Background: MicroRNA-138 deregulation is a common event in cancer.
Results: Our study describes a microRNA regulatory module consisting of tumor suppressor miR-138 and proto-oncogene FOSL1.
Conclusion: The deregulation of miR-138-FOSL1 regulatory module may play an important role in cancer initiation and progression.
Significance: Our results suggest that microRNAs target both canonical and non-canonical targeting sites located in all areas of mRNA molecules.
Keywords: AP1 Transcription Factor, Epithelial Mesenchymal Transition, MicroRNA, RNA Interference (RNAi), Tumor Suppressor Gene, FOSL1, microRNA Target Prediction, MicroRNA-138, Squamous Cell Carcinoma
Abstract
MicroRNA-138 is one of the most frequently down-regulated microRNAs in cancer. We recently identified 51 candidate targets of microRNA-138 (Jiang, L., Dai, Y., Liu, X., Wang, C., Wang, A., Chen, Z., Heidbreder, C. E., Kolokythas, A., and Zhou, X. (2011) Hum. Genet. 129, 189–197). Among these candidates, Fos-like antigen 1 (FOSL1) is a member of Fos gene family and is a known proto-oncogene. In this study, we first confirmed the microRNA-138-mediated down-regulation of FOSL1 in squamous cell carcinoma cell lines. We then demonstrated the effect of this microRNA-138-FOSL1 regulatory module on downstream genes (homolog of Snail 2 (Snai2) expression and the Snai2-mediated repression of E-cadherin expression), as well as its contributions to tumorigenesis. The microRNA-138-directed recruitment of FOSL1 mRNA to the RNAi-induced silencing complex was confirmed by a ribonucleoprotein-immunoprecipitation assay. Three canonical and three high affinity non-canonical microRNA-138 (miR-138) targeting sites were identified on the FOSL1 mRNA: one in the 5′-UTR, three overlapping sites in the coding sequences, and two overlapping sites in the 3′-UTR. The direct targeting of miR-138 to these sites was confirmed using luciferase reporter gene assays. In summary, we describe an important microRNA regulatory module, which may play an important role in cancer initiation and progression. Our results also provide evidence that microRNAs target both canonical and non-canonical targeting sites located in all areas of the mRNA molecule (e.g. 5′-UTR, coding sequences, and 3′-UTR).
Introduction
MicroRNA deregulation is a frequent event in cancer. Several microRNAs have been functionally classified as proto-oncogenes or tumor suppressors. Deregulation (e.g. overexpression or loss of expression) of these “cancerous” microRNAs plays critical roles in tumor initiation and progression (1, 2). The deregulation of miR-138 4 has been previously observed in a number of cancer types, including head and neck squamous cell carcinoma (HNSCC) (3, 4), thyroid cancer (5, 6), and lung cancer (7). Two miR-138 precursor genes, termed pre-miR-138-1 and pre-miR-138-2, were initially identified in the mouse genome (8), and their human homologs were mapped to chromosome 3p21.33 and 16q13, respectively. Interestingly, loss of heterozygosity at both chromosome loci has been frequently observed in HNSCC (9–11). Our recent study demonstrated that a reduced miR-138 level is associated with enhanced cell growth and invasion in HNSCC (12, 13). Potential roles of miR-138 deregulation in chemoresistance have also been suggested (14–16).
