Background: Membrane repair requires the assembly of multiple proteins to orchestrate the repair process.
Results: A complex between three membrane repair proteins was assembled to explore their arrangement.
Conclusion: A novel asymmetric complex is formed involving all three proteins.
Significance: This is the first evidence of an asymmetric multiprotein complex that might control membrane repair.
Keywords: Annexin, Calcium-binding Proteins, Membrane Fusion, Protein Structure, Spectroscopy
Abstract
Membrane repair is mediated by multiprotein complexes, such as that formed between the dimeric EF-hand protein S100A10, the calcium- and phospholipid-binding protein annexin A2, the enlargeosome protein AHNAK, and members of the transmembrane ferlin family. Although interactions between these proteins have been shown, little is known about their structural arrangement and mechanisms of formation. In this work, we used a non-covalent complex between S100A10 and the N terminus of annexin A2 (residues 1–15) and a designed hybrid protein (A10A2), where S100A10 is linked in tandem to the N-terminal region of annexin A2, to explore the binding region, stoichiometry, and affinity with a synthetic peptide from the C terminus of AHNAK. Using multiple biophysical methods, we identified a novel asymmetric arrangement between a single AHNAK peptide and the A10A2 dimer. The AHNAK peptide was shown to require the annexin A2 N terminus, indicating that the AHNAK binding site comprises regions on both S100A10 and annexin proteins. NMR spectroscopy was used to show that the AHNAK binding surface comprised residues from helix IV in S100A10 and the C-terminal portion from the annexin A2 peptide. This novel surface maps to the exposed side of helices IV and IV′ of the S100 dimeric structure, a region not identified in any previous S100 target protein structures. The results provide the first structural details of the ternary S100A10 protein complex required for membrane repair.
Introduction
The fusion of cellular phospholipid membranes is an important step during membrane repair, vesicular trafficking, and exocytosis. A requisite for plasma membrane repair is calcium-regulated exocytosis, controlled by a number of proteins that coordinate resealing of the membrane. One exocytotic compartment implicated in membrane fusion is the enlargeosome, characterized by the large scaffolding protein AHNAK, that relocates to the plasma membrane immediately following membrane damage (1). In addition to AHNAK, calcium sensors, such as members from the synaptotagmin, ferlin (dysferlin), and annexin protein families, are involved in membrane repair. Upon membrane rupture, these sensor proteins respond to the calcium influx by localizing near the inner membrane surface. For example, annexins A1 and A2 have been implicated in skeletal muscle cell repair based on their calcium-dependent interaction and localization with dysferlin near the plasma membrane (2), whereas others, such as the SNARE protein VAMP2, have been shown to co-localize and interact with annexin A2 and S100A10 during exocytosis in chromaffin cells (3). Recent proteomic analyses have identified ∼115 proteins in the dysferlin complex, including calpain 3, annexin A2, and AHNAK (4, 5), that might be responsible for the repair process. These studies indicate that large protein complexes are probably needed to orchestrate the repair process and restore the integrity of the membrane.
One potential multiprotein repair complex includes S100A10, annexin A2, AHNAK, and actin identified in enlargeosomes. S100A10 is a member of the S100 protein family of dimeric, EF-hand proteins. Three-dimensional structures of several S100 proteins have revealed a calcium-induced conformational change in these molecules (6, 7) that modulates their interactions with other proteins. S100A10 is a unique member of the family that has lost its ability to accommodate calcium ions due to alterations in both of its calcium-binding loops. Consequently, S100A10 recruits annexins A1 and A2 (8, 9) to form tight heterotetrameric complexes in a calcium-insensitive manner with respect to the EF-hand protein. In vivo and in vitro studies show that annexin A2 is a central protein involved in membrane transport events, such as endo-/exocytosis (10–20). Members of the annexin family form a group of highly helical proteins containing a core domain composed of four (annexins A1–A5, A7–A11, and A13) or eight (annexin A6) conserved structural repeat sequences (I–IV), each about 70–75 residues in length, and a short N-terminal region (21, 22). Calcium binding to the annexins promotes their localization and association with phospholipid-containing membranes through the convex surface of the core domain (23). In the case of annexins A1 and A2, calcium binding is proposed to lead to the release of the previously buried N terminus to an exposed position available for interaction with S100A10 protein (24). Thus, an S100A10 dimer can coordinate a pair of annexin A2 proteins, potentially bridging adjacent phospholipid membranes into close proximity during a membrane fusion event (24). As with S100A10 and annexin A2, the enlargeosome protein AHNAK is also trafficked to the plasma membrane in response to calcium flux (25). Recruitment of AHNAK to the plasma membrane by S100A10 in complex with annexin A2 is also proposed to maintain the proper cytoarchitecture of epithelial cells (26). In vitro and in vivo experiments have shown that the C-terminal domain of AHNAK (27–29) interacts with S100A10 while bound to the N terminus of annexin A2 (26, 30).
There is little structural information about the arrangement and mechanisms used by multiprotein complexes, such as those involved in membrane repair. Interestingly, for S100A10, similar higher order complexes involving TRPV5/6, plasminogen, and the serotonin receptor have also been observed. Most of these complexes require the presence of annexin A2 (26–28, 30, 31), indicating that the S100A10-annexin A2 heterotetrameric complex might act as a scaffold to recruit additional proteins. In this work, we have used peptide array analysis to uncover the binding site(s) for the S100A10-annexin A2 complex with the C-terminal region of AHNAK. We provide evidence for a novel asymmetric interaction between the S100A10-annexin A2 dimer and AHNAK that requires contributions from both the S100A10 protein and the N terminus of annexin A2 and utilizes a surface not previously observed in any other S100 target protein complex. Overall, our work indicates a novel ternary arrangement of S100A10, annexin A2, and AHNAK that provides the first structural model of a multiprotein assemblage required for membrane repair.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors
DNA fragments encoding either rabbit S100A10 (Dr. Michael P. Walsh, University of Calgary, Alberta, Canada) or the S100A10-annexin A2 (A10A2)2 hybrid protein (32) were contained in pGEX-6P-1 vectors. QuikChange site-directed mutagenesis (33) was performed to convert the cysteine codon (Cys82) to a serine in both plasmids using forward (5′-GCCGGCCTCACCATTGCATCCAATGACTATTTTGTAGTGC-3′) and reverse (5′-GCACTACAAAATAGTCATTGGATGCAATGGTGAGGCCGGC-3′) primers. PCR products were treated with DpnI (34) restriction enzyme for 1 h at 37 °C to digest the methylated template DNA and transformed into E. coli strain JM-109. The single point mutations in the resulting vectors were confirmed by DNA sequencing.
