Abstract
Expression of the clusterin (CLU) gene results in the synthesis of a conventional secretory isoform set (pre- and mature secretory clusterin proteins, psCLU/sCLU), as well as another set of intracellular isoforms, appearing in the cytoplasm (pre-nuclear CLU, pnCLU) and in the nucleus as an ∼55-kDa mature nuclear clusterin (nCLU) form. These two isoform sets have opposing cell functions: pro-survival and pro-death, respectively. Although much is known about the regulation and function of sCLU as a pro-survival factor, the regulation and function of endogenous nCLU in cell death are relatively unexplored. Here, we show that depletion of endogenous nCLU protein using siRNA specific to its truncated mRNA increased clonogenic survival of ionizing radiation (IR)-exposed cells. nCLU-mediated apoptosis was Bax-dependent, and lethality correlated with accumulation of mature nCLU protein. nCLU accumulation was regulated by CRM1 because binding between CRM1 and nCLU proteins was significantly diminished by leptomycin B (LMB), and nuclear levels of nCLU protein were significantly enhanced by LMB and IR co-treatment. Moreover, LMB treatment significantly enhanced IR-induced nCLU-mediated cell death responses. Importantly, bax−/− and bax−/−/bak−/− double knock-out cells were resistant to nCLU-mediated cell death, whereas bak−/− or wild-type bax+/+/bak+/+ cells were hypersensitive. The regulation of nCLU by CRM1 nuclear export/import may explain recent clinical results showing that highly malignant tumors have lost the ability to accumulate nCLU levels, thereby avoiding growth inhibition and cell death.
Keywords: Apoptosis, Nuclear Pore, Nuclear Translocation, Nuclear Transport, Radiation Inactivation, Bax, Clusterin, Crm1
Introduction
Clusterin (CLU)3 is a stress-induced protein implicated in apoptosis, complement-mediated cell lysis, lipid transport, central nervous system disorders, glomerulonephritis, and atherosclerosis (1). Elevated CLU mRNA levels are observed in many cancers, including those of breast, esophagus, and colon (1–3). Induction of CLU mRNA and its secreted protein forms (psCLU, sCLU) are commonly associated with apoptosis in the heart, breast, prostate, brain, lung, liver, kidney, pancreas, and retina after various cell stresses (4–14). Elevated sCLU levels were noted in regressing rat ventral prostate following castration (15).
Two different sets of CLU protein isoforms have been described in mammalian cells, a conventional secreted isoform set (pre-secretetory and secretory, psCLU and sCLU, respectively) that has pro-survival functions, and a cytoplasmic/nuclear (pnCLU/nCLU) isoform set that has pro-apoptotic cell death functions. Pre-secretory CLU is synthesized by translation from the first AUG codon of full-length CLU mRNA, which directs the synthesis of a leader peptide targeting the protein to the endoplasmic reticulum. The mature secretory (sCLU) protein is ultimately secreted from cells as a heavily glycosylated and proteolytically cleaved 80-kDa protein (16). Overexpression of sCLU in human prostate cancer cells results in drug resistance and cytoprotection against a variety of cytotoxic agents that induce apoptosis (13, 17, 18). CLU functions as an extracellular chaperone that binds hydrophobic regions of partially unfolded proteins and via an ATP-independent mechanism. It inhibits protein aggregation and precipitation, otherwise caused by physical or chemical stresses (e.g. heat, oxidative reduction) (19). Thus, sCLU was classified as a functional homolog of the small heat shock proteins (20, 21). Depletion of sCLU protein levels using siRNA specific to exon II caused dramatic increases in the radiosensitivity of transfected MCF-7 breast cancer cells (22). Similar results were reported for various chemotherapeutic agents (5, 12). The efficacy of siRNA specific to sCLU was enhanced further by nanoparticle micelle delivery, enhancing strategies for improving tumor-selective radiotherapies as well as chemotherapies (11). Thus, sCLU is a general pro-survival factor in most cells after stress, acting to clear cell debris from traumatized tissue. The anti-apoptotic function of sCLU was attributed to its ability to bind, sequester, and prevent the movement of the pro-apoptotic Bax protein into mitochondria (14, 23).