We previously identified 194 genes that were significantly down-regulated by miR-138 (based on microarray analysis on HNSCC cells that were ectopically transfected with miR-138) (17). Bioinformatics analysis identified miR-138 targeting sites in the 3′-UTR of 51 out of 194 genes. Among these candidates, FOSL1 is one of the most significantly down-regulated genes upon ectopic transfection of miR-138 into the HNSCC cell line. FOSL1 is a member of Fos gene family and an important proto-oncogene that has been implicated in the initiation and progression of various types of cancers. This represents an important microRNA regulatory module consisting of a candidate tumor suppressor microRNA-138 and a proto-oncogene FOSL1. However, only one miR-138 targeting site was predicted in a non-conserved region in the 3′-UTR of the FOSL1 mRNA based on six commonly used bioinformatics tools (4-way PicTar, 5-way PicTar, TargetScanS, TargetScanHuman 5.1, the miRanda database through the Memorial Sloan-Kettering Cancer Center, and miRanda at miRBase). This renders FOSL1 a “weak candidate” for miR-138-mediated post-transcriptional regulation and is not consistent with the wet laboratory-based observation. It is worth knowing that the microRNA targeting prediction tools described above are limited to the 3′-UTR of the mRNA sequence and do not consider possible non-canonical targeting sites (e.g. allowing for G:U wobble base pairing). The present study is motivated by this lack of consistency between the experimental observations and the bioinformatics prediction using currently available tools, and we aim to functionally test the microRNA-138-FOSL1 regulatory module.
MATERIALS AND METHODS
Cell Culture and Transfection
Human HNSCC cell lines (UM1, 1386Ln, and 686Tu) used in this study were maintained in DMEM/F12 supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C in a humidified incubator containing 5% CO2. For functional analysis, hsa-miR-138 mimic and control microRNA mimic (cel-miR-67 from Caenorhabditis elegans) (Dharmacon), locked nucleic acid knockdown probe specific to miR-138 (anti-miR-138 LNA), negative control LNA (Scramble-miR control) (Exiqon), and gene-specific siRNAs (On-TargetPlus SMARTpool, Dharmacon) were transfected into cells using DharmaFECT Transfection Reagent 1 as described previously (12, 13).
Western Blot Analysis
Western blots were performed as described previously (13) using antibodies specific for FOSL1, Fos, FosB, Snai2 (Cell Signaling), E-cadherin (BD Biosciences), FOSL2, and β-actin (Sigma-Aldrich).
Fluorescent Immunocytochemical Analysis
Immunofluorescence analysis was performed as described previously (13). In brief, cells were cultured on eight-chamber glass slides (BD Biosciences), fixed, permeabilized, and blocked as described. The slides were incubated with primary antibodies against FOSL1 (1:100) (Cell Signaling) or E-cadherin (1:100) (BD Biosciences) and then incubated with a FITC-conjugated anti-rabbit IgG antibody (1:50, Santa Cruz Biotechnology) or Alexa Fluor 594 goat anti-mouse IgG antibody (1: 400, Invitrogen). The slides were mounted with ProLong Gold antifade reagent containing DAPI (Invitrogen) and examined with a fluorescence microscope (Carl Zeiss).
Dual-Luciferase Reporter Assay
Two reporter constructs were used to test the effect of miR-138 on gene promoters containing activator protein 1 (AP-1) sites. The first reporter construct was obtained from SABiosciences (Cignal AP1 reporter kit, a Dual-Luciferase assay kit), which is a luciferase construct containing a synthetic promoter with multiple AP-1 sites. The second reporter construct contains a 1155-bp fragment of the Snai2 promoter that contains a previously identified AP-1 site (18). The Snai2 promoter construct was a gift from Dr. Shuang Huang at the Department of Biochemistry and Molecular Biology, Georgia Health Sciences University. The reporter constructs and the pRL-TK vector (Promega) were co-transfected using Lipofectamine 2000 (Invitrogen). The Dual-Luciferase assays were then performed as described previously (13) using a GloMax 20/20 luminometer (Promega). Experiments were performed in quadruplicate.
The luciferase reporter gene construct containing the miR-138 targeting site from the 5′-UTR of FOSL1 (E1) was created by cloning a 40-bp fragment (positions 142–181, NM_005438, containing the miRNA-138 binding site E1) into the NcoI site on the 5′-UTR of the luciferase gene in the pGL3-Control firefly luciferase reporter vector (Promega). The luciferase reporter gene constructs containing miR-138 targeting sites from the CDs and 3′-UTR of FOSL1 were created by cloning a 71-bp fragment from the CDs (positions 713–783, NM_005438, containing the miRNA-138 binding sites E2, E3, and E4), and a 58-bp fragment from the 3′-UTR (positions 1389–1446, NM_005438, containing the miRNA-138 binding sites E5 and E6) of the FOSL1, respectively, into the XbaI site on the 3′-UTR of the luciferase gene in the pGL3-Control firefly luciferase reporter vector (Promega). The corresponding mutant constructs were created by replacing the seed regions (positions 2–8) of the miR-138 binding sites with 5′-TTTTTTT-3′. All constructs were verified by sequencing. The reporter constructs and the pRL-TK vector (Promega) were co-transfected using Lipofectamine 2000 (Invitrogen). The luciferase activities were then determined as described previously (13) using a GloMax 20/20 luminometer (Promega). Experiments were performed in quadruplicate.