Expression and Purification of S100A10 and A10A2
All experiments used either S100A10C82S or S100A10C82S-annexin A2 hybrid proteins, which are simply referred to as S100A10 and A10A2 in this work. Unlabeled and uniformly 15N,13C-labeled rabbit GST-S100A10 and GST-A10A2 were overexpressed in the BL21-CodonPlus(DE3)-RIL Escherichia coli strain as described previously (32) using 1 liter of 2× YT or M9 minimal medium supplemented with 1 g of 15NH4Cl and 2 g of [13C6]glucose as the sole nitrogen and carbon sources, respectively. Briefly, the cultures were grown at 37 °C in the presence of ampicillin (100 μg/ml) to a density of (A600) 0.8 absorbance units. Expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside and allowed to continue for another 4 or 8 h with constant shaking at 37 °C. Cells were harvested by centrifugation at 6,000 rpm for 15 min, lysed by French pressure at 20,000 p.s.i., and centrifuged at 38,000 rpm for 90 min. Proteins were purified using 2 × 5-ml GSTrap FF columns (GE Healthcare) connected in series at 4 °C and equilibrated in PBS buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.3). The GST-S100A10 or GST-A10A2 proteins were eluted using 50 mm Tris and 20 mm reduced glutathione at pH 8.0. Fractions containing the protein were pooled and dialyzed against PreScission protease cleavage buffer (50 mm Tris, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, pH 7.0) overnight. S100A10 or A10A2 was cleaved from the GST protein using 150 units of PreScission protease for 48 h. The cleaved protein was applied to a 5-ml GSTrap HP column in PBS buffer, and the flow-through fractions were collected. MALDI-TOF mass data for unlabeled S100A10 (MWcalc = 11,598.6; MWobs = 11,599.4) and A10A2 (MWcalc = 13,733.8; MWobs = 13,733.5) confirmed the protein identities.
Fluorescent Labeling of S100A10 and A10A2
Freshly reduced S100A10 or A10A2 was loaded onto a pre-equilibrated Sephadex G-25 PD-10 column (GE Healthcare) in 20 mm Tris-HCl and 1 mm EDTA, pH 7.0, in order to remove excess DTT in the protein samples. Acrylodan or Alexa Fluor 680 maleimide (Invitrogen) was dissolved in acetonitrile to a concentration of 100 mm. The acrylodan or Alexa Fluor 680 solution (100 μl) was added to 1 ml of 80 μm S100A10 or A10A2, and the reaction continued for 2 h at room temperature until quenched with the addition of 5 mm DTT. The unreacted dye was removed by chromatography through a Sephadex G-25 PD-10 column, which was preequilibrated in TBS buffer (50 mm Tris-HCl, 120 mm NaCl, pH 7.4). The protein samples were exhaustively dialyzed against 20 mm Tris-HCl, 120 mm NaCl, 1 mm DTT, pH 7.4, to eliminate all noncovalently linked dye. Electrospray ionization-MS indicated that a single dye molecule was covalently linked to each protein.
Peptide Array Experiments
Peptide arrays of AHNAK were produced with an Auto-Spot Robot ASP222 (Amimed) on nitrocellulose membranes using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. This array contained 330 spots of 18-residue peptides that shifted by 3 residues through the C-terminal sequence of AHNAK (residues 4884–5890). A10A2 was uniformly labeled at Cys61 with Alexa Fluor 680 maleimide as described above. Membranes were hydrated using anhydrous methanol and then washed three times with a buffer containing TBS + 0.05% Tween for 10 min at room temperature. Subsequently, membranes were incubated with TBS + 0.05% Tween and skim milk blocking buffer for 1 h, followed by a final rinse for 10 min with TBS + 0.05% Tween wash buffer. Alexa-A10A2 (0.2 μm) in TBS + 0.05% Tween and skim milk was used to probe each array for 2 h. The array was then rinsed with TBS + 0.05% Tween buffer three times for 10 min each. To eliminate background fluorescence, the array was treated three times with a solution of 8 m urea, 1% SDS, and 0.5% β-mercaptoethanol, pH 7.0, in a sonication bath at 40 °C and washed with 10% acetic acid, 50% ethanol, and 40% distilled and deionized H2O. The spots on the array were visualized using Odyssey infrared imaging system (LI-COR Biosciences) at 700 nm.
Peptide Synthesis
The AHNAK5 (GKVTFPKMKIPKFTFSGREL) and annexin A2 (STVHEILSKLSLEGD) peptides were purchased from Bio Basic Inc. (Toronto, Canada). The N termini of the peptides were acetylated, whereas the C termini were amidated. The synthesized peptides were purified by C18 reversed-phase HPLC and lyophilized. The MALDI-TOF mass data for the acetylated AHNAK5 (MWcalc = 2353.8; MWobs = 2352.5) and annexin A2 (MWcalc = 1669.8; MWobs = 1668.6) confirmed the peptide identities.
Mass Spectrometry
Mass spectrometry analyses were carried out using a quadrupole time-of-flight mass spectrometer (Q-TOF, Micromass) equipped with a nanoelectrospray (nano-ES) source. Samples were loaded on a gold-coated capillary (Protona) with the tip manually opened to produce an orifice of ∼10 μm. Positive ES was performed at a capillary voltage of 1.5–2 kV and cone voltage of 50 V. CID experiments were carried out with collision energy of 80 V using argon as the collision gas. Average molecular masses were calculated using Mass Lynx 4.0 (Micromass).
Fluorescence Spectroscopy
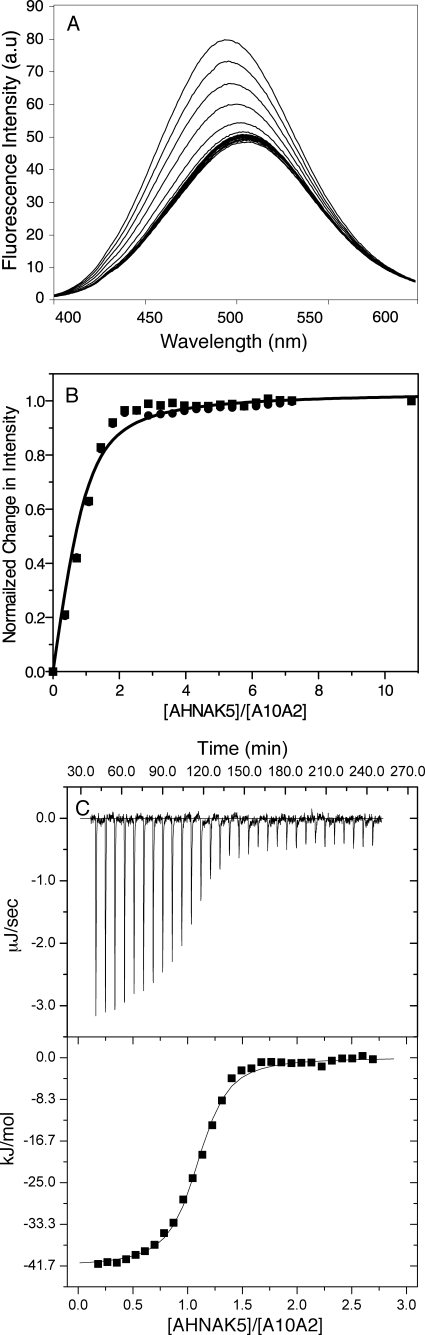
A10A2 was purified and labeled with acrylodan as described. The concentrations of the protein stock solutions were determined using the Bradford protein assay and triplicate amino acid analysis (Advanced Protein Technology Centre, Toronto, Canada). Fluorescence experiments were conducted in duplicate using acrylodan-labeled A10A2 in 20 mm Tris-HCl, 120 mm NaCl, and 1 mm DTT at pH 7.4 (3 ml). A Fluorolog-3 steady-state fluorometer (Horiba Scientific) was used for all measurements. Solutions were excited at 375 nm, and emission was monitored between 400 and 600 nm using an emission band pass of 1 nm and an integration time of 1 s.