In contrast, a pro-death intracellular (pnCLU) or nuclear CLU (nCLU) isoform set has also been described (8, 10). Numerous groups have reported accumulation of nCLU in the nuclei of stress-induced cells undergoing apoptotic cell death (4, 8, 9, 10). We previously showed that pnCLU was translated in human cells from an alternatively spliced nCLU mRNA, created by direct splicing of exons I and III (8). This splicing event eliminated exon II that encoded the first AUG start codon and the endoplasmic reticulum-targeting signal peptide present in sCLU mRNA (8). Translation from this truncated nCLU mRNA, using a second in-frame AUG codon in exon III, produced an ∼49-kDa nCLU protein located in the cytoplasm. Unlike sCLU, the mature ∼55-kDa nCLU co-immunoprecipitated with Ku70 after cell stress (e.g. after IR exposure) and its C-terminal region contained a functional nuclear localization sequence (NLS) and pro-death coiled-coil domain (8, 17). Overexpression of nCLU, but not NLS-mutated nCLU, nor nCLU mutated in its Ku70 binding domain, induced apoptotic cell death.
The regulation and roles of nCLU protein before and after stress have not been examined thoroughly. We showed previously that in log phase MCF-7 cells endogenous nCLU protein was located predominantly in the cytosol (17), apparently sequestered or excluded from the nucleus. Exposure to high doses of IR (>1 Gy; ≥LD50) (8, 17) or cytotoxic doses of TGF-β1 (10) triggered accumulation of nCLU in the nuclei of exposed cells. Overexpression of nCLU in MCF-7 cells acted as a pro-death signal, inhibiting cell growth, inducing G1 cell cycle arrest responses, stimulating apoptosis, and resulting in dramatic losses in clonogenic survival (8, 17). Thus, with two apparently functional NLSs in pnCLU, it was not clear how pnCLU was regulated in its basal state, where it was sequestered in the cytosol. It was also unclear how nCLU accumulated after cytotoxic cell stress responses (i.e. 1 Gy) in the nuclei of irradiated cells scheduled to undergo cell death (apoptosis).
Here, we report that endogenous nCLU is a major pro-death factor, affecting the radiosensitivity of cancer cells. Its subcellular localization is regulated by a defined NLS (8) and the CRM1 nuclear transport/exportin protein. Binding between nCLU and CRM1 was inhibited by leptomycin B (LMB), which significantly enhanced nCLU accumulation and cell death of both untreated and cell stress (IR)-exposed cancer cells. Specific nCLU protein knockdown using a splice-specific siRNA significantly spared cell death and enhanced long term clonogenic survival of IR-treated cells. Furthermore, bax expression was critical to cell death mediated by mature nCLU because genetically deficient cells lacking bax expression, as well as nCLU-resistant bax-defective cells, were resistant to nCLU-induced cell death. Because two recent clinical reports showed that aggressive esophageal, breast, and colon tumors have significantly lowered overall basal nCLU levels and nuclear accumulation with concomitantly high levels of sCLU (1, 2), data presented in this paper may offer potential insights into resistance mechanisms. Thus, CRM1 activity may correlate with aggressiveness in tumors as otherwise regulatory mechanisms that control expression and intracellular regulation of nCLU and its growth-suppressive responses may be abrogated.
EXPERIMENTAL PROCEDURES
Chemicals and Tissue Culture
LMB was purchased from the Sigma. Human osteosarcoma U-2 OS cells were purchased from the American Tissue Culture Collection (Manassas, VA). MCF-7 human breast cancer cells were grown in RPMI 1640 medium (Invitrogen) as described (16). Wild-type, bax−/−/bak+/+, bax+/+/bak−/−, and bax−/−/bak−/− mouse embryonic fibroblasts (MEFs) were generously provided by Dr. Craig Thompson (Memorial Sloan Kettering) and grown as described (18).
siRNAs
A double strand siRNA 23-oligomer was made against a nCLU-specific sequence (see Fig. 1A) that spanned the exon I/exon III junction. siRNA-nCLU and scrambled siRNA (siRNA-Scr) (0.02-μmol scale) were synthesized by Dharmacon (Lafayette, CO). Mock transfections using transfection reagent alone were performed as controls.
