Background: ESCRT proteins catalyze membrane budding and fission away from the cytosol.
Results: The ESCRT-III protein CHMP2B polymerizes into tubular helical structures deforming the plasma membrane.
Conclusion: CHMP2B, not only mediates recruitment of the ESCRT-dissociating ATPase VPS4, as proposed previously, but also molds membranes.
Significance: ESCRT-III polymerize into a novel kind of membrane deforming filament distinct of actin and tubulin.
Keywords: Electron Microscopy (EM), Endosomes, Membrane, Scaffold Proteins, Trafficking, Cryo-EM, Membrane Remodeling, Endosomal Sorting Complexes Required For Transport, Protein Polymers
Abstract
The endosomal sorting complexes required for transport (ESCRT-0-III) allow membrane budding and fission away from the cytosol. This machinery is used during multivesicular endosome biogenesis, cytokinesis, and budding of some enveloped viruses. Membrane fission is catalyzed by ESCRT-III complexes made of polymers of charged multivesicular body proteins (CHMPs) and by the AAA-type ATPase VPS4. How and which of the ESCRT-III subunits sustain membrane fission from the cytoplasmic surface remain uncertain. In vitro, CHMP2 and CHMP3 recombinant proteins polymerize into tubular helical structures, which were hypothesized to drive vesicle fission. However, this model awaits the demonstration that such structures exist and can deform membranes in cellulo. Here, we show that depletion of VPS4 induces specific accumulation of endogenous CHMP2B at the plasma membrane. Unlike other CHMPs, overexpressed full-length CHMP2B polymerizes into long, rigid tubes that protrude out of the cell. CHMP4s relocalize at the base of the tubes, the formation of which depends on VPS4. Cryo-EM of the CHMP2B membrane tubes demonstrates that CHMP2B polymerizes into a tightly packed helical lattice, in close association with the inner leaflet of the membrane tube. This association is tight enough to deform the lipid bilayer in cases where the tubular CHMP2B helix varies in diameter or is closed by domes. Thus, our observation that CHMP2B polymerization scaffolds membranes in vivo represents a first step toward demonstrating its structural role during outward membrane deformation.
Introduction
Studies in yeast have revealed that protein complexes called ESCRTs (endosome-sorting complexes required for transport) play a pivotal role in intraluminal vesicle budding inside endosomes, an essential step for the sorting of receptors toward degradation (1–3). The particularity of ESCRT complexes is to induce membrane vesiculation with a topology that is inverted compared with coated vesicles budding from a donor membrane and released into the cytosol. Four ESCRT complexes have been characterized so far: ESCRT-0, -I, -II, and -III. ESCRTs 0, I, and II are recruited to membranes as complexes (4). In contrast, ESCRT-III assembles sequentially and transiently at membranes, and requires VPS4 for disassembly (5). Yeast ESCRT-III is composed of seven highly related proteins. Each of them has one to three homologs in mammals, which are grouped into eight families designated CHMP1–7 (charged multivesicular body proteins) and IST1 (increased sodium tolerance-1). In yeast, four ESCRT-III proteins essential for vesicle release have been shown to be recruited on endosomal membranes in the following order: two Vps20p subunits (CHMP6) recruited by ESCRT-II to nucleate the assembly of two Snf7p (CHMP4A-C) spirals, which are capped by Vps24p (CHMP3) and Vps2p (CHMP2A and -B) (6–8). The latter recruits VPS4 (9) (10), to catalyze disassembly of ESCRT-III (11–14). In vitro, yeast ESCRT-I and II are sufficient to form vesicles on giant unilamellar vesicles and ESCRT-III induces membrane fission (15).
In mammalian cells, ESCRT complexes also play a role at the plasma membrane during cytokinesis (16–19) and budding of some enveloped viruses (20–25). Although all ESCRT-III members and VPS4 are required during cell divison (17), siRNA knockdown experiments indicate that only one of the two CHMP2, and one of the three CHMP4, isoforms and their direct interaction are absolutely required for virus egress (26).
Little direct evidence exists so far revealing how mammalian ESCRT-III complexes mold membranes in vivo. ESCRT-III proteins are small helical assemblies that are auto-inhibited in the cytosol and become activated upon interaction with a membrane (12, 27–31). One common theme between CHMP proteins is their capacity to polymerize in vitro. CHMP1A polymerizes into large tubes, IST1 assembles into curled sheets (29), whereas CHMP4B associates into arrays of loose circular filaments (32). In contrast, CHMP3 and CHMP2A polymerize into helical tubular structures (12, 29) that can be closed at one end to form dome-like structures. It has been hypothesized that the tight interaction of the dome with the membrane at the neck of the budding vesicle might lead to the closing of the neck and spontaneous membrane fission (33). In contrast to these structures observed in vitro, CHMP4B overexpression in cultured cells led to its assembly into circular filaments at the plasma membrane. Membrane tubes of unknown composition were emanating from the plasma membrane when CHMP4B was overexpressed together with catalytically inactive VPS4 (34). Recent evidence from HIV-1 budding suggests that although CHMP4 isoforms assemble within the neck, they do not suffice to induce membrane fission in the absence of CHMP2A or -B. This suggests that CHMP2 may be the minimal fission machinery (26) requiring VPS4 to catalyze fission and virion release (35), consistent with the sequential recruitment of ESCRTs and VPS4 to the HIV-1 budding site (36).
Here, we report that down-regulating VPS4 leads to accumulation of endogenous CHMP2B at the plasma membrane, indicating its presence in ESCRT-III complexes formed transiently at the plasma membrane. When overexpressed, CHMP2B is recruited to the plasma membrane, where it forms long protrusions, partly depending on its capacity to bind VPS4. The capacity of CHMP2B to make plasma membrane protrusions is not shared by other CHMPs, and neither CHMP2A, CHMP3, nor CHMP4 isoforms colocalize with CHMP2B inside tubes. However, CHMP4A and -B tend to relocalize at the base of CHMP2B tubes. Cryo-electron microscopy of membrane tubes shed into the cell culture supernatant demonstrates that CHMP2B polymerizes into a tightly packed helical polymer intimately associated with the inner leaflet of the bilayer. Our results show for the first time that CHMP proteins can form tightly packed helical structures in vivo that associate with cellular membranes. Second, such structures vary in diameter to induce membrane constriction, which indirectly supports the dome model for membrane fission (33). Finally, our data demonstrate that in mammalian cells, CHMP2B is not only an adaptor protein for the recruitment of VPS4 as demonstrated in yeast but also directly plays a structural role in membrane scaffolding.
EXPERIMENTAL PROCEDURES
DNA Construct
The wild-type human CHMPs (2A, 3, 4A, 4B, and 4C) cDNAs were cloned from HeLa cells by RT-PCR, in frame with a C-terminal FLAG inserted into reverse primers (supplemental Table S1). CHMP2B, CHMP2Bintron5-FLAG, and CHMP2B-FLAG, CHMP2BL207D/L210D-FLAG and CHMP2BL4D/F5D-FLAG were generated by PCR using plasmids containing HA-CHMP2B or GFP-VPS4B as templates, introducing Stop codon or point mutations inside primers where needed. (All PCR primers used in this study are described in supplemental Table S1.) PCR fragments were inserted into the pcDNA3.1 vector using a TOPO directional cloning kit (Invitrogen).
