Background: Disc degeneration is characterized by elevated levels of cytokines, TNF-α and IL-1β and an increase in aggrecan-rich matrix degradation.
Results: Cytokine-dependent syndecan-4 expression regulate ADAMTS-5/aggrecanse2 activation in disc cells.
Conclusion: Syndecan-4 may play a key role in pathogenesis of degenerative disc disease.
Significance: Syndecan-4 may offer a potential therapeutic target for controlling progression of degenerative disc disease.
Keywords: ADAM ADAMTS, Connective Tissue, Interleukin, NF-kappa B (NF-κB), Tumor Necrosis Factor (TNF), Aggrecan, Arthritis, Degenerative Disc Disease
Abstract
Elevated levels of TNF-α, IL-1β and a resultant increase in ADAMTS (a disintegrin-like and metalloprotease with thrombospondin type I motifs) expression is seen during disc degeneration. However, if these pro-inflammatory cytokines control ADAMTS activity is not definitively known. The goal of the investigation was to study if TNF-α and IL-1β regulate syndecan-4 (SDC4) expression, and if SDC4 was responsible for promoting aggrecan degradation through controlling ADAMTS activity in nucleus pulposus cells of the intervertebral disc. Cytokine treatment increased SDC4 expression and promoter activity. Use of inhibitor, SM7368 and co-transfections with IκBα, RelA/p50 showed that NF-κΒ regulated both basal and cytokine-dependent SDC4 transcription. SDC4 promoter harboring RelA binding site mutation was unresponsive to the cytokines. Moreover, cytokines failed to increase SDC4 promoter activity in RelA-null cells. Cytokines increased ADAMTS-4/5 expression and aggrecan degradation and promoted SDC4 interaction with ADAMTS-5. Treatment with heparinase-III and p-nitrophenyl-β-d-xylopyranoside (PNPX), an inhibitor of heparan sulfate synthesis and transfection with SDC4-shRNA partially blocked cytokine mediated aggrecan degradation. Analysis of human tissues showed increased aggrecan degradation with a concomitant increase in SDC4 and ADAMTS-5 protein expression with severity of disc disease. Likewise, SDC4, TNF-α, IL-1β, ADAMTS-4, and ADAMTS-5 mRNA expression increased in degenerate tissues. We conclude that in nucleus pulposus, TNF-α and IL-1β regulate SDC4 expression, which plays a key role in pathogenesis of degenerative disc disease by promoting aggrecan degradation by ADAMTS-5.
Introduction
The intervertebral disc is a specialized tissue that permits rotation as well as flexure and extension of the human spine. It consists of an outer ligamentous annulus fibrosus that encloses gel-like nucleus pulposus. Cells in the nucleus pulposus are derived from the embryonic notochord and often mistakenly considered as chondrocytes, another matrix-rich skeletal tissue. The superior and inferior boundaries of the intervertebral disc are formed by the cartilage endplates. Blood vessels infiltrate the superficial region of the endplates and the outer third of annulus fibrosus, but do not enter the nucleus pulposus making this tissue hypoxic (1, 2). Cells in the nucleus pulposus secrete a complex extracellular matrix that primarily contains the hydrophilic proteoglycans such as aggrecan that is substituted with numerous negatively charged chondroitin sulfate chains. In its fully sulfated form, the molecule exhibits a high negative charge density and when hydrated assumes a linear configuration. Bound to the aggrecan core protein, and associated with hyaluronic acid, the chondroitin sulfate chains form a giant polydispersed supramolecular structure. The high osmotic pressure of the aggregate contains the biomechanical forces applied to the spine (3). Surprisingly, while the importance of proteoglycan secretion and function has been discussed by many investigators, molecular mechanisms that control aggrecan turnover, are not well understood. Intervertebral disc degeneration, linked to persistent back pain and the development of disc herniation, is characterized by profound anatomical and biologic changes that include a decrease in cell number and a simultaneous increase in matrix degradation resulting in a reduction in disc height (4). Elevated levels of pro-inflammatory cytokines and other inflammatory mediators have been reported to be present in degenerate discs including tumor necrosis factor alpha (TNF-α), interleukin-1beta (IL-1β) and IL-6, IL-8, prostaglandin E2 (PGE2) (5–9). During disc degeneration and herniation, in addition to infiltrating immune cells, resident nucleus pulposus and annulus fibrosus cells produce high levels of cytokines such as TNF-α and IL-1β (8, 10). Animal studies suggest that TNF-α may elicit radicular pain associated with long-term sensitization and pathological changes in dorsal root ganglion (DRG) neurons (11). Moreover, TNF-α and IL-1β stimulate production of NGF, BDNF and VEGF, molecules associated with nerve ingrowth and angiogenesis by nucleus pulposus cells (12). Importantly, both TNF-α and IL-1β upregulate expression of matrix degrading matrix metalloproteinases (MMPs2 and two major aggrecanases (ADAMTS-4 and ADAMTS-5) by nucleus pulposus cells (8, 13–15). Noteworthy, in osteoarthritis, a degenerative joint disease, aggrecan cleavage by ADAMTS-5 is important for the breakdown of cartilage matrix (16, 17). A recent study indicates that syndecan-4 (SDC4) is required for ADAMTS-5 activation in chondrocytes during osteoarthritis (18), while, syndecan-1 has been shown to activate ADAMTS-4 (19). Whether, syndecans are required for ADAMTS in embryologically distinct nucleus pulposus tissue and whether activation is TNF-α and IL-1β dependent is as yet unknown.
The major objective of the investigation was to examine the mechanism that regulates expression of SDC4 by nucleus pulposus cells and to elucidate its role in ADAMTS activation. We show for the first time that TNF-α and IL-1β increased SDC4 expression in NF-κB dependent fashion. Moreover, SDC4 was required for the full activity of ADAMTS-5. Unlike chondrocytes, our results show that SDC4-dependent ADAMTS-5 activation did not require MMP-3 activity suggesting that this regulation is unique to nucleus pulposus cells. From this perspective, TNF-α, IL-1β, and SDC4 and ADAMTS-5 form a regulatory loop that controls aggrecan turnover in nucleus pulposus cells.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
Human SDC4 wild type (−895/+40 bp) and a NF-κB mutant promoter constructs are provided by Dr. Jitendra Gautam, University of Virginia (20). Expression plasmids pCMX-IκBM (#12330), RelA/p65 (#20012), p50 (#20018), developed by Dr. Inder Verma, RelB (#20017) developed by Dr.Thomas Gilmore and c-Rel (27253) developed by Dr. Stephen Smale were obtained from Addgene. Lenti-viral ShRNA plasmids targeting rat SDC4 were purchased from Genecoepia. The vector pRL-TK (Promega) containing Renilla luciferase gene was used as an internal transfection control. The amount of transfected plasmid, the pre-transfection period after seeding, and the post-transfection period before harvesting, have been optimized for rat nucleus pulposus cells using pSV β-galactosidase plasmid (Promega) (21). p65-null and wild type cells were a kind gift from Dr. Denis Guttridge, University of Ohio, Columbus. Antibodies that recognize aggrecan degradation in intraglobular domain (anti-NITEGE and anti-G1) were a gift from Dr. Peter Roughley. Shriners Hospital, Toronto. SDC4, ADAMTS4/5, and ARGSVIL antibodies were from Abcam. p-p65, p65, and p50 antibodies were from Cell Signaling. β-Tubulin and GAPDH antibodies were purchased from DSHB and Novus Biologicals, respectively. TNF-α and IL-1β were purchased from Peprotech. Heparinase III was purchased from Sigma-Aldrich. p-Nitrophenyl-β-d-xylopyranoside (PNPX) and NF-κB transcriptional activity inhibitor SM7368 was purchased from EMD Chemicals.