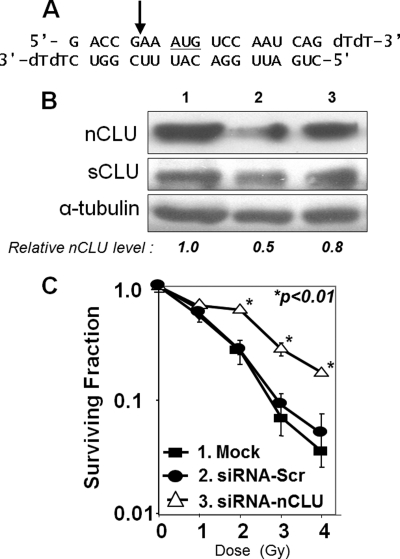
FIGURE 1.
nCLU knockdown creates IR-resistant MCF-7 cells. A, sequence of double strand siRNA-nCLU. The AUG translational start site for nCLU protein is underlined. The arrow indicates the exon I/III junction that results from nCLU-specific alternative splicing (8). B, changes in nCLU and sCLU protein levels in MCF-7 cells 60 h after transfection with siRNA-nCLU. MCF-7 cells were transfected as follows: lane 1, mock-transfected cells; lane 2, siRNA-nCLU, siRNA selective for nCLU and the exon I/III junction (arrow, A); and lane 3, siRNA-Scr, siRNA 21-oligomer generated to a nonspecific sequence in the human genome. nCLU, sCLU, and α-tubulin steady-state protein levels were monitored by Western blot analyses. The blot shown is representative of experiments performed three or more times. C, MCF-7 cells transfected with siRNA-nCLU and resistant to IR. MCF-7 cells were transfected with siRNA-nCLU (open triangles), siRNA-Scr (filled circles), and mock-siRNA (filled squares) as described under “Experimental Procedures.” Colony forming assays were performed on transfectants as described under “Experimental Procedures.” An increase in the surviving fraction of siRNA-nCLU-transfected MCF-7 cells after exposure to 2–4 Gy versus mock- or scramble-siRNA-transfected cells was noted (p < 0.01).
Intracellular Localization Studies Using Confocal Analyses
The human nCLU open reading frame (ORF) was cloned by RT-PCR from MCF-7 cells, inserted in-frame with hrGFP into the SmaI site of the phrGFP-N1 vector (Stratagene), and verified by DNA sequencing. All transfections were performed using Lipofectamine PlusTM as described by the manufacturer (Invitrogen). For confocal microscopy, MCF-7 cells were plated onto coverslips in 6-well dishes at 2 × 105 cells/well and transfected with 1 μg of plasmid DNA/well. After transfection (24 h), cells were treated with IR (5 Gy) using a 137Cs radiation source (17). LMB (10 or 20 ng/ml) was added 1 h prior to IR exposure and 24–48 h after treatment. IR-exposed and/or LMB-treated cells were fixed 48 h after treatment with ice-cold methanol for 10 min at 20 °C, rinsed twice with PBS, and treated with RNase A (1 μg/ml in PBS) for 20 min at 37 °C. Cells were mounted in propidium iodide (PI)-containing VectashieldTM mounting medium (Vector Laboratories, Burlingame, CA), and microscopy was performed using an MRC 1024 confocal laser scanning microscope (Bio-Rad). Photomicrograph z-sections were taken at ×1000 magnification. For quantification, at least 200 GFP-positive cells were visually screened for intracellular distribution of hrGFP-nCLU protein and its co-localization with PI-stained nuclei. The number of cells with nuclear-only, cytoplasmic and nuclear, or cytoplasmic-only intercellular localization of GFP-fused proteins was counted manually. Percentages means ± S.D. of cells in each population were then calculated. All experiments were performed three times in triplicate, and statistical significance (p values) was evaluated by Student's t tests.
