Background: Endoparasitoid wasps inject venom proteins at oviposition to alter host immunity.
Results: The venom of Leptopilina boulardi, but not of a related species, contains an active extracellular SOD. Recombinant SODs inhibit the Drosophila host phenoloxidase activity.
Conclusion: An extracellular SOD is secreted and active in an insect fluid.
Significance: SODs may be used as immune suppressive virulence factors by parasitoid wasps.
Keywords: Drosophila, Insect Immunity, Invertebrates, Superoxide Dismutase (SOD), Virulence Factors, Parasitoid Wasp, Phenoloxidase
Abstract
Endoparasitoid wasps inject venom proteins with their eggs to protect them from the host immune response and ensure successful parasitism. Here we report identification of Cu,Zn superoxide dismutase (SOD) transcripts for both intracellular SOD1 and extracellular SOD3 in the venom apparatus of two Leptopilina species, parasitoids of Drosophila. Leptopilina SODs show sequence and structure similarity to human SODs, but phylogenetic analyses indicate that the extracellular SODs are more related to cytoplasmic vertebrate SODs than to extracellular SODs, a feature shared by predicted insect extracellular SODs. We demonstrate that L. boulardi SOD3 is indeed secreted and active as monomeric glycosylated forms in venom. Our results also evidence quantitative variation in SOD3 venom contents between closely related parasitoid species, as sod3 is 100-fold less expressed in Leptopilina heterotoma venom apparatus and no protein and SOD activity are detected in its venom. Leptopilina recombinant SOD3s as well as a mammalian SOD in vitro inhibit the Drosophila phenoloxidase activity in a dose-dependent manner, demonstrating that SODs may interfere with the Drosophila melanization process and, therefore, with production of cytotoxic compounds. Although the recombinant L. boulardi SOD3 quantity needed to observe this effect precludes a systemic effect of the wasp venom SOD3, it is still consistent with a local action at oviposition. This work provides the first demonstration that insect extracellular SODs are indeed secreted and active in an insect fluid and can be used as virulence factors to counteract the host immune response, a strategy largely used by bacterial and fungal pathogens but also protozoan parasites during infection.
Introduction
Endoparasitoid wasps are insects that lay eggs in arthropods, generally other insects, eventually killing them as the result of their development. To ensure successful parasitism, they inject with the egg various components, including venom factors that alter the host immunity and/or manipulate its development and physiology (1, 2). The typical host immune defense against parasitoids is the formation of a multicellular melanized capsule around the egg, which requires the coordination of cellular and humoral components (3, 4). After egg parasite recognition, host hemocytes proliferate, become adhesive, attach and spread at the surface of the egg (5). In the meantime, a cascade involving several serine proteases is triggered, leading to the cleavage of prophenoloxidase (proPO)2 into active phenoloxidase (PO), which catalyzes oxidation of phenolic compounds that polymerize to form melanin (4, 6). This melanization pathway involves oxidoreduction reactions and generates diverse cytotoxic molecules including reactive oxygen (ROS) and nitrogen species that have been suggested to play a major role in the death of the parasitoid egg (7–9).
Until now, a limited number of proteins have been identified and characterized from parasitoid venom fluids, and only a few of them have been demonstrated to interfere with host immune defenses (10). Known immune-suppressive proteins target either host hemocytes or the PO cascade (10), as shown for Leptopilina boulardi, a parasitoid of Drosophila species of the melanogaster subgroup (2). This species is likely the best known parasitoid model regarding venom virulence factors, and it is also an important model for understanding intraspecific variation of virulence with the occurrence of two well characterized strains, ISm and ISy (for review, see Ref. 11). The venom of ISm females, a highly virulent strain against Drosophila melanogaster, contains a RhoGAP protein that is required for parasitism success (12). This factor targets Drosophila lamellocytes, the main capsule-forming hemocytes, and induces changes in their morphology by inactivating the RhoGTPases Rac1 and Rac2, both required for successful encapsulation (13–17). L. boulardi ISy females differ from ISm females in their ability to suppress immune defenses of both D. melanogaster and Drosophila yakuba hosts but depending on the host resistance genotype (11, 18). The main identified effect of ISy female venom is the inhibition of the host serine protease cascade leading to the proPO-PO conversion thanks to the action of a serpin, a specific serine protease inhibitor (19).
Although it has been suggested that L. boulardi ISm venom might interfere with the oxidation pathways during the melanization process (20), no parasitoid-injected protein had ever been described that could prevent the deleterious effects of the production of ROS or reactive nitrogen species. This is quite surprising as a well known virulence strategy employed by numerous bacterial and fungal pathogens, including pathogens of insects as well as protozoan parasites, is the detoxification of reactive species by antioxidant enzymes, mainly superoxide dismutases (SODs) (21–26). SODs are one of the most important cellular enzymatic defenses against the detrimental ROS generated by the aerobic metabolism. These ubiquitous metalloenzymes catalyze the dismutation of the superoxide anion, O2˙̄, to hydrogen peroxide H2O2, which in turn is converted into H2O by catalase and peroxidases (for review, see Ref. 27). In mammals, three distinct SODs have been identified and characterized that have a similar enzymatic function, although they differ in their metal cofactor requirements and/or cellular compartmentalization (for review, see Ref. 28). The cytoplasmic Cu,Zn-SOD (SOD1) is primarily localized in cytoplasm and also found in the mitochondrial inter-membrane space (29). The extracellular Cu,Zn-SOD (SOD3) is similar in structure to SOD1 but contains an N-terminal signal peptide and a C-terminal region involved in extracellular matrix binding (30). Finally, the MnSOD (SOD2), which is very different in protein sequence and structure from Cu,Zn-SODs, is restricted to the inner matrix of mitochondria (31).
While studying candidate virulence proteins in Leptopilina wasp venom, we identified predicted cytoplasmic and extracellular Cu,Zn-SODs from the two strains of L. boulardi as well as from L. heterotoma, a more generalist species than L. boulardi. The extracellular SOD gene is specifically overexpressed in the venom apparatus of L. boulardi compared with the remnant body, in contrast to what was observed for L. heterotoma. The corresponding extracellular SOD protein as well as SOD activity has been detected in L. boulardi venom only, and SOD identity was confirmed by mass spectrometry. In vitro assays using a recombinant protein show a direct inhibitory effect of the L. boulardi extracellular SOD on D. melanogaster PO activity.
To our knowledge this work is the first demonstration that insect-predicted extracellular SODs are indeed secreted. It is also the first report of the presence and specific overproduction of a SOD protein in parasitoid wasp venom and of its in vitro effect on host immune components. We suggest that parasitoid venom SODs likely have a role in parasitism success either directly, by interfering with the local host melanization pathway, or indirectly by protecting other virulence factors from ROS damages during their storage in the wasp venom reservoir.
EXPERIMENTAL PROCEDURES
Biological Material
The origin of L. boulardi ISy (Lby; Gif stock 486) and ISm (Lbm; Gif stock 431) lines has been previously described (32). Briefly, they derive from single females collected, respectively, in Brazzaville (Congo) and Nasrallah (Tunisia). The L. heterotoma (Lh) strain originates from the south of France (Gotheron). All parasitoid lines were reared at 25 °C on a susceptible D. melanogaster strain (Gif stock 1333). The wasp venom apparatus that can be easily isolated consists of a unique long gland connected to a reservoir (2) that can be separated and processed independently.
Cloning and Molecular Analysis of Leptopilina Cu,Zn-SODs
Total RNA was extracted from 100 venom apparatus (venom gland + reservoir) collected from Lbm, Lby, or Lh using TRIzol reagent (Invitrogen). Three cDNA libraries were constructed using 1 μg of RNA and the Creator SMART cDNA Library Construction kit (Clontech) as previously described (19). Among the 120 clones sequenced from each library and analyzed by BLAST against the NCBI non-redundant databases, three sequences had very high similarities with Cu,Zn-SODs. They were named LbmSOD3, LbySOD3, and LhSOD1 according to the presence or the absence of a putative peptide signal predicted using the CBS prediction servers.