For expression in bacteria, cDNA fragments derived from human full-length CHMP2B (wt CHMP2B and mutant L4D/F5D), were subcloned in frame with a C-terminal His6 tag into the expression vector pETM-13, using NcoI and XhoI. Proteins were produced in Escherichia coli strain BL21 codon plus (Invitrogen).
GPF-AMSH was a generous gift from Sylvie Urbé. pCaggs GFP-VPS4B and pCaggs GFP-VPS4E235Q were described previously (37).
Reagents and Antibodies
Polyclonal antibodies against the C-terminal part of CHMP2B were purchased from Abcam (ab33174). Polyclonal anti-CHMP4A (H-52) and CHMP4B (C-12) were obtained from Santa Cruz Biotechnology. Polyclonal anti-CHMP4B (ab105767) was from Abcam. Polyclonal antibodies against VPS4A and B were a kind gift from W. I. Sundquist (University of Utah). Monoclonal and polyclonal anti-FLAG antibodies were obtained from Sigma-Aldrich. Rabbit polyclonal antibody against β-tubulin was a generous gift from A. Andrieux (Inserm U836, Grenoble, France). Secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 594, Alexa 594-WGA, and Texas Red-X-phalloidin were obtained from Molecular Probe.
Cell Cultures and Transfections
For immunofluorescence, HeLa cells were seeded onto sterile glass coverslips in six-well plates (105 cells per well) in DMEM (Invitrogen) containing 10% FBS (Sigma), and 2 mm l-glutamine (Sigma). Cells were transfected 24 h later with 1 μg of DNA and 6 μl of jetPEI transfection reagent (Polyplus) and mixed in a final volume of 200 μl of 150 mm NaCl. For cotransfection, 1 μg of CHMP2B containing plasmid was mixed with 500 ng of plasmids encoding the other proteins. Cells were fixed and processed for immunostaining 24 or 36 h after transfection. For actin destabilization, cells were incubated 1 h with 1 μm cytochalasin D or with 0.2 mg/ml latrunculin A (Sigma).
siRNA Transfection
HeLa cells transfected with a mixture containing 10 nm of each of the previously described siRNA duplexes (17) (VPS4A, CCGAGAAGCUGAAGGAUUAtt; VPS4B, CCAAAGAAGCACUGAAAGAtt) using Lipofectamine RNAi MAX (Invitrogen). Cells were transfected again after 24 h and fixed 24 h later. Control cells were transfected with 20 nm of siRNA duplexes against LucGL2 (CGUACGCGGAAUACUUCGAtt). The extent of down-regulation was estimated by Western blot analysis of cell lysates.
Western Immunoblotting
Cells were lysed in radioimmune precipitation assay buffer containing protease inhibitors (Complete, Roche Molecular). Proteins (20 μg of total protein per lane) were separated on SDS-polyacrylamide gels (10%) and analyzed by immunoblotting with the indicated antibodies.
Immunofluorescence
Cells were fixed and immunostained as described in Ref. 39. In brief, cells were fixed with 4% paraformaldehyde. After three washes in PBS, cells were permeabilized with PBS containing 0.5% Triton X-100 in blocking solution (PBS containing 5% goat pre-immune serum) for 30 min. Primary antibodies diluted in blocking solution were then incubated for 1 h. After washing in PBS, cells were incubated for 1 h with secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 594, Alexa Fluor 594-WGA, or Alexa Fluor 488-phalloidin. After three washes in PBS, coverslips were rinsed in water and mounted in Mowiol.
For staining of F-actin, Texas Red-X phalloidin was incubated together with the secondary antibodies. For delineating cellular membranes, live cells were washed in HBSS, incubated 10 min at 4 °C with Alexa Fluor 494-WGA (5 μg/ml in Hank's Buffered salt solution) and washed in PBS before fixation.
Liposome Flotation Assay
Lipsomes were made of synthetic 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine and 1,2-dioleoyl-sn-glycero-3-phosphoserine from Avanti Polar Lipids. Lipids were mixed homogeneously in chloroform to achieve a 8:2 molar ratio. Dried thin lipid films were obtained by evaporation. Lipids were hydrated in 20 mm HEPES, pH 7, to a final concentration of 2.5 mg/ml. The translucent solution was then extruded 15 times through a 200-nm polycarbonate membrane (Avanti Polar Lipids). The average diameter of the liposomes was estimated to range between 200 to 400 nm by dynamic light scattering measurement and electron microscopy
Proteins and large unilamellar vesicles were incubated overnight. The solution was mixed with an equal volume of 80% (w/v) sucrose in 20 mm HEPES, pH 7, resulting in a final sucrose concentration of 40% (w/v) with a final large unilamellar vesicle concentration of 0.625 mg/ml. 300 μl of this solution was overlaid with 100 μl of 30, 20, and 10% (w/v) sucrose in no-salt buffer. The gradient was then subjected to ultracentrifugation at 190,000 × g for 6 h at 4 °C. After ultracentrifugation, gradients were separated into seven fractions. Sucrose concentration of each fraction was determined by refractometry; one-third of each fraction was analyzed by SDS-PAGE.
Confocal Microscopy
Images of fluorescent cells were acquired with a Zeiss LSM-710 laser scanning confocal microscope with a 63× Plan-Apochromat objective (numerical aperture 1.4). Laser power was adjusted to maximize the dynamic range of each sample. For dual-color samples, the adjustable spectral window of fluorescence collection was set for each channel with a singly colored control sample, so that cross-contamination between channels was avoided.
Because CHMP2B tubes protrude out of the cell in a large Z range, stacks of optical sections were acquired with the required electronic magnification. Image files were processed with ImageJ software. Except when mentioned, images represent maximum intensity projections.
CHMP2B Tube Purification
For isolation of CHMP2B tubes, HeLa cells were seeded in 10-cm culture dishes (∼106 cells/well). The culture medium was the same as mentioned above, except that FBS was previously cleared by 60,000 × g centrifugation to remove potential contaminants. Cells were transfected 24 h later with 3 μg of DNA and 18 μl of jetPEI transfection reagent (Polyplus transfection) and mixed in a final volume of 600 μl of 150 mm NaCl. Culture media harvested 36 h after transfection were centrifuged twice at 300 × g for 5 min to remove debris and then at 30,000 × g (1 h) to pellet membranes. The resulting pellet (P1) was washed once with HBS (150 mm NaCl, 20 mm HEPES, pH 7.4), resuspended and incubated for 30 min at 4 °C in HBS containing 1 mm AMPPnP6 (Sigma), CompleteTM protease inhibitor (Roche Applied Science), and 1% Triton X-100. Half of this suspension was centrifuged at 20,000 × g for 30 min, and the pellet (P2) was resuspended in the same volume as S2. Identical volumes of P1, S2, and P2 were analyzed by Western blot together with the cell lysate (20 μg of proteins estimated using BCA protein assay reagent (Pierce).
