIGURE 2.
Preferential binding of PAK3 with Nck2 in vitro and in vivo. A, stronger interaction between PAK3 and Nck2 compared with Nck1. COS-7 cells were co-transfected with plasmids coding for HA-tagged Nck1 or Nck2 with FLAG-PAK3-WT. Nck proteins were HA-immunoprecipitated (IP(HA)). The co-immunoprecipitated PAK3 protein was revealed by anti-FLAG blotting (upper panel). Equal PAK3 expression levels in transfected cells and equal amount of immunoprecipitated Nck proteins were controlled by Western blotting on TCL (lower panel) and immunoprecipitated proteins (IP-HA, WB HA, middle panel), respectively. B, quantification of the amount of PAK3 precipitated with Nck adaptors from the experiments illustrated in A. Comparison with Student's t test: ***, p < 0.001, n = 4. Error bars indicate the S.E. C, PAK3 co-immunoprecipitates with Nck2 but not with Nck1 from mouse brain. E18 or adult brain lysates (10 mg), analyzed by Western blot in Fig. 1F, were immunoprecipitated with preimmune or immune Nck1 or Nck2 sera. Immunoprecipitated Nck proteins were revealed with the pan-Nck antibodies (WB pan Nck). Associated PAK3 proteins were revealed by Western blotting using PAK3 antibodies (WB PAK3). D, three PAK3 proteins bearing the mental retardation mutation A365E, R419Stop, and R67C co-immunoprecipitate with Nck2 with an apparent normal efficiency. COS-7 cells were co-transfected with plasmids coding for HA-tagged PAK3 mutants with FLAG-Nck2. The co-immunoprecipitated HA Nck2 protein was revealed by anti-FLAG blotting (upper panel). Equal Nck2 expression levels in transfected cells and amount of HA-immunoprecipitated PAK3 proteins were controlled by Western blotting on TCL (lower panel) and immunoprecipitated proteins (IP-HA, WB HA, middle panel). Data shown in the A, C, and D are representative of a typical experiment.
PAK3 Binds Nck2 Independently of Its Kinase Activity
The main functions of PAK1 are dependent on its cycling activity that is mostly linked to its subcellular location (17–19). Following its activation at the membrane, PAK1 autophosphorylates and dissociates from the Nck1 complex and then returns to the cytoplasm where it may be dephosphorylated and deactivated (18, 19). To analyze whether the interactions of PAK3 with Nck adaptors are dependent on the kinase activity as described for PAK1, we compared the affinity precipitation of wild-type, kinase dead (K297L), or constitutively active (T421E) variants with the GST-immobilized second SH3 domain of Nck1 or Nck2 (Fig. 3A). We found that the three PAK3 proteins (wild-type, kinase dead, or constitutively active) were equally affinity-precipitated by the second SH3 domain of Nck1 (relative binding compared with wild-type, kinase dead, 89.0 ± 18.0; constitutively active, 91.1 ± 23.6) and of Nck2 (relative binding compared with wild-type, kinase dead, 92.9.± 22.1, constitutively active, 115.1 ± 19.5) (Fig. 3B).
To analyze whether the in vivo interactions of PAK3 with Nck adaptors are regulated by active GTPases, COS-7 cells were co-transfected either with the FLAG-tagged PAK3 wild-type protein or with the kinase-dead mutant (PAK3-K297L), together with HA-Nck1 (Fig. 3C) or HA-Nck2 (Fig. 3D), and PAK3 proteins were activated by co-transfecting the constitutively active GFP-Cdc42-V12 GTPase or the inactive GFP-Cdc42-N17 mutant as a negative control. Surprisingly, we found that the Cdc42-activated PAK3 protein co-immunoprecipitated with Nck2 and even with Nck1 (Fig. 3, C and D). The presence of a kinase-dead mutation in PAK3 did not modify this interaction. Furthermore, the co-expression of the dominant negative N17 form of Cdc42 did not further increase co-immunoprecipitation of PAK3 with Nck adaptors. Thus, in sharp contrast with PAK1, PAK3 binds the Nck adaptors independently of its activation state and kinase activity.
Serine 20 Residue of PAK3 Is Not a Phosphorylated Site
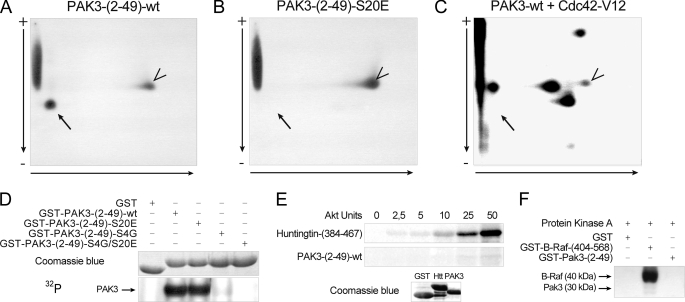
The activation-dependent autophosphorylation of PAK1 serine 21 (white arrow, Fig. 1B) has been shown previously to be responsible for suppression of the PAK1/Nck1 interaction upon PAK1 activation (18). This serine residue lies at the extremity of the highly conserved PPXPPXRXXS consensus sequence for binding to the Nck SH3 domain (18) and is conserved among all PAK proteins of group I, in invertebrates and vertebrates (Fig. 1B). Interestingly, the sequence of the surrounding proline-rich domain is PAK isoform-specific, and the PAK3 amino acid sequence of this domain is totally conserved among all vertebrates (Fig. 1C). We further investigated whether the homologous PAK3 serine 20 residue is indeed an autophosphorylation site. As the serine 20 residue is located in a strong RXXS consensus sequence and is likely to be phosphorylated in vitro by several kinases, such as CaMKII, we used recombinant CaMKII to identify the phosphorylated peptide containing the serine 20. We generated a GST-fused recombinant peptide corresponding to the N-terminal extremity (amino acids 2–49) of PAK3 and tested it in an in vitro kinase assay (supplemental data 1). We observed that the N-terminal part of PAK3 is phosphorylated in vitro by recombinant CaMKII. We then generated a nonphosphorylatable mutated form by the substitution of the serine 20 residue. The tryptic maps of the wild-type (Fig. 4A) and S20E (Fig. 4B) recombinant N-terminal peptides phosphorylated in vitro confirm that the serine 20 residue in the tryptic MNS20NNR peptide is phosphorylated by CaMKII in vitro. Two-dimensional mapping of the autophosphorylated full-length PAK3 protein (Fig. 4C), expressed in COS-7 cells and activated by recombinant Cdc42-V12 protein, indicated multiple autophosphorylated sites in the PAK3 protein, whereas in the absence of active Cdc42, PAK3 does not present any autophosphorylation, as observed previously (24, 25). However, no spot corresponding to the Ser-20-containing peptide (black arrow) is observed in this map (Fig. 4C). The two-dimensional map obtained with the full-length PAK3-S20E mutant gave a similar pattern than the wild-type protein confirming that the Ser-20-containing peptide was not phosphorylated. This was also confirmed by co-migration of peptides from the GST-PAK3-(2–49)-WT sample mixed with those obtained from the autophosphorylated wild-type PAK3 (data not shown). These experiments indicate that the serine 20 residue is not autophosphorylated upon PAK3 activation.