To obtain the corresponding sequences LbmSOD1, LbySOD1, and LhSOD3, RNA isolated from the venom apparatus of Lbm, Lby, or Lh was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad). PCR were performed using the GoTaq DNA Polymerase (Promega) and the primer pairs designed from LhSOD1 and LbmSOD3/LbySOD3 sequences (supplemental Table 1). Amplified fragments were cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced.
Pairwise sequence comparisons were performed using the EMBOSS program Needle at EMBL-EBI. Multiple amino acid sequence alignment was performed using MUSCLE (33), and phylogenetic analysis was conducted using PhyML (34) at The Laboratory of Informatics, Robotics, and Microelectronics (LIRMM). Molecular modeling of Leptopilina Cu,Zn-SODs was performed using the Phyre server (35). Visualization of structures, structural alignments, and root mean square deviations were done using PyMOL. N-Glycosylation sites were predicted using the CBS prediction servers.
Semiquantitative and Quantitative RT-PCR
RNA isolated from dissected Lby, Lbm, or Lh venom apparatus or from the remnant female bodies (without venom apparatus) were reverse-transcribed as described above. PCRs were carried out with primers common to LbmSOD1/LbySOD1, common to LbmSOD3/LbySOD3, specific to LhSOD1, and specific to LhSOD3 (supplemental Table 1). Semi-qRT-PCR experiments were performed for 28 or 33 cycles. PCR products were then separated and quantified using the GelEval software (FrogDance Software). qRT-PCR experiments were carried out on an Opticon monitor 2 (Bio-Rad) using the Absolute qPCR SYBR MasterMix Plus (Eurogentec). PCR conditions were 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 30 s. Each reaction was performed in triplicate, and the mean of three independent biological replicates was calculated. All data were normalized using the ITS2 ribosomal sequence (Internal Transcribed Spacer 2; supplemental Table 1) and analyzed by a Student's t test (R software package; The R Project for Statistical Computing).
Production of Recombinant Proteins
For production of the thioredoxin (Trx) fusion proteins Trx-LbmSOD3, Trx-LbySOD3, and Trx-LhSOD3, cDNA fragments corresponding to the mature proteins were amplified by RT-PCR from RNA isolated from Lbm, Lby, and Lh venom apparatus, respectively. Amplified fragments were cloned into pET-32a(+) vector (Novagen), and recombinant plasmids were transformed in competent Origami (DE3) Escherichia coli cells. Production and purification of Trx-LbmSOD3, Trx-LbySOD3, Trx-LhSOD3, and Trx tag alone were performed according to manufacturer's instructions.
Immunoblotting Experiments
For dot blot experiments, serial 1:3 dilutions of recombinant Trx-LbmSOD3, Trx-LbySOD3, Trx-LhSOD3, and of Trx tag, starting from 1 μg of protein, were spotted onto a nitrocellulose membrane. After blocking with TBS-Tween, 2% milk, the membrane was probed overnight at 4 °C with a Homo sapiens SOD1 rabbit polyclonal antibody (1:1000, ab13498, Abcam). After washing and a 2-h incubation at room temperature with a goat anti-rabbit IgG horseradish peroxidase conjugate (1:25000, Sigma), chemiluminescence signal detection was performed (Immobilon Western, Millipore). Relative spot intensities were quantified using GeneSnap and GeneTools (Syngene).
For all gel and Western blot experiments, venom reservoirs were dissected in insect Ringer, and residual tissues were removed by centrifugation 2 min at 500 × g. For SDS-PAGE comparison, the content of 10 reservoirs was separated on a 12.5% polyacrylamide gel before transfer. For reducing and non-reducing conditions, 60 reservoirs were dissected in 40 μl of Ringer, and 20 μl were mixed with sample buffer and heated either in presence or absence of 2.5% 2-mercaptoethanol before SDS-PAGE and transfer. For native PAGE comparison, the contents of 20 reservoirs were mixed (50/50, v/v) with sample buffer without SDS and 2-mercaptoethanol and separated on a 7.5% native PAGE. The gel was soaked 15 min in SDS-PAGE running buffer before transfer. All membranes were probed as described above.
N-Glycosylation Assay
The content of 10 Lbm reservoirs was collected in 50 mm sodium phosphate buffer (pH 7.5) before supplementation with 0.2% SDS and 0.5% 2-mercaptoethanol. The sample was heated at 95 °C for 10 min, cooled on ice for 2 min, and incubated with 1 unit of N-glycosidase F (Roche Applied Science) overnight at 37 °C. The reaction was stopped at 95 °C for 5 min. The extent of carbohydrate removal was determined by Western blot analysis as described above.
Mass Spectrometry
Silver-stained bands cut from a 6–16% SDS-PAGE gel were destained, reduced, and alkylated with iodoacetamide and incubated overnight at 37 °C with 12.5 ng/μl trypsin (Sigma). The solution was dried, reconstituted with 0.1% formic acid, and sonicated for 10 min. Generated peptides were sequenced by nano-LC-tandem mass spectrometry (MS/MS) (Q-TOF Ultima with a nano-electrospray ionization source, Waters/Micromass) in data-dependent acquisition (DDA) mode using the five most intense parent ions. Peptides were loaded on a C18 column (XBridge™ BEH130 3.5 μm, 75-μm ×150-mm, Waters) and eluted with a 5–60% linear gradient at a flow rate of 180 nl/min over 90 min (buffer A: water/acetonitrile (98:2, v/v) and 0.1% formic acid; buffer B: water/acetonitrile (20:80, v/v) and 0.1% formic acid). MS/MS data analysis was performed with Mascot software using Leptopilina Cu,Zn-SOD sequences and the following parameters: trypsin specificity, two missed cleavages, carbamidomethyl cysteine and oxidation of methionine as variable modifications, and 0.2-Da mass tolerance on both precursor and fragment ions. To validate SOD identification, only matches with peptides with a score above the identity threshold of 13 (p < 0.005) and containing at least five consecutive y or b ions were considered.
SOD Activity Assays
SOD activity was detected by zymography after 7.5% non-denaturing, non-reducing electrophoresis at 100 V at 4 °C. For recombinant Trx-LbmSOD3, Trx-LbySOD3, Trx-LhSOD3, and Trx tag, 3 μg of proteins were loaded. For venom assays, the content of 25 venom reservoirs was loaded. SOD activity was visualized as inhibition of the reduction of NBT according to Beauchamp and Fridovich (36). Upon illumination, achromatic bands indicate zones of SOD activity. SOD spectrophotometric assays were performed using the SOD determination kit (Sigma). Assays were performed using 20 and 200 ng of recombinant protein or Trx as a control, 200 ng of BtSOD, a commercial SOD from bovine erythrocytes (Sigma), and protein extracts from 20 Lbm, Lby, and Lh reservoirs. Inhibition assays were performed with 2 mm KCN. p values were generated by ANOVA followed by pairwise t tests (R software package).