Titrations of A10A2 (167 nm) with the AHNAK peptide were carried out at room temperature in a stirred cell holder. A solution of AHNAK peptide was added in 3-μl increments using a calibrated 10-μl Hamilton syringe followed by three 30-μl additions to ensure protein saturation. After each AHNAK addition, the sample was stirred for 2 min before scanning. The dissociation constant (35) was determined by plotting the normalized change in fluorescence (ΔF) intensity, monitored at 500 nm for acrylodan-labeled A10A2 (Pt), as a function of AHNAK concentration (Lt). Data were fit using GraphPad Prism5 to Equations 1 and 2, where ΔFmax is the maximum normalized change in fluorescence, and [PL] is the concentration of the A10A2-AHNAK complex.
 |
NMR Titration Experiments
All NMR experiments were acquired at 35 °C on a Varian INOVA 600-MHz spectrometer equipped with a pulse field gradient triple resonance probe. NMR samples of uniformly 15N,13C-labeled A10A2 (143 ± 6 μm) were prepared in 10% D2O, 20 mm MOPS, 1 mm EDTA, 1 mm DTT, 50 mm arginine, 50 mm glutamic acid, and 100 mm NaCl buffer, pH 7.0, using 2,2-dimethyl-2-silapentanesulfonic acid as an internal standard. A10A2 and AHNAK concentrations were determined by triplicate amino acid analysis (Advanced Protein Technology Centre, Toronto, Canada). AHNAK peptide was added from a stock solution (3320 ± 16.4 μm) to give a final AHNAK concentration of 584.6 μm. Samples were equilibrated for 15 min after each peptide addition. 1H-15N HSQC spectra were collected using carrier frequencies of 4.699 (1H) and 114.0 ppm (15N) and spectral widths of 8000.0 and 1700.0 Hz, respectively. All data were processed using NMRPipe and NMRDraw (36) and analyzed by NMRView (37). Intensities of peaks undergoing slow exchange were measured after each addition and fit to Equation 1 using GraphPad Prism5.
Isothermal Titration Calorimetry
The association constant of the AHNAK5 peptide to A10A2 was determined by measuring the heat of reaction during the titration of the peptide into the protein solution. Experiments were performed using a Microcal VP-ITC microcalorimeter (Microcal Inc., Northampton, MA). For the titration, a sample of 15 μm dimeric A10A2 in 20 mm Pipes (pH 7.2), 50 mm NaCl, 0.2 mm EDTA, and 1 mm TCEP was exhaustively dialyzed at 4 °C, and the AHNAK peptide was dissolved in the last dialysis buffer. Protein and peptide solutions were degassed under vacuum prior to each titration. Titrations consisted of 5-μl injections of 300 μm AHNAK5 peptide into a 1.43-ml cell containing 15 μm A10A2 protein (dimer concentration) at 35 °C. Heats of dilution were measured in a separate experiment in which the peptide was injected into the buffer alone. This heat of dilution corresponded to the average of data points after saturation in the AHNAK5-A10A2 titration and was subtracted before performing curve fitting. Base-line correction was completed by manually selecting the peak areas for integration. The dissociation constant (Kd) was obtained by fitting the data to a one-site model using Microcal Origin software. Protein and peptide concentrations were determined from triplicate amino acid analysis (Amino Acid Analysis Facility, Hospital for Sick Children, Toronto). Measurements were done in duplicate with similar results.
RESULTS
The crystal structures of calcium-S100A11 and S100A10 bound to the N-terminal regions of the phospholipid-binding proteins annexin A1 (9) and annexin A2 (8), respectively, illustrate the heterotetrameric nature of these complexes. The symmetric arrangement and stoichiometry of these complexes are characteristic of every S100 protein-target peptide structure examined to date, including calcium-bound S100B in complex with peptides from the C-terminal region of p53 (38), the N-terminal regulatory domain from the Ndr kinase (39), and the actin-capping protein CapZ (TRTK12) (40–42); calcium-bound S100A1 in complex with peptides from the cytosolic region of the ryanodine receptor (43) and CapZ (44); and calcium-bound S100A6 in complex with the C terminus of the Siah-1-interacting protein (45). These structures show that some variation in the target peptide binding site exists (46), although a symmetric arrangement is always maintained, comprising two S100 protomers and two target peptides. Structural information on the architecture of higher order complexes, such as those involving S100A10, annexin A2, and the AHNAK, are unknown and would be important to understand how this group of proteins assembles to participate in the cell membrane repair process. To examine how S100A10 and annexin A2 recruit AHNAK, we used two S100A10 complexes. The first utilized a non-covalent complex between S100A10 and a 15-residue N-terminal peptide from annexin A2. In addition, we used a hybrid protein complex (A10A2) in which S100A10 was linked in tandem to the N-terminal 15 residues of annexin A2, separated by a 9-residue spacer sequence. Previous work has shown that the A10A2 hybrid protein has an association and structure similar to those of the non-covalent S100A10-annexin A2 peptide complex (32).
A10A2 Binds to Multiple Consensus Regions on AHNAK
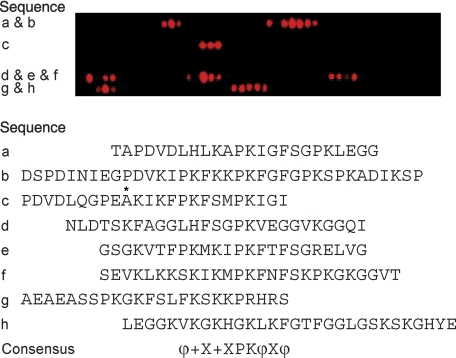
To identify the AHNAK residues involved in the interaction with annexin A2-bound S100A10 complex, Alexa Fluor 680-labeled A10A2 hybrid (Alexa-A10A2) protein was used in two separate peptide array experiments covering the C terminus of AHNAK (residues 4884–5890). This approach identifies the sequence-dependent but structurally independent interactions between A10A2 and a series of 18-residue peptides spanning the C-terminal sequence of AHNAK. Overlapping sequences in the arrays that fluoresced upon Alexa-A10A2 binding were used to identify the AHNAK binding sites (Fig. 1). Blotting of the arrays with Alexa-A10A2 revealed that A10A2 associates with eight distinct regions within the C terminus of AHNAK. Using the Blocks Server (available on the World Wide Web) to analyze these regions, a 10-residue consensus sequence was identified for AHNAK that could best be represented as ϕ+X+XPKϕXϕ (where X is variable, ϕ represents a hydrophobic residue; and + represents a positively charged residue).
FIGURE 1.
Mapping the A10A2 binding site on the C-terminal region of AHNAK by peptide array analysis. A representative peptide array of the Asp4884–Glu5890 region of AHNAK was synthesized on a cellulose membrane. Each spot contained an 18-residue peptide shifted by 3 residues from its predecessor until the C terminus was reached. The array was probed with Alexa Fluor 680-A10A2 and imaged at 700 nm. The sequences of AHNAK that showed the best interaction with A10A2 are listed beside and below the array: sequence a (Thr4917–Gly4940), sequence b (Asp4959–Pro4994), sequence c (Pro5220–Ile5243), sequence d (Asn5607–Ile5633), sequence e (Gly5652–Gly5675), sequence f (Ser5703–Thr5729), sequence g (Ala5757–Ser5780), and sequence h (Leu5808–Glu5837). These sequences were used to define a consensus sequence showing positively charged (+), hydrophobic (ϕ), and variable (X) residues using the Blocks Server (available on the World Wide Web).