Electron Microscopy
Immuno-EM Analysis of CHMP2b-expressing Cells
Cells were fixed with 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 m phosphate buffer, pH 7.3, during 2 h. Cells were then gently detached using a cell scraper, centrifuged at 1200 rpm during 5 min and embedded in 10% gelatin. The cell pellet was then cut into 1 mm3 pieces. These samples were cryoprotected during 4 h in 2.3 m sucrose and frozen in liquid nitrogen. Ultrathin cryosections of 40 nm were made at −120 °C using an ultra-cryo-microtome (Leica-Reichert) and retrieved with a 1:1 solution of 2.3 m sucrose and 2% methylcellulose according to the Tokuyasu protocol (40). Cryosections were first incubated with primary polyclonal anti-FLAG antibody (Sigma) and revealed with protein A-gold conjugate (cell microscopy center, Utrecht, The Netherlands). Double immunogold labeling was performed sequentially using 15-nm protein A gold to recognize anti-CHMP4A and 10-nm protein A gold to detect CHMP2B. Labeled cryosections were viewed at 80 kV with a 1200EX JEOL TEM microscope, and images were acquired with a digital camera (Veleta, SIS, Olympus).
Negative Staining
P1 pellet obtained from the culture media of ten 100-mm dishes of HeLa cells transfected with CHMP2B-FLAG were resuspended in 80 μl of HBS. 6 μl of this sample was incubated during 10 min on an EM grid coated with Formvar and carbon. Grids were then quickly blotted and washed three times in water before staining with 1% uranyl acetate during 1 min, followed by blotting to remove excess liquid. For immunolabeling, samples were permeabilized with Triton X-100 1% during 20 min, and labeling was performed after extensive washing with water using an anti-CHMP2B antibody against the whole protein. Protein A gold (Cell Microscopy Center, Utrecht, The Netherlands) was used to reveal the antibody. Finally, grids were stained with 1% uranyl acetate, pH 4, during 1 min. Grids were viewed under a transmission electron microscope (JEOL 1200EX), and images were acquired with a digital camera (Veleta, Soft Imaging Solutions, Olympus).
Cryo-EM
P1 pellets obtained from the culture medium of thirty 100-mm dishes were resuspended in 40 μl of HBS. 4 ml of sample were loaded onto a Quantifoil R2/1 holey grid (Quantifoil Micro Tools GmbH), blotted for 1–2 s to remove the excess liquid, and then rapidly plunged into liquid ethane cooled using liquid nitrogen. The frozen grid was transferred into an FEI Tecnai Polara electron microscope. Images were taken under low dose conditions at 300 kV (<10 e−/Å2) and with a nominal magnification of 45,000 and a defocus between 2 and 3 μm using a CCD camera (GATAN Ultrascan 4000).
Two-dimensional Average
Two-dimensional averaging of particles was performed by selecting a total of 35 boxes (900 × 900 pixels) equally spaced along one tube using the x3d program (41) and averaging them together after cross-correlation using SPIDER (42).
RESULTS
CHMP2B Localizes to Plasma Membrane upon VPS4 Knockdown
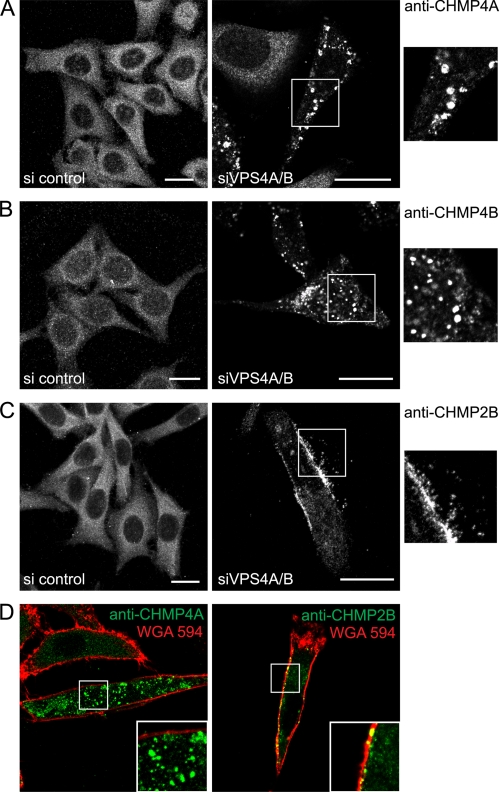
Immunostaining of CHMP4A, -B, and -2B revealed that all endogenous proteins were distributed homogeneously throughout the cytoplasm. siRNA knockdowns of VPS4A and VPS4B (supplemental Fig. S2A) led to a relocalization of cytosolic CHMP4A and CHMP4B to intracellular patches, possibly reflecting their accumulation at the surface of endosomes (Fig. 1, A and B). In contrast, similar depletion of VPS4 led to relocalization of endogenous CHMP2B from the cytoplasm into patches, which were associated with the plasma membrane in some cells (Fig. 1, C and D, and supplemental Fig. S2B). This suggests that VPS4 is required to dissociate plasma membrane-based ESCRT-III complexes containing CHMP2B and thus intimately controls ESCRT-III activity at the plasma membrane.
FIGURE 1.
VPS4A and -B down-regulation induces plasma membrane accumulation of endogenous CHMP2B. HeLa cells were depleted of VPS4A and VPS4B using a mixture of siRNA against both proteins (right panels). Control cells were transfected with an irrelevant siRNA (left panels). The confocal section of control cells or of cells depleted of VPS4 was immunostained with antibodies against CHMP4A (A), CHMP4B (B), or CHMP2B (C). Enlargements of the boxed regions are shown on the right. D, confocal sections of VPS4-depleted cells treated with Alexa Fluor 594-WGA to delineate membranes. Cells were immunostained using anti-CHMP4A (left panel) or anti-CHMP2B (right panel). Scale bars, 20 μm.
Expression of Wild-type CHMP2B Induces Cell Surface Protrusions
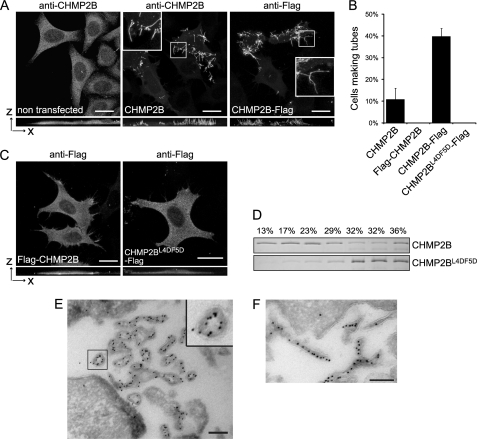
Overexpressed CHMP2B was distributed homogeneously throughout the cytoplasm. However, in 10% of cells, the cytoplasmic staining was partially or entirely lost, and CHMP2B was relocated to the plasma membranes, producing long protrusions pointing out into the culture medium (Fig. 2A). Some protrusions had a length of up to several tens of microns. Fusion of a FLAG peptide to the C-terminal part of CHMP2B increased the capacity of the protein to assemble into surface protrusions, which were detected in ∼40% of CHMP2B-FLAG-positive cells (Fig. 2, A and B). Projection in the x-z plane (Fig. 2A) and three-dimensional reconstruction (supplemental Movies S1 and S2) of cells expressing CHMP2B or CHMP2B-FLAG show that the tubes protrude on the sides and above the cell and are rarely attached to the cell culture substrate. In some cases, however, CHMP2B protrusions detached from the cell surface were observed lying on the cell culture substrate.