Ribonucleoprotein-IP (RIP-IP) Assay and Quantitative RT-PCR Analysis
RIP-IP assays were performed as described previously with minor modifications (19, 20). In brief, cells were co-transfected with a pIRESneo-FLAG/HA-Ago2 expression vector (Addgene plasmid 10822, Addgene Inc.) and hsa-miR-138 mimic or non-targeting microRNA mimic (Dharmacon). 48 h after transfection, cells were washed and lysed in radioimmune precipitation buffer (Sigma) containing 10% proteinase inhibitor mixture (Sigma), 1 mm PMSF (Fluka), and 100 units/ml SUPERase·In (Ambion) followed by a 15-s sonication for three times on ice. The samples were then subjected to centrifugation for 30 min at 14,000 rpm, and the supernatants were collected. A fraction of the whole cell lysate was saved for Western blot and RNA isolation, and the remaining lysate was subjected to immunoprecipitation (IP) using anti-FLAG M2 affinity gel (Sigma). RNA from the whole cell lysate and the RIP-IP fraction was extracted with QIAzol and purified by miRNeasy mini kit (Qiagen).
The relative levels of the FOSL1 mRNA were examined using a quantitative two-step RT-PCR assay as described previously (21) with specific primer sets for the 5′-UTR (sense, 5′-CAGCCGTGTACCCCGCAGAG-3′; antisense, 5′-CGAAGTCTCGGAACATGCCCGG-3′), the CD region (sense, 5′-CCAAGGAGGGGGACACAGGCA-3′; antisense, 5′-CAAGGTACAGGGCGGCAGGG-3′), and the 3′-UTR (sense, 5′-TGCCACTTTACCCACCTAGAACACT-3′; antisense, 5′-CCTGGTCCAATCACCTGCTGCT-3′). The specificity of PCR reactions was confirmed by melting curve analysis. The relative expression level was computed using the 2−ΔΔCt analysis method, where actin was used as an internal reference (22). The relative enrichment of mRNA in the RIP-IP fractions was computed based on the ratio of relative mRNA levels in the RIP-IP fractions and the relative mRNA levels in the whole cell lysates as described previously (19).
RESULTS AND DISCUSSION
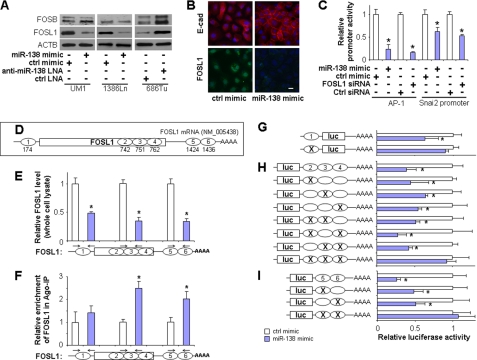
Our previous study suggested that restoring miR-138 in the HNSCC cell line UM1 led to a significant reduction of FOSL1 mRNA (17). As illustrated in Fig. 1A, we confirmed that ectopic transfection of miR-138 led to significant down-regulation of FOSL1 at the protein level in UM1 and 1386Ln cells, a previously established HNSCC cell line with a low miR-138 level (12). Knockdown of miR-138 in 686Tu cells (HNSCC cell line with high miR-138) by anti-miR-138 LNA enhanced FOSL1 expression. The miR-138-induced changes in FOSL1 expression were also confirmed at the mRNA level by quantitative RT-PCR (supplemental Fig. 1). We also examined the expression of FOS, FOSB, and FOSL2, three additional members of the Fos gene family (Fig. 1A). Although FOS and FOSL2 were not expressed to any significant extent under basal conditions, a slight decrease and an apparent increase of FOSB levels were observed in 1386Ln cells treated with miR-138 mimic and in 686Tu cells treated with anti-miR-138 LNA, respectively. As shown in Fig. 1B and supplemental Fig. 