Examination of the sequences that interact with A10A2 allowed for the identification of some general trends. For example, sequences b, e, and f show the highest sequence similarity, whereas the most intense binding at any one spot was observed within sequences b and e (4968–4985 and 5652–5669). Because the residues of the conserved motif for these sequences are identical, the differences in the binding intensities for A10A2 could indicate that the variable residues in the motif or residues on the periphery may be important for the interaction. Sequences a, d, and g showed weaker interactions with the A10A2 protein, probably due to one or more substitutions within the consensus region. For example, sequences a and d have a histidine at the first position instead of lysine, whereas sequences d and g have a small non-polar residue (G/S) at the last position in place of a larger hydrophobe. Sequence c is unusual due to the presence of two possible consensus sequences, the first as shown in Fig. 1 and an alternate sequence starting at Ala5229, 5 residues prior to the indicated consensus sequence.
In general, these results agree with a previous study in which the S100A10-annexin A2 binding region within the C terminus of AHNAK was mapped utilizing a series of GST-AHNAK deletion mutants by pull-down assays (30). A 20-residue region in the C terminus of AHNAK corresponding to residues 5654–5673 (lying within sequence e, residues 5652–5675) was shown to be sufficient for binding to the S100A10-annexin A2 complex. Further deletions from the N- or C-terminal extremity of this 20-residue peptide abolished the interaction with the S100A10-annexin A2 complex, confirming that these residues are crucial for the interaction (30).
Unique Interactions of AHNAK with the S100A10-Annexin A2 Complex
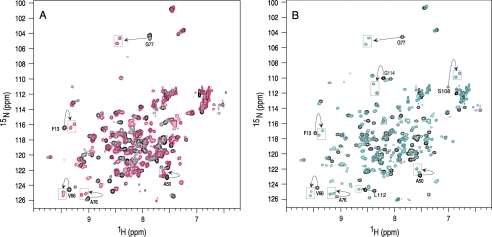
A 20-residue AHNAK peptide (5654–5673; AHNAK5) that displayed a strong interaction with A10A2 based on the peptide array experiments and was consistent with GST-AHNAK pull-down assays (30) was synthesized and used in NMR titration experiments. Initially, the AHNAK5 peptide was titrated into a non-covalent complex formed between 15N-labeled S100A10 and unlabeled annexin A2 peptides (residues 1–15) (Fig. 2A). In the absence of AHNAK5, a single resonance was observed in the 1H-15N HSQC spectrum for each residue in S100A10, consistent with the symmetric structure of the S100A10-annexin A2 complex, where only the 15N-labeled S100A10 is visible. Titration of unlabeled AHNAK5 peptide to this solution led to the gradual disappearance of many of the S100A10 resonances in the S100A10-annexin A2 complex and the reappearance of new peaks, indicative of a slow exchange binding process. Remarkably, the 1H-15N HSQC spectrum of the 15N-labeled S100A10 in the presence of annexin A2 and AHNAK5 resulted in an increased number of peaks that did not change even in the presence of a 4-fold excess of AHNAK5 peptide. In addition, many of the new resonances appeared in pairs, with each peak having ∼50% of the intensity of the original peak (Fig. 2A). For example, Phe13, Ala50, Val66, Ala76, and Gly77 in 15N-labeled S100A10 all appeared as two peaks in the 1H-15N HSQC spectrum of S100A10-annexin A2 in complex with AHNAK5.
FIGURE 2.
An overlay of 1H-15N HSQC spectra of annexin A2-bound S100A10 or A10A2 hybrid protein complexes bound to the AHNAK5 peptide. A, 1H-15N HSQC spectrum of 0.5 mm 15N-labeled S100A10 (protomer concentration) bound to 0.5 mm unlabeled annexin A2 peptide (Ac-STVHEILSLKQLEGD) (black) and in complex with 0.25 mm AHNAK5 (Ac-GKVTFPKMKIPKFTFSGREL) (pink). B, 1H-15N HSQC spectrum of 0.5 mm uniformly 15N-labeled A10A2 hybrid protein (protomer) (black) and in complex with 0.25 mm AHNAK5 peptide (cyan). Peaks that exhibited obvious multiplicity in the AHNAK5 complex are labeled according to their one-letter amino acid code and residue number. Spectra were collected on a Varian Inova 600-MHz spectrometer at 35 °C in 90% H2O, 10% D2O at pH 7.0.
The A10A2 hybrid protein has been shown to have a similar structure and interactions as the non-covalent S100A10 complex with N-terminal annexin A2 peptides (32). The A10A2 complex also offers the ability to examine the binding effects of AHNAK5 on annexin A2 using NMR titration experiments because it is also 15N-labeled in the A10A2 hybrid. As expected, the 1H-15N HSQC spectrum of A10A2 (Fig. 2B) is very similar to that of the S100A10-annexin A2 complex (Fig. 2A), indicating that the structures and interactions between the S100A10 and N terminus of annexin A2 are similar in the two complexes (32). In addition, because the annexin A2 portion of A10A2 is also 15N-labeled, resonances from this segment of the hybrid protein (i.e. Leu112 and Gly114) are clearly visible in the spectrum. Similar to the S100A10-annexin A2 peptide complex, the addition of AHNAK5 to the 15N-labeled A10A2 protein led to the disappearance of peaks from S100A10 and the appearance of new resonances on the slow exchange time scale. Upon saturation, the 1H-15N HSQC spectrum of 15N-labeled A10A2 bound to unlabeled AHNAK5 (Fig. 2B) bore a strong resemblance to that of 15N-labeled S100A10 in complex with both unlabeled annexin A2 and AHNAK5 peptides (Fig. 2A). This included an increased number of peaks in the A10A2 spectrum, where several resonances from the S100A10 protein appeared as pairs (Phe13, Ala50, Val66, Ala76, and Gly77). Interestingly, resonances from the annexin A2 portion of A10A2 also exhibited peak doubling upon the addition of AHNAK5 (Leu112 and Gly114), indicating that binding of AHNAK5 also affected the annexin A2 portion of the hybrid protein.
The increased number of resonances and multiplicity of some peaks in both the S100A10-annexin A2 and A10A2 complexes with AHNAK5 are in contrast to 1H-15N HSQC spectra of all other S100-target peptide complexes, including calcium-bound S100B with CapZ, p53, or NDR kinase; S100A6 with Siah1-interacting protein; and S100A1 in complex with CapZ or ryanodine receptor (38–41, 43–45). NMR spectra from these complexes all display a similar number of resonances in the presence or absence of target, indicating that a symmetric relationship in the dimer is maintained. This is borne out in three-dimensional structures of the complexes. In order to account for the unique spectral observations made for the S100A10-annexin A2 complex with AHNAK5, several models were considered, including displacement of annexin A2 by AHNAK5, multiple orientations of AHNAK5 binding, and asymmetric binding of a single AHNAK5 peptide to the S100A10-annexin A2 complex.