FIGURE 2.
Overexpression of CHMP2B or CHMP2B-FLAG induces formation of long cell surface protrusions in which they concentrate. HeLa cells were transfected with the indicated plasmids and immunostained 36 h later. Except otherwise stated, photographs presented in all figures are maximal intensity projections of confocal image stacks. In A and C, bottom panels represent projections in the x-z plane. Bars, 20 μm. A, CHMP2B immunostaining of cells transfected with the empty vector reveals a homogeneous cytoplasmic localization of the protein. Overexpression of CHMP2B and CHMP2B-FLAG induces the formation of cell surface protrusions in which CHMP2B concentrates. Note the absence of cytoplasmic staining in tube forming cells. B, percentage of transfected cells displaying tubes as revealed with anti-CHMP2B antibodies. For each condition, 200 CHMP2B-expressing cells were counted. Data from three independent experiments are plotted. Standard error bars are shown. C, FLAG-CHMP2B and CHMP2BL4D/F5D-FLAG do not induce surface protrusions. D, flotation experiments demonstrate that mutations in Leu-4–Phe-5 impair the capacity of CHMP2B to associate with membranes in vitro. Purified recombinant full-length CHMP2B and CHMP2BL4D/F5D were mixed with liposomes. Liposomes were floated on a sucrose gradient, fractions were run on SDS-PAGE, and proteins were revealed by Coomassie staining. E and F, FLAG immunogold labeling of cells overexpressing CHMP2B-FLAG. Cross-sections of tubes reveal the presence of the C-terminal FLAG lining the lumen of a hollow, electron-dense structure closely associated with the membrane of tubes; longitudinal sections reveal the presence of CHMP2B along the entire length of the tubes. Scale bars, 200 nm.
Immunoelectron microscopy of CHMP2B-FLAG-expressing cells demonstrated the presence of tubular, electron dense structures covered by a single membrane (Fig. 2, E and F). The hollow electron dense structures were labeled with FLAG antibodies, revealing the presence of CHMP2B along the entire length of the tubes (Fig. 2F). In tube cross-sections, FLAG immunolabeling was seen in the lumen of the tubes indicating that the C-terminal part of CHMP2B points toward the interior of the tubular structure (Fig. 2E).
Expression of CHMP2B also induced the formation of tubes in HEK 293T cells (data not shown). CHMP2B expressed in post-mitotic neurons also deformed the membrane into protrusions in which it accumulates (supplemental Fig. S3A). This demonstrates that CHMP2B protrusions do not result from cell retraction occurring during cell migration or division.
N-terminal End of CHMP2B Is Required to Deform Plasma Membrane
In contrast to CHMP2B-FLAG and wild-type CHMP2B, N-terminally FLAG-tagged CHMP2B (FLAG-CHMP2B) remained cytoplasmic and failed to induce growth of surface protrusions (Fig. 2, B and C). Similarly, an HA tag fused to the N terminus blocked the capacity of CHMP2B to induce membrane tubulation (data not shown). Western blot analysis showed similar levels of expression of CHMP2B, FLAG-CHMP2B, or CHMP2B-Flag (supplemental Fig. S3B), demonstrating that the inability of N-terminally tagged versions of CHMP2B to induce tubule formation is not due to differences in their expression level.
Based on these observations, we reasoned that the N-terminal part of the protein might be crucial for deforming membranes. The CHMP2B N-terminal eight amino acids are conserved throughout the CHMP family. In particular Leu-4 and Phe-5 of CHMP2B, followed by a cluster of positively charged residues (Lys and Arg) are conserved within all CHMPs (supplemental Fig. S1B). Expression of the double mutant L4D/F5D (CHMP2BL4D/F5D-FLAG) did not deform the plasma membrane into protrusions (Fig. 2, B and C). To test whether these mutations affect membrane binding, a liposome flotation assay was employed using purified recombinant CHMP2B and CHMP2BL4D/F5D. The presence of CHMP2B in the upper fractions demonstrated the capacity of the protein to interact with liposomes (Fig. 2D). In contrast, most CHMP2BL4D/F5D stayed in the bottom fraction, demonstrating that the L4D/F5D mutation impairs CHMP2B lipid bilayer interaction in vitro. These data confirm the requirement of an intact N terminus for CHMP2B function.
Co-expression of CHMP4 with CHMP2B Affects Capacity of CHMP2B to Induce Membrane Tubulation
Because formation of CHMP2B cell protrusions depends on an intact N terminus, we re-examined the expression of C-terminally FLAG-tagged CHMP2A, CHMP3, and the CHMP4A, -B, and -C isoforms. None of these induced membrane protrusions when expressed alone. Although CHMP2A and CHMP3 polymerize into tubes in vitro, no such structures could be observed in vivo upon co-expression of CHMP2A-FLAG and CHMP3-FLAG (data not shown).
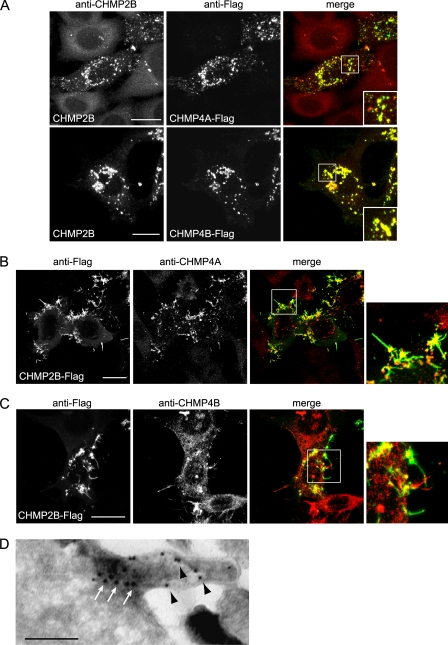
CHMP4 co-expressed with CHMP2B abolished CHMP2B protrusions and led to relocalization of CHMP2B to CHMP4-containing structures (Fig. 3A). In contrast, neither CHMP2A nor CHMP3 abolished formation of CHMP2B protrusions (data not shown). These observations thus suggest that CHMP4 isoforms regulate the capacity of CHMP2B to polymerize and deform the plasma membrane.
FIGURE 3.