2, the miR-138-mediated down-regulation of FOSL1 was also confirmed by immunofluorescence analysis in UM1 cells. Furthermore, an apparent inverted correlation of miR-138 and FOSL1 levels was observed in HNSCC tissue samples (supplemental Fig. 3). The reduction of FOSL1 level was accompanied by enhanced expression of E-cadherin (Fig. 1B and supplemental Fig. 4). These miR-138-mediated expression changes were accompanied by functional changes at the cellular level. Although miR-138- or siRNA-mediated down-regulation of FOSL1 was accompanied by reduced proliferation, cell migration, and invasion and enhanced apoptosis, the anti-miR-138 LNA-mediated up-regulation of FOSL1 led to enhanced proliferation, reduced apoptosis, and enhanced cell migration and invasion (supplemental Fig. 5). This finding is in agreement with our previous observation (12) and is in agreement with the notion that FOSL1 is essential to proliferation, apoptosis, cell motility, invasion, and metastasis.
FIGURE 1.
miR-138 direct targeting of FOSL1 mRNA. A, Western blot analyses were performed to examine the effects of miR-138 on FOS, FOSB, FOSL1, and FOSL2 gene expression at the protein level on UM1 and 1386Ln cells treated with either miR-138 mimic or negative control mimic (ctrl mimic) and 686Tu cells treated with either anti-miR-138 LNA or negative control LNA (ctrl LNA). B, the expression changes in FOSL1 and E-cadherin (E-cad) were measured by fluorescent immunocytochemical analysis in UM1 cells treated with either miR-138 mimic or negative control mimic (green, FOSL1; red, E-cadherin; blue, DAPI nuclear staining; scale bar = 5 μm). C, Dual-Luciferase reporter assays were performed to test the effect of miR-138 and FOSL1 on gene promoter activities using constructs containing a synthetic promoter containing multiple AP-1 sites and the SNAI2 gene promoter. D, the relative location of six predicted miR-138 targeting sequences located in the FOSL1 mRNA. The numbers under the diagram indicate the bp locations of the starts of the seed sequences in the FOSL1 mRNA. RIP-IP assays were performed to co-immunoprecipitate the Ago2 complexes from cells transfected with either miR-138 mimic or negative control mimic. E, quantitative RT-PCR assays specific to the 5′-UTR, CDs, and 3′-UTR were performed on RNA samples isolated from the whole cell lysates to quantify the cellular FOSL1 mRNA levels. F, quantitative RT-PCR assays specific to the 5′-UTR, CDs, and 3′-UTR were performed on RNA samples isolated from the Ago2 co-IP (Ago-IP) fractions to measure the relative enrichment of the FOSL1 mRNA. G–I, Dual-Luciferase reporter assays were performed to test the interaction of miR-138 and its targeting sequences in the 5′-UTR (G), CDs (H), and 3′-UTR (I) of the FOSL1 mRNA using constructs containing the predicted targeting sequences and mutated targeting sequences. Data represent at least three independent experiments with similar results. Error bars indicate S.E. *, p < 0.05. luc, luciferase.