FIGURE 4.
Serine 20 residue of PAK3 is not a regulatory site. A–C, two-dimensional chromatography of PAK3 peptides obtained from recombinant GST-PAK3-(2–49) protein (A) or GST-PAK3-(2–49)-S20E protein (B), which were phosphorylated in vitro by CaMKII, before analysis of the tryptic phosphopeptides by two-dimensional mapping. C, two-dimensional mapping of full-length activated wild-type PAK3 protein. HA-PAK3-transfected COS-7 cell lysate was incubated with recombinant GST-Cdc42-V12 before immunoprecipitation. The precipitate was then subjected to a kinase assay with radioactive ATP. Vertical arrows indicate the electrophoretic direction and horizontal arrows the chromatography migration. Black arrow indicates the serine 20-corresponding peptide, and the arrowheads show an uncharacterized phosphorylated peptide. D, phosphorylation of mutated PAK3-N-terminal proteins. The serine residues of the PAK3-N-terminal proteins were successively mutated and tested in a CaMKII kinase assay, as described above. E, Akt phosphorylation of PAK3-N-terminal and huntingtin as positive control. huntingtin and PAK3 recombinant peptides, visualized with Coomassie Blue staining (lower panel), were tested in an Akt kinase assay (upper panel). F, PKA phosphorylation of PAK3-N-terminal and B-Raf as positive control. B-Raf and PAK3 recombinant peptides were tested in a PKA kinase assay. All data shown are representative of a typical experiment from three independent experiments.
However, the mutation of serine 20 of PAK3 is not sufficient to totally abrogate N-terminal phosphorylation suggesting the presence of other phosphorylation sites targeted by CaMKII (Fig. 4, B and D). To identify these other sites, we have mutated all the putative phosphorylated sites of the N-terminal portion of PAK3 (supplemental data 2) and tested their phosphorylation by CaMKII. The single mutation of the serine 4 residue is sufficient to dramatically decrease the phosphorylation level of PAK3 (Fig. 4D). However, this serine 4 residue present in rodents corresponds to a glycine residue in primates (Fig. 1C). PAK3 N-terminal peptide with a glycine 4 residue, thus corresponding to a human like peptide sequence, does not appear to be phosphorylated by CaMKII, suggesting that CaMKII does not regulate PAK3/Nck2 interaction in primates. We also observed using similar approaches that the serine 20 residue of PAK3 is not phosphorylated by Akt (Fig. 4E) or PKA (Fig. 4F), which were previously described as regulatory kinases for the PAK1 serine 21 residue (19, 37). Altogether, these data suggest that the Akt, CaMKII, and PKA kinases do not regulate the PAK3/Nck2 interaction and that the PAK3-Nck2 complex displays unique properties compared with the complex of PAK1 with Nck1.
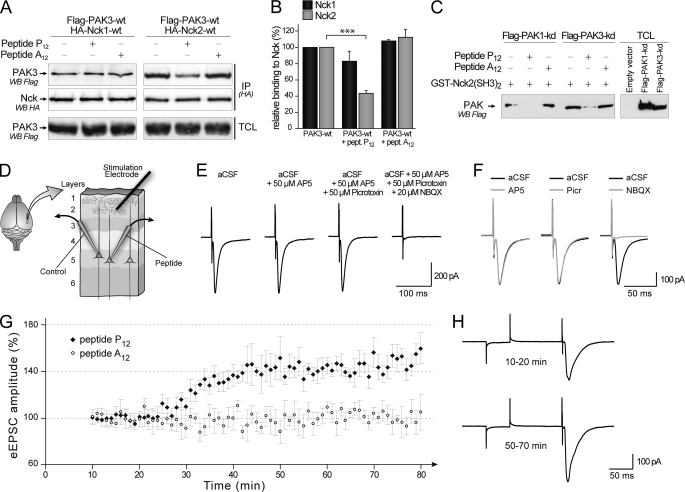
PAK-Nck2 Uncoupling Peptide P12 Increases eEPSC Amplitude in Perfused Neurons of Acute Cortical Slices
The role of the PAK1/Nck1 interaction in cell migration, cell spreading, atherogenesis, and angiogenesis was proved in several experimental models using inhibitory peptides (17, 19, 38, 39). So we tested whether a PAK3 homologous peptide from the first proline-rich region would inhibit PAK3/Nck interaction. We designed and synthesized a P12 inhibitory peptide, homologous to the previously used PAK1 peptide, and corresponding to the 15-amino acid peptide 8EEKPPAPPLRMNSNN22 near the N-terminal extremity of the wild-type PAK3 protein (underlined in the sequence of the PAK3-N-terminal region, in Fig. 1B). We also produced a mutated version of the P12 peptide that contained a substitution of the proline 12 by alanine (A12 peptide, 8EEKPAAPPLRMNSNN22) to be used as a negative control. The inhibitory effects of these two peptides were checked in co-immunoprecipitation assays of PAK3 performed with either Nck1 or Nck2, and in the presence of the P12 or the A12 peptide. As shown in Fig. 5A, the presence of the P12 peptide during immunoprecipitation strongly decreased the recovery of PAK3 with Nck2 but not with Nck1. Quantification (Fig. 5B) confirmed that the P12 peptide inhibits PAK3 binding to Nck2 (43.4 ± 6.2%, p = 0.0004) but not significantly to Nck1 (83.0 ± 21.2%, p = 0.2988), whereas the A12 peptide did not significantly change PAK3 binding with Nck2 or Nck1 (108.3 ± 2.1 and 112.4 ± 16.1%, respectively). However, because this P12 peptide should interact with the second SH3 domain of Nck2, we checked whether this interaction affects the PAK1-Nck2 complex. By co-immunoprecipitation (data not shown) and pulldown assay (Fig. 5C), we observed that addition of the P12 peptide to cell lysate strongly decreases the binding of both PAK1 and PAK3 to the SH3 domain of Nck2. The control peptide A12 does not have any effect on PAK1 and PAK3 interactions with Nck2. Thus, the mimetic peptide P12 prevents the interaction between Nck2 and the partners of its second SH3 domain, in particular PAK1 and PAK3. Once the mimetic peptide specificity was confirmed toward Nck2, we could use this peptide to clarify the role of the Nck2 complex in neuronal signaling. To do so, the uncoupling P12 or the mutated A12 peptide was applied through the patch clamp pipette into layer 5 pyramidal neurons while performing whole-cell recordings of evoked excitatory post-synaptic currents (eEPSCs) in visual cortical slices of juvenile rats (Fig. 5D). The time course of peptide diffusion in layer 5 pyramidal neurons was estimated prior to electrophysiological recordings by filling pyramidal cells through whole-cell patches with pipettes containing a 3000-Da