Relationship between CHMP4- and CHMP2B-induced protrusions. HeLa cells were transfected with the indicated plasmids and immunostained 36 h later. A, CHMP4A-FLAG or CHMP4B-FLAG, co-expressed with similar amounts of CHMP2B, block the capacity of CHMP2B to form protrusions. Note that the relocalization of CHMP2B inside CHMP4A or CHMP4B patches. In cells expressing CHMP2B-FLAG only, immunostaining with anti-CHMP4A (B) or anti-CHMP4B (C) antibodies reveals the relocalization of endogenous CHMP4A and CHMP4B at the base of CHMP2B tubes. The right panels show enlargement of the boxed regions in the merged panels. In all cases, scale bars are 20 μm. D, double immunogold labeling of a tube emanating from a cell overexpressing CHMP2B-FLAG. Anti-FLAG staining (10-nm beads, black arrowheads) reveals the presence of CHMP2B along the entire length of the tube; anti CHMP4A (15-nm beads, white arrows) reveals CHMP4A labeling restricted at the base of the tube. Scale bars, 200 nm.
Immunostaining with antibodies against CHMP4A and CHMP4B of CHMP2B-overexpressing cells revealed that the endogenous proteins were both recruited at the base of CHMP2B protrusions (Fig. 3, B and C). This localization was confirmed using double immunogold labeling of CHMP2B-FLAG expressing cells with anti-FLAG and anti CHMP4A antibodies: CHMP4A was confined to a dense core material at the base of the tube whereas CHMP2B was distributed along the entire length (Fig. 3D).
CHMP2B Membrane Tube Formation Is Regulated by VPS4
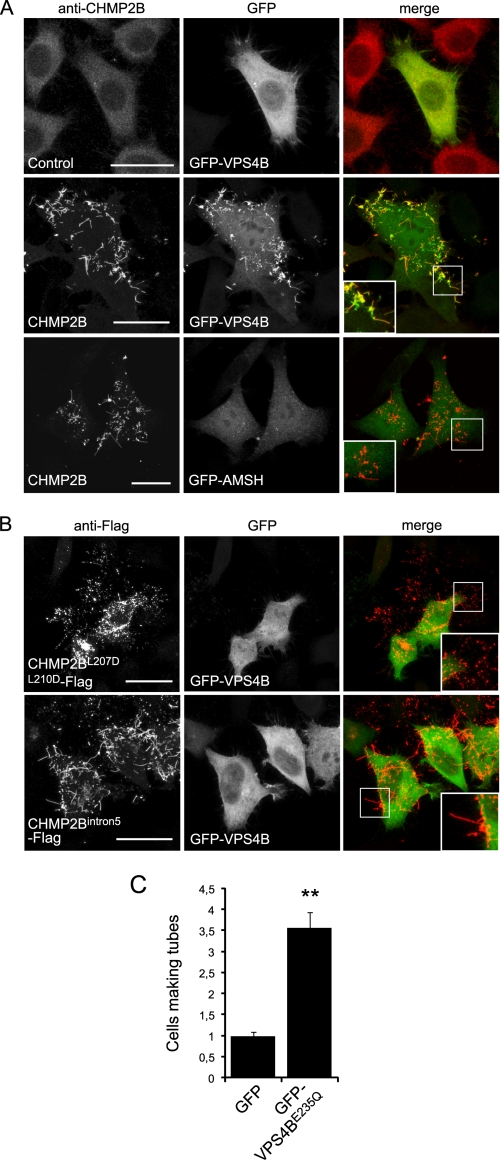
The C-terminal MIT domain-interacting motif (MIM) of CHMP2B recruits the ATPase VPS4 (9). Accordingly, we found that GFP-VPS4B decorated the entire length of CHMP2B-containing protrusions (Fig. 4A). In contrast, the ubiquitin hydrolase GFP-AMSH, which binds to the MIMs of CHMP1A, -B, -2A, and -3 but not that of CHMP2B (28, 43) was absent from CHMP2B protrusions (Fig. 4A). This further indicates that none of these proteins take part in the making of CHMP2B protrusions.
FIGURE 4.
Relationship between VPS4 and CHMP2B protrusions. HeLa cells were co-transfected with the indicated plasmids and observed 36 h later. A, in cells transfected with the control plasmid together with GFP-VPS4, VPS4 is distributed homogeneously in the cytoplasm. In cells expressing both GFP-VPS4B and CHMP2B, VPS4 is mainly present in CHMP2B tubes. In contrast, AMSH is not recruited in CHMP2B protrusions made by cells expressing both GFP-AMSH and CHMP2B. B, CHMP2BL207D/L210D-FLAG, mutated in the MIM domain, accumulates in plasma membrane patches but does not induce protrusions. CHMP2Bintron5, which lacks the C-terminal α6 helix containing the MIM domain, forms everting tubes. GFP-VPS4 is recruited neither in CHMP2BL207D/L210D-FLAG patches nor in CHMP2Bintron5-FLAG containing tubes. C, the catalytically inactive form of VPS4B (GFP-VPS4BE235Q) increases the proportion of cells displaying CHMP2B protrusions: HeLa cells were transfected with CHMP2B together with GFP-VPS4BE235Q and observed 24 h later. For each condition, 200 cells from three independent experiments were counted. Standard error bars are shown. Mann-Whitney statistical analysis was used (p < 0.0011). Scale bars, 20 μm. **, p < 0.0011.
We next tested whether VPS4 binding to CHMP2B is necessary for its membrane tubulating activity. Here, we used a CHMP2B double mutation L207D/L210D (CHMP2BL207D/L210D-FLAG), which impairs VPS4B recruitment (supplemental Fig. S1) (9). CHMP2BL207D/L210D-FLAG did not co-localize with GFP-VPS4B, confirming its inability to bind the ATPase (Fig. 4B). Unexpectedly, CHMP2BL207D/L210D-FLAG was concentrated inside patches but almost never induced tube formation. This observation suggests that VPS4 is required for the formation or integrity of CHMP2B tubes. However, a CHMP2B mutant (CHMP2Bintron5), which lacks part of the C terminus, including the MIM domain, formed patches at the plasma membrane like CHMP2BL207D/L210D-FLAG but nevertheless polymerized efficiently into membrane tubes (Fig. 4B). GFP-VPS4B was not detected inside tubes made of CHMP2Bintron5-FLAG. We conclude that interaction of VPS4 to the C-terminal MIM domain is required for full-length CHMP2B to form membrane tubes. This was further supported by the fact that co-expressing catalytically inactive VPS4B (VPS4E235Q) with CHMP2B, strikingly increased the number of cells making tubes compared with cells expressing CHMP2B only (Fig. 4C). The requirement for the VPS4-CHMP2B interaction can be overcome by deletion of the C-terminal part, which is thought to release autoinhibition as in the case of CHMP2Bintron5. Thus, VPS4 interaction might be necessary to release the autoinhibition in full-length CHMP2B, a process that is required to induce CHMP membrane association and polymerization. However, it is not necessary to form or maintain the structures of the protrusions.