Isolation of Nucleus Pulposus Cells and Treatments
Rat and human nucleus pulposus cells were isolated using a method reported earlier by Risbud et al. (21). Nucleus pulposus cells were maintained in Dulbecco's modified Eagles medium (DMEM) and 10% fetal bovine serum (FBS) supplemented with antibiotics. To investigate effect of cytokines, cells were treated with IL-1β (10 ng/ml) and TNF-α (50–100 ng/ml) for 4–24 h. In some experiments, cells were treated with heparinase III (1 units/ml) or PNPX (1 mm) 24 h before addition of cytokines.
Human Tissue Collection and Grading
Lumbar disc tissues were collected as surgical waste from individuals undergoing elective spinal surgical procedures (average age 54 years, ranging from 38–82 years). In line with Thomas Jefferson University's Institutional Review Board guidelines, informed consent for sample collection was obtained from each patient. Assessment of the disease state was performed using the Pfirrmann grading (22).
Immunohistological Studies
Freshly isolated spines or whole embryos were immediately fixed in 4% paraformaldehyde in PBS and then embedded in paraffin. Transverse and coronal sections, 6–8 μm in thickness, were deparaffinized in xylene, rehydrated through graded ethanol and some sections stained with alcian blue, eosin, and hematoxylin. For localizing SDC4, sections were incubated with the anti-SDC4 antibody in 2% bovine serum albumin in PBS at a dilution of 1:200 at 4 °C overnight. After thoroughly washing the sections, the bound primary antibody was incubated with Alexa fluor-488 conjugated anti-rabbit secondary antibody (Invitrogen), at a dilution of 1:200 for 45 min at room temperature. Sections were visualized using a fluorescence microscope (Nikon, Japan).
Real-time RT-PCR Analysis
Following treatment, total RNA was extracted from nucleus pulposus cells using RNAeasy mini columns (Qiagen). For human samples, total RNA was isolated from 100 to 300 mg of nucleus pulposus tissue. Tissue was homogenized in Trizol (Invitrogen) on ice using Omni TH Homogenizer (Omni International). Following Trizol extraction, RNA was passed through the RNA easy mini columns. Before elution from the column, RNA was treated with RNase free DNase I. 2 μg of total DNA free-RNA was used to synthesize cDNA using SuperScipt III cDNA synthesis kit (Invitrogen). Reactions were set up in triplicate in 96-well plate using 1 μl cDNA with Fast SYBR Green PCR Master Mix (Applied Biosystems) to which gene-specific forward and reverse PCR primers were added. Each set of samples included a template-free control. PCR reactions were performed in a StepOnePlus real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. All the primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Immunofluorescence Microscopy
Cells were plated in flat bottom 96-well plates (5 × 103/well) and treated with TNF-α or IL-1β for 1–24 h. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton-X 100 in PBS for 10 min, blocked with PBS containing 5% FBS, and incubated with antibodies against SDC4 (1:200), p65, p-p65, and p50 (1:200), at 4 °C overnight. As a negative control, cells were reacted with isotype IgG under similar conditions. After washing, the cells were incubated with Alexa fluor-488 conjugated anti-rabbit secondary antibody (Invitrogen), at a dilution of 1:100 for 1 h at room temperature. Cells were imaged using a laser scanning confocal microscope using 20×/0.4 LCPlanFl objective (Olympus Fluoview, Japan).
Protein Extraction, Immunoprecipitation, and Western Blotting
Cells were placed on ice immediately following treatment and washed with ice-cold HBSS. For SDC4 IP we used a commercial kit and followed manufacturer's instructions (Pierce Crosslink IP kit). All the wash buffers and final re-suspension buffer included 1× protease inhibitor mixture (Roche), NaF (5 mm) and Na3VO4 (200 μm). For detecting aggrecan neoepitopes, protein lysates were pretreated with chondroitinase ABC (0.1 unit/ml, Sigma) for 1–6 h at 37 °C. Immunoprecipitated or total cell proteins were resolved on 8–12% SDS-polyacrylamide gels and transferred by electroblotting to PVDF membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBST (50 mm Tris, pH 7.6, 150 mm NaCl, 0.1% Tween 20) and incubated overnight at 4 °C in 3% nonfat dry milk in TBST with the anti-SDC4, anti-ADAMTS5, anti-ARGSVIL, anti-NITEGE, anti-G1, or anti-p65, Pp65, or p50 antibody all at a dilution of 1:1000. Immunolabeling was detected using the ECL reagent (Amersham Biosciences).
Transfections and Dual Luciferase Assay
Cells were transferred to 24-well plates at a density of 4 × 104 cells/well 1 day before transfection. To investigate the effect of NF-κB on SDC4 promoter activity, cells were cotransfected with 100–300 ng of IκBM or p65 or p50 or p65 plus p50 with or without appropriate backbone vector and 350 ng of SDC4 reporter and 350 ng of pRL-TK plasmid. In some experiments, cells were transfected with 500 ng of SDC4 reporter plasmids with 500 ng pRL-TK plasmid. Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent. 48 h after trasfection, the cells were harvested and a Dual-LuciferaseTM reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were carried out using a luminometer (TD-20/20, Turner Designs, CA). At least three independent transfections were performed, and all analyses were carried out in triplicate.
Lentiviral Particle Production and Viral Transduction
HEK 293T cells were seeded in 10-cm plates (1.3 × 106 cells/plate) in DMEM with 10% heat-inactivated FBS 2 days before transfection. Cells were transfected with 2.5 μg of Sh-SDC4-Ctr, sh-SDC4 plasmids along with 1.875 μg psPAX2 and 0.625 μg pMD2.G. After 16 h, transfection media was removed and replaced with DMEM with 5% heat-inactivated FBS and penicillin-streptomycin. Lentiviral particles were harvested at 48 and 60 h post-transfection. Nucleus pulposus cells were plated in DMEM with 5% heat-inactivated FBS 1 day before transduction. Cells in 10 cm plates were transduced with 5 ml of conditioned media containing viral particles along with 6 μg/ml polybrene. After 24 h, conditioned media was removed and replaced with DMEM with 5% heat-inactivated FBS. Cells were harvested for protein extraction 5 days after viral transduction.