dextran-FITC, a fluorescent polymer whose molecular mass is similar to that of the P12 (1666 Da) peptide. The full-length of the apical dendrite of pyramidal cells (from the soma to the layer 1) showed a green fluorescence signal 15–20 min after membrane break-in (data not shown). Next, layer 5 pyramidal neurons were held at −80 mV (i.e. the reversal potential for evoked inhibition under our experimental conditions), and eEPSCs were recorded every minute for more than 1 h. A typical control eEPSC, evoked by layer 2/3 stimulation, is shown in Fig. 5E (aCSF panel). The corresponding current wave did not change after bath application of AP5 and picrotoxin, the selective blockers of NMDA and GABAA receptors, respectively. However, bath application of 2,3-dihydroxy-6-nitro-7-sulfamoylbenzo(F)quinoxaline, a potent AMPA receptor antagonist, suppressed the inward current of the evoked response (Fig. 5, E and F), showing that the eEPSCs evoked under our experimental conditions are strictly due to AMPA receptor activation, mainly on the apical dendrite of pyramidal neurons (30). We found that the perfusion of the uncoupling P12 peptide in pyramidal neurons produced a robust and persistent increase in eEPSCs amplitude (Fig. 5, G and H). This increase in eEPSCs amplitude started 20–30 min after the beginning of the perfusion (i.e. after breaking-in cell membrane) and reached a plateau after 40 min (+41.8% ± 7.7% between 50 and 70 min, p < 0.001, n = 4) (Fig. 5, G, black squares, and H). This delay is consistent with the estimated peptide diffusion time course in the apical dendrite of the recorded neuron, as mentioned above. In contrast, the perfusion of the mutated A12 peptide failed to produce any change in eEPSC amplitude (Fig. 5G, open dots), thus strongly suggesting that the wild-type proline-rich peptide P12 specifically modifies evoked excitatory post-synaptic currents through the inhibition of Nck2 signaling.
FIGURE 5.
Post-synaptic inhibition of the PAKs/Nck2 interaction by perfusion of the inhibitory P12 peptide induced an increase in evoked EPSC. A, P12 peptide decreased the amount of co-immunoprecipitated PAK3 protein with Nck2 but not with Nck1. COS-7 cells were co-transfected with plasmids coding for FLAG-PAK3-WT and HA-tagged Nck1 (left lanes) or Nck2 (right lanes). Nck proteins were HA-immunoprecipitated (IP(HA)) in the absence or the presence of 1 mm of the P12 or the A12 peptide. Nck proteins are visualized after immunoblotting (middle panel). The co-immunoprecipitated PAK3 protein was revealed by anti-FLAG blotting (upper panel). PAK3 expression levels in TCLs were controlled as indicated (lower panel). B, quantification of amount of precipitated PAK3 protein with Nck adaptors illustrated in A (n = 3). C, pulldown assay of PAK1 or PAK3 proteins with the GST-(SH3)2 domain of Nck2 in presence of the P12 competing peptide or its A12 control peptide. Lysates of COS-7 cells transfected with FLAG-PAK1 or PAK3-kinase-dead expression vector were incubated 5 min without or with 250 μm of the P12 peptide or the A12 peptide before the PAK proteins were pulled down with the recombinant GST protein (20 μg). Precipitated proteins (left lanes) and expressed proteins in TCL (right lanes) were revealed by FLAG Western blot (WB). D, schematic representation of simultaneous patch clamp recordings. Pyramidal neurons were targeted in layer 5 of parasagittal slices of juvenile rat primary visual cortex. Two simultaneous whole-cell patch clamp recordings were then performed using two pipettes, one of them containing the P12 or A12 peptide, whereas the second consists of control recording. Extracellular electrical stimulations were applied in layer 2/3, on a vertical axis set at 50–70 μm from the recorded neuron apical dendrite. Evoked EPSCs (eEPSCs) were recorded under voltage clamp at a holding potential of −80 mV. Recordings and analysis involving P12 or A12 peptides were performed under blind conditions. E, characterization of the AMPA-mediated evoked excitatory responses in layer 5 neurons. The aCSF panel represents the typical current response obtained under control conditions after the brief extracellular stimulation (as indicated by the corresponding stimulation artifact). Bath application of NMDA (50 μm AP5) or GABAA (50 μm picrotoxin) receptor blockers had no impact on the evoked EPSC, whereas AMPA receptor blockade 2,3-dihydroxy-6-nitro-7-sulfamoylbenzo(F)quinoxaline (NBQX, 20 μm) suppressed the response. Current responses represented in this panel are means of three consecutive individual traces. This experiment was replicated three times in different slices. F, superposition of the control eESPC (aCSF condition, black trace) with the eEPSCs recorded under the three different conditions shown in E (gray traces). G, perfusion of the P12 peptide through the patch clamp pipette increases the amplitude of eEPSCs recorded in layer 5 pyramidal neurons by roughly 40% (black square). In contrast, recordings performed with the noninteracting peptide A12 (open dots) showed unmodified amplitudes during more than 1 h (n = 3 cells per condition, p < 0.001; Mann Whitney test). Note that the control recordings (performed without any peptide in the patch clamp pipette, see E) were stable for all cells included in the data set and are not illustrated in this figure. H, significant representation of eEPSC obtained with P12 peptide-containing recording pipette at two different times, during an early step between 10 and 20 min (upper trace) and during a later phase between 50 and 70 min (lower trace).