Asymmetric Binding of AHNAK to the A10A2 Dimer
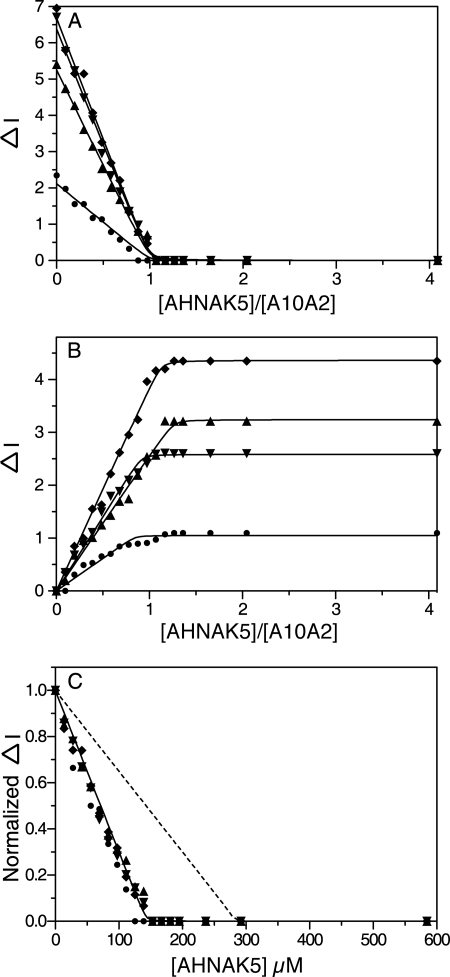
Titrations of 15N-labeled S100A10 with AHNAK5 in the absence of annexin A2 peptide did not show an interaction between S100A10 and AHNAK5 and were prone to precipitation (data not shown). This result indicates that both S100A10 and annexin A2 are required for the recruitment of AHNAK5 to the ternary complex. Analysis of the NMR experiments using 15N-labeled A10A2 titrated with unlabeled AHNAK5 peptide were carried out to investigate the stoichiometry and affinity of the interaction. The change in the peak intensities of residues from a variety of locations in S100A10 and annexin A2 moieties (Gly40, Val51, and Gly77 in S100A10 and Leu112 in annexin A2) were plotted as a function of AHNAK5 and A10A2 concentrations (Fig. 3). The data show that all of the peaks have nearly linear decreases for their intensities in 15N-labeled A10A2 (Fig. 3A) and concomitant increases in the peak intensities in the AHNAK5 complex (Fig. 3B). The inflection point for the data sets occurs near a stoichiometry of one AHNAK5 peptide to one A10A2 dimer. Fitting for this interaction indicated a Kd of <1 μm, although a precise determination was not possible by this method due to the high A10A2 protein concentrations used in the NMR experiments. The data were also fit for the interaction of two AHNAK5 peptides per A10A2 dimer, similar to other S100-target peptide complexes that display symmetric binding (Fig. 3C). This resulted in obvious disagreement with the data, indicating the 2:1 AHNAK/A10A2 (dimer) model was incorrect.
FIGURE 3.
Stoichiometry of the AHNAK5 peptide with A10A2 measured from 1H-15N HSQC experiments. Aliquots of the unlabeled AHNAK5 peptide were added to a solution of 143 μm 15N,13C-labeled A10A2 (dimer concentration), and the resulting changes in peak intensity were monitored. The binding between AHNAK5 and A10A2 is shown as the plot of decrease (A) and increase (B) in peak intensities for Gly40 (●), Val51 (♦), and Gly77 (▴) in S100A10 and Leu112 (▾) in annexin A2 as a function of the AHNAK5/A10A2 ratio. C, the normalized changes in peak intensities are shown as a function of AHNAK5 concentration. Data sets for Gly40 (●), Val51 (♦), Gly77 (▴), and Leu112 (▾) were globally fit for 1:1 (solid line) and 2:1 (dashed line) AHNAK5/A10A2 stoichiometries, indicating that the dissociation constant (Kd) was <1 μm. Data fitting is described under “Experimental Procedures.”
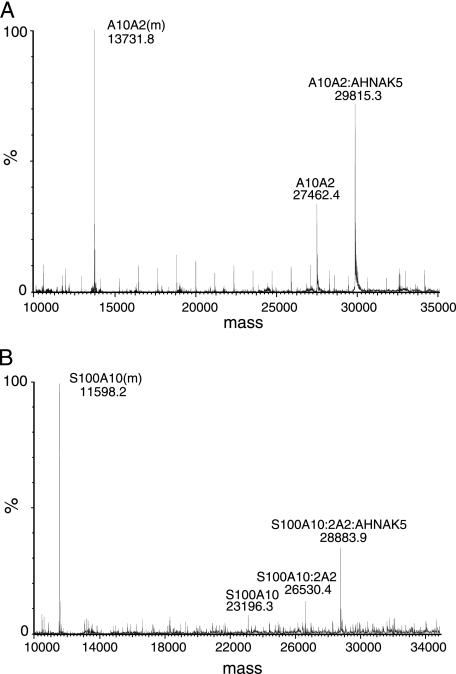
To provide further evidence for a 1:1 AHNAK/A10A2 (dimer) stoichiometry and to show that both proteins are required for the recruitment of AHNAK5, a mixture of A10A2 and AHNAK5 was subjected to non-denaturing ES-MS (Fig. 4) (47). These data showed three major peaks in the spectrum corresponding to the A10A2 protomer (MWobs = 13,731.8, MWcalc = 13,733.8), A10A2 dimer (MWobs = 27,462.4, MWcalc = 27,467.6), and the A10A2 dimer in complex with a single AHNAK5 peptide (MWobs = 29,815.5, MWcalc = 29,820.4). There was no evidence for a peak at 32,173.2 in the spectrum that would be expected for the mass of the A10A2 dimer in complex with two AHNAK5 peptides. This result corroborates the NMR titration experiments that show that only one AHNAK5 peptide is bound to the A10A2 dimer. In separate experiments, S100A10 was mixed with both the annexin A2 and AHNAK5 peptides (Fig. 4B). A representative ES-MS spectrum showed peaks representing the S100A10 protomer (MWobs = 11,598.2, MWcalc 11,598.6), S100A10 dimer (MWobs = 23,196.3, MWcalc = 23,197.2), and S100A10 dimer bound to two annexin A2 peptides (MWobs = 26530.4, MWcalc = 26,531.4). There was little evidence for a peak corresponding to an S100A10 protomer bound to a single annexin A2 peptide (MWcalc = 132,666.8), in agreement with the crystal structure that shows that the annexin A2 binding site comprises regions from both subunits of the S100A10 dimer (8). The second most intense peak in the ES-MS spectrum corresponded to the S100A10 dimer in complex with two annexin A2 peptides and a single AHNAK5 peptide (MWobs = 28,883.9, MWcalc = 28,884.2). Repeated experiments showed no evidence of complexes with masses corresponding to the S100A10 dimer complexed with either one or two AHNAK5 peptides (MWcalc = 25,549.9 and 27,901.9). Experiments using the A10A2 hybrid protein and the non-covalent complex of S100A10 with the N-terminal peptide from annexin A2 only show AHNAK5 interaction with dimeric S100A10 having bound annexin A2. This indicates that the S100A10-annexin A2 complex is a prerequisite for AHNAK interaction, in agreement with NMR experiments that showed no evidence for interaction of AHNAK5 with S100A10 in the absence of annexin A2.
FIGURE 4.
Mass spectra showing complexes of S100A10, annexin A2 peptide, and AHNAK5 peptides. A, A10A2 hybrid protein mixed with the AHNAK5 peptide, showing masses attributed to the A10A2 protomer (A10A2(m)), A10A2 dimer, and A10A2 complex with a single AHNAK5 peptide. B, S100A10 mixed with annexin A2 and AHNAK5 peptides, showing masses corresponding to the S100A10 protomer (S100A10(m)), S100A10 dimer, S100A10 dimer in complex with two annexin A2 peptides (S100A10:2A2), and S100A10 dimer in complex with two annexin A2 peptides and a single AHNAK5 peptide(S100A10:2A2:AHNAK5). Spectra were obtained under non-denaturing conditions as described under “Experimental Procedures.”