Statistical Analysis
All measurements were performed in triplicate, data are presented as mean ± S.E. Differences between groups were analyzed by the Student's t test and ANOVA; *, p < 0.05.
RESULTS
Expression of SDC4 and Its Regulation by Cytokines in the Nucleus Pulpous Cells
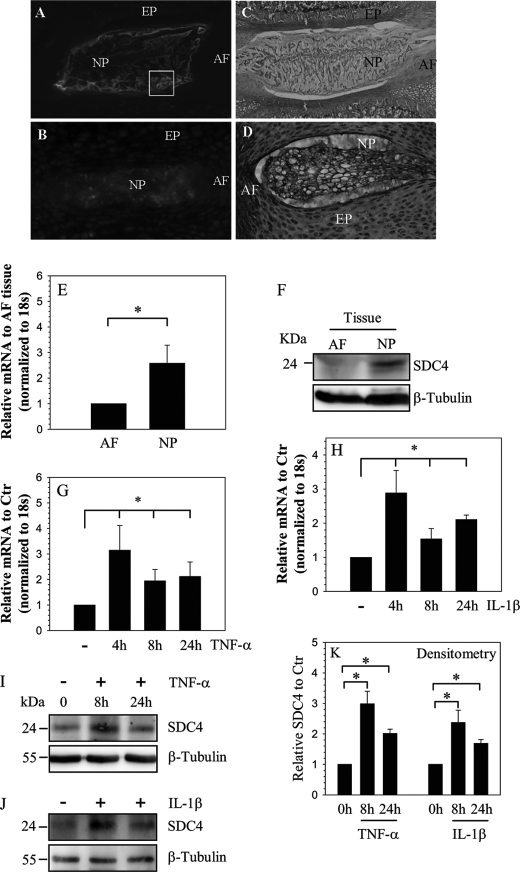
Saggital sections of skeletally mature (Fig. 1A) and neonatal (Fig. 1B) rat discs were stained with an antibody to SDC4 or with hemotoxylin, eosin, and alcian blue (Fig. 1, C and D). Fig. 1, A and B indicate that SDC4 is robustly expressed by cells of the nucleus pulposus and weakly expressed by cells of the annulus fibrosus and cartilaginous endplate. In skeletally mature animals, nucleus pulposus shows a strong cell surface SDC4 staining (see box, Fig. 1A). In the embryonic nucleus pulposus, staining is localized to the cytosol as well as to the pericellular matrix (Fig. 1B). Expression of SDC4 in mature rat tissues was studied using real-time RT-PCR and Western blot analysis. Expression level of SDC4 mRNA in nucleus pulposus is significantly higher than annulus fibrosus cells (Fig. 1E). Fig. 1F shows that nucleus pulposus tissue express a 24–26 kDa band representing the full-length SDC4; again expression is prominent in nucleus pulposus than annulus fibrosus. To explore the premise that cytokines concerned with disc degeneration-regulated SDC4 expression, nucleus pulposus cells were treated with TNF-α and IL-1β and expression of SDC4 analyzed. Fig. 1, G and H show that treatment with both TNF-α and IL-1β results in increased SDC4 mRNA levels in nucleus pulposus cells. In addition, we studied expression of SDC4 in nucleus pulposus cells using Western blot analysis. Cytokine treatment results in elevated SDC4 protein expression (Fig. 1, I–K).
FIGURE 1.
Expression and cytokine dependence of SDC4 in NP cells. Sagittal sections of the intervertebral disc of a mature (A, C) and a neonatal rat (B, D). Sections were treated with SDC4 antibody (A, B), or counterstained with hemotoxylin, eosin, and alcian blue (C and D). Cell surface staining of SDC4 was evident in NP cells in mature disc (box in A), SDC4 was expressed at a higher level in NP of the compared with the AF. Mag. × 4–10). E, real-time RT-PCR analysis shows significantly higher expression of SDC4 in NP than AF tissue. F, Western blot analysis of SDC4 expression in AF and NP tissue. Note, the expression of the 24–26 kDa SDC4 band, higher expression of SDC4 in NP compared with AF tissue. G and H, real-time RT-PCR analysis of SDC4 expression by nucleus pulposus cells treated with TNF-α (G) and IL-1β (H) up to 24 h. There was increased expression at 4 h which continued until 24 h. I and J, Western blot analysis showing increased expression of SDC4 by NP cells following TNF-α (I) and IL-1β (J) treatment. K, densitometric analysis of three blots each for experiment described in I and J above. β-Tubulin was used as a loading control and to calculate relative expression levels. Cytokine treatment significantly increased SDC4 expression. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
TNF-α and IL-1β Promotes NF-κB Signaling in the Nucleus Pulposus Cells
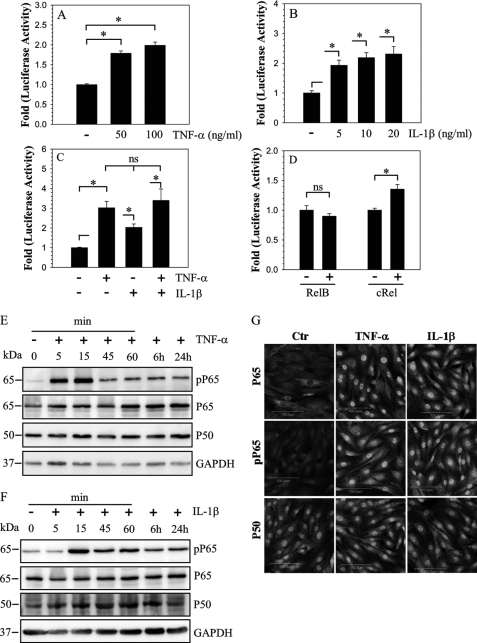
To investigate the cytokine mediated regulation of SDC4 expression, we examined the effect of IL-1β and TNF-α on SDC4 promoter activity in nucleus pulposus cells. TNF-α treatment results in a dose dependent increase in SDC4 promoter activity (Fig. 2A). Similarly, an induction in promoter activity is seen following treatment with IL-1β (Fig. 2B). However, no additive or synergistic effects of TNF-α and IL-1β, are seen on the promoter activity (Fig. 2C). To determine if NF-κB signaling was required for the regulation of SDC4 expression in nucleus pulposus cells, we first measured SDC4 promoter activity in presence of NF-κB subunits, RelB and c-Rel. A smaller increase in SDC4 promoter activity is seen with only cRel (Fig. 2D). We next evaluated status of NF-κB subunits, RelA/p65 and p50 following treatment with TNF-α (Fig. 2E) and IL-1β (Fig. 2F). Fig. 2, E and F show that following treatment, there is a rapid increase in phospho-p65 levels. Activation is maximal at 15–30 min and declines rapidly thereafter, still staying elevated until 24 h. On the other hand, levels of total p65 as well as p50 did not show appreciable increase. Immunofluorescence analysis shows an increase in the nuclear accumulation of both phosphorylated and total p65 (Fig. 2G). Interestingly, p50 shows strong nuclear localization in both untreated and treated cells but level of expression remained similar.