PO Activity Measurements
Phenoloxidase activity was assayed using a method modified from Colinet et al. (19). For each replicate the hemolymph of three D. melanogaster 96-h aged-larvae was collected in 10 μl of cacodylate buffer (10 mm cacodylic acid, 5 mm CaCl2, pH 7). After 15 min of incubation at 21 °C during which proPO is converted into PO, 10 μl of cacodylate buffer containing 0.003–3 μg of the recombinant protein, 3 μg of BtSOD, or 3–45 μg of venom proteins (estimated using Bradford assay; Coo Protein Assay, Interchim) was added. Samples were incubated for 15 min at 21 °C before the addition of the substrate (180 μl of 1 mg/ml dopamine hydrochloride in cacodylate buffer). After 1 h at 21 °C, PO activity was estimated spectrophotometrically at 490 nm by detecting the quantity of dopachrome formed. Hemolymph samples in which 10 μl of cacodylate buffer alone or containing 1-phenyl-2-thiourea (PTU, final concentration 0.01%), a PO inhibitor, were added after the initial 15-min incubation period were used as controls. To determine whether recombinant Leptopilina Cu,Zn-SODs affect the PO activity itself or the proPO to PO activation by serine proteases, absorbance values were compared between hemolymph samples collected in the presence of recombinant proteins or in which the recombinant proteins were added after the 15-min incubation period that follows hemolymph collection. Hemolymph samples collected in PTU (0.01%), in which PTU was added after the 15-min incubation period or collected in cacodylate buffer alone, were used as controls. Three replicates were made for each treatment. p values were generated by ANOVA followed by pairwise t tests (R software package).
RESULTS
Identification of Cu,Zn-SOD Sequences from Leptopilina Venom Apparatus
Among the 120 clones sequenced from each venom apparatus cDNA library of L. boulardi ISm (Lbm), L. boulardi ISy (Lby), and L. heterotoma (Lh), full-length sequences with high similarity to Cu,Zn-SODs were identified. Sequence comparisons and prediction of a peptide signal in L. boulardi ISm and ISy Cu,Zn-SOD transcripts strongly suggested that they were both extracellular SODs. They were thus named LbmSOD3 and LbySOD3, according to the human nomenclature. The L. heterotoma sequence had no predicted peptide signal, suggesting that it was a cytoplasmic Cu,Zn-SOD, and it was named LhSOD1. The sequences of L. boulardi LbmSOD1 and LbySOD1 and of L. heterotoma LhSOD3 were further obtained by RT-PCR using specific primers designed from the homolog sequences (supplemental Table 1).
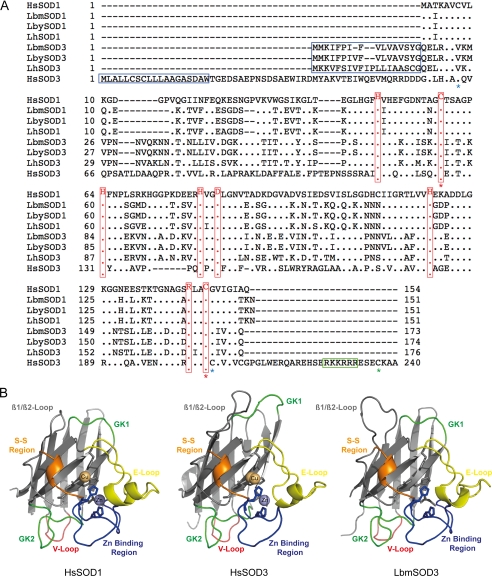
The predicted Leptopilina Cu,Zn-SODs are proteins of 151 amino acids for SOD1 and 173–176 amino acids for SOD3 (Table 1 and Fig. 1A). The estimated molecular mass of the mature proteins is 15.5 kDa for SOD1 and 16.4–16.6 kDa for SOD3, but 2 and 3 N-glycosylation sites are predicted for Leptopilina SOD3 and SOD1, respectively (Table 1). Cu,Zn-SOD sequences are nearly identical between L. boulardi strains and very similar between L. boulardi and L. heterotoma, with LhSOD3 and LhSOD1 closely related to extracellular and cytoplasmic L. boulardi Cu,Zn-SODs, respectively (Table 2 and Fig. 1A). As a whole, extracellular SODs are clearly more divergent from one another than are cytoplasmic SODs (Table 2). Multiple sequence alignment with the fully characterized H. sapiens SOD1 and SOD3 indicates that Leptopilina SODs contain all the conserved residues essential for dismutation, metal binding, and stability (28), suggesting that they are active Cu,Zn-SODs (Fig. 1A). In addition, a strong conservation of key structural elements is shown by superposition of the predicted three-dimensional models for Leptopilina proteins with the solved H. sapiens SOD structures (37) (see Fig. 1B, illustrated for LbmSOD3 only).
TABLE 1.
Main characteristics of Cu,ZnSOD sequences identified in L. boulardi ISm, L. boulardi ISy, and L. heterotoma venom apparatus
| LbmSOD1 | LbySOD1 | LhSOD1 | LbmSOD3 | LbySOD3 | LhSOD3 | |
|---|---|---|---|---|---|---|
| ORF (bp) | 456 | 456 | 456 | 522 | 525 | 531 |
| Protein (aa) | 151 | 151 | 151 | 173 | 174 | 176 |
| Signal peptide | No | No | No | Yes | Yes | Yes |
| Molecular mass (kDa) | 15.5 | 15.5 | 15.5 | 16.4 | 16.4 | 16.6 |
| N-Glycosylation | 2 | 2 | 2 | 3 | 3 | 3 |
FIGURE 1.
Sequence and structure conservation of Cu,Zn-SOD sequences. A, multiple sequence alignment of Leptopilina and H. sapiens (SOD1, P00441; SOD3, P08294) Cu,Zn-SODs. Residues identical to H. sapiens SOD1 (HsSOD1) sequence are illustrated by a dot. The signal peptide is indicated by a blue box. Functional residues are highlighted in red (28). Red stars, positions of HsSOD3 Cys-107—Cys-189 involved in the Cu,Zn-SOD-conserved intrasubunit disulfide bridge; blue stars, positions of the Cys45-Cys190 involved in the HsSOD3 intrasubunit disulfide bridge. Green star, position of the HsSOD3 Cys-219 residue involved in the intersubunit disulfide bridge; green box, heparin binding region. B, comparison of the predicted tertiary structure of LbmSOD3 with the solved HsSOD1 structure 1PU0 and the solved HsSOD3 structure 2JLP. β-Strands are displayed as flat arrows, and helices are displayed as ribbons. Metal-liganding residues are shown as sticks. Key structural elements are color-coded: E-Loop, Electrostatic loop; GK1 and GK2, Greek key loops 1 and 2; S-S, disulfide region; V-loop, variable loop (28).
TABLE 2.