Fluorescence spectroscopy and isothermal calorimetry were used to calculate the binding affinity of AHNAK5 to the A10A2 complex and S100A10. Because AHNAK5, A10A2, and S100A10 do not possess a good intrinsic fluorescence probe, S100A10 and the A10A2 hybrid proteins were covalently labeled with acrylodan at Cys61 in the second EF-hand loop. Initial fluorescence experiments using acrylodan-labeled S100A10 in the absence of the annexin A2 peptide showed less than 12% change in fluorescence and little wavelength shift, suggestive of a poor interaction. In contrast, fluorescence experiments showed that acrylodan-labeled A10A2 had a fluorescence maximum near 495 nm (Fig. 5A) that decreased in intensity and gradually shifted to 500 nm upon the addition of AHNAK5, indicative of a much tighter interaction. The normalized change in fluorescence intensity for this titration was plotted as a function of the AHNAK5/A10A2 (dimer) ratio (Fig. 5B). Global fitting of duplicate data sets with a 1:1 ligand-binding curve yielded a dissociation constant of 35 ± 4 nm. Duplicate experiments were also conducted using isothermal titration calorimetry (Fig. 5C) as an additional measure to determine binding affinity. The titration of AHNAK5 into a solution of A10A2 yielded a exothermic peak profile having a Kd = 28 ± 2 nm for the binding of a single AHNAK5 peptide to the A10A2 dimer.
FIGURE 5.
Interaction of AHNAK5 with the A10A2 hybrid protein monitored by fluorescence spectroscopy and isothermal titration calorimetry. A, fluorescence spectra of acrylodan-A10A2 (167 nm) showing the change in acrylodan fluorescence with increasing AHNAK5 peptide concentration. B, binding curve for acrylodan-labeled A10A2 titrated with the AHNAK5 peptide showing the normalized change in fluorescence measured at 500 nm as a function of AHNAK5 concentration. Data were collected from duplicate titrations, and curves were fit globally with a 1:1 ligand binding function (solid line). C, isothermal titration calorimetry experiment for AHNAK5 peptide binding to A10A2. The spectra show the base line-corrected calorimetric titration data for 30 injections of AHNAK5 into a cell containing a solution of A10A2 (top) and the derived binding isotherm for the experiment obtained by integrating the area of each peak after each injection (bottom). The solid line represents the best fit of the data to a single binding site with n = 1.02. The fit yields Kd = 28 ± 2 nm, ΔH = −41.9 ± 0.4 kJ/mol (binding enthalpy), and ΔS = 8.4 J/mol (entropy change).
Unique Surface for AHNAK Binding to A10A2
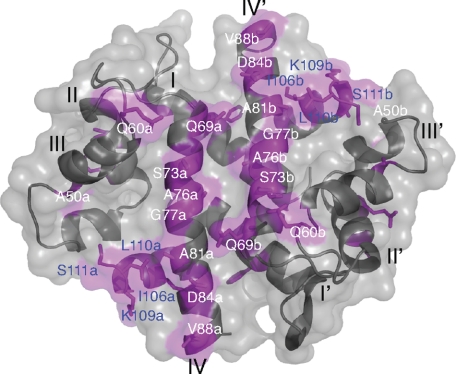
The initial backbone assignment and 1H-15N HSQC spectra of A10A2 in the absence and presence of AHNAK5 (Fig. 2B) were used to identify the binding surface for AHNAK on the S100A10-annexin A2 complex. In this analysis, we used peaks that exhibited the largest difference in pairs of chemical shifts (i.e. the difference between “doubled” peaks) in the AHNAK complex as an indicator of AHNAK binding. This was used instead on the traditional chemical shift perturbation because some pairs of peaks were such that one peak exhibited a much larger shift than the other in the complex, and our assignment was unable to distinguish asymmetric halves of the dimeric structure. Using this analysis, the residues in the A10A2 complex with AHNAK5 that showed the largest difference were mapped to the surface of the S100A10-annexin A2 structure (8). This yielded a contiguous interaction surface that included the surfaces of the central crossing portions of helix IV and IV′ (Ser73a,b, Ala76a,b, Gly77a,b, and Ala81a,b) and the extreme C termini of the annexin A2 peptides (Ile106a,b, Lys109a,b, Leu110a,b, and Ser111a,b).
DISCUSSION
Proteomic studies have indicated that large numbers of proteins are involved in membrane repair processes (4, 5). Included in this group is the EF-hand protein S100A10, the phospholipid-binding protein annexin A2, and the enlargeosome protein AHNAK, all proposed to participate in a multiprotein complex near the plasma membrane. The current work provides the first details of the architecture for a ternary complex between these three proteins.
Evidence for a Scaffolding Role of the AHNAK C Terminus
Peptide array experiments show eight potential binding regions within the C-terminal domain of AHNAK for the S100A10-annexin A2 hybrid protein (A10A2). Not surprisingly, these sequences all have some similarity, forming a consensus sequence, ϕ+X+XPKϕXϕ. The repeating nature of these sequences is not unique to the C terminus of AHNAK. The central region of the protein includes 24 repeat motifs (165 residues each), suggested to adopt a series of β-propeller structures similar to RCC1 (29, 48). The nature of the AHNAK consensus sequence differs from other S100 binding motifs, such as those found in the N terminus of annexins A1 and A2 ((A/V)XXϕLXXϕXϕ) and CapZ ((R/K)(L/I)XWXXIL). The lack of a 3–4-hydrophobic residue spacing in the AHNAK sequence suggests that it does not have amphipathic character similar to the α-helical structure found in the annexin-binding sequences. Similarly, a central proline residue would not favor helix formation upon binding. This is confirmed by the positive entropy change observed in calorimetry experiments, which suggests that a coil-helix transition in AHNAK does not occur upon binding to A10A2. The possibility for eight binding regions in the C terminus of AHNAK for the S100A10-annexin A2 complex is consistent with a scaffolding role for AHNAK. In principle, this would allow up to eight S100A10-annexin A2 heterotetramers to assemble on the C terminus. Because binding of the complex to the phospholipid surface is governed by calcium binding to the annexin moiety, this would allow a highly cooperative association of the complex with the membrane. Despite this potential, other experiments have shown that only the Gly5654–Leu5673 region within the C terminus of AHNAK displays a strong interaction with the S100A10-annexin A2 (30). This could indicate that other sequences identified in the peptide array are masked by the AHNAK tertiary structure. Similarly, the Gly5654–Leu5673 region had the strongest binding spot in the peptide array, suggesting that it could have a much stronger affinity than the other sequences.
An Asymmetric S100A10-Annexin A2-AHNAK Complex
A representative sequence (AHNAK5) identified from the peptide array was used to identify the arrangement of the S100A10-annexin A2-AHNAK ternary complex. Using both the A10A2 hybrid protein and complex of S100A10 with an N-terminal peptide of the annexin A2, a single AHNAK5 peptide was found to coordinate in an asymmetric fashion with the S100A2-annexin A2 heterotetramer. Although it is a ternary complex, an asymmetric arrangement within an S100-target protein complex has not been observed previously. For example, multiple S100-peptide complexes, such as those between S100B and CapZ (40–42), p53 (38), or Ndr kinase (39); between S100A1 and ryanodine receptor (43) and CapZ (44); and between S100A6 and Siah1-interacting protein (45) all display symmetric arrangements binding two target proteins per S100 dimer. In the S100A10-annexin A2 complex, the addition of AHNAK acts to break the symmetry of the heterotetramer. In contrast, the EF-hand calcium-binding protein calmodulin can form asymmetric complexes with the dimeric targets glutamate decarboxylase and Cdc42-WASP. In these cases, binding of calcium-activated calmodulin to the symmetric, dimeric arrangements of glutamate decarboxylase and Cdc42-WASP results in an overall asymmetric arrangement essential for enzyme activity (49–51).