Generation of the C-terminal 120-Amino Acid Residue Fragment of nCLU (C120) and HA-tagged C120 Plasmids
Using specific primers, either the C120 cDNA (primers used: C120-HindIII forward, 5′-CGAATTCGCGGAAGCTTCATGTCTGTGGACT-3′ and RevBamHIstop, 3′-ATCAGATGGATCCTTATCACTCCTCCCGGTGCTTTTTGC-5′) or a C120 cDNA containing an HA tag at its 5′-end (primers used: HA-C120-HindIII forward, 5′-AACCCAAAAAAAGAAGCTTGTATGGCT-3′ and RevBamHIstop, 3′-ATCAGATGGATCCTTATCACTCCTCCCGGTGCTTTTTGC-5′) were amplified from the original pACT2-C120 vector (13). The cDNA of interest was excised using HindIII and BamHI and subcloned directly into pcDNA3.1/Myc-His+, which contains a Geneticin resistance gene (Invitrogen). Expression from pACT2-C120 produces a polypeptide with a theoretical and actual molecular mass of ∼16.3 kDa, whereas the HA-tagged HA-C120 vector expressed a molecular mass of ∼19.5 kDa. Correct cloning was verified by double strand DNA sequencing, and expression was verified using 12% SDS-PAGE analyses.
Isolation of nCLU-resistant Cells
Resistance to nCLU-mediated cell death was explored using C120 forced overexpression in U-2 OS osteosarcoma cells. Resistant cell lines that expressed varying levels of C120-related nCLU proteins were isolated. Briefly, cells were transfected with pcDNA3.1 vector alone, C120, or HA-C120 plasmids using Lipofectamine 2000TM. Transient overexpression and cell responses were monitored 48 h after transfection, as well as at various times thereafter as described (17). To isolate cell lines that stably overexpress C120 or HA-C120 polypeptides, transfectants were selected in Complete medium containing 800 μg/ml Geneticin, beginning 48 h after transfection. To avoid clonal variation, pools of transduced cells were isolated and analyzed. All transfectants were tested for C120 or HA-C120 polypeptide expression by immunoblotting using the H330 antibody (Santa Cruz Biotechnology). Apoptotic rates were quantified by cytoplasmic histone-associated DNA fragmentation at 48 h after transfection using the Cell Death Detection ELISAPLUS photometric enzyme-immunoassay method (Roche Applied Science). Colonies and transfectant cell number were counted after growing an equal starting number of cells for 11 days in Geneticin-containing medium. Colonies were tested for nCLU expression, as well as bax−/− expression because loss of Bax expression was predicted to confer resistance based on nCLU-mediated cell death responses in Bax-deficient MEFs.
Transfections and Colony Forming Ability Assays
Log phase MCF-7 cells (5 × 105 cells/60-mm dish) were mock-transfected or transfected with siRNA-nCLU or siRNA-Scr (5 μg/dish) using Lipofectamine PlusTM. After transfection (48 h), cells were trypsinized and plated onto 60-mm dishes (500 cells/dish) in triplicate. Cells were exposed to varying doses of IR as indicated, and 10 days later colonies were fixed and stained with crystal violet (16), and colonies containing ≥100 normal appearing cells counted. Normal-appearing colonies in each treatment were normalized by plating efficiency of mock-transfected cells, and means of surviving fractions ± S.D. were calculated and graphed on semi-log scales. All experiments were performed three times and evaluated by Student's t tests.
Antibodies and Western Blot Analyses
Cells were harvested either in radioimmuneprecipitation assay buffer for whole cell extracts or in a hypotonic/hypertonic buffers for cytoplasmic/nuclear fractionation after various treatments as indicated. Whole cell extracts (50 μg of total protein) were separated using SDS-PAGE. nCLU levels were assessed by Western blot analyses using the H-330 anti-nCLU antibody (Santa Cruz Biotechnology). Variations in loading were assessed by α-tubulin levels using an appropriate antibody (EMD Biosciences, San Diego, CA).
All samples were adjusted to equal protein amounts by Bradford (Bio-Rad) assays, and proteins were resolved by SDS-PAGE followed by immunoblot analyses. Antibodies against human CLU (sc-6419), nCLU (H330), p53 (sc-126), Bcl-2 (sc-509), Bax (sc-493), and GAPDH (sc-25778) were purchased from Santa Cruz Biotechnology. Equal protein loading was verified by GAPDH levels. All Western blots shown were representative of experiments performed at least three times.