FIGURE 2.
Modulation of SDC4 promoter activity and NF-κB signaling by cytokines in NP cells. Treatment with both TNF-α (A) and IL-1β (B) induces activity of SDC4 promoter in NP cells. C, a combined treatment of TNF-α and IL-1β shows no additive or synergistic effect on induction of SDC4 promoter activity over individual cytokine treatments. D, NP cells were co-transfected with RelB or cRel and SDC4 promoter activity measured. Only cRel has a small inductive effect on SDC4 promoter activity. Values shown are mean ± S.E., of three independent experiments; *, p < 0.05. E and F, Western blot analysis of NF-κB signaling proteins, p65 and p50 following treatment of NP cells with TNF-α (E) and IL-1β (F). Cytokine treatment induced phosphorylation of p65 within first 15 min and levels remained elevated after 24 h. No appreciable change in expression of p65 and p50 was seen. G, immunofluorescent analysis of NP cells treated with TNF-α and IL-1β. Cells showed increased nuclear localization of both phospho-p65 and p65 whereas p50 expression and localization did not change.
NF-κB Signaling Controls SDC4 Transcription in the Nucleus Pulposus Cells
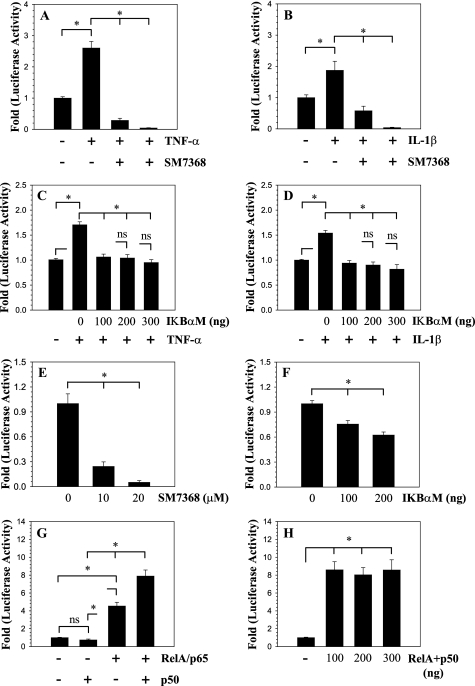
To investigate the role of NF-κB in the transcriptional regulation of SDC4 expression, we performed loss of function studies. When cells are treated with the NF-κB inhibitor SM7368, both TNF-α and IL-1β mediated induction in SDC4 promoter activity is completely inhibited (Fig. 3, A and B). To further explore the mechanism of regulation, we co-transfected nucleus pulposus cells with a plasmid expressing IκBαM, a negative regulator of NF-κB signaling. It was found that suppression of NF-κB activity blocks cytokine-mediated induction in SDC4 promoter activity (Fig. 3, C and D). Moreover, suppression of NF-κB signaling either by inhibitor treatment (Fig. 3E) or IKBαM (Fig. 3F), in absence of exogenous cytokines, results in a significant decrease in basal activity of the SDC4 promoter. To confirm that SDC4 promoter activity is responsive to NF-κB signaling, we co-transfected nucleus pulposus cells with plasmids encoding p65/RelA and/or p50 (Fig. 3, G and H). Transfection with p65/RelA significantly increases SDC4 promoter activity. Moreover, activation is pronounced when both p65 and p50 are present, while p50 alone has no effect on the promoter activity (Fig. 3G). The stimulatory effect of p65 with p50 on SDC4 promoter activity is such that when the amount of plasmids is decreased from 300 to 100 ng the level of induction remained constant (Fig. 3H).
FIGURE 3.
Effect of NF-κB on SDC4 promoter activity in NP cells. A and B, SDC4 promoter activity was measured following TNF-α (A) and IL-1β (B) treatment of NP cells with or without NF-κB signaling inhibitor SM7368. Inhibitor treatment completely abolished cytokine mediate promoter induction. C and D, cells were co-transfected with IκBαM and promoter activity was measured following TNF-α (C) and IL-1β (D) treatment. Inhibition of NF-κB signaling resulted in significant blocking of cytokine dependent induction in SDC4 promoter activity. E, treatment of NP cells with inhibitor SM7368 or F, co-transfection with IKBαM, in absence of exogenous cytokines resulted in suppression of basal SDC4 promoter activity. G, NP cells were co-transfected with RelA/p65 and/or p50 and SDC4 promoter activity measured. Co-transfection with p65 alone but not p50 resulted in increased promoter activity. When p65 and p50 are added together they elicited a higher induction in activity than p65 alone. H, note the significant induction in SDC4 reporter activity even when transfected with 100 ng of total RelA and p50 plasmids. No further induction in activity was seen when plasmid concentration was increased till 300 ng. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05.
NF-κB Interaction with SDC4 Promoter Controls Its Activity in the Nucleus Pulposus
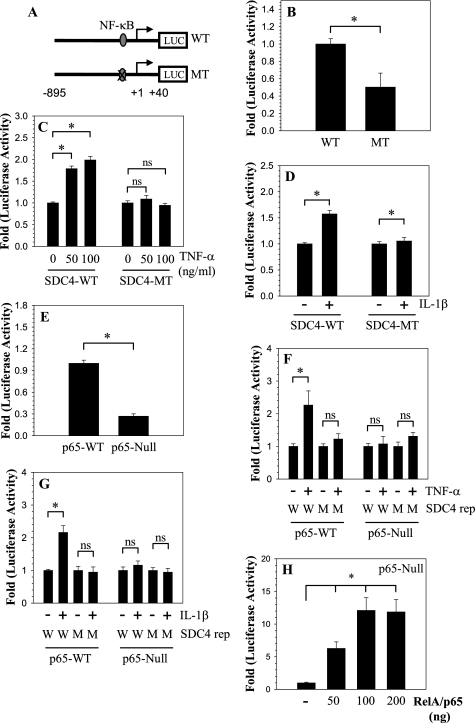
Analysis of human SDC4 promoter showed that there is a conserved RelA binding site GGGGAATTCC within first kb of the promoter at −97/−88 bp (Fig. 4A). We evaluated the requirement of the NF-κB responsive element using a reporter containing mutation at this site (Fig. 4A). Compared with the wild type reporter, the mutant reporter exhibits decreased basal activity in nucleus pulposus cells (Fig. 4B). We then examined the effect of TNF-α and IL-1β on the activity of the mutant reporter (Fig. 4, C and D). Results clearly show that unlike the wild type reporter, the mutation at the RelA binding site does not change the activity of the reporter. To further confirm the role of RelA in SDC4 regulation, and to determine if this mechanism is cell type-specific, we measured SDC4 promoter activity in RelA null and wild type embryonic fibroblasts. Compared with wild type cells, the basal activity of the SDC4 promoter is significantly lower in RelA-null cells (Fig. 4E). Moreover, similar to nucleus pulposus cells, only in wild type cells, the promoter activity is inducible by both cytokines (Fig. 4, F and G). As expected, the mutant SDC4 promoter is unresponsive to cytokines in both wild type and null cells (Fig. 4, F and G). Moreover, when co-transfected with RelA, SDC4 promoter activity is significantly induced in RelA-null cells (Fig. 4H).