In the A10A2-AHNAK5 complex, significant changes in chemical shift and peak doubling occur mainly for residues in helix IV of the S100A10 (Ser73, Ala76, Gly77, and Ala81) as well as the N terminus of the annexin A2 (Ser111, Leu112, and Gly114) peptides. In the S100A10-annexin A2 structure (8), helices IV and IV′ from S100A10 form a portion of the dimer interface oriented roughly 180° from each other on one face of the protein The two annexin A2 peptides in the heterotetramer are both oriented such that their C termini lie on the same side of the complex as helices IV and IV′ (Fig. 6). Thus, the interaction surface of AHNAK5 with S100A10-annexin A2 is consistent with binding that utilizes one face of the dimeric protein comprising helices IV and IV′ and the adjacent annexin C termini (Fig. 6). This surface for AHNAK is unique compared with other S100 target protein complexes. For example, complexes between Ca2+-S100B and p53 or Ndr kinase (38, 39) and between Ca2+-S100A6 and Siah1-interacting protein (45) utilize the cleft between helices III and IV (III′ and IV′) to recruit these proteins. This is the region that undergoes a large conformational change upon calcium binding. In these structures, only the extreme C terminus of helix IV (IV′) is involved in the interaction with the target protein. In contrast, the AHNAK surface utilizes residues in the center of helix IV, in addition to the bound annexin sequence that the other S100 complexes do not possess. Because only a single AHNAK5 peptide interacts with the S100A10-annexin A2 complex, it is envisioned that the AHNAK species will lie across the top of helices IV and IV′ in the complex and interact on either side with the protruding C termini of the bound annexin species.
FIGURE 6.
Binding surface for AHNAK5 on the S100A10-annexin A2 complex. Residues that exhibited the largest differences in chemical shift for pairs of peaks, in the bound AHNAK5 complex with A10A2 hybrid protein, were mapped to the surface of S100A10-annexin A2 derived from its three-dimensional crystal structure (8). Residues that showed the largest changes are shown in magenta and labeled in S100A10 (white) and annexin A2 (blue). Each residue label is followed by either a or b to denote the asymmetric relationship of the dimer caused by AHNAK5 binding. Helices are labeled I–IV and I′–IV′ for each protomer.
The tight binding between AHNAK5 and S100A10-annexin A2 (∼30 nm) requires initial formation of the heterotetramer because experiments lacking the annexin A2 peptide yielded little or no observable interaction. This observation is in excellent agreement with yeast triple-hybrid and in vitro binding assays that show that both S100A10 and annexin A2 are required for strong association with the C-terminal domain of AHNAK (30). This requirement indicates that there are two calcium-regulated steps for AHNAK delivery to the membrane surface, both mediated by annexin A2. The first is the extrusion of N terminus of annexin A2 upon calcium binding to the protein core domain, allowing formation of the heterotetramer and subsequent recruitment of AHNAK. The second step is the localization of the entire complex to the phospholipid surface through calcium bridging between annexin A2 and the membrane. It is interesting that several other proteins have also shown an association with the S100A10-annexin A2 heterotetramer, including TPRV5/6 (27, 28), TASK-1 (52, 53), and NS3 (54). Although the calcium regulation of these interactions is less well known, it is possible that the asymmetric interaction proposed here for AHNAK is a general feature for trafficking of these proteins to the cell membrane.
S100A10, annexin A2, and AHNAK have been identified as binding partners of dysferlin, a central protein in the membrane repair complex (55). This multiprotein complex is proposed to facilitate wound repair of damaged epithelial, auditory, and muscle cells upon extracellular calcium influx. The association of annexins A1 and A2 with a dysferlin-containing vesicle and damaged plasma membranes (56–58) is calcium-dependent. The C terminus of AHNAK has also been shown to interact with the N-terminal C2A domain of dysferlin (amino acids 2–130) (55). Further, proteomic analyses have identified both annexin A2 and AHNAK as members of the dysferlin complex (4, 5). Consequently, it is possible that the interaction between S100A10, annexin A2, and dysferlin is mediated through AHNAK, the largest protein within the complex. The identification of a ternary asymmetric complex between S100A10-annexin A2 and AHNAK in this work provides an initial framework for understanding this intricate assembly of these membrane repair proteins.
Acknowledgments
We thank Paula Pittock, Suya Liu, and Dr. Gilles Lajoie in the University of Western Ontario Biological Mass Spectrometry Laboratory for expertise in acquiring and analyzing mass spectral data and Dr. Brian Dempsey for critical reading of the manuscript. We also thank Dr. Shawn Li for help and advice regarding the peptide arrays used in this work.
This work was supported by grant FRN #95320 from the Canadian Institutes of Health Research (to G. S. S.) and the Canada Research Chairs Program (to G. S. S.) and an Ontario Graduate Student Scholarship (to A. R.).
- A10A2
- S100A10-annexin A2 hybrid protein
- AHNAK5
- peptide comprising AHNAK residues Gly5654–Leu5673
- acrylodan
- 6-acryloyl-2-dimethylaminonaphthalene
- HSQC
- heteronuclear single quantum coherence
- ES
- electrospray.
REFERENCES
- 1. Zhang Z. Q., Wietgrefe S. W., Li Q., Shore M. D., Duan L., Reilly C., Lifson J. D., Haase A. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5640–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lennon N. J., Kho A., Bacskai B. J., Perlmutter S. L., Hyman B. T., Brown R. H., Jr. (2003) J. Biol. Chem. 278, 50466–50473 [DOI] [PubMed] [Google Scholar]
- 3. Umbrecht-Jenck E., Demais V., Calco V., Bailly Y., Bader M. F., Chasserot-Golaz S. (2010) Traffic 11, 958–971 [DOI] [PubMed] [Google Scholar]
- 4. de Morrée A., Hensbergen P. J., van Haagen H. H., Dragan I., Deelder A. M., 't Hoen P. A., Frants R. R., van der Maarel S. M. (2010) PLoS One 5, e13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cacciottolo M., Belcastro V., Laval S., Bushby K., di Bernardo D., Nigro V. (2011) J. Biol. Chem. 286, 5404–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rintala-Dempsey A. C., Santamaria-Kisiel L., Liao Y., Lajoie G., Shaw G. S. (2006) Biochemistry 45, 14695–14705 [DOI] [PubMed] [Google Scholar]
- 7. Smith S. P., Shaw G. S. (1998) Biochem. Cell Biol. 76, 324–333 [DOI] [PubMed] [Google Scholar]
- 8. Réty S., Sopkova J., Renouard M., Osterloh D., Gerke V., Tabaries S., Russo-Marie F., Lewit-Bentley A. (1999) Nat. Struct. Biol. 6, 89–95 [DOI] [PubMed] [Google Scholar]