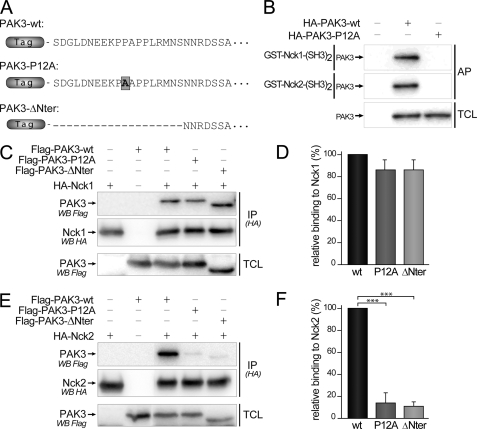
Mutation of the PAK3 Proline 12 Residue Targets the Interaction with Nck2 but Not with Nck1
Mutations of the second proline residue (12PPAPPMRNTS21)in the first proline-rich domain of PAK1 (black arrow in Fig. 1B), and its homologous residue in Drosophila D-PAK protein, abolished PAK/Nck interaction and suppressed PAK functions (16, 32). To investigate the role of the interaction between PAK3 and Nck, we constructed PAK3 mutants in which the homologous proline 12 residue (bold black on gray square, in Fig. 6A) was mutated into alanine (PAK3-P12A) or the first 20 N-terminal amino acids encompassing the proline-rich domain of PAK3 were deleted (PAK3-ΔNter) (Fig. 6A). First, pulldown assays were performed with the second SH3 domains of Nck1 and Nck2 (Fig. 6B), with WT or P12A mutated proteins expressed in COS-7 cells. The P12A mutation totally abolishes the precipitation of the Nck adaptors. We also observed that the P12A mutation suppresses the interaction between PAK3 and Nck1 in a two-hybrid assay, a result in total agreement with the previously published results obtained with Drosophila homologs (16) as well as with human genes (supplemental data 3)(32). We then performed co-immunoprecipitation assays in transfected COS-7 cells expressing these different FLAG-PAK3 proteins and either HA-Nck1 (Fig. 6C) or HA-Nck2 (Fig. 6E) proteins. The HA-tagged Nck proteins were immunoprecipitated and the FLAG-tagged PAK3 proteins revealed by Western blotting. We observed that introducing the P12A mutation or the small N-terminal deletion had differential effects on Nck1 and Nck2 binding. Whereas both mutations induced only a nonsignificant decrease in binding with Nck1 (−13.7 ± 9.1 and −14.5 ± 9.4% for P12A and ΔNter mutants, respectively) (Fig. 6D), both mutations induced a strong decrease of the interaction between PAK3 and Nck2 (−86.4 ± 9.3 and −89.2 ± 4.4%, for P12A and ΔNter proteins, respectively) (Fig. 6F). Among several studies analyzing these complexes (16–19), we report here the first experimental data showing that the mutation of the proline 12 residue suppresses co-immunoprecipitation between a PAK protein and a Nck adaptor. Because the Nck proteins are the only known partners of the PAK first proline-rich domain, the P12A mutant of PAK3 is a highly selective tool to study the implication of PAK3/Nck2 in biological processes.
FIGURE 6.
Mutation of the proline 12 residue and deletion of the N-terminal part of PAK3 suppress co-immunoprecipitation with Nck2 but not with Nck1. A, amino acid sequences of the N-terminal extremity of mouse PAK3 protein and the two mutated proteins used in this study. B, P12A mutation impairs the affinity precipitation of PAK3 protein with the second SH3 domains of Nck1 and Nck2 fused to GST. C, mutation of the proline 12 in PAK3 does not significantly change the interaction with Nck1. FLAG-PAK3 wild-type (wt), with the mutation P12A (P12A) or deleted for the first 20 amino acids (ΔNter) were co-expressed with HA-Nck1 in COS-7 cells. Nck1 protein was HA-immunoprecipitated and the presence of PAK3 in precipitated complexes (IP(HA)) and in TCLs was analyzed by Western blotting (WB) with FLAG antibodies. The amount of Nck1 precipitated proteins was controlled by anti-HA immunoblotting. D, relative binding of PAK3 proteins to Nck1, quantified from three independent experiments illustrated from C and averaged. E, mutation of the proline 12 in PAK3 abolished the interaction with Nck2. PAK3 proteins were co-expressed with HA-Nck2 in COS-7 cells. Nck2 protein was HA-immunoprecipitated and the presence of PAK3 in precipitated complexes (IP(HA)) and in TCL was analyzed by Western blotting with FLAG antibodies. The amount of Nck2 precipitated proteins was controlled by anti-HA immunoblotting. F, relative binding of PAK3 proteins to Nck2, quantified from four independent experiments illustrated from E. Comparison with Student's t test: ***, p < 0.001, n = 3. Errors bars indicate the S.E.
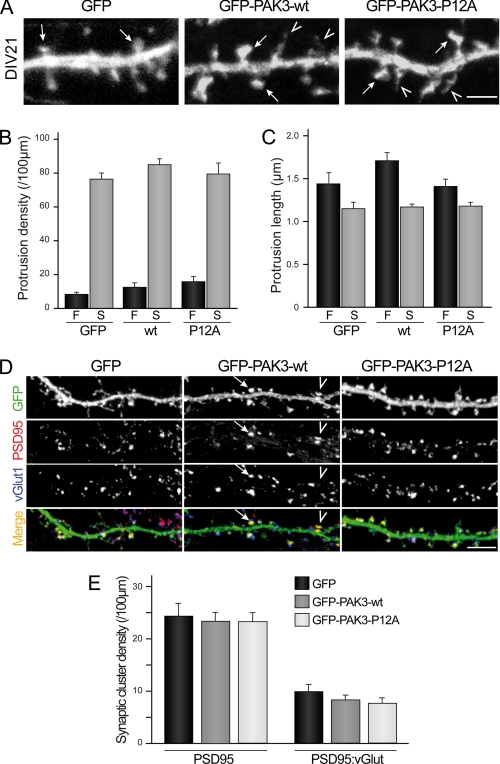
PAK3 Acts on Spinogenesis and Synaptogenesis Independently of Its Interaction with Nck2
Because PAK3 was described to regulate dendritic spine formation (8, 9), we studied the role of the PAK3/Nck2 interaction on dendritic spine maturation. PAK3-WT and -P12A expression vectors were transfected in mouse hippocampal neurons at DIV18 and neurons were fixed 3 days later. A morphological study of spine differentiation was carried out, and protrusions were classified into the following two categories as illustrated on Fig. 7A: filopodia defined as long protrusions without head or widening, and spines defined as protrusions with a mushroom-like head or an enlargement at the tip. No significant difference of the protrusion density was observed between the two constructs, compared with the GFP control (Fig. 7B). Moreover, the average length of protrusions was not significantly modified in P12A mutant-expressing neurons when compared with PAK3-WT or GFP-transfected neurons (Fig. 7C). We also analyzed protrusion density in hippocampal neurons at an early stage of differentiation, after 14 days of culture in vitro differentiation, and did not find any difference between control, WT, and P12A-expressing neurons (data not shown). To investigate the involvement of the PAK3-Nck2 complex in synaptogenesis, we analyzed the post-synaptic structure of DIV21-transfected neurons, by imaging the PSD-95 clusters, a key scaffold protein of the post-synaptic density of excitatory synapses (40) and the pre-synaptic contact using vGLUT1 as a marker (27). Interestingly, we found that expression of PAK3-WT and PAK3-P12A proteins does not modify the density of PSD-95 clusters in dendritic spines (Fig. 7E, left histogram). In addition, the density of the synapses defined as PSD-95:vGLUT1 double-positive clusters were unaltered by expression of any PAK3 constructs (Fig. 7E, right histogram). Altogether, these results strongly suggest that the interaction between PAK3 and Nck2 does not control either the pathway involved in spine morphogenesis or synapse formation.