FIGURE 4.
NF-κB regulation of SDC4 expression. A, schematic of different SDC4 reporter constructs (WT: wild type; MT: mutant RelA). B, cells were transfected with WT and MT SDC4 reporter constructs and basal activity measured. Mutant promoter exhibits significantly lower basal activity than the wild type promoter. C and D, cells transfected with WT and MT SDC4 reporter constructs were treated with C, TNF-α and D, IL-1β. Only the WT SDC4 promoter fragment with elicited an increase in activity. E, SDC4 promoter activity in MEFs isolated from RelA WT and null mice. There was a 70% decrease in basal SDC4 reporter activity in the RelA-null cells. F and G, RelA WT and null cells transfected with WT and MT SDC4 reporter constructs and treated with F, TNF-α and G, IL-1β. Wild type cells show increase in only WT reporter activity. While null cells do not show increase either WT or MT promoter. H, RelA-null cells were transfected with plasmid expressing RelA and SDC4 promoter activity was measured. A dose-dependent increase in SDC4 reporter activity was seen, at a concentration higher than 200 ng of RelA no further increase in activity was seen. Values shown are mean ± S.E. from three independent experiments, *, p < 0.05.
TNF-α and IL-1β Promote ADAMTS Activity and SDC4 Interaction with ADAMTS-5 in the Nucleus Pulposus Cells
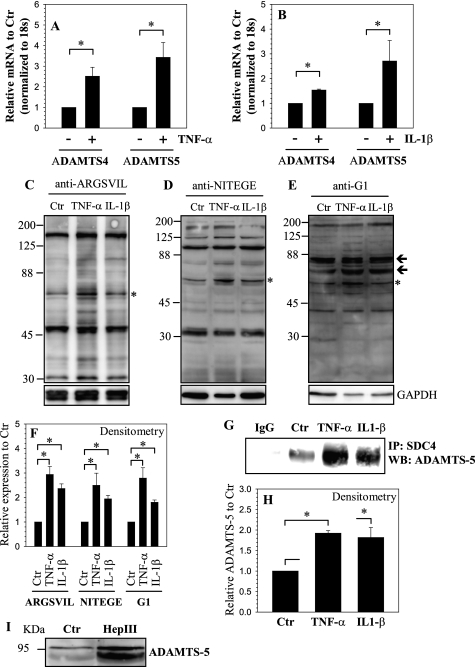
Next, we examined the effect of cytokines IL-1β and TNF-α on ADAMTS expression in nucleus pulposus cells. Fig. 5, A and B shows that treatment increases mRNA expression of both ADAMTS-4/aggrecanase1 and ADAMTS-5/aggrecanase2 (Fig. 5, A and B). To determine if cytokines promoted ADAMTS activity, we measured generation of aggrecan fragments using neoepitope recognizing anti-ARGSVIL (Fig. 5C), anti-NITEGE (Fig. 5D), and anti-G1 (Fig. 5E) antibodies. In addition to a 65–70 kDa fragment corresponding to the primary neoepitope, a few high molecular weight fragments can be recognized by all the three antibodies (Fig. 5, C–E). A significant increase in aggrecan neoepitope generation is detected when cells were treated with cytokines compared with untreated cells (Fig. 5F). We used IP-Western blot to determine if ADAMTS proteins interacted with SDC4. Pull down of SDC4 resulted in co-precipitation of ADAMTS-5, but not ADAMTS-4 (Fig. 5G). Fig. 5, G and H indicates that there is an increase in association between SDC4 and ADAMTS-5 in cells treated with either TNF-α or IL-1β. To understand if heparan sulfate side chains played a role in SDC4 interaction with ADAMTS-5, we treated cells with heparinase III and measured levels of secreted ADAMTS-5. Fig. 5I shows that heparinase III treatment results in increased levels of unprocessed ADAMTS-5 in conditioned medium.
FIGURE 5.
Regulation of ADAMTS expression and activity by TNF-α and IL-1β. Real-time RT-PCR analysis of cells treated with A, TNF-α and B, IL-1β show increase in expression of ADAMTS-4 and ADAMTS-5. C–E, Western blot analysis to measure aggrecan degradation using neoepitope antibodies C, anti-ARGSVIL D, anti-NITEGE E, anti-G1. A prominent increase in degradation products corresponding to particular epitopes (marked by asterisk and arrows). F, densitometric analysis of three blots each for experiment described in C, D, and E above. GAPDH was used as a loading control and to calculate relative expression. Increase in degradation products corresponding to epitopes marked by asterisk in C–E was seen by the cytokine treatment. G, immunoprecipitation (IP) of SDC4 from control and cytokine-treated cells followed by a Western blot using ADAMTS-5 antibody shows increased association between SDC4 and ADAMTS-5 in treated cells. IP using an isotype IgG in place of SDC4 antibody showed absence of ADAMTS-5 co-precipitation. H, densitometric analysis of three blots for experiment described in G above shows increased co-precipitation of SDC4 and ADAMTS-5 in cytokine-treated NP cells. I, treatment of NP cells with heparinase (Hep) III resulted in increased shedding of pro-ADAMTS-5 in the medium. Values shown in quantitative experiments are mean ± S.E. from three independent experiments, *, p < 0.05.