Relationship between CHMP2B and Actors of Membrane Deformation
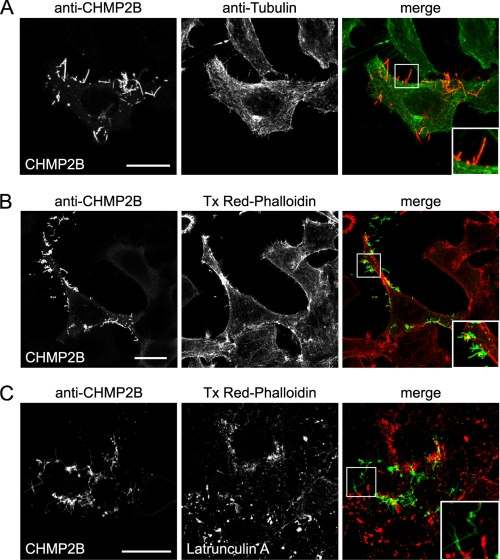
Classically, actin and tubulin underlie plasma membrane protrusions. No immunostaining of β-tubulin was detected inside CHMP2B tubes (Fig. 5A). Absence of microtubules inside CHMP2B protrusions was also obvious from electron microscopy data (Fig. 2E). Thus, CHMP2B-induced protusions are not related to microvilli or other protrusions made by microtubules.
FIGURE 5.
HeLa cells CHMP2B protrusions do not contain actin or tubulin. Immunostaining with anti-β-tubulin antibody (A) and staining with Texas Red (Tx Red)-phalloidin (B) of HeLa cells overexpressing CHMP2B demonstrate the absence of tubulin and F-actin in CHMP2B tubes. C, latrunculin A treatment (0.1 μg/ml; 60 min) induces depolymerization of actin but does not collapse CHMP2B tubes.
F-actin stained by phalloidin was almost never present inside CHMP2B protrusions, demonstrating that it is not necessary to keep them intact (Fig. 5B). The lack of a structural role of actin for maintaining CHMP2B protrusions was confirmed by the fact that latrunculin A (Fig. 5C) or cytochalasin D (data not shown), which dramatically disrupted the actin cytoskeleton, did not affect their integrity. Our observations thus demonstrate that CHMP2B assembly can scaffold plasma membrane tubes independently of actin and tubulin.
Membrane Tubes Made of CHMP2B Are Resistant to Detergent
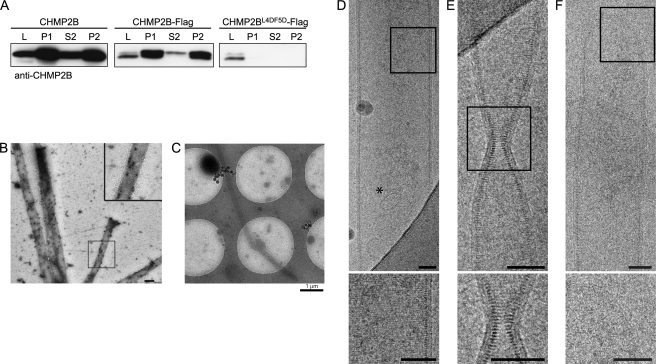
Because CHMP2B protrusions are often found shed from HeLa cells, we collected them from the culture medium of CHMP2B-expressing cells. After low speed centrifugation, media were centrifuged at 30,000 × g. The pelleted material (P1) was solubilized in 1% Triton X-100 and centrifuged at 20,000 × g (P2 and S2). Western blot analysis demonstrated the presence of large amounts of CHMP2B in the P1 pellets, the vast majority remaining insoluble in Triton X-100 (Fig. 6A). This is in good agreement with the known resistance of ESCRT-III complexes to non-ionic detergents. No CHMP2B immunopositive material was recovered from P1 or P2 pellets prepared from cell supernatants of cells transfected with CHMP2BL4D/F5D-FLAG in accordance with the inability of the mutant to make tubes.
FIGURE 6.
Characterization of tubes pelleted from culture media of CHMP2B-expressing cells. A, Western blotting analysis using anti-CHMP2B of culture media of CHMP2B, CHMP2B-FLAG, and CHMP2BL4D/F5D-FLAG expressing cells. L, cell lysates; P1, 30,000 × g pellet of culture media. P2 and S2, P1 pellets solubilized in 1% Triton X-100 were centrifuged at 20,000 × g: most of the CHMP2B-containing material present in culture media is resistant to detergent extraction. No CHMP2B pelletable material could be recovered from media of CHMP2BL4D/F5D-FLAG-expressing cells. B, P1 pellets of culture medium of CHMP2B-FLAG-expressing cells contain tubes made up of CHMP2B. P1 pellets were fixed, permeabilized, and immunolabeled with anti-CHMP2B antibodies revealed by protein A gold (10 nm). C–F, cryo-EM analysis of P1 pellets prepared from culture medium from CHMP2B-FLAG-expressing cells reveals the structure of CHMP2B tubes. C, low magnification shows tubes with a length reaching at least 8 μm. D, striations perpendicular to the longitudinal axis can be seen across the width of the tube. An asterisk indicates a 50-nm vesicle inside of the tube. E, constriction of a tube with no change in the helix pitch. F, dome closing one end of a tube. The inner leaflet of the membrane is closely associated with the CHMP2B protein lattice. Scale bars, 50 nm.
EM examination of P1 pellets negatively stained with uranyl acetate demonstrated the presence of numerous rigid tubes surrounded by plasma membrane (data not shown). Their diameter ranged from 70 to 350 nm. No such tubes could be seen in similar supernatants of untransfected cells. Immunogold labeling performed after permeabilization revealed the presence of CHMP2B along the entire length of the tubes (Fig. 6B).
Cryo-EM Observation Reveals CHMP2B Polymerized into Tightly Packed Helical Structure
We next observed CHMP2B tubes present in P1 pellets using cryo-EM to better characterize their structure (Fig. 6, C–F, and supplemental Fig. S4). In these conditions, the diameter of the tubes ranged from 100 to 400 nm. Regular striations perpendicular to the longitudinal axis of the tube could be revealed. The hollow nature of the tubes was demonstrated as they were sometimes filled with small vesicles (Fig. 6D). Regardless of the diameter of the tubes, the striations of CHMP2B tubes were regular (Fig. 6, D–F, and supplemental Fig. S4) and spaced by a 32 Å gap as determined by Fourier transform of an image, indicating a helical structure with a pitch of 32 Å (supplemental Fig. S4). The CHMP2B protein lattice was tightly linked to the internal leaflet of the membrane derived from the plasma membrane. In some cases, the tube was constricted for example from a diameter of ∼80 to ∼16 nm. Because the lateral striation remains well visible, constriction is most likely induced by gradually reducing the number of CHMP protomers per helical turn (Fig. 6E). Sometimes tubes had a succession of two uniform segments with different diameters (data not shown) or presented closed ends with continuous membrane (Fig. 6F and supplemental Fig. S4). These analyses show that CHMP2B assembles into a helical polymer to deform the membrane into tubes with variable diameters.