FIGURE 7.
PAK3 acts on spinogenesis and synaptogenesis independently of its interaction with Nck2. A, P12A mutation did not affect spine maturation. Hippocampal neurons were transfected either with GFP as control or GFP-PAK3-WT or GFP-PAK3-P12A constructs at DIV18 and fixed at DIV21. Confocal images showed mature spines (arrows) and filopodia (arrowheads) at DIV21. Scale bar represents 2 μm. B and C, quantitative analysis of protrusion density or length. No significant difference for filopodia (F) or spines (S) between the three expressed constructs was observed. Data are means ± S.E. of measurements obtained from 8 to 11 cells (length of dendrites analyzed: 50–250 μm/cells; 1030–1800 protrusions analyzed). D, P12A mutation did not affect synaptic density. Hippocampal neurons were transfected either with GFP (1st panels) as control, GFP-PAK3-WT (2nd panels) or GFP-PAK3-P12A (3rd panels) constructs at DIV18, fixed at DIV21, and immunolabeled for PSD95 as postsynaptic marker and vGLUT1 as presynaptic marker. Confocal images showed GFP proteins in green, PSD95 in red, and vGLUT1 in blue. PSD95-positive spines were identified from green and red images (arrowhead), and synapses were identified as PSD95-positive spines with adjacent staining of vGLUT1 (arrow). Scale bar, 5 μm. E, quantitative analysis of synaptic cluster density. No significant difference was observed in PSD95-positive spine (left histogram) or synapse (right histogram) density between the three conditions. Data are means ± S.E. of measurements obtained from 15 to 17 neurons (length of dendrites analyzed: 50–250 μm/cell; 680–820 PSD-95 clusters and 230–330 synapses analyzed).
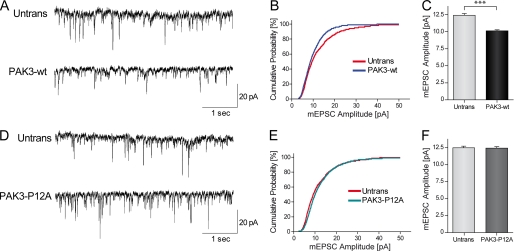
PAK3 Expression Decreases mEPSC Amplitude through Nck2 Interaction in Hippocampal Neurons
We next investigated the functional outcome of PAK3 or P12A expression on synaptic transmission. Thus, whole-cell recordings of spontaneous miniature excitatory post-synaptic currents (mEPSCs) were performed in DIV18 hippocampal neurons, transiently transfected with either GFP-PAK3-WT or GFP-PAK3-P12A plasmids. During each set of experiments, control recordings were performed on nonfluorescent untransfected neurons surrounding the targeted transfected cell on the same coverslip. Transfected and untransfected neurons showed similar resting potential and membrane input resistance (data not shown). Postsynaptic neurons were held at −60 mV to record only AMPA receptor-mediated mEPSCs. In both PAK3-WT and PAK3-P12A conditions, quantification of mEPSCs frequency showed no significant modification, thus ruling out any impact of PAK3 transfection on presynaptic release (Fig. 8, A and B, and data not shown). However, we did find that the expression of the wild-type PAK3 protein produced a decreased amplitude (−17.0 ± 0.2%, p < 0.001, n = 15) of the averaged mEPSCs compared with untransfected neurons (Fig. 8, A–C), although the expression of the PAK3-P12A protein, which does not interact with Nck2, did not change the average amplitude of mEPSCs (untransfected, 12.38 ± 0.24 pA, 1500 events on 15 neurons; WT, 10.28 ± 0.17 pA, 1300 events on 13 neurons; P12A, 11,90 ± 0.24 pA, 1000 events on 10 neurons) (Fig. 8, D–F). This suggests that PAK3 overexpression down-regulates excitatory post-synaptic currents, by a mechanism dependent upon its interaction with Nck2.
FIGURE 8.
Expression of PAK3-WT down-regulates mEPSC amplitudes in hippocampal neurons through its interaction with Nck2. A, representative sample traces of 5 s recording of mEPSCs in untransfected or GFP-PAK3-WT transfected neurons. Scale bars, 20 pA and 1 s. B, cumulative distribution plots of the mEPSC amplitude, assembled from the first 100 events from each cell recorded. C, quantification of mean mEPSC amplitude (mean ± S.E.). n = 10–15 cells per condition; ***, p < 0.001, Mann Whitney test. D–F, sample traces, cumulative distribution and quantification of mEPSC amplitude (means ± S.E.) recorded in untransfected neurons and in neurons transfected with GFP-PAK3-P12A construct showed no difference. n = 15 cells per condition.
DISCUSSION
Mutations of the pak3 gene are responsible for mental retardation, indicating that PAK3 has specific synaptic functions that, when lost or altered, cannot be compensated for by the two other PAK proteins of the group I. These unique functions of PAK3 in neuronal signaling have been largely unexplored (4). The amino acid sequence identity of PAK3 compared with the two other members of PAK group I is strong (41), but the level of identity drops drastically within the N-terminal region of these kinases. Indeed, many of the first 60 amino acids of human PAK3 are unique to PAK3 (53 and 49% identity with PAK1 and PAK2, respectively) and are highly conserved among PAK3s in the vertebrate phylum. Thus the NH2 extremity of PAK3 may support its specific functions, in particular through interaction with Nck adaptors, the only known partners of the N-terminal part of PAKs (5), because the second proline-rich sequence PP42NP localized in the N-terminal part of PAK1 that links with Grb2 is not conserved in PAK3 (42). The absence of PAK3 interaction with Grb2 was verified by a two-hybrid assay (data not shown). We showed that PAK3 is more closely associated with Nck2 than with Nck1 in brain co-immunoprecipitates, and we demonstrated a higher affinity of PAK3 for Nck2 than for Nck1. This is a new specific feature of PAK3, because PAK1 has been shown to interact similarly with the two adaptors (43).