SDC4 Controls TNF-α- and IL-1β-dependent Increase in ADAMTS-5 Activity
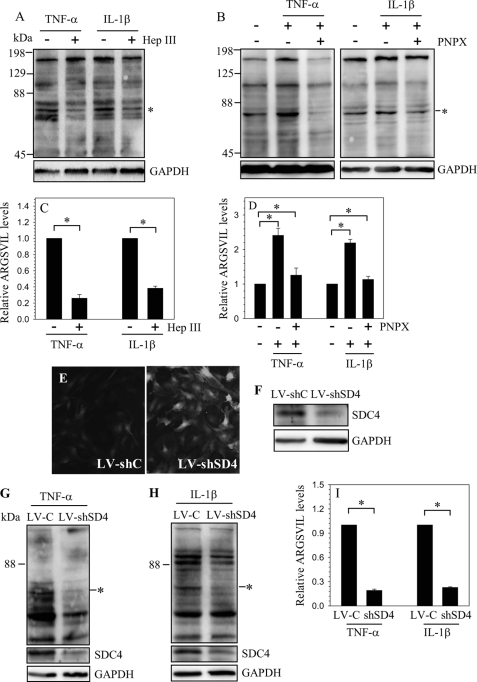
We then determined if SDC4 plays a role in cytokine induced aggrecanase activity in nucleus pulposus cells. When cytokines are added to cells pretreated with heparinase III, generation of aggrecan fragments is decreased (Fig. 6, A and B). A similar phenomenon is observed when cells are pretreated with PNPX, a known inhibitor of heparan sulfate synthesis (Fig. 6, C and D). To further elucidate this observation, we suppressed SDC4 expression in nucleus pulposus cells by transduction with lentivirus expressing SDC4-shRNA. Fig. 6E shows that there is a robust GFP expression by the virally transduced cells, indicating high level of transduction efficiency and transgene expression. Moreover, Fig. 6F shows that in nucleus pulposus cells transduced with ShRNA there is a significant decrease in SDC4 protein expression. Furthermore, when silenced cells were treated with TNF-α (Fig. 6, G and I) as well as IL-1β (Fig. 6, H and I), a significant decrease in aggrecan fragmentation is seen. We then investigated if MMP-3 played a role in cytokine and SDC4-dependent activation of ADAMTS-5. Treatment of nucleus pulposus cells with TNF-α and IL-1β increases mRNA and protein expression of MMP-3 (supplemental Fig. S1, A and B). In addition, we observed that SDC4 is required for cytokine dependent MMP-3 induction (supplemental Fig. S1C). To further explore role of MMP-3 in cytokine dependent ADAMTS function, rat as well as human nucleus pulposus cells were treated with cytokines together with a specific MMP-3 inhibitor, NNGH. Suppression of MMP-3 did not affect cytokine dependent aggrecan degradation in either rat (supplemental Fig. S1D) or human nucleus pulposus cells (supplemental Fig. S1E).
FIGURE 6.
Regulation of ADAMTS activity by SDC4. Western blot analysis of aggrecan neoepitope (ARGSVIL) in cells treated with cytokines with or without A, Heparinase (Hep) III or B, PNPX. A prominent decrease in aggrecan neoepitope formation is seen when cells are treated with either heparinase or PNPX. C and D, densitometric analysis of three blots each for experiment described in A and B, respectively. GAPDH was used as a loading control and to calculate relative expression. A significant decrease in aggrecan neoepitope formation is seen in cells treated with either heparinase or PNPX. E, immunofluorescent detection of GFP in NP cells transduced with lentivirus co-expressing GFP and control ShRNA (LV-shC) and lentivirus co-expressing GFP and SDC4 shRNA (LV-shSD4) shows high transduction efficiency. Magnification ×20. F, Western blot analysis of cells infected with LV-shC and LV-shSD4. The expression of SDC4 was suppressed by SDC4 shRNA compared with cells transduced with control lentivirus. G and H, Western blot analysis of aggrecan neoepitope (ARGSVIL) in LV-shC and LV-shSD4 transduced cells treated with G, TNF-α and H, IL-1β. I, densitometric analysis of three blots each for experiment described in G and H. Note that aggrecan neoepitope generation is significantly reduced following cytokine treatment in SDC4-silenced cells compared with controls. Values shown in quantitative experiments are mean ± S.E. from three independent experiments, *, p < 0.05.
Degenerate Human Disc Tissues Show Elevated SDC4 and ADAMTS-5 Expression and Increase in Aggrecan Degradation
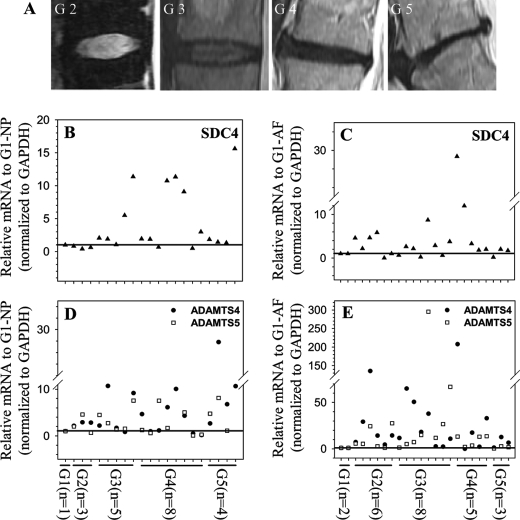
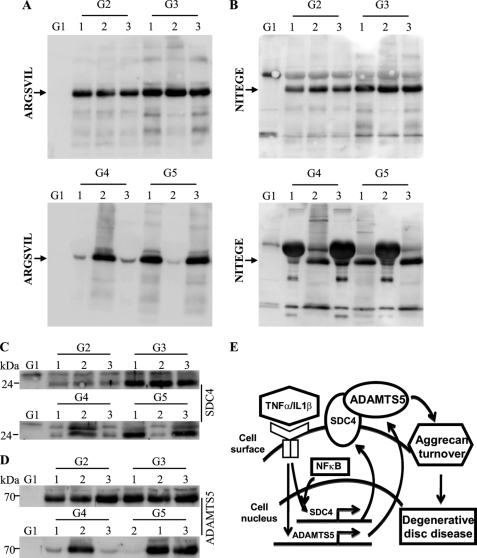
Next, using MRI graded degenerate human nucleus pulposus and annulus fibrosus tissues (Fig. 7A), we evaluated the expression of SDC4, ADAMTS-4, ADAMTS-5 (Fig. 7, B–E), TNF-α, and IL-1β (supplemental Fig. S2). Real-time RT-PCR analysis shows that there is an increase in expression of SDC4 mRNA in human degenerate nucleus pulposus (Fig. 7B) as well as annulus fibrosus (Fig. 7C) samples when compared with the normal control. Similarly, ADAMTS-4 and -5 expression is also responsive to degeneration with higher level of expression at the later stages (stage 3 and stage 4) (Fig. 7, D and E). Moreover, the level of both TNF-α (supplemental Fig. S2A) and IL-1β (supplemental Fig. S2B) transcripts exhibit a similar pattern in degenerate tissues. Finally, we measured aggrecan degradation in human nucleus pulposus tissue samples using anti-G1 neopeptide antibodies. By staining the electrophoresed protein extracts with Coomassie Blue tissue protein integrity and equal protein loading was ensured (supplemental Fig. S2C). While the normal sample showed no evidence of aggrecan degradation, anti-ARGSVIL (Fig. 8A) and anti-NITEGE (Fig. 8B) antibodies indicated that there is a high level of accumulation of G1 neoepitopes in the degenerate samples. Moreover, samples that evidenced increased accumulation of aggrecan neoepitopes also displayed an elevated level of SDC4 (Fig. 8C) as well as ADAMTS-5 (Fig. 8D) protein. Not surprisingly, there is a considerable patient-to-patient variation within the same grade.
FIGURE 7.