Sequences identity (%)/similarity (%) between Leptopilina and H. sapiens (Hs) Cu,Zn-SODs (SOD1, NP_000445; SOD3, P08294) (amino acid sequence alignment using the EMBOSS program Needle)
| LbmSOD3 | LbySOD3 | LhSOD3 | LbmSOD1 | LbySOD1 | LhSOD1 | HsSOD1 | |
|---|---|---|---|---|---|---|---|
| LbySOD3 | 98.9/98.9 | ||||||
| LhSOD3 | 73.3/88.1 | 73.9/88.6 | |||||
| LbmSOD1 | 44.6/56.0 | 44.3/55.7 | 45.5/55.1 | ||||
| LbySOD1 | 44.6/56.0 | 44.3/55.7 | 45.5/55.1 | 100/100 | |||
| LhSOD1 | 44.0/54.9 | 43.8/54.5 | 43.8/53.9 | 95.4/98.7 | 95.4/98.7 | ||
| HsSOD1 | 47.7/58.5 | 47.5/58.2 | 47.5/56.5 | 65.8/74.2 | 65.8/74.2 | 64.5/74.8 | |
| HsSOD3 | 25.5/37.7 | 25.0/35.9 | 24.1/37.3 | 23.5/30.8 | 23.5/30.8 | 24.0/31.1 | 25.1/34.4 |
Leptopilina SOD3s Are Closely Related to Human SOD1
Interestingly, sequence comparisons of Leptopilina extracellular SODs with human ones clearly show that they are more related to the human intracellular SOD1 than to the human extracellular SOD3 (Table 2 and Fig. 1A). More specifically, the Leptopilina SOD3 signal peptide differs from that of human SOD3, and Leptopilina SOD3 lack the heparin binding C-terminal part of human SOD3 involved in the binding to the cellular matrix (30). In addition, Leptopilina and human SOD3 differ in the number of disulfide bridges. The human SOD3 contains two intrasubunit disulfide bonds: Cys-107—Cys-189, also found in human SOD1 and predicted for Leptopilina SOD1s, and Cys-45—Cys-190, unique to human SOD3 (38, 39). The specific C-terminal heparin binding region of human SOD3 also contains a cysteine (Cys-219) implicated in an additional intersubunit disulfide bridge, necessary for the formation of the human SOD3 tetramer (38, 39). Only the disulfide bridge geometrically similar to Cys-107—Cys-189 could be predicted for Leptopilina SOD3, as a Val and a Gly are found at positions corresponding to Cys-45 and Cys-190 (Fig. 1A), and the absence of the C-terminal region precludes the formation of the intersubunit disulfide bridge. Finally, comparisons of solved human and Leptopilina-predicted SOD1 and SOD3 structures confirmed that Leptopilina extracellular SODs are more different from human SOD3 (with root mean square deviations ranging from 0.839 to 0.851 Å for 116 to 127 α-carbon atoms) than from human SOD1 (with root mean square deviations ranging from 0.448 to 0.449 Å for 125 α-carbon atoms; Table 3). Among key structural elements, the Greek key loop 1 (GK1, Fig. 1B), involved in SOD stability (39), is longer in human SOD3 only.
TABLE 3.
Structural alignments between Leptopilina and H. sapiens (SOD1, 1PU0; SOD3, 2JLP) Cu,ZnSODs based on the root mean square deviations (Å) using PyMOL
Values in parentheses correspond to number of aligned α-carbon atoms.
| LbmSOD3 | LbySOD3 | LhSOD3 | LbmSOD1 | LbySOD1 | LhSODI | HsSODI | |
|---|---|---|---|---|---|---|---|
| HsSOD1 | 0.449 (125) | 0.449 (125) | 0.448 (125) | 0.420 (132) | 0.420 (132) | 0.440 (133) | |
| HsSOD3 | 0.839 (127) | 0.839 (127) | 0.851 (116) | 0.881 (108) | 0.881 (108) | 1.090 (100) | 0.615 (110) |
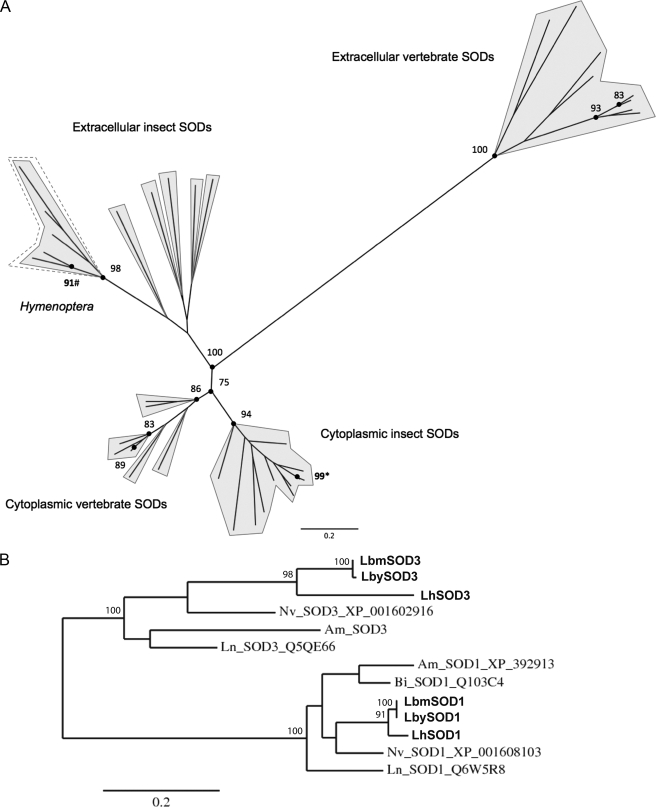
Phylogenetic analysis with selected vertebrate and insect Cu,Zn-SOD sequences (Fig. 2A) and with available hymenopteran Cu,Zn-SOD sequences (Fig. 2B) confirms that Leptopilina SOD3 clusters with predicted extracellular insect Cu,Zn-SODs, whereas Leptopilina SOD1 groups with cytoplasmic Cu,Zn-SODs (Fig. 2, A and B). In agreement with sequence and structure comparisons, extracellular vertebrate and insect Cu,Zn-SOD sequences form largely separated clusters, insect extracellular Cu,Zn-SODs being more closely related to cytoplasmic vertebrate Cu,Zn-SODs than to extracellular ones (Fig. 2A).
FIGURE 2.
Maximum-likelihood phylogenetic trees using vertebrate and insect Cu,Zn-SOD sequences (A) and using hymenopteran Cu,Zn-SOD sequences (B) (SOD3, extracellular SOD; SOD1, cytoplasmic SOD). The dashed line delineates hymenopteran extracellular Cu,Zn-SODs. Numbers at corresponding nodes are bootstrap support values (500 bootstrap replicates). Numbers followed by asterisks (*) are bootstrap values for Leptopilina SOD1 nodes. Numbers followed with number symbols (#) are bootstrap values for Leptopilina SOD3 nodes. Only bootstrap support values ≥ 75% are shown. Am, Apis mellifera; Bi, Bombus ignitus; Lb, L. boulardi; Ln, Lasius niger; Nv, Nasonia vitripennis.
The Extracellular Cu,Zn-SOD Gene Is Overexpressed in L. boulardi Venom Apparatus
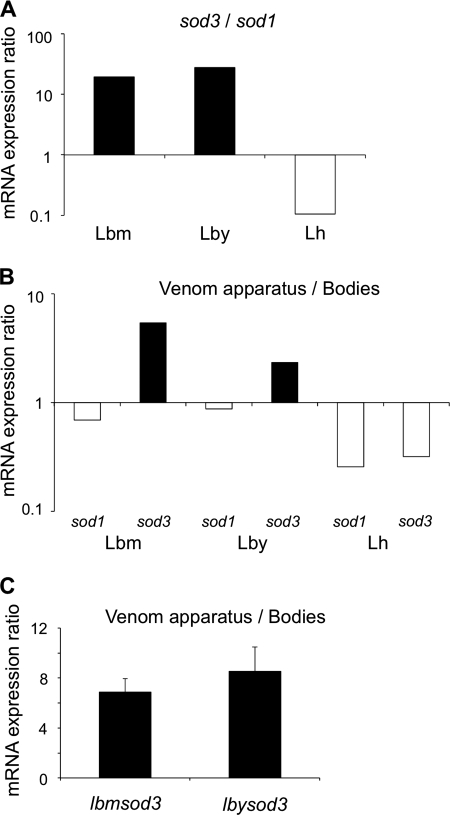
The relative abundance of Cu,Zn-SOD transcripts in venom apparatus of Leptopilina wasps was estimated by semi-qRT-PCR using ITS2 as a reference gene. After 28 and 33 cycles, L. boulardi lbmsod3 and lbysod3 genes were found 20 and 30 times more expressed in venom apparatus than lbmsod1 and lbysod1, whereas in L. heterotoma, lhsod1 was 10 times more expressed in venom tissues than lhsod3 (Fig. 3A, shown for 28 cycles). Overall, the expression level of sod3 in venom apparatus was ∼100 times higher in L. boulardi than in L. heterotoma, whereas expression of sod1 was only twice higher. Using a similar approach, lbmsod3 and lbysod3 were found more expressed in venom-producing tissues than in the remnant body in contrast to lhsod3 and all cytoplasmic Cu,Zn-SOD genes (Fig. 3B, shown for 28 cycles). qRT-PCR results confirmed that extracellular Cu,Zn-SODs gene expression in L. boulardi venom apparatus is 7–9 times higher than in the remnant body (Fig. 3C), with no significant difference between lbmsod3 and lbysod3 (Student's t test: t = −0.6117, df = 3, p = 0.584). All these results demonstrate that only L. boulardi wasps specifically overexpress extracellular Cu,Zn-SOD genes in venom-producing tissues.