DISCUSSION
ESCRT-III catalyzes membrane budding and fission processes during Multi Vesicule Body biogenesis, cytokinesis, and enveloped virus budding (25, 44, 45). In yeast, sequential assembly of VPS20p (CHMP6), Snf7p (CHMP4), Vps24p (CHMP3), and Vps2p (CHMP2) is required for function, whereas SNF7p was suggested to constitute the driving force of membrane fission (6–8). The importance of Snf7 was further highlighted by giant unilamellar vesicles budding assays showing that vesicles can be released into the lumen of the giant unilamellar vesicles by yeast ESCRT-III Vps20p (CHMP6), Snf7p (CHMP4), and Vps24p (CHMP3) (15). However, the mammalian system might differ in that CHMP4 still plays a crucial role but requires in addition CHMP2 to catalyze membrane fission: although CHMP4 isoforms are recruited to the interior of the membrane neck of budding HIV-1 virions, virions are not released in the absence of both CHMP2 isoforms (26). Furthermore, VPS4 is recruited last and before virion release (35, 36). Similarly, CHMP4 in conjunction with CHMP2A have been implicated in constriction and membrane fission at the cytokinetic midbody (19, 26, 46). Thus, both CHMP4 and CHMP2 play important roles in HIV-1 release and cytokinesis.
Here, we show that overexpression of intact CHMP2B is sufficient to induce the formation of membrane tubes protruding from the cell surface. The capacity of CHMP2B to make cell protrusions and to bind to membranes in vitro depends on an intact N-terminal end, suggesting that this domain participates in the regulation of the interaction between the lattice and the inner membrane leaflet. Even though membrane recruitment of and deformation by CHMP2B were easily observed in the present experiments, this property has escaped detection in previous studies. This can be explained by the fact that all the other studies used tagging of the protein at the N terminus, which clearly abolishes its capacity to form plasma membrane tubes.
Even if CHMP proteins vary extensively in their primary sequence, they share a common folded structure with an N-terminal basic domain interacting with the C-terminal acidic regulatory domain. Because CHMPs are cytosolic, they require an activation step to polymerize on membranes (12, 27–29). The autoinhibition is thought to be lifted by displacement of the C-terminal region from the N-terminal core, which subsequently permits polymerization at membranes. The fact that wild-type CHMP2B is able to form tubes indicates that the protein needs to be activated prior to polymerization at the plasma membrane. The AAA-ATPase VPS4 could be part of this activation through binding to the C-terminal MIM domain of CHMP2B, thus releasing autoinhibition. A similar mechanism has already been described for activation of CHMP3 by AMSH (28). Indeed, mutations in the VPS4 binding site of CHMP2B prevent membrane tube formation. Furthermore, co-expression with catalytically inactive VPS4B increases the propensity of CHMP2B to polymerize into tubes. In humans, dominant mutations in CHMP2B cause frontotemporal dementia (47–49). One mutation generates a distinct aberrant transcript, CHMP2Bintron5, which encodes a protein lacking the α6 helix, thus rendering the protein constitutively active. CHMP2Bintron5 mutant induces tube formation, demonstrating that the lack of VPS4 binding can be overcome by the absence of the C-terminal regulatory domain.
On the other hand, the ATPase activity of VPS4 is known to regulate the dissociation of ESCRT-III complexes (11, 13, 14). Accordingly, down-regulation of VPS4A and -B revealed the accumulation of cytoplasmic CHMP4A-containing ESCRT-III complexes. In contrast, the same down-regulation led to preferential accumulation of endogenous CHMP2B in plasma membrane patches. This suggests that ESCRT-III complexes forming transiently at the plasma membrane are major sites of CHMP2B polymerization. Altogether, our results suggest that VPS4 plays a dual regulatory role: binding to CHMP2B would structurally favor polymerization into tubes, whereas its ATPase activity depolymerizes plasma membrane ESCRT-III complexes. CHMP2Bintron5 polymerization would escape control by VPS4 potentially contributing to its pathogenic activity (39).
A remarkable finding was the recruitment of endogenous CHMP2B in VPS4 knocked down cells at the plasma membrane, which correlates well with the fact that the overexpressed protein makes tubes only at the cell surface. This preferential recruitment of CHMP2B at the plasma membrane could explain our recent observation in neurons of its involvement in the shaping of dendritic spines, which are small protrusions corresponding to post synaptic parts of synapses (39). It is also in good agreement with the crucial role played by CHMP2s in HIV-1 virus egress (26).
None of the CHMPs tested so far, including the closest homolog CHMP2A, polymerized into tubes or copolymerized with CHMP2B on the plasma membrane, although they all tended to concentrate at their base. However, using co-transfection experiments, only CHMP4 isoforms had a drastic effect in blocking CHMP2B-tubulating activity. The dominant negative effect of CHMP4 on CHMP2B tube formation may be explained by sequestering of CHMP2B by the overexpressed CHMP4. On the other hand, the presence of CHMP4 at the base of the tubes suggests that it also plays a positive role in that it sets the stage for CHMP2B membrane polymerization. Similarly, CHMP2 isoforms have no effect on HIV-1 budding in the absence of CHMP4 isoforms consistent with the absence of CHMP2 in the membrane necks of budding virions in the absence of CHMP4 (26).
Recruitment of FLAG-CHMP4B at the plasma membrane has already been observed using “deep etched” electron microscopy. In this case, the protein polymerized into filaments, which self-associated into circular arrays underneath the plasma membrane (34). Expression of C-terminal deleted CHMP4A or -B or co-expression of the full-length protein together with an ATPase-dead version of VPS4 (VPS4BE235Q) led to the appearance of buds and tubes pointing out of the cells. The identity of the CHMPs making these hollow tubes was not defined, but our results open the possibility that they represent CHMP2B polymers.
Cryo-EM analyses of isolated membrane tubes demonstrated the presence of a protein lattice in close association with the inner leaflet of the tube membrane. Immunogold labeling indicates that the protein lattice is made of CHMP2B, confirming the immunofluorescence staining of CHMP2B along the entire length of cellular membrane tubes. The CHMP2B protein lattice present in the tubes displays a lateral striation that is produced by helical symmetry; thus, tube formation most likely followed the biophysical principle of membrane buckling as proposed for ESCRT-III polymer assembly (38). The pitch of the helix was determined by cryo-EM to be 32 Å. Furthermore, immunogold labeling revealed that the arrangement of CHMP2B within the helix must be such that the C-terminal part points inside the lumen. These characteristics have already been observed in the case of CHMP2A/CHMP3 tubular structures assembled in vitro in the absence of membranes (12), indicating common structural principles for CHMP polymerization. The CHMP2B tubes varied in diameter, and the CHMP2B lattice was able to constrict the membrane tube down to a diameter of 16 nm; furthermore, tube ends are closed and often reveal dome-like structures covered with continuous membrane. The induced membrane constriction and the observation of tubes with dome-like structural ends tightly covered by membrane indirectly support the following membrane fission model: the building up of a dome-like CHMP protein lattice attracts and bends membranes sufficiently to induce neck thinning, leading to spontaneous membrane fission (33). Furthermore, our observations are in agreement with the tomography data that present spiral CHMP2A tubes at the cytokinetic midbody with a diameter of 17 nm (46). Thus, both CHMP2A and CHMP2B can constrict membranes to narrow diameters. Because none of the other CHMPs have yet been shown to induce membrane constriction, it is tempting to speculate that CHMP2 polymers catalyze the final membrane fission step, as suggested for HIV-1 budding (26), by providing sufficient membrane bending energy that leads to close apposition of the membranes within the membrane neck of a virus or vesicle or within the midbody structure.