- 9. Rety S., Osterloh D., Arie J. P., Tabaries S., Seeman J., Russo-Marie F., Gerke V., Lewit-Bentley A. (2000) Structure Fold. Des. 8, 175–184 [DOI] [PubMed] [Google Scholar]
- 10. Emans N., Gorvel J. P., Walter C., Gerke V., Kellner R., Griffiths G., Gruenberg J. (1993) J. Cell Biol. 120, 1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harder T., Kellner R., Parton R. G., Gruenberg J. (1997) Mol. Biol. Cell 8, 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jost M., Zeuschner D., Seemann J., Weber K., Gerke V. (1997) J. Cell Sci. 110, 221–228 [DOI] [PubMed] [Google Scholar]
- 13. Diakonova M., Gerke V., Ernst J., Liautard J. P., van der Vusse G., Griffiths G. (1997) J. Cell Sci. 110, 1199–1213 [DOI] [PubMed] [Google Scholar]
- 14. Merrifield C. J., Rescher U., Almers W., Proust J., Gerke V., Sechi A. S., Moss S. E. (2001) Curr. Biol. 11, 1136–1141 [DOI] [PubMed] [Google Scholar]
- 15. Sarafian T., Pradel L. A., Henry J. P., Aunis D., Bader M. F. (1991) J. Cell Biol. 114, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harder T., Gerke V. (1993) J. Cell Biol. 123, 1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knop M., Aareskjold E., Bode G., Gerke V. (2004) EMBO J. 23, 2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ayala-Sanmartin J., Zibouche M., Illien F., Vincent M., Gallay J. (2008) Biochim. Biophys. Acta 1778, 472–482 [DOI] [PubMed] [Google Scholar]
- 19. Illien F., Finet S., Lambert O., Ayala-Sanmartin J. (2010) Biochim. Biophys. Acta. 1798, 1790–1796 [DOI] [PubMed] [Google Scholar]
- 20. Zibouche M., Vincent M., Illien F., Gallay J., Ayala-Sanmartin J. (2008) J. Biol. Chem. 283, 22121–22127 [DOI] [PubMed] [Google Scholar]
- 21. Rosengarth A., Gerke V., Luecke H. (2001) J. Mol. Biol. 306, 489–498 [DOI] [PubMed] [Google Scholar]
- 22. Rosengarth A., Luecke H. (2003) J. Mol. Biol. 326, 1317–1325 [DOI] [PubMed] [Google Scholar]
- 23. Swairjo M. A., Concha N. O., Kaetzel M. A., Dedman J. R., Seaton B. A. (1995) Nat. Struct. Biol. 2, 968–974 [DOI] [PubMed] [Google Scholar]
- 24. Gerke V., Moss S. E. (2002) Physiol. Rev. 82, 331–371 [DOI] [PubMed] [Google Scholar]
- 25. Kouno M., Kondoh G., Horie K., Komazawa N., Ishii N., Takahashi Y., Takeda J., Hashimoto T. (2004) J. Invest. Dermatol. 123, 700–707 [DOI] [PubMed] [Google Scholar]
- 26. Benaud C., Gentil B. J., Assard N., Court M., Garin J., Delphin C., Baudier J. (2004) J. Cell Biol. 164, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van de Graaf S. F., Hoenderop J. G., Gkika D., Lamers D., Prenen J., Rescher U., Gerke V., Staub O., Nilius B., Bindels R. J. (2003) EMBO J. 22, 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borthwick L. A., Neal A., Hobson L., Gerke V., Robson L., Muimo R. (2008) Cell Calcium 44, 147–157 [DOI] [PubMed] [Google Scholar]
- 29. Shtivelman E., Bishop J. M. (1993) J. Cell Biol. 120, 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Seranno S., Benaud C., Assard N., Khediri S., Gerke V., Baudier J., Delphin C. (2006) J. Biol. Chem. 281, 35030–35038 [DOI] [PubMed] [Google Scholar]
- 31. MacLeod T. J., Kwon M., Filipenko N. R., Waisman D. M. (2003) J. Biol. Chem. 278, 25577–25584 [DOI] [PubMed] [Google Scholar]
- 32. Rezvanpour A., Phillips J. M., Shaw G. S. (2009) Protein Sci. 18, 2528–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang W., Malcolm B. A. (1999) BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 34. Nebgen D. R., Inoue H., Sabsay B., Wei K., Ho C. S., Veis A. (1999) J. Dent. Res. 78, 1484–1494 [DOI] [PubMed] [Google Scholar]
- 35. Morigasaki S., Li F., Kawai A., Yamazaki K., Sikdar D., Hibino Y., Hiraga K. (2000) Biochem. Biophys. Res. Commun. 273, 261–266 [DOI] [PubMed] [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37. Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 38. Rustandi R. R., Baldisseri D. M., Weber D. J. (2000) Nat. Struct. Biol. 7, 570–574 [DOI] [PubMed] [Google Scholar]
- 39. Bhattacharya S., Large E., Heizmann C. W., Hemmings B., Chazin W. J. (2003) Biochemistry 42, 14416–14426 [DOI] [PubMed] [Google Scholar]
- 40. Inman K. G., Yang R., Rustandi R. R., Miller K. E., Baldisseri D. M., Weber D. J. (2002) J. Mol. Biol. 324, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 41. McClintock K. A., Shaw G. S. (2003) J. Biol. Chem. 278, 6251–6257 [DOI] [PubMed] [Google Scholar]
- 42. Charpentier T. H., Thompson L. E., Liriano M. A., Varney K. M., Wilder P. T., Pozharski E., Toth E. A., Weber D. J. (2010) J. Mol. Biol. 396, 1227–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wright N. T., Prosser B. L., Varney K. M., Zimmer D. B., Schneider M. F., Weber D. J. (2008) J. Biol. Chem. 283, 26676–26683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright N. T., Cannon B. R., Wilder P. T., Morgan M. T., Varney K. M., Zimmer D. B., Weber D. J. (2009) J. Mol. Biol. 386, 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee Y. T., Dimitrova Y. N., Schneider G., Ridenour W. B., Bhattacharya S., Soss S. E., Caprioli R. M., Filipek A., Chazin W. J. (2008) Biochemistry 47, 10921–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rezvanpour A., Shaw G. S. (2009) Gen. Physiol. Biophys. 28, F39–F46 [PubMed] [Google Scholar]
- 47. Loo J. A. (2000) Int. J. Mass Spectrom. 200, 175–186 [Google Scholar]
- 48. Shtivelman E., Cohen F. E., Bishop J. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5472–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yap K. L., Yuan T., Mal T. K., Vogel H. J., Ikura M. (2003) J. Mol. Biol. 328, 193–204 [DOI] [PubMed] [Google Scholar]
- 50. Yuan T., Vogel H. J. (1998) J. Biol. Chem. 273, 30328–30335 [DOI] [PubMed] [Google Scholar]
- 51. Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. (2000) Nature 404, 151–158 [DOI] [PubMed] [Google Scholar]
- 52. Girard C., Tinel N., Terrenoire C., Romey G., Lazdunski M., Borsotto M. (2002) EMBO J. 21, 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Renigunta V., Yuan H., Zuzarte M., Rinné S., Koch A., Wischmeyer E., Schlichthörl G., Gao Y., Karschin A., Jacob R., Schwappach B., Daut J., Preisig-Müller R. (2006) Traffic 7, 168–181 [DOI] [PubMed] [Google Scholar]
- 54. Beaton A. R., Rodriguez J., Reddy Y. K., Roy P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13154–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang Y., Laval S. H., van Remoortere A., Baudier J., Benaud C., Anderson L. V., Straub V., Deelder A., Frants R. R., den Dunnen J. T., Bushby K., van der Maarel S. M. (2007) FASEB J. 21, 732–742 [DOI] [PubMed] [Google Scholar]
- 56. Doherty K. R., McNally E. M. (2003) Trends Mol. Med. 9, 327–330 [DOI] [PubMed] [Google Scholar]
- 57. Glover L., Brown R. H., Jr. (2007) Traffic 8, 785–794 [DOI] [PubMed] [Google Scholar]
- 58. Han R., Campbell K. P. (2007) Curr. Opin. Cell Biol. 19, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]