SDC4 and ADAMTS-4/5 mRNA expression in human degenerate NP and AF tissues. A, representative MRI images of patients with degenerative disc disease. (From left, Grade II,III,IV,V). B–E, real-time RT-PCR analysis of SDC4 (B and C) and ADAMTS4/5 (D and E) mRNA expression in multiple human NP (B and D) and AF (C and E) tissue samples. Samples from multiple different patients were used for each degenerative grade. Compared with normal control, with increasing disc degeneration there is a relative increase in the expression of SDC4 and ADAMTS mRNA. Mean expression of normal controls (c) was set at 1.0 and shown by a horizontal line.
FIGURE 8.
Increased aggrecan degradation is correlated with SDC4 and ADAMTS-5 protein levels in degenerate human NP tissues. A and B, Western blot analysis of ADAMTS generated aggrecan neoepitopes in human degenerate NP tissue samples using A, anti-ARGSVIL and B, anti-NITEGE antibodies. Both ARGSVIL and NITEGE neoepitope levels are significantly higher in all the degenerate NP samples compared with control tissue. Control sample showed undetectable levels of these aggrecan degradation products. Samples from three different patients were used for each degenerative grade. C, SDC4 and D, ADAMTS-5 expression levels in corresponding human degenerate NP samples. Degenerate samples that exhibit high level of aggrecan neoepitope accumulation also showed a high expression of both SDC4 and ADAMTS-5. Expression of SDC4 and ADAMTS-5 was very low in control sample. E, a proposed model of relationship between inflammatory cytokines (TNF-α, IL-1β), SDC4, and ADAMTS-5 in NP cells. During degeneration high levels of inflammatory cytokines TNF-α and IL-1β transcriptionally induce SDC4 expression through NF-κB signaling pathway. Increased level of SDC4 binds and anchors ADAMTS-5 on cell surface for processing and results in its activation. Active ADAMTS-5 cleaves aggrecan matrix resulting in loss of water binding capacity and disc height.
DISCUSSION
The experiments described in this investigation demonstrated for the first time that expression of SDC4, a key heparan sulfate proteoglycan concerned with activation of many growth factors and MMPs, was regulated by the inflammatory cytokines TNF-α and IL-1β. Our studies also revealed that the cytokine-mediated expression of SDC4 in nucleus pulposus cells was NF-κB dependent. A second major observation was that the by regulating ADAMTS-4 and -5 expression and activity, these inflammatory cytokines controlled aggrecan turnover. Importantly, binding to SDC4 was required for ADAMTS-5 activation and thus this cell surface heparan sulfate proteoglycan indirectly regulated aggrecan turnover in nucleus pulposus cells. In this way, TNF-α and IL-1β compromise the water binding capacity and biomechanical properties of the intervertebral disc and thereby promote the degenerative state.
We showed for the first time that SDC4 was expressed in tissues of the intervertebral disc. The differential expression by cells of the nucleus pulposus and annulus fibrosus may indicate a tissue specific role for SDC4 that may possibly reflect the distinct embryological origins of the two tissues. Although SDC4 has a number of diverse physiological functions, the mechanism of regulation of its expression is not completely understood. The study clearly showed that the treatment of nucleus pulposus cells with TNF-α and IL-1β induced SDC4 expression and promoter activity indicating that regulation was at the transcript level. There is some evidence to indicate that SDC4 expression by H. pylori and TLR agonists may be dependent on NF-κB signaling (20). Results of the studies described herein suggest that this regulatory mechanism is conserved. We showed that both cytokines promoted NF-κB signaling and by binding to the motif between −97/−88 bp, RelA significantly influenced SDC4 expression. Importantly, regardless of the presence of cytokines, suppression of NF-κB activity blocked SDC4 promoter activity, indicating that this transcription factor also controlled basal gene expression. Further support for this finding was the failure of cytokines to increase the activity of a NF-κB mutant reporter. Moreover, cells from RelA-null mice displayed decreased basal SDC4 promoter activity and failed to induce reporter activity even when treated with cytokines. Together, the results of these functional studies indicated that by controlling the expression of SDC4, NF-κB and especially RelA signaling may modulate the activity of several heparan sulfate proteoglycan-dependent growth factors and proteinases in nucleus pulposus cells.
The stimulatory effect of TNF-α and IL-1β on ADAMTS-4 and -5 mRNA expression confirmed and extended earlier studies of nucleus pulposus cells (8, 13, 14). Concerning enzymatic function, it has been shown that SDC1 contributed to ADAMTS-4 activation through interaction with its heparan sulfate and chondroitin sulfate side chains (19). Similarly, in a recent study it was reported that SDC4 plays a role in ADAMTS-5 activation and may contribute to the pathogenesis of osteoarthritis (18). Our results showed that in nucleus pulposus cells, SDC4 interacted selectively with ADAMTS-5, but not ADAMTS-4 and that this heparan sulfate mediated interaction was required to anchor pro-ADAMTS-5 on the cells surface. Moreover, enzymatic removal of heparan sulfate as well as blocking its synthesis clearly indicated that ADAMTS-dependent aggrecan fragmentation was partially dependent on the presence of heparan sulfate. Partial blockage of aggrecan cleavage in cytokine treated SDC4 silenced nucleus pulposus cells lent further support for the role of this molecule in ADAMTS-5 activation. Based on these findings it was concluded that in nucleus pulposus cells, SDC4 is required for cell surface anchoring required for processing and functional activation of ADAMTS-5.
In mouse and human chondrocytes, in addition to SDC4, MMP-3 is required for activation of ADAMTS-5 (18). Moreover, cytokine dependent MMP-3 expression is dependent on SDC4 (18). Of interest, was the observation that unlike chondrocytes, MMP-3 inhibition did not decrease aggrecan turnover in rat and human nucleus pulpous cells indicating cell type specificity. Moreover, inhibition of ERK signaling, a pathway that connects SDC4 to MMP-3 expression in chondrocytes had no effect on aggrecan degradation in cells of the nucleus pulposus (not shown). Based on these findings, it is not unreasonable to assume that in nucleus pulposus cells, MMP-3 activation plays little or no role in cytokine-dependent SDC4-mediated ADAMTS-5 activation.
In human as well as animal models of degenerative disc disease there are increased levels of inflammatory cytokine such as TNF-α and IL-1β; our studies confirmed and extended these earlier findings (7, 8, 23–28). Likewise, in both mouse and human articular cartilage, cytokines and SDC4 expression is elevated during osteoarthritis (18). While the number of control disc tissue samples were very limited (due to practical difficulties in acquiring normal MRI graded human discs), measurement of both mRNA and protein expression levels suggested that as the tissues became degenerate, there was a rise in expression of SDC4, ADAMTS-5 as well as accumulation of high level of ADAMTS generated aggrecan fragments. These results suggested a strong correlation among the expression of SDC4 and ADAMTS-5 and ADAMTS enzymatic activity. Noteworthy, high level of expression of SDC4 in the annulus fibrosus contrasted with the very low levels found in healthy human and rat discs. This observation suggested that during degeneration, in both the nucleus pulposus and annulus fibrosus aggrecan undergoes SCD4-dependent ADAMTS-mediated degradation.