FOSL1 exerts its proto-oncogene function by dimerizing with proteins of the Jun family to form the AP-1 complex, a transcription factor that controls critical cellular processes including differentiation, proliferation, and apoptosis. To assess the functional relevance of miR-138-mediated down-regulation of FOSL1, we tested the effect of miR-138 on gene promoters containing AP-1 sites (a synthetic promoter containing multiple AP-1 sites and the Snai2 gene promoter, which contains a previously described AP-1 binding site (18)). Overexpression of Snai2 is frequently observed in HNSCC and is associated with epithelial-to-mesenchymal transition (EMT) and metastasis (23). As shown in Fig. 1C, significant reductions in promoter activities were observed for both promoters when cells were treated with miR-138 mimics. Similar results were observed when cells were treated with siRNA specific against FOSL1. The miR-138-mediated down-regulation of Snai2 was further confirmed at the protein level (supplemental Fig. 4). These findings are consistent with our recent observation that miR-138 regulates EMT, in part, by regulating Snai2 expression, which in turn controls E-cadherin expression (24). However, no miR-138 targeting site was identified in the Snai2 mRNA. Taken together, our results suggest a novel paradigm in which miR-138-mediated Snai2 down-regulation is achieved through down-regulation of FOSL1, which in turn leads to the reduced promoter activity of the Snai2 gene. The reduction of Snai2, a transcription repressor, then leads to enhanced E-cadherin expression, which prevents EMT and cancer progression.
Using commonly available bioinformatics tools (4-way PicTar, 5-way PicTar, TargetScanS, TargetScanHuman 5.1, the miRanda database through the Memorial Sloan-Kettering Cancer Center, and miRanda at miRBase), a canonical miR-138 targeting site was identified in the 3′-UTR of the FOSL1 mRNA in a region that is not highly conserved (17). Because several microRNA targeting sites have recently been reported in the 5′-UTR (25–27) and CDs (24, 28–30), we extended our search to the entire mRNA molecule. Two additional canonical targeting sites were located in the CDs. When G:U wobble base pairing was considered, three additional high affinity (based on predicted minimum free energy) non-canonical targeting sites were identified: one each in the 5′-UTR, CDs, and 3′-UTR of the FOSL1 mRNA (supplemental Fig. 6). As such, a total of six miR-138 targeting sites were identified (Table 1 and Fig. 1D), one in the 5′-UTR, three in the CDs, and two in the 3′-UTR of the FOSL1 mRNA. It is worth noting that the three miR-138 targeting sites located in CDs are partially overlapped, and the two sites in the 3′-UTR are also overlapped (supplemental Fig. 7). This provided us with a unique opportunity to test microRNA targeting sites located in different regions of the mRNA molecule.
TABLE 1.
Predicted hsa-miR-138 targeting sites on FOLS1 mRNA
The accession number for the FOSL1 mRNA sequences used in this analysis is NM_005438.3.
| Positiona | Location | Adjacent siteb | Seed seq. c | Typed | Predicted mfee |
|---|---|---|---|---|---|
| 174 | 5′-UTR | CgCCAGC | Non-canonical | −25.1 kcal/mol | |
| 742 | CDS | Yes | uACCAGu | Non-canonical | −23.7 kcal/mol |
| 751 | CDS | Yes | CACCAGC | Canonical | −26.3 kcal/mol |
| 762 | CDS | Yes | CACCAGC | Canonical | −25.4 kcal/mol |
| 1424 | 3′-UTR | Yes | CACCAGC | Canonical | −22.6 kcal/mol |
| 1436 | 3′-UTR | Yes | ugCCAGC | Non-canonical | −21.8 kcal/mol |
a The bp location of the start of the seed sequence in the FOSL1 mRNA sequence.
b If the distance between the start positions of two seed sequences is less than 23 bp (the length of miR-138 molecule), then these targeting sites are defined as adjacent sites.
c The seed sequences (seq.) are identified from FOSL1 mRNA as 7-mers that are reverse complementary to the sequence from the 2 to 8 positions of hsa-miR-138. The canonical bases are identified by uppercase letters, and the non-canonical bases are identified by lowercase letters.
d The canonical targeting site is defined by the presence of seed sequence CACCAGC in the mRNA molecule. The non-canonical targeting site allows G:U wobble base pairing. The seed sequence on the mRNA molecule allows the substitution of C by U and the substitution of A by G.
e The minimum free energy (mfe) for the binding of hsa-miR-138 to the targeting sequence in FOSL1 mRNA is predicted using the RNAhybrid program (32). An mfe cut-off of −20.0 kcal/mol was used.