FIGURE 3.
Expression levels of Leptopilina Cu,Zn-SOD genes. A, relative expression ratio of extracellular to cytoplasmic Cu,Zn-SOD genes (logarithmic scale) in Lbm, Lby, and Lh venom apparatus after 28 cycles of semi-qRT-PCR. B, relative expression ratio of Leptopilina Cu,Zn-SOD genes (logarithmic scale) in venom apparatus compared with the remnant bodies after 28 cycles of semi-qRT-PCR. C, relative expression level (qRT-PCR) in venom-producing tissues compared with the remnant bodies for lbmsod3 and lbysod3 genes.
SOD3 Proteins Are Secreted in Different Amounts in L. boulardi and L. heterotoma Venom
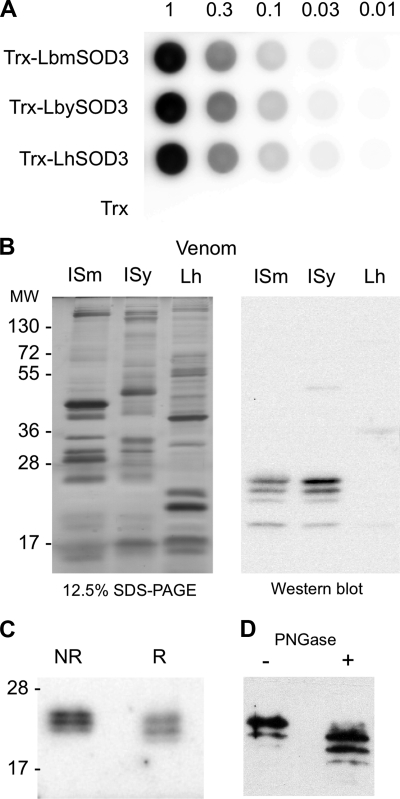
To confirm that predicted extracellular Cu,Zn-SODs are secreted in venom, we performed immunoblotting using a polyclonal antibody against H. sapiens SOD1, which recognizes the recombinant Trx-fusion proteins LbmSOD3, LbySOD3, and LhSOD3 with the same efficiency (Fig. 4A). On a Western blot with venom from 10 female reservoirs, 4 bands of different intensity ranging from 19 to 25 kDa were detected for Lbm and Lby, whereas only a very faint band at 19 kDa was obtained for Lh (Fig. 4B). By comparing signal intensities between Lbm and Lby venom extracts and dilutions of recombinant LbmSOD3s, we estimated the total quantity of SOD as ∼10 ng per reservoir for both L. boulardi strains (results not shown).
FIGURE 4.
Immunoblotting experiments using a H. sapiens SOD1 polyclonal antibody. A, dot blot experiment on serial dilutions starting with 1 μg of the recombinant proteins Trx-LbmSOD3, Trx-LbySOD3, Trx-LhSOD3, and Trx tag alone. B, 12.5% SDS-PAGE (left) and an equivalent Western blot (right) on protein extracts from 10 reservoirs of Lbm, Lby, and Lh. C, Western blot on protein extracts from 30 Lbm venom reservoirs in non-reducing (NR) and reducing (R) conditions. D, glycosylation profile analysis of Leptopilina Cu,Zn-SOD using Western blot on extracts from 10 L. boulardi ISm reservoirs either untreated (−) or treated (+) with N-glycosidase F (PNGase). Molecular mass is in kDa.
To ascertain the identity of the proteins detected in Lbm and Lby venom, we performed mass spectrometry on gel bands cut at the corresponding position on an SDS-PAGE gel of venom proteins. For both strains, we found peptides specifically matching with the extracellular Cu,Zn-SOD (Table 4), demonstrating that this protein is indeed secreted in L. boulardi venom. By contrast, no peptides corresponding to SOD3 were found in L. heterotoma venom. Some peptides were also found that matched SOD1 in L. boulardi and L. heterotoma, possibly because of the presence of contaminants originating from cell leakage during venom collection. All these results show that the SOD antibody signal in L. boulardi venom essentially corresponds to detection of SOD3.
TABLE 4.
Cu,ZnSOD peptides identified by mass spectrometry in L. boulardi and L. heterotoma venom
| Organism | Protein | Peptides |
|---|---|---|
| Lbm | LbmSOD1 | LACGVIGITKN |
| LbmSOD3 | GDISKGCISTGK | |
| HVGDLGNVIANK | ||
| AFVVHEK | ||
| MVPNNVQK | ||
| Lby | LbySOD1 | GTVFFEESGDSVK |
| LACGVIGITKN | ||
| LbySOD3 | HVGDLGNVIANK (2 bands) | |
| GNTSLSLETGDAGDR | ||
| Lh | LhSOD1 | GNNNIIGR |
| LACGVIGITK | ||
| TLVVHGDPDDLGK | ||
| GTVFFEEAGDSVK |
In mammals, Cu,Zn-SODs are classically found in SDS-PAGE as dimers or tetramers even in presence of a reducing agent (28). In contrast, in L. boulardi, Cu,Zn-SOD migration profiles were similar between reducing and non-reducing conditions (Fig. 4D), which suggests that LbSOD3 is mainly under the monomeric form in venom. Besides, treatment of L. boulardi ISm venom extracts with N-glycosidase F induced a shift in size for the strongest reactive bands, suggesting the presence of N-glycosylations, although deglycosylation might be incomplete (Fig. 4C). The 19–25-kDa bands thus likely correspond to SOD3 monomers with post-translational modifications (Fig. 4B).
Cu,Zn-SODs Are Active in the Venom of L. boulardi
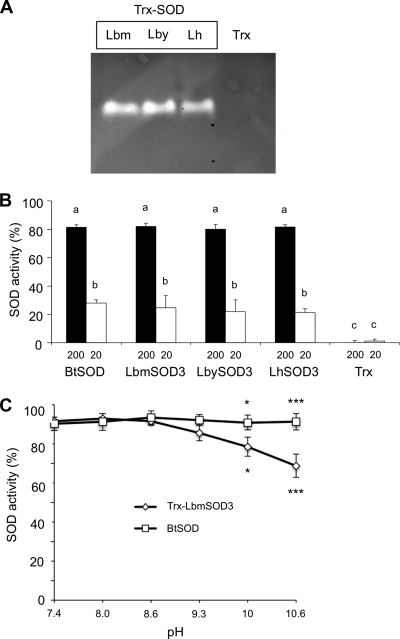
SOD zymography and spectrophotometric assays clearly demonstrated that the recombinant Trx-LbmSOD3, Trx-LbySOD3, and Trx-LhSOD3 proteins were all active (Fig. 5A). The level of activity was similar between these proteins and with that of BtSOD, a commercial SOD from bovine erythrocytes (Fig. 5B; ANOVA: df = 9, F = 67.238, p > 0.9). All Trx-SODs and BtSOD activities significantly decreased with the concentration of the protein (Fig. 5B; ANOVA: df = 9, F = 67.238, p < 0.001), and there was no activity for the Trx tag alone. In contrast with that of BtSOD, the activity of Trx-LbmSOD3 decreased at pH 10 and above (Fig. 5C; ANOVA: df = 11, F = 11.954, p < 0.05).
FIGURE 5.