In addition to the transcriptional induction of SDC4 by TNF- α and IL-1β, its expression has also been shown to respond to a variety of other stimuli, including hypoxia (29). It is therefore likely that in the degenerate disc, in addition to the elevated levels of TNF-α and IL-1β, confounding factors in the avascular, hypoxic microenvironment may also contribute to the raised levels of SDC4. From a functional perspective, we conclude that basal expression of SDC4 is possibly required for maintenance of activities of several heparan sulfate-dependent growth factors in the healthy intervertebral disc. On the other hand, in the degenerate disc, cytokine-induced SDC4 expression raised ADAMTS-5 activity in conjunction with other catabolic proteins including MMPs may drive the degradation of the aggrecan-rich matrix (Fig. 8E).
Supplementary Material
This work was supported, in whole or in part, by Grants R01-AR050087 and R01-AR055655 from the National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2.
- MMP
- matrix metalloproteinase
- ADAMTS
- A disintegrin-like and metalloprotease with thrombospondin type I motifs
- PNPX
- p-nitrophenyl-β-d-xylopyranoside
- SDC
- Syndecan
- NP
- nucleus pulposus cells
- AF
- annulus fibrosus.
REFERENCES
- 1. Feng H., Danfelter M., Stromqvist B., Heinegard D. (2006) J Bone Joint Surg Am 88, Suppl. 2, 25–29 [DOI] [PubMed] [Google Scholar]
- 2. Setton L.A., Chen J. (2006) J. Bone Joint Surg. Am. 88, Suppl. 2, 52–57 [DOI] [PubMed] [Google Scholar]
- 3. Ng L, Grodzinsky A.J., Patwari P., Sandy J., Plaas A., Ortiz C. (2003) J. Struct. Biol. 143, 242–257 [DOI] [PubMed] [Google Scholar]
- 4. Roberts S., Evans H., Trivedi J., Menage J. (2006) J. Bone Joint Surg. Am. 88, Suppl. 2, 10–14 [DOI] [PubMed] [Google Scholar]
- 5. Miljkovic D., Trajkovic V. (2004) Cytokine Growth Factor Rev. 15, 21–32 [DOI] [PubMed] [Google Scholar]
- 6. Shamji M. F., Setton L. A., Jarvis W., So S., Chen J., Jing L., Bullock R., Isaacs R. E., Brown C., Richardson W. J. (2010) Arthritis Rheum. 62, 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weiler C., Nerlich A. G., Bachmeier B. E., Boos N. (2005) Spine 30, 44–53 [DOI] [PubMed] [Google Scholar]
- 8. Le Maitre C. L., Freemont A. J., Hoyland J. A. (2005) Arthritis Res. Ther. 7, 732–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kang J. D., Stefanovic-Racic M., McIntyre L. A., Georgescu H. I., Evans C. H. (1997) Spine 22, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 10. Lee S., Moon C. S., Sul D., Lee J., Bae M., Hong Y., Lee M., Choi S., Derby R., Kim B. J., Kim J., Yoon J. S., Wolfer L., Kim J., Wang J., Hwang S. W., Lee S.H. (2009) Clin Biochem. 42, 1504–1511 [DOI] [PubMed] [Google Scholar]
- 11. Yamashita M., Ohtori S., Koshi T, Inoue G., Yamauchi K., Suzuki M., Takahashi K. (2008) Spine 33, 1836–1842 [DOI] [PubMed] [Google Scholar]
- 12. Lee J. M., Song J. Y., Baek M., Jung H. Y., Kang H., Han I. B., Kwon Y. D., Shin D. E. (2011) J. Orthop. Res. 29, 265–269 [DOI] [PubMed] [Google Scholar]
- 13. Séguin C. A., Pilliar R. M., Roughley P. J., Kandel R..A. (2005) Spine 30, 1940–1948 [DOI] [PubMed] [Google Scholar]
- 14. Séguin C. A., Bojarski M., Pilliar R. M., Roughley P. J., Kandel R. A. (2006) Matrix Biol. 25, 409–418 [DOI] [PubMed] [Google Scholar]
- 15. Millward-Sadler S. J., Costello P. W., Freemont A. J., Hoyland J. A. (2009) Arthritis Res. Ther. 11, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glasson S. S., Askew R., Sheppard B., Carito B., Blanchet T., Ma H. L., Flannery C. R., Peluso D., Kanki K., Yang Z., Majumdar M. K., Morris E. A. (2005) Nature 434, 644–648 [DOI] [PubMed] [Google Scholar]
- 17. Stanton H., Rogerson F. M., East C. J., Golub S. B., Lawlor K. E., Meeker C. T., Little C. B., Last K., Farmer P. J., Campbell I. K., Fourie A. M., Fosang A. J. (2005) Nature 434, 648–652 [DOI] [PubMed] [Google Scholar]
- 18. Echtermeyer F., Bertrand J., Dreier R., Meinecke I., Neugebauer K., Fuerst M., Lee Y. J., Song Y. W., Herzog C., Theilmeier G., Pap T. (2009) Nat. Med. 15, 1072–1076 [DOI] [PubMed] [Google Scholar]
- 19. Gao G., Plaas A., Thompson V. P., Jin S., Zuo F., Sandy J. D. (2004) J. Biol. Chem. 279, 10042–10051 [DOI] [PubMed] [Google Scholar]
- 20. Smith M. F., Jr., Novotny J., Carl V. S., Comeau L. D. (2006) Glycobiology 16, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., Shapiro I. M. (2006) J. Cell. Biochem. 98, 152–159 [DOI] [PubMed] [Google Scholar]
- 22. Pfirrmann C. W., Metzdorf A., Zanetti M., Hodler J., Boos N. (2001) Spine 26, 1873–1878 [DOI] [PubMed] [Google Scholar]
- 23. Le Maitre C. L., Hoyland J. A., Freemont A. J. (2007) Arthritis Res. Ther. 9, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bachmeier B. E., Nerlich A. G., Weiler C., Paesold G., Jochum M., Boos N. (2007) Ann. N.Y. Acad. Sci. 1096, 44–54 [DOI] [PubMed] [Google Scholar]
- 25. Omlor G. W., Lorenz H., Engelleiter K., Richter W., Carstens C., Kroeber M. W., Guehring T. (2006) J. Orthop. Res. 24, 385–392 [DOI] [PubMed] [Google Scholar]
- 26. Ulrich J. A., Liebenberg E. C., Thuillier D. U., Lotz J. C. (2007) Spine 32, 2812–2819 [DOI] [PubMed] [Google Scholar]
- 27. Barbir A., Godburn K. E., Michalek A. J., Lai A., Monsey R. D., Iatridis J. C. (2011) Spine 36, 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haschtmann D., Stoyanov J. V., Gédet P., Ferguson S. J. (2008) Eur. Spine J. 17, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Asplund A., Ostergren-Lundén G., Camejo G., Stillemark-Billton P., Bondjers G. (2009) J. Leukoc. Biol. 86, 381–388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.