We then analyzed the molecular mechanisms of PAK3 interaction with Nck2, by comparing them with those described for the PAK-Nck1 complex (13, 14, 18, 19, 32). Our results indicate that PAK3 interacts directly with Nck1 and Nck2 because the full-length PAK3 protein precipitates with the second SH3 domain of Nck1 and Nck2 in pulldown assays, and reciprocally, the amino part of PAK3 (the first 49 amino acids) fused to GST precipitates with Nck1. So, the main interaction is a direct binding between the second SH3 domain of Nck1 and Nck2 and the amino extremity of PAK3. These results do not differentiate PAK3 from PAK1, and similar interactions are widely observed with all PAK and Nck orthologs from Drosophila to human. Analyzing the molecular bases of the PAK3-Nck complex, we unexpectedly found major differences between Nck1 and Nck2 (see recapitulative Table in supplemental data 4). We observed that the P12A mutation in PAK3 does not abolish the co-immunoprecipitation between PAK3 and Nck1, whereas it disrupts the PAK3-Nck2 complex. In contrast, in pulldown assays, the P12A mutation suppresses the interaction with both Nck1 and Nck2. We also confirmed that the P12A mutation suppresses the interaction between PAK3 and Nck1 in a two-hybrid assay, as described previously for Drosophila homologs (16) as well as for human genes (32). The co-immunoprecipitation of the P12A mutant with Nck1 was also confirmed using a PAK3 mutant deleted for the first 20 amino acids corresponding to the Nck-binding domain because this deletion suppresses the interaction with Nck2 but not with Nck1. Moreover, the mimetic peptide displays a differential effect on Nck1 and Nck2, inhibiting only the interaction with the latter. Thus, the mutation of the second proline residue P12, the deletion of the first 20 amino acids, and the inhibitory mimetic peptide, all showed a minor effect on PAK3/Nck1 co-immunoprecipitation but a strong effect on recovery of the PAK3-Nck2 complex, indicating that interactions of PAK3 with the two adaptors are different. To explain these results, we hypothesize that the interaction between PAK3 and Nck1 may involve a third partner consolidating this interaction or may implicate other interacting domains of PAK3 and Nck1. As an example, it was shown that although the second SH3 domain of Nck1 plays an important role in the binding with PAK1, the presence of the third SH3 domain increases the interaction (5). However, another type of interaction may exist as recently described for the atypical association of the Nck adaptors with Robo, a binding that may involve multiple SH3 domains (44). The fact that the full-length PAK3-P12A mutant does not interact in the two-hybrid assay with the full-length Nck1 protein is an argument strengthening the hypothesis of a third partner. Another unique property of PAK3 is that PAK3 binds Nck independently of its kinase activity thus demonstrating that regulation of this complex is different from that characterized for PAK1. Indeed the PAK3 serine 20, which is a key residue for the regulation of the PAK1/Nck1 interaction (18), is neither autophosphorylated upon kinase activation nor is it a substrate for Akt and PKA. Whether the PAK3/Nck2 interaction is regulated is still unknown, and several mechanisms may be considered. For instance, the observed dissociation of the PAK3-Nck2 complex with the mimetic phosphorylated serine 20 residue suggests that a kinase may phosphorylate this crucial serine 20 residue. But the upstream asparagine 19 residue, which is conserved in all vertebrate PAK3 proteins, is a bad peptide consensus for almost all kinases. So we hypothesize that this interaction is not regulated by phosphorylation of this site. Another mechanism of regulation may target the amount of PAK3 at synapses. Indeed previous data effectively demonstrated the regulation of the pak3 gene expression during long term potentiation (45). Another attractive option may be the involvement of the GIT-PIX complex that controls membrane localization of PAK (46) and that plays an important role in synapse formation (9). To date, there is no evidence of cross-talk between Nck and PIX signaling (for example see Ref 47). Finally, another hypothesis may be that this interaction is constitutive, as has been described for the Nck1-DCC complex (48).
Whereas invalidation of Nck1 or Nck2 genes has uncovered a large redundancy of both protein functions (49, 50), several recent data also demonstrate independent roles for each adaptor (33, 44), although the involvement of a specific PAK isoform has not been previously described. Nck2 plays specific roles in cellular signaling and also in neuronal differentiation and in dendritic spine formation (22, 23, 44, 51). When we analyzed the biological role of the PAK3-Nck2 complex during dendritic spine formation and maturation, we did not find any significant effect when the PAK3-P12A mutant or the wild-type protein were expressed in hippocampal neurons, at an early stage of dendritic spine formation (DIV7,) or later during spine maturation (DIV18). A first possibility could be that effects due to disruption of the PAK3-Nck2 complex may be undetectable in spine and synapse differentiation assays. Alternatively, the PAK3/Nck2 pathway may regulate synaptic plasticity independently of spine morphogenesis, an hypothesis in agreement with the normal dendritic spine structure in PAK3 knock-out mice (7). The two kinase-dead mutations associated with mental retardation have no effect on the interaction suggesting that plasticity defect due to these mutations is not directly related to the absence of the PAK3-Nck2 complex. However, the slight decrease observed in the binding of the R67C mutant with Nck2 may be responsible for neuronal anomalies and be related to the severity of the mental retardation in this family (36).