Mature microRNAs form stable complexes with Argonaute proteins (such as Ago2), the core of the RNAi-induced silencing complex (RISC). The microRNA then directs RISC to bind to the mRNA molecules containing specific targeting sequences and results in translational repression and/or enhanced mRNA degradation. To test the miR-138-directed RISC binding to FOSL1 mRNA, an RIP-IP assay was performed to determine whether FOSL1 mRNA fragments were enriched in the Ago2 co-IP fraction. As shown in Fig. 1E, a significant reduction of FOSL1 mRNA was observed in the whole cell lysates based on quantitative RT-PCR (assays specific to the 5′-UTR, CDS, and 3′-UTR of FOSL1) in cells transfected with miR-138 mimic as compared with cells treated with control mimic. As shown in Fig. 1F, in the Ago2 co-IP fractions, significant enrichments of the fragments representing the CDs and the 3′-UTR of the FOSL1 mRNA were observed in cells treated with miR-138 mimic. Apparent enrichment of the 5′-UTR fragment was also observed; however, the difference was not statistically significant. As a control, we also tested miR-138-mediated enrichment of RHOC mRNA, a known miR-138 targeting gene (13), in the Ago2 co-IP fractions. As shown in supplemental Fig. 8, a significant reduction of RHOC mRNA was observed in the total cell lysate, and a significant enrichment of RHOC mRNA was observed in the Ago2-IP fraction. This is in agreement with our previous findings (13).
To further confirm that miR-138 directly interacts with these predicted miR-138 targeting sites, Dual-Luciferase reporter assays were performed using constructs containing these targeting sites from the 5′-UTR, CDs, and 3′-UTR (Fig. 1, G, H, and I, respectively). As illustrated in Fig. 1G, when cells were transfected with miR-138, the luciferase activities of the construct containing the miR-138 targeting site from the 5′-UTR of the FOSL1 was significantly reduced as compared with the cells transfected with negative control. When the seed region of this targeting site was mutated, the miR-138 effect on luciferase was abolished. As shown in Fig. 1H, for the construct containing three miR-138 targeting sites from the CDs of the FOSL1, when cells were transfected with miR-138, the luciferase activities were significantly reduced as compared with the cells transfected with negative control. When the seed region(s) from one or two of the targeting sites were mutated, the miR-138-mediated reduction in luciferase activity was still observed. When the seed regions of all three targeting sites were mutated, the miR-138 effect on the luciferase activity was abolished. Similar results were observed for miR-138 targeting sites located in the 3′-UTR (Fig. 1I). When cells were treated with the miR-138 mimic, the luciferase activity was significantly reduced as compared with the cells transfected with negative control. When the seed region from one of the two targeting sites was mutated, the effect of miR-138 on luciferase activity was still retained. When the seed regions from both targeting sites were mutated, the miR-138 effect on the luciferase activity was abolished. As controls, we also tested the effects of miR-138 on the 3′-UTR of RHOC gene, which contains a known miR-138 targeting site (13), and the 3′-UTR of IGF1R gene, which has no miR-138 targeting site (but multiple miR-7 targeting sites) (31). As shown in supplemental Fig. 9, although a significant reduction in luciferase activity from construct containing the 3′-UTR of RHOC was observed when cells were treated with miR-138 mimic as compared with cells treated with negative control, no significant change was observed with the construct containing the 3′-UTR of IGF1R mRNA. However, when the cells were treated with miR-7 mimic, the luciferase activity from construct containing the 3′-UTR of IGF1R was significantly reduced. Taken together, our results provide solid evidence to support an important microRNA regulatory module consisting of a candidate tumor suppressor microRNA (miR-138) and a proto-oncogene (FOSL1).