SOD activity assays with recombinant proteins. A, SOD zymography assay using 3 μg of recombinant proteins Trx-LbmSOD3, Trx-LbySOD3, and Trx-LhSOD3. B, SOD spectrophotometric assay using 200 and 20 ng of BtSOD and of Trx-LbmSOD3, Trx-LbySOD3, and Trx-LhSOD3. Trx alone is used as a control. Different letters above the bars designate significantly different values. C, effect of pH on activity performed with 200 ng of BtSOD and LbmSOD3. Mean values (±S.E.) are given for each category (three replicates). The asterisk indicates statistically significant differences (*, p < 0.05; ***, p < 0.001).
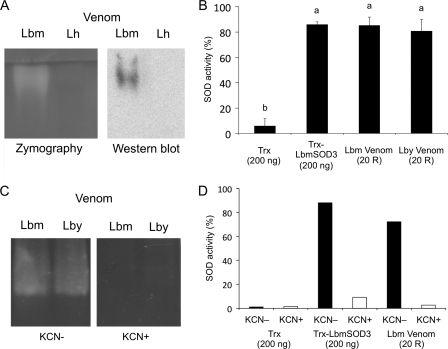
Using zymographic assays, a SOD activity was detected in the venom of both L. boulardi strains (Fig. 6, A and C) at a position corresponding to the signal observed on Western blot under native conditions (Fig. 6A, not shown for Lby). In contrast, no activity was observed in L. heterotoma venom (Fig. 6A). Spectrophotometric assays confirmed that the venom of L. boulardi strains has a similar SOD activity (Fig. 6B; ANOVA: df = 3, F = 113.94, p = 1). In addition, venom samples, which contain 200 ng of SOD, had a SOD activity equivalent to that of 200 ng of recombinant LbmSOD3, suggesting that recombinant and venom SOD3 have a similar specific activity (Fig. 6B; ANOVA: df = 3, F = 113.94, p = 1). Finally, a complete inhibition was observed when adding KCN both in zymography (Fig. 6C) and spectrophotometric assays (Fig. 6D, not shown for Lby), indicating that all venom activity is due to a Cu,Zn-SOD activity.
FIGURE 6.
SOD activity assays with venom. A, SOD zymography assay and Western blot experiment in native conditions with extracts of 25 Lbm and Lh venom reservoirs. B, SOD spectrophotometric assay using 200 ng of Trx-LbmSOD3 and protein extracts of 20 Lbm, Lby, and Lh venom reservoirs (20 R). Different letters above the bars designate significantly different values. C, SOD zymography assay using extracts of 25 Lbm and Lby venom reservoirs with (KCN+) and without KCN (KCN−). D, SOD spectrophotometric assay using 200 ng of Trx-LbmSOD3 and extracts of 20 Lbm reservoirs (20 R) with (KCN+) and without KCN (KCN−). Trx tag alone was used as a control.
Cu,Zn-SODs Inhibit PO Activity in D. melanogaster
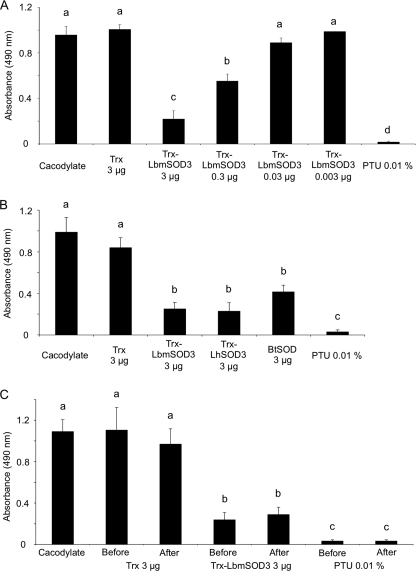
To test whether Leptopilina extracellular Cu,Zn-SODs may interfere with the host PO cascade, PO assays were conducted on Drosophila hemolymph samples in the presence of recombinant Trx-LbmSOD3 or Trx tag alone as a control (Fig. 7A). The PO activity was decreased by 70% using 3 μg of recombinant protein and by 40% using 0.3 μg (Fig. 6A; ANOVA: df = 6, F = 49.334, p < 0.01), whereas the addition of lower concentrations (0.03 and 0.003 μg) had no inhibitory effect as Trx tag or buffer alone (Fig. 7A, p > 0.5). The same inhibition level was observed with 3 μg of Trx-LbySOD3, of Trx-LhSOD3, and, interestingly, of BtSOD (Fig. 7B; ANOVA: df = 5; F = 19.3058, p < 0.01). As expected, PO activity was totally inhibited by PTU.
FIGURE 7.
Effect of Leptopilina recombinant SOD3 and venom on Drosophila PO activity. PO activity was measured as absorbance at 490 nm for A, D. melanogaster larval hemolymph in presence of buffer alone, Trx-LbmSOD3 (3–0.003 μg), Trx-tag alone (3 μg), or PO inhibitor (PTU). B, D. melanogaster larval hemolymph in the presence of buffer alone, 3 μg of Trx-LbmSOD3, Trx-LhSOD3, BtSOD or Trx tag alone, or PTU. C, D. melanogaster larval hemolymph in presence of Trx-LhSOD3 (3 μg), Trx-tag alone (3 μg), or PTU added either before or after PO activation. Mean values (±S.E.) are given for each category (three replicates). Different letters above the bars designate significantly different values.
To investigate whether the reduction of absorbance reflected the inhibition of proPO to PO conversion through action on the serine protease cascade or the inhibition of the PO activity itself, we performed parallel experiments in which Trx-LbmSOD3 protein, Trx tag alone, or PTU was added either before hemolymph collection or after proPO to PO conversion (Fig. 7C). The inhibitory effect of PTU and of Trx-LbmSOD3 did not differ whether they were added before or after PO activation (Fig. 7C; ANOVA: df = 6, F = 18.4594, p = 1), thus demonstrating that recombinant SODs do not affect the PO activation cascade but PO activity itself.
Because of the presence of a serpin in L. boulardi ISy venom, which inhibits proPO to PO conversion (19), PO assays with L. boulardi ISm and ISy venom were performed by adding venom in conditions where the proPO was already converted into PO. Assays were performed using venom quantities containing from 0.04 to 0.6 μg of SOD. However, no significant effect was detected, even when using an amount of venom estimated to contain twice the minimal amount of recombinant SOD3 (0.3 μg) that induced a detectable effect on PO activity (data not shown).
DISCUSSION
In contrast to mammals, insects have long been thought to produce only the cytoplasmic and mitochondrial SODs. The presence of extracellular SODs was questioned only recently with the description of two highly divergent Cu,Zn-SOD sequences in the ant Lasius niger, one of them containing a signal peptide (40). Subsequent database searches and phylogenetic analyses then revealed that insect genomes, including that of D. melanogaster, contain sequences corresponding to the two distinct Cu,Zn-SOD families related to mammalian SOD1 and SOD3 (40, 41). However, although predicted by in silico analyses, occurrence of extracellular Cu,Zn-SOD in insects had never been proven in vivo.
In this work we demonstrate that insects produce and secrete extracellular Cu,Zn-SODs. Indeed, having identified Cu,Zn-SOD transcripts containing a signal peptide from the venom apparatus of two Leptopilina wasps, parasitoids of Drosophila, we confirmed the presence of the corresponding secreted SOD3 protein in L. boulardi venom using Western blot and mass spectrometry identification. In accordance with the conservation of predicted essential residues and key structural elements, this SOD3 protein is active, and its activity is specific of Cu,Zn-SODs.