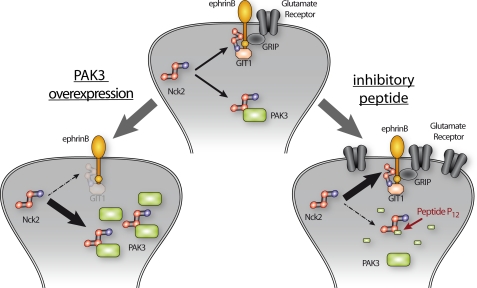
We report here two independent observations that indicate that PAKs, and in particular PAK3, are involved in a down-regulation of post-synaptic excitatory currents. Perfusion of an inhibitory peptide in cortical pyramidal neurons increases the evoked post-synaptic response, and conversely overexpression of PAK3 in hippocampal neurons induces a decrease of spontaneous excitatory postsynaptic currents. Interestingly, transgenic expression of the auto-inhibitory domain of PAK and pak1/pak3 double gene invalidation has been shown to induce an increase of excitatory synaptic transmission (6, 52) also indicating that PAKs down-regulate synaptic transmission. Together, these data strongly suggest that PAK3 down-regulates excitatory currents through its interaction with Nck2. These results raise the question as to how this complex acts on the dynamics of excitatory synaptic transmission. One possibility may be that PAK3 targets the ephrinB signaling through its interaction with Nck2 whose recruitment to the C-terminal phosphotyrosine of ephrinB after the Eph/ephrinB engagement initiates reverse signaling (53). In particular, the ephrinB/Nck2 association permits synaptic recruitment of the scaffold protein GIT1 to promote spine morphogenesis and synapse formation (22). An interesting hypothesis may be that the PAK3/Nck2 interaction regulates glutamate receptor dynamics at excitatory synapses, through regulation of signaling complexes involving EphrinB, GIT, and other synaptic proteins (54), either by titration of Nck2 or by interfering with Nck2 function at synapse In this model (Fig. 9), overexpression of PAK3 is responsible for Nck2 titration and induces a decrease of the Nck2-dependent stabilization of glutamate receptors. Conversely, perfusion of the P12 inhibitory peptide decreases the cytoplasmic binding of Nck2 to PAK3, releasing Nck2 and allowing an Nck2-dependent stabilization of glutamate receptors at the postsynaptic level, and leading to an increase in synaptic transmission. Interestingly, a similar model in which Nck sequesters PAK1 to the cytoplasm was recently proposed in prolactin signaling (55). In conclusion, we have shown that PAK3 forms a signaling complex with the Nck2/Grb4 adaptor, which is involved in the down-regulation of post-synaptic excitatory currents.
FIGURE 9.
Model of regulation of synaptic transmission by PAK3. Nck2 regulates glutamate receptor stabilization (center diagram). Overexpression of PAK3 titrates Nck2 and induces a decrease of the Nck2-dependent stabilization of glutamate receptors (left diagram). Conversely, perfusion of an inhibitory peptide inhibits the Nck2-PAK3 complex, releasing Nck2 and allowing a Nck2-dependent stabilization of glutamate receptors (right diagram).
Supplementary Material
Acknowledgments
We thank Wei Li and Bruce Mayer for Nck plasmids, Fréderic Saudou and Sandrine Humbert for Huntingtin plasmids, Martine Latta-Mahieu for the DA-PAK1-kd plasmid, and Salah El Mestikawy for the vGLUT1 rabbit antibody. We also thank Annie Lamouroux for the help with quantitative-PCR; Jacques Camonis for the help with two-hybrid assay; Julie Perroy for advice in mouse neuronal cultures; Sandrine Poëa-Guyon for help in confocal imaging, and Gérard Sadoc for the electrophysiological recording software. We thank Seana O'Regan for helpful comments on the manuscript.
This work was supported in part by Agence Nationale de la Recherche Grant ANR-MNP-2009, the Fondation Jérôme Lejeune, and by a fellowship from Association pour la Recherche sur le Cancer (to E. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- PAK
- p21-activated kinase
- TCL
- total cell lysate
- DIV
- days in vitro
- aCSF
- artificial cerebrospinal fluid
- eEPSC
- evoked excitatory postsynaptic current
- mEPSC
- miniature excitatory postsynaptic current
- SH
- Src homology.
REFERENCES
- 1. Ropers H. H. (2008) Curr. Opin. Genet. Dev. 18, 241–250 [DOI] [PubMed] [Google Scholar]
- 2. Chelly J., Khelfaoui M., Francis F., Chérif B., Bienvenu T. (2006) Eur. J. Hum. Genet. 14, 701–713 [DOI] [PubMed] [Google Scholar]
- 3. Allen K. M., Gleeson J. G., Bagrodia S., Partington M. W., MacMillan J. C., Cerione R. A., Mulley J. C., Walsh C. A. (1998) Nat. Genet. 20, 25–30 [DOI] [PubMed] [Google Scholar]
- 4. Kreis P., Barnier J. V. (2009) Cell. Signal. 21, 384–393 [DOI] [PubMed] [Google Scholar]
- 5. Bokoch G. M. (2003) Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 6. Huang W., Zhou Z., Asrar S., Henkelman M., Xie W., Jia Z. (2011) Mol. Cell. Biol. 31, 388–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meng J., Meng Y., Hanna A., Janus C., Jia Z. (2005) J. Neurosci. 25, 6641–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boda B., Alberi S., Nikonenko I., Node-Langlois R., Jourdain P., Moosmayer M., Parisi-Jourdain L., Muller D. (2004) J. Neurosci. 24, 10816–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H., Webb D. J., Asmussen H., Niu S., Horwitz A. F. (2005) J. Neurosci. 25, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kreis P., Thévenot E., Rousseau V., Boda B., Muller D., Barnier J. V. (2007) J. Biol. Chem. 282, 21497–21506 [DOI] [PubMed] [Google Scholar]
- 11. Boda B., Nikonenko I., Alberi S., Muller D. (2006) Mol. Neurobiol. 34, 67–80 [DOI] [PubMed] [Google Scholar]
- 12. Bagrodia S., Taylor S. J., Creasy C. L., Chernoff J., Cerione R. A. (1995) J. Biol. Chem. 270, 22731–22737 [DOI] [PubMed] [Google Scholar]
- 13. Bokoch G. M., Wang Y., Bohl B. P., Sells M. A., Quilliam L. A., Knaus U. G. (1996) J. Biol. Chem. 271, 25746–25749 [DOI] [PubMed] [Google Scholar]
- 14. Lu W., Katz S., Gupta R., Mayer B. J. (1997) Curr. Biol. 7, 85–94 [DOI] [PubMed] [Google Scholar]
- 15. Buday L., Wunderlich L., Tamás P. (2002) Cell. Signal. 14, 723–731 [DOI] [PubMed] [Google Scholar]