Although our results provide strong evidence supporting the miR-138-FOSL1 regulatory module, it is possible that other members of the Fos gene family may also be regulated by miR-138. As shown in supplemental Table 1, in addition to FOSL1, miR-138 targeting sites were also identified on mRNAs of FOS, FOSB, and FOSL2. It is worth noting that two canonical and seven non-canonical miR-138 targeting sites were identified in the CDs of the FOSB mRNA, and three additional non-canonical sites were identified in the 3′-UTR of the FOSB mRNA. This is consistent with the observation that ectopic transfection of miR-138 led to reduced FOSB expression in 1386Ln cells, and knockdown of miR-138 led to an apparent increase in FOSB levels in 686Tu cells (Fig. 1A).
Although our results suggest that both canonical and non-canonical targeting sites located in all areas of the mRNA molecule (e.g. 5′-UTR, CDs, 3′-UTR) contribute to miR-138-mediated post-transcriptional regulation of the FOSL1 gene (and potentially also the FOSB gene), more studies will be needed to test whether this is true for other target genes. We previously identified 194 genes that were significantly down-regulated by miR-138 (17). As shown in supplemental Table 2, significant enrichment of canonical microR-138 targeting sites was observed in the 5′-UTR, CDs, and 3′-UTR of the genes that were regulated by miR-138. Furthermore, significant enrichment of non-canonical miR-138 targeting sites was observed in the 3′-UTR of the genes that were regulated by miR-138. Apparent enrichment of non-canonical miR-138 targeting sites was also observed in the 5′-UTR, but the difference is not statistically significant. These observations support our hypothesis that both canonical and non-canonical microRNA targeting sites and the microRNA targeting sites located in all areas of the mRNA molecule (e.g. 5′-UTR, CDs, 3′-UTR) may contribute to microRNA-mediated post-transcriptional regulation.
In summary, we demonstrate an important microRNA regulatory module consisting of candidate tumor suppressor miR-138 and proto-oncogene FOSL1. The deregulation of the miR-138-FOSL1 regulatory module may contribute significantly to the initiation and progression of cancer, such as HNSCC. Our results also provide evidence that microRNAs target both canonical and non-canonical targeting sites located in all areas of the mRNA molecule (e.g. 5′-UTR, CDs, 3′-UTR). More studies will be needed to fully explore the precise molecular mechanisms that govern the microRNA-mediated post-transcriptional regulation of the target genes.
Supplementary Material
Acknowledgments
We thank Dr. Shuang Huang from Georgia Health Sciences University for providing us with the Snai2 promoter luciferase construct. We thank Dr. Thomas Tuschl from Rockefeller University for providing us with the pIRESneo-FLAG/HA-Ago2 expression vector through Addgene Inc. We thank Dr. Wendy Cerny for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant Public Health Service Grants CA135992, CA139596, and DE014847 (to X. Z.). This work was also supported by supplementary funding from the UIC Center for Clinical and Translational Science (CCTS) (Grant UL1RR029879).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S9.
- miR-138
- microRNA-138
- pre-miR-138
- precursor miRNA-138
- miRNA
- microRNA
- HNSCC
- head and neck squamous cell carcinoma
- RISC
- RNAi-induced silencing complex
- CD
- coding sequence
- RIP-IP
- ribonucleoprotein-IP
- co-IP
- co-immunoprecipitation
- AP-1
- activator protein 1
- EMT
- epithelial-to-mesenchymal transition
- mfe
- minimum free energy
- LNA
- locked nucleic acid.