Supplementary Material
Acknowledgments
We acknowledge the support by the Grenoble Parternship for Structural Biology and the GIS-IBISA platform for access to the Polara microscope. We thank W.I. Sundquist and S. Urbé for generous gift of reagents, D. Béal for technical assistance, F. Hemming for critical reading of the manuscript and all members of the lab for helpful suggestions.
This work was supported by INSERM, CNRS, Université Joseph Fourier, Fondation France Alzheimer, and ANR (Grant 08-Blanc-0271, to W. W., R. S., and G. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S4, and Movies S1 and S2.
- AMPPnP
- 5′-adenylyl-β,γ-imidodiphosphate
- MIM
- MIT domain-interacting motif
- MIT
- Microtubule Interacting and Transport domain
- VPS4
- Vacuolar Protein Sorting homolog 4
- AMSH
- Associated Molecule with the Src homology domain of STAM
- WGA
- Wheat Germ Agglutinin.
REFERENCES
- 1. Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. (2002) Dev. Cell 3, 283–289 [DOI] [PubMed] [Google Scholar]
- 2. Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. (2002) Dev. Cell 3, 271–282 [DOI] [PubMed] [Google Scholar]
- 3. Katzmann D. J., Babst M., Emr S. D. (2001) Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 4. Hurley J. H. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M. (2010) Mol. Biol. Cell 21, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Teis D., Saksena S., Emr S. D. (2008) Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 7. Teis D., Saksena S., Judson B. L., Emr S. D. (2010) EMBO J. 29, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009) Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. (2007) Nature 449, 740–744 [DOI] [PubMed] [Google Scholar]
- 10. Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. (2007) Nature 449, 735–739 [DOI] [PubMed] [Google Scholar]
- 11. Lin Y., Kimpler L. A., Naismith T. V., Lauer J. M., Hanson P. I. (2005) J. Biol. Chem. 280, 12799–12809 [DOI] [PubMed] [Google Scholar]
- 12. Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. (2008) Science 321, 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babst M., Sato T. K., Banta L. M., Emr S. D. (1997) EMBO J. 16, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Babst M., Wendland B., Estepa E. J., Emr S. D. (1998) EMBO J. 17, 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wollert T., Hurley J. H. (2010) Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carlton J. G., Martin-Serrano J. (2007) Science 316, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 17. Morita E., Colf L. A., Karren M. A., Sandrin V., Rodesch C. K., Sundquist W. I. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12889–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. (2007) EMBO J. 26, 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elia N., Sougrat R., Spurlin T. A., Hurley J. H., Lippincott-Schwartz J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Usami Y., Popov S., Popova E., Inoue M., Weissenhorn W. G., Göttlinger H. (2009) Biochem. Soc. Trans. 37, 181–184 [DOI] [PubMed] [Google Scholar]
- 21. Demirov D. G., Freed E. O. (2004) Virus Res. 106, 87–102 [DOI] [PubMed] [Google Scholar]
- 22. Morita E., Sundquist W. I. (2004) Annu. Rev. Cell Dev. Biol. 20, 395–425 [DOI] [PubMed] [Google Scholar]
- 23. Bieniasz P. D. (2009) Cell Host Microbe 5, 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin-Serrano J., Zang T., Bieniasz P. D. (2003) J. Virol. 77, 4794–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin-Serrano J., Neil S. J. (2011) Nat. Rev. Microbiol. 9, 519–531 [DOI] [PubMed] [Google Scholar]
- 26. Morita E., Sandrin V., McCullough J., Katsuyama A., Baci Hamilton I., Sundquist W. I. (2011) Cell Host Microbe 9, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muzioł T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H., Weissenhorn W. (2006) Dev. Cell 10, 821–830 [DOI] [PubMed] [Google Scholar]
- 28. Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G., Göttlinger H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W. M., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I. (2009) Nat. Struct. Mol. Biol. 16, 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lata S., Roessle M., Solomons J., Jamin M., Gottlinger H. G., Svergun D. I., Weissenhorn W. (2008) J. Mol. Biol. 378, 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shim S., Kimpler L. A., Hanson P. I. (2007) Traffic 8, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 32. Pires R., Hartlieb B., Signor L., Schoehn G., Lata S., Roessle M., Moriscot C., Popov S., Hinz A., Jamin M., Boyer V., Sadoul R., Forest E., Svergun D. I., Göttlinger H. G., Weissenhorn W. (2009) Structure 17, 843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fabrikant G., Lata S., Riches J. D., Briggs J. A., Weissenhorn W., Kozlov M. M. (2009) PLoS Comput. Biol. 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanson P. I., Roth R., Lin Y., Heuser J. E. (2008) J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baumgärtel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P. W., Kräusslich H. G., Bräuchle C., Müller B., Lamb D. C. (2011) Nat. Cell Biol. 13, 469–474 [DOI] [PubMed] [Google Scholar]
- 36. Jouvenet N., Zhadina M., Bieniasz P. D., Simon S. M. (2011) Nat. Cell Biol. 13, 394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mahul-Mellier A. L., Strappazzon F., Chatellard-Causse C., Blot B., Béal D., Torch S., Hemming F., Petiot A., Verna J. M., Fraboulet S., Sadoul R. (2009) Biochem. Soc. Trans. 37, 200–203 [DOI] [PubMed] [Google Scholar]
- 38. Lenz M., Crow D. J., Joanny J. F. (2009) Phys. Rev. Lett. 103, 038101 [DOI] [PubMed] [Google Scholar]
- 39. Belly A., Bodon G., Blot B., Bouron A., Sadoul R., Goldberg Y. (2010) J. Cell Sci. 123, 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liou W., Geuze H. J., Slot J. W. (1996) Histochem. Cell Biol. 106, 41–58 [DOI] [PubMed] [Google Scholar]
- 41. Conway J. F., Steven A. C. (1999) J. Struct. Biol. 128, 106–118 [DOI] [PubMed] [Google Scholar]
- 42. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 43. Agromayor M., Martin-Serrano J. (2006) J. Biol. Chem. 281, 23083–23091 [DOI] [PubMed] [Google Scholar]
- 44. Hurley J. H., Hanson P. I. (2010) Nat. Rev. Mol. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peel S., Macheboeuf P., Martinelli N., Weissenhorn W. (2011) Trends Biochem. Sci. 36, 199–210 [DOI] [PubMed] [Google Scholar]
- 46. Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., Gerlich D. W. (2011) Science 331, 1616–1620 [DOI] [PubMed] [Google Scholar]
- 47. Skibinski G., Parkinson N. J., Brown J. M., Chakrabarti L., Lloyd S. L., Hummerich H., Nielsen J. E., Hodges J. R., Spillantini M. G., Thusgaard T., Brandner S., Brun A., Rossor M. N., Gade A., Johannsen P., Sørensen S. A., Gydesen S., Fisher E. M., Collinge J. (2005) Nat. Genet. 37, 806–808 [DOI] [PubMed] [Google Scholar]
- 48. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007) J. Cell Biol. 179, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee J. A., Beigneux A., Ahmad S. T., Young S. G., Gao F. B. (2007) Curr. Biol. 17, 1561–1567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.