- 16. Hing H., Xiao J., Harden N., Lim L., Zipursky S. L. (1999) Cell 97, 853–863 [DOI] [PubMed] [Google Scholar]
- 17. Fryer B. H., Wang C., Vedantam S., Zhou G. L., Jin S., Fletcher L., Simon M. C., Field J. (2006) J. Biol. Chem. 281, 11487–11495 [DOI] [PubMed] [Google Scholar]
- 18. Zhao Z. S., Manser E., Lim L. (2000) Mol. Cell. Biol. 20, 3906–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou G. L., Zhuo Y., King C. C., Fryer B. H., Bokoch G. M., Field J. (2003) Mol. Cell. Biol. 23, 8058–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guan S., Chen M., Woodley D., Li W. (2007) Mol. Cell. Biol. 27, 6001–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pilpel Y., Segal M. (2005) J. Neurochem. 95, 1401–1410 [DOI] [PubMed] [Google Scholar]
- 22. Segura I., Essmann C. L., Weinges S., Acker-Palmer A. (2007) Nat. Neurosci. 10, 301–310 [DOI] [PubMed] [Google Scholar]
- 23. Xu N. J., Henkemeyer M. (2009) Nat. Neurosci. 12, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rousseau V., Goupille O., Morin N., Barnier J. V. (2003) J. Biol. Chem. 278, 3912–3920 [DOI] [PubMed] [Google Scholar]
- 25. Kreis P., Rousseau V., Thévenot E., Combeau G., Barnier J. V. (2008) J. Neurochem. 106, 1184–1197 [DOI] [PubMed] [Google Scholar]
- 26. König S., Guibert B., Morice C., Vernier P., Barnier J. V. (2001) C. R. Acad. Sci. III 324, 673–681 [DOI] [PubMed] [Google Scholar]
- 27. Herzog E., Bellenchi G. C., Gras C., Bernard V., Ravassard P., Bedet C., Gasnier B., Giros B., El Mestikawy S. (2001) J. Neurosci. 21, RC181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moreau A. W., Amar M., Le Roux N., Morel N., Fossier P. (2010) Cereb. Cortex 20, 456–467 [DOI] [PubMed] [Google Scholar]
- 29. Le Roux N., Amar M., Baux G., Fossier P. (2006) Eur. J. Neurosci. 24, 3507–3518 [DOI] [PubMed] [Google Scholar]
- 30. Le Roux N., Amar M., Moreau A., Baux G., Fossier P. (2008) Eur. J. Neurosci. 27, 3244–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seeburg D. P., Feliu-Mojer M., Gaiottino J., Pak D. T., Sheng M. (2008) Neuron 58, 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Galisteo M. L., Chernoff J., Su Y. C., Skolnik E. Y., Schlessinger J. (1996) J. Biol. Chem. 271, 20997–21000 [DOI] [PubMed] [Google Scholar]
- 33. Guan S., Fan J., Han A., Chen M., Woodley D. T., Li W. (2009) J. Invest Dermatol. 129, 1909–1920 [DOI] [PubMed] [Google Scholar]
- 34. Cahoy J. D., Emery B., Kaushal A., Foo L. C., Zamanian J. L., Christopherson K. S., Xing Y., Lubischer J. L., Krieg P. A., Krupenko S. A., Thompson W. J., Barres B. A. (2008) J. Neurosci. 28, 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T. M., Chin M. C., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H. W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K. R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Yuan X. F., Zhang B., Zwingman T. A., Jones A. R. (2007) Nature 445, 168–176 [DOI] [PubMed] [Google Scholar]
- 36. Bienvenu T., des Portes V., McDonell N., Carrié A., Zemni R., Couvert P., Ropers H. H., Moraine C., van Bokhoven H., Fryns J. P., Allen K., Walsh C. A., Boué J., Kahn A., Chelly J., Beldjord C. (2000) Am. J. Med. Genet 93, 294–298 [DOI] [PubMed] [Google Scholar]
- 37. Howe A. K., Juliano R. L. (2000) Nat. Cell Biol. 2, 593–600 [DOI] [PubMed] [Google Scholar]
- 38. Kiosses W. B., Hood J., Yang S., Gerritsen M. E., Cheresh D. A., Alderson N., Schwartz M. A. (2002) Circ. Res. 90, 697–702 [DOI] [PubMed] [Google Scholar]
- 39. Orr A. W., Stockton R., Simmers M. B., Sanders J. M., Sarembock I. J., Blackman B. R., Schwartz M. A. (2007) J. Cell Biol. 176, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee H. W., Kim Y., Han K., Kim H., Kim E. (2010) J. Neurosci. 30, 5508–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arias-Romero L. E., Chernoff J. (2008) Biol. Cell 100, 97–108 [DOI] [PubMed] [Google Scholar]
- 42. Puto L. A., Pestonjamasp K., King C. C., Bokoch G. M. (2003) J. Biol. Chem. 278, 9388–9393 [DOI] [PubMed] [Google Scholar]
- 43. Braverman L. E., Quilliam L. A. (1999) J. Biol. Chem. 274, 5542–5549 [DOI] [PubMed] [Google Scholar]
- 44. Round J. E., Sun H. (2011) Mol. Cell. Neurosci. 47, 265–273 [DOI] [PubMed] [Google Scholar]
- 45. Boda B., Mas C., Muller D. (2002) Neuroscience 114, 13–17 [DOI] [PubMed] [Google Scholar]
- 46. Brown M. C., West K. A., Turner C. E. (2002) Mol. Biol. Cell 13, 1550–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ku G. M., Yablonski D., Manser E., Lim L., Weiss A. (2001) EMBO J. 20, 457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bladt F., Aippersbach E., Gelkop S., Strasser G. A., Nash P., Tafuri A., Gertler F. B., Pawson T. (2003) Mol. Cell. Biol. 23, 4586–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fawcett J. P., Georgiou J., Ruston J., Bladt F., Sherman A., Warner N., Saab B. J., Scott R., Roder J. C., Pawson T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20973–20978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li X., Meriane M., Triki I., Shekarabi M., Kennedy T. E., Larose L., Lamarche-Vane N. (2002) J. Biol. Chem. 277, 37788–37797 [DOI] [PubMed] [Google Scholar]
- 51. Tu Y., Li F., Wu C. (1998) Mol. Biol. Cell 9, 3367–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hayashi M. L., Choi S. Y., Rao B. S., Jung H. Y., Lee H. K., Zhang D., Chattarji S., Kirkwood A., Tonegawa S. (2004) Neuron 42, 773–787 [DOI] [PubMed] [Google Scholar]
- 53. Cowan C. A., Henkemeyer M. (2001) Nature 413, 174–179 [DOI] [PubMed] [Google Scholar]
- 54. Essmann C. L., Martinez E., Geiger J. C., Zimmer M., Traut M. H., Stein V., Klein R., Acker-Palmer A. (2008) Nat. Neurosci. 11, 1035–1043 [DOI] [PubMed] [Google Scholar]
- 55. Tao J., Oladimeji P., Rider L., Diakonova M. (2011) Mol. Endocrinol. 25, 1565–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.