REFERENCES
- 1. Esquela-Kerscher A., Slack F. J. (2006) Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 2. Calin G. A., Croce C. M. (2006) Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 3. Kozaki K., Imoto I., Mogi S., Omura K., Inazawa J. (2008) Cancer Res. 68, 2094–2105 [DOI] [PubMed] [Google Scholar]
- 4. Wong T. S., Liu X. B., Wong B. Y., Ng R. W., Yuen A. P., Wei W. I. (2008) Clin. Cancer Res. 14, 2588–2592 [DOI] [PubMed] [Google Scholar]
- 5. Mitomo S., Maesawa C., Ogasawara S., Iwaya T., Shibazaki M., Yashima-Abo A., Kotani K., Oikawa H., Sakurai E., Izutsu N., Kato K., Komatsu H., Ikeda K., Wakabayashi G., Masuda T. (2008) Cancer Sci. 99, 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yip L., Kelly L., Shuai Y., Armstrong M. J., Nikiforov Y. E., Carty S. E., Nikiforova M. N. (2011) Ann. Surg. Oncol. 18, 2035–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seike M., Goto A., Okano T., Bowman E. D., Schetter A. J., Horikawa I., Mathe E. A., Jen J., Yang P., Sugimura H., Gemma A., Kudoh S., Croce C. M., Harris C. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Obernosterer G., Leuschner P. J., Alenius M., Martinez J. (2006) RNA 12, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piccinin S., Gasparotto D., Vukosavljevic T., Barzan L., Sulfaro S., Maestro R., Boiocchi M. (1998) Br. J. Cancer 78, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hogg R. P., Honorio S., Martinez A., Agathanggelou A., Dallol A., Fullwood P., Weichselbaum R., Kuo M. J., Maher E. R., Latif F. (2002) Eur. J. Cancer 38, 1585–1592 [DOI] [PubMed] [Google Scholar]
- 11. Wang X., Gleich L., Pavelic Z. P., Li Y. Q., Gale N., Hunt S., Gluckman J. L., Stambrook P. J. (1999) Int. J. Oncol. 14, 557–561 [DOI] [PubMed] [Google Scholar]
- 12. Liu X., Jiang L., Wang A., Yu J., Shi F., Zhou X. (2009) Cancer Lett. 286, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang L., Liu X., Kolokythas A., Yu J., Wang A., Heidbreder C. E., Shi F., Zhou X. (2010) Int. J. Cancer 127, 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y., Huang J. W., Li M., Cavenee W. K., Mitchell P. S., Zhou X., Tewari M., Furnari F. B., Taniguchi T. (2011) Mol. Cancer Res. 9, 1100–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q., Zhong M., Liu W., Li J., Huang J., Zheng L. (2011) Exp. Lung Res. 37, 427–434 [DOI] [PubMed] [Google Scholar]
- 16. Zhao X., Yang L., Hu J., Ruan J. (2010) Leuk. Res. 34, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 17. Jiang L., Dai Y., Liu X., Wang C., Wang A., Chen Z., Heidbreder C. E., Kolokythas A., Zhou X. (2011) Hum. Genet. 129, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H., Zhu G., Li Y., Padia R. N., Dong Z., Pan Z. K., Liu K., Huang S. (2009) Cancer Res. 69, 9228–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamura Y., Inloes J. B., Katagiri T., Kobayashi T. (2011) Mol. Cell. Biol. 31, 3019–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendrickson D. G., Hogan D. J., Herschlag D., Ferrell J. E., Brown P. O. (2008) PLoS One 3, e2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou X., Temam S., Oh M., Pungpravat N., Huang B. L., Mao L., Wong D. T. (2006) Neoplasia 8, 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 23. Wang C., Liu X., Huang H., Ma H., Cai W., Hou J., Huang L., Dai Y., Yu T., Zhou X. (August 16, 2011) Int. J. Cancer 10.1002/ijc.26226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X., Wang C., Chen Z., Jin Y., Wang Y., Kolokythas A., Dai Y., Zhou X. (July 19, 2011) Biochem. J., 10.1042/BJ20111006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moretti F., Thermann R., Hentze M. W. (2010) RNA 16, 2493–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005) Science 309, 1577–1581 [DOI] [PubMed] [Google Scholar]
- 27. Lytle J. R., Yario T. A., Steitz J. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao J. M., Lu H. (2011) J. Biol. Chem. 286, 33901–33909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin W., Shi Y., Zhao B., Yao C., Jin L., Ma J., Jin Y. (2010) PLoS One 5, e9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schnall-Levin M., Rissland O. S., Johnston W. K., Perrimon N., Bartel D. P., Berger B. (2011) Genome Res. 21, 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang L., Liu X., Chen Z., Jin Y., Heidbreder C. E., Kolokythas A., Wang A., Dai Y., Zhou X. (2010) Biochem. J. 432, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.