Interestingly, Leptopilina SOD3s are more similar in sequence and structure to human SOD1 than to human SOD3, and they lack in particular the human SOD3 motif involved in binding of the protein to the cellular matrix (30). Moreover, our results suggest that Leptopilina venom SOD3s are active as monomers, in agreement with the absence of the intersubunit disulfide bridge and of one of the two intrasubunit disulfide bridges found in human SOD3. Phylogenetic analyses show that not only the Leptopilina but all the predicted insect SOD3s are more closely related to vertebrate SOD1 than SOD3 sequences, thus suggesting a rapid divergence of the extracellular vertebrate Cu,Zn-SOD sequences.
The characterization of Leptopilina SOD3 is the first example of secretion of an active extracellular SOD in hymenopteran venom despite recent proteomic analyses from different species, including parasitoids wasps (42–45). This questions the potential role of SOD3 in L. boulardi, which, as most endoparasitoids, inject at oviposition venom toxins that among other effects counteract the host immune response. Production of ROS is part of this response, as evidenced for D. melanogaster (7–9), and the detoxification of ROS is known to be part of virulence mechanisms. SODs, for instance, are used as immune-suppressive toxins by a large number of pathogens, including eukaryotic protozoan parasites (21–26).
A likely role of L. boulardi venom SOD3 in parasitism success is supported by the specific overexpression of the sod3 gene in the venom apparatus compared with other tissues, as shown for most parasitoid virulence factors (43–52), including two major venom toxins of L. boulardi (17, 19). In L. heterotoma, the SOD3 protein is hardly detected in venom, and the venom contains no measurable Cu,Zn-SOD-specific activity. This suggests that SOD3 is not used as a virulence factor by L. heterotoma as it is by L. boulardi, thus evidencing interspecific variation in venom toxins in Leptopilina parasitoids despite phylogenetic proximity and the use of similar hosts. We recently showed that the quantitative difference in the LbGAP venom toxin was at the origin of the intraspecific variation of virulence between L. boulardi ISm and ISy lines (12). No such variation was found for SOD3 proteins neither in sequence or in venom quantity and activity, which seems to preclude a role of SODs in intraspecific variation of L. boulardi virulence.
Interestingly, the recombinant L. boulardi SOD3, but also a purified mammalian SOD, have inhibitory effects on D. melanogaster PO activity in vitro in a dose-dependent manner. We thus hypothesized that L. boulardi injection of the SOD3 protein in Drosophila could result in a modification of enzymatic equilibria through dismutation of the superoxide anion O2˙̄ that could disturb the host melanization pathway. However, no significant effect of L. boulardi ISm nor ISy venom on PO activity was observed in vitro that could correspond to the SOD effect. This was not due to quantitative differences as the venom sample used in the assays contained a SOD3 quantity higher than the SOD3 recombinant protein amount that yielded 40% inhibition of PO activity. However, the very high quantity of venom used (corresponding to the content of 60 reservoirs) concentrated in the small volume of the assay might have disturbed the PO assay that is very sensitive to salt and/or pH modifications (53). In any case, the SOD3 quantity in one unique female reservoir was estimated to be 30 times smaller than the minimal amount of recombinant SOD3 shown to inhibit Drosophila hemolymph PO activity in vitro.
The SOD3 effect could also be through direct detoxification of O2˙̄, which is produced at high levels by the host during the melanization process (7). Although O2˙̄ is not cytotoxic against the parasitoid (7), it likely plays a critical role in Drosophila cellular immunity by initiating the formation of .OH, the most reactive free radical in biological systems, through the reduction of some metal ions (7, 9).
If a systemic effect of injection of SOD3 in the host, whether directly on the superoxide anion or on PO activity, is rather unlikely due to its low quantity in venom, this does not preclude a local effect nearby the deposited parasitoid egg. After recognition, the egg is indeed rapidly covered by host hemocytes that likely produce killing oxygen radicals (54). Also, lamellocytes, the main forming-capsule hemocytes, specifically express the PO3 gene (55, 56), and a deposit of melanin, indicating PO activity, is observed at the surface of the parasitoid egg (3, 57, 58).
Finally, a possible function of Leptopilina venom SODs might be the protection from oxidative stress of venom apparatus tissues and/or venom proteins during storage. Protection of venom gland tissues by the SOD1 protein was suggested, for instance, in the honeybee (42) and can be hypothesized as well in Leptopilina parasitoids. However, as the SOD3 protein is only secreted in venom and in a high amount in L. boulardi, its role in protecting venom components would suggest a specific need of this species that remains to be evidenced.
In summary, our results demonstrate that insects indeed produce and secrete extracellular SODs and that venom-secreted SODs are likely used as virulence factors by parasitoid wasps as they are by pathogens of mammals and insects. The exact role of the venom SOD in parasitism success nevertheless remains an open question that might be explored in the future through development of promising methods, such as RNAi, for parasitoids (59).
Supplementary Material
Acknowledgments
We are grateful to Christian Rebuf and Didier Crochard for technical assistance, to Didier Herouart for help with SOD zymography assays, and to Hugo Mathé-Hubert for help with statistical analyses.
This work was supported by Grants CLIMEVOL (ANR-08-BLAN-0231) and PARATOXOSE (ANR-09-BLAN-0243-01) from the French National Agency for Research and grants from the Department of Plant Health and Environment from the French National Institute for Agricultural Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- proPO
- prophenoloxidase
- Lbm
- L. boulardi ISm
- Lby
- L. boulardi ISy
- Lh
- L. heterotoma
- PO
- phenoloxidase
- ROS
- reactive oxygen species
- SOD
- superoxide dismutase
- Bt
- Bos taurus
- Trx
- thioredoxin
- PTU
- 1-phenyl-2-thiourea
- qRT
- quantitative real-time
- ANOVA
- analysis of variance
- df
- degrees of freedom
- F
- F ratio.
REFERENCES
- 1. Godfray H. (1994) Parasitoids: Behavioral and Evolutionary Ecology, Princeton University Press, Princeton, NJ [Google Scholar]
- 2. Poirié M., Carton Y., Dubuffet A. (2009) C. R. Biol. 332, 311–320 [DOI] [PubMed] [Google Scholar]
- 3. Carton Y., Poirié M., Nappi A. J. (2008) Insect Sci. 15, 67–87 [Google Scholar]
- 4. Nappi A. J. (2010) Invert. Surv. J. 7, 198–210 [Google Scholar]
- 5. Strand M. (2008) Insect Sci. 15, 1–14 [Google Scholar]
- 6. Cerenius L., Lee B. L., Söderhäll K. (2008) Trends Immunol. 29, 263–271 [DOI] [PubMed] [Google Scholar]
- 7. Nappi A. J., Vass E., Frey F., Carton Y. (1995) Eur. J. Cell Biol. 68, 450–456 [PubMed] [Google Scholar]
- 8. Nappi A. J., Christensen B. M. (2005) Insect Biochem. Mol. Biol. 35, 443–459 [DOI] [PubMed] [Google Scholar]
- 9. Nappi A., Poirié M., Carton Y. (2009) Adv. Parasitol. 70, 99–121 [DOI] [PubMed] [Google Scholar]
- 10. Asgari S., Rivers D. B. (2011) Annu. Rev. Entomol. 56, 313–335 [DOI] [PubMed] [Google Scholar]
- 11. Dubuffet A., Colinet D., Anselme C., Dupas S., Carton Y., Poirié M. (2009) Adv. Parasitol. 70, 147–188 [DOI] [PubMed] [Google Scholar]
- 12. Colinet D., Schmitz A., Cazes D., Gatti J. L., Poirié M. (2010) PLoS Pathog. 6, e1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Labrosse C., Eslin P., Doury G., Drezen J. M., Poirié M. (2005) J. Insect Physiol. 51, 161–170 [DOI] [PubMed] [Google Scholar]
- 14. Labrosse C., Stasiak K., Lesobre J., Grangeia A., Huguet E., Drezen J. M., Poirié M. (2005) Insect Biochem. Mol. Biol. 35, 93–103 [DOI] [PubMed] [Google Scholar]
- 15. Williams M. J., Ando I., Hultmark D. (2005) Genes Cells 10, 813–823 [DOI] [PubMed] [Google Scholar]
- 16. Williams M. J., Wiklund M. L., Wikman S., Hultmark D. (2006) J. Cell Sci. 119, 2015–2024 [DOI] [PubMed] [Google Scholar]
- 17. Colinet D., Schmitz A., Depoix D., Crochard D., Poirié M. (2007) PLoS Pathog. 3, e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dupas S., Carton Y., Poirié M. (2003) Heredity 90, 84–89 [DOI] [PubMed] [Google Scholar]
- 19. Colinet D., Dubuffet A., Cazes D., Moreau S., Drezen J. M., Poirié M. (2009) Dev. Comp. Immunol. 33, 681–689 [DOI] [PubMed] [Google Scholar]
- 20. Kohler L. J., Carton Y., Mastore M., Nappi A. J. (2007) Arch. Insect Biochem. Physiol. 66, 64–75 [DOI] [PubMed] [Google Scholar]
- 21. Lynch M., Kuramitsu H. (2000) Microbes Infect. 2, 1245–1255 [DOI] [PubMed] [Google Scholar]
- 22. Hwang C. S., Rhie G. E., Oh J. H., Huh W. K., Yim H. S., Kang S. O. (2002) Microbiology 148, 3705–3713 [DOI] [PubMed] [Google Scholar]
- 23. Ghosh S., Goswami S., Adhya S. (2003) Biochem. J. 369, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Narasipura S. D., Ault J. G., Behr M. J., Chaturvedi V., Chaturvedi S. (2003) Mol. Microbiol. 47, 1681–1694 [DOI] [PubMed] [Google Scholar]
- 25. Ammendola S., Pasquali P., Pacello F., Rotilio G., Castor M., Libby S. J., Figueroa-Bossi N., Bossi L., Fang F. C., Battistoni A. (2008) J. Biol. Chem. 283, 13688–13699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie X. Q., Wang J., Huang B. F., Ying S. H., Feng M. G. (2010) Appl. Microbiol. Biotechnol. 86, 1543–1553 [DOI] [PubMed] [Google Scholar]
- 27. Fridovich I. (1995) Annu. Rev. Biochem. 64, 97–112 [DOI] [PubMed] [Google Scholar]
- 28. Perry J. J., Shin D. S., Getzoff E. D., Tainer J. A. (2010) Biochim. Biophys. Acta 1804, 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okado-Matsumoto A., Fridovich I. (2001) J. Biol. Chem. 276, 38388–38393 [DOI] [PubMed] [Google Scholar]
- 30. Stenlund P., Lindberg M. J., Tibell L. A. (2002) Biochemistry 41, 3168–3175 [DOI] [PubMed] [Google Scholar]
- 31. Weisiger R. A., Fridovich I. (1973) J. Biol. Chem. 248, 4793–4796 [PubMed] [Google Scholar]
- 32. Dupas S., Frey F., Carton Y. (1998) J. Hered. 89, 306–311 [DOI] [PubMed] [Google Scholar]
- 33. Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guindon S., Gascuel O. (2003) Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 35. Kelley L. A., Sternberg M. J. E. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 36. Beauchamp C., Fridovich I. (1971) Anal. Biochem. 44, 276–287 [DOI] [PubMed] [Google Scholar]
- 37. DiDonato M., Craig L., Huff M. E., Thayer M. M., Cardoso R. M., Kassmann C. J., Lo T. P., Bruns C. K., Powers E. T., Kelly J. W., Getzoff E. D., Tainer J. A. (2003) J. Mol. Biol. 332, 601–615 [DOI] [PubMed] [Google Scholar]
- 38. Petersen S. V., Oury T. D., Valnickova Z., Thogersen I. B., Hojrup P., Crapo J. D., Enghild J. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13875–13880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Antonyuk S. V., Strange R. W., Marklund S. L., Hasnain S. S. (2009) J. Mol. Biol. 388, 310–326 [DOI] [PubMed] [Google Scholar]
- 40. Parker J. D., Parker K. M., Keller L. (2004) Insect Mol. Biol. 13, 587–594 [DOI] [PubMed] [Google Scholar]
- 41. Corona M., Robinson G. E. (2006) Insect Mol. Biol. 15, 687–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peiren N., de Graaf D. C., Vanrobaeys F., Danneels E. L., Devreese B., Van Beeumen J., Jacobs F. J. (2008) Toxicon 52, 72–83 [DOI] [PubMed] [Google Scholar]
- 43. de Graaf D. C., Aerts M., Brunain M., Desjardins C. A., Jacobs F. J., Werren J. H., Devreese B. (2010) Insect Mol. Biol. 19, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vincent B., Kaeslin M., Roth T., Heller M., Poulain J., Cousserans F., Schaller J., Poirié M., Lanzrein B., Drezen J. M., Moreau S. J. M. (2010) BMC Genomics 11, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu J. Y., Fang Q., Wang L., Hu C., Ye G. Y. (2010) Arch. Insect Biochem. Physiol. 75, 28–44 [DOI] [PubMed] [Google Scholar]
- 46. Asgari S., Reineke A., Beck M., Schmidt O. (2002) Insect Mol. Biol. 11, 477–485 [DOI] [PubMed] [Google Scholar]
- 47. Crawford A. M., Brauning R., Smolenski G., Ferguson C., Barton D., Wheeler T. T., McCulloch A. (2008) Insect Mol. Biol. 17, 313–324 [DOI] [PubMed] [Google Scholar]
- 48. Parkinson N., Smith I., Weaver R., Edwards J. P. (2001) Insect Biochem. Mol. Biol. 31, 57–63 [DOI] [PubMed] [Google Scholar]
- 49. Price D. R., Bell H. A., Hinchliffe G., Fitches E., Weaver R., Gatehouse J. A. (2009) Insect Mol. Biol. 18, 195–202 [DOI] [PubMed] [Google Scholar]
- 50. Vinchon S., Moreau S. J., Drezen J. M., Prévost G., Cherqui A. (2010) Insect Biochem. Mol. Biol. 40, 38–48 [DOI] [PubMed] [Google Scholar]
- 51. Zhang G., Schmidt O., Asgari S. (2006) Dev. Comp. Immunol. 30, 756–764 [DOI] [PubMed] [Google Scholar]
- 52. Zhu J. Y., Ye G. Y., Hu C. (2008) Toxicon 51, 1391–1399 [DOI] [PubMed] [Google Scholar]
- 53. Zufelato M. S., Lourenço A. P., Simões Z. L., Jorge J. A., Bitondi M. M. G. (2004) Insect Biochem. Mol. Biol. 34, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 54. Nappi A. J., Vass E. (1998) J. Parasitol. 84, 1150–1157 [PubMed] [Google Scholar]
- 55. Irving P., Ubeda J. M., Doucet D., Troxler L., Lagueux M., Zachary D., Hoffmann J. A., Hetru C., Meister M. (2005) Cell. Microbiol. 7, 335–350 [DOI] [PubMed] [Google Scholar]
- 56. Nam H. J., Jang I. H., Asano T., Lee W. J. (2008) Mol. Cells 26, 606–610 [PubMed] [Google Scholar]
- 57. Cerenius L., Kawabata S., Lee B. L., Nonaka M., Söderhäll K. (2010) Trends Biochem. Sci. 35, 575–583 [DOI] [PubMed] [Google Scholar]
- 58. Reumer A., Van Loy T., Schoofs L. (2010) Invert. Surv. J. 7, 32-44 [Google Scholar]
- 59. Werren J. H., Loehlin D. W., Giebel J. D. (2009) CSH Protoc. 2009, pdb.prot5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.