Co-immunoprecipitations
MCF-7 cells were exposed to 5 Gy and/or LMB as specified, harvested in lysis buffer as described (24). Each extract (∼1 mg of total protein) was added to 4 μg of either the exportin/CRM1 monoclonal antibody (BD Biosciences Pharmingen) or normal mouse IgG (Santa Cruz Biotechnology) and incubated overnight at 4 °C. Protein G-agarose beads (Pierce) were added ∼12 h later and incubated for 3 h. Beads were collected by centrifugation (500 × g), washed in PBS, and boiled in 2×SDS-PAGE loading buffer. Total cell extracts (50 μg) were separated by SDS-PAGE along with immunoprecipitates. Western blotting was performed as indicated using anti-nCLU H330 (Santa Cruz Biotechnology) or anti-exportin/CRM1 monoclonal (BD Biosciences Pharmingen) antibodies at 1:2000 and 1:3000 dilutions, respectively.
Cell Death Assays
Apoptotic/necrotic cell populations were assessed by determining the percentage of sub-G1/G0 populations using flow cytometry (EPICS XL-MCC; Beckman Coulter) as described (25). Experiments were repeated three times in triplicate, and means ± S.E. were determined and graphed using SigmaPlot v.4.0 (SPSS, San Rafael, CA). Cell responses (×-fold changes in apoptosis, percentage of colony forming ability, or changes in overall cell number) were monitored for comparison with corresponding controls. Statistical significance was evaluated using one-way analyses of variance (ANOVA), and significant differences at p values <0.05 are indicated on the graphs by asterisks.
RESULTS
nCLU Knockdown Protects Cells from IR Lethality
To establish a role for nCLU in IR-induced lethality, we specifically knocked down nCLU protein expression using a 21-mer siRNA specific for the exon I/III splicing junction (siRNA-nCLU, Fig. 1A) present in truncated nCLU mRNA, but not in full-length CLU mRNA. Western blot analyses indicated that the steady-state level of nCLU was ∼2-fold lower in siRNA-nCLU transfectants (lane 2) than in mock-transfected (lane 1) or scrambled siRNA-transfected (siRNA-Scr, lane 3) MCF-7 cells, 60 h after transfection (Fig. 1B). In contrast, sCLU levels (generated from expression from full-length CLU mRNA) were not significantly altered (p > 0.5) by siRNA-nCLU or siRNA-Scr transfection. Importantly, siRNA-nCLU MCF-7 transfectants showed a significant ∼3–4-fold increase in survival after IR exposures of 2–4 Gy compared with mock- or siRNA-Scr transfectants (Fig. 1C). Transfection with siRNA-Scr did not affect the plating efficiency of MCF-7 cells, the survival of cells post-IR, and had minimal effects on endogenous nCLU protein levels. In contrast, siRNA knockdown of sCLU in MCF-7 cells enhanced IR lethality (22). Thus, nCLU is a pro-death factor, whereas sCLU is a pro-survival factor in the same cells.
Endogenous nCLU Accumulates in the Nuclei of IR-exposed Cells
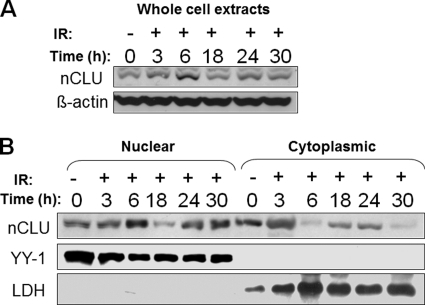
We previously showed significant accumulation of mature ∼55-kDa nCLU protein in the nuclei of IR-exposed MCF-7 cells (17). To investigate the kinetics of this translocation further, time course studies of nCLU intracellular localization in MCF-7 cells before and after 5 Gy were performed. In control cells, roughly equivalent levels of nCLU were noted in the cytoplasm and nucleus (Fig. 2B). Western blot analyses showed an increase in total nCLU protein of ∼3-fold, beginning at 3 h and peaking at 6 h after 5 Gy (Fig. 2A). In fractionation studies, we noted a 2–3-fold increase in cytoplasmic levels at 3 h after irradiation and detected a dramatic shift (3–5-fold increases in separate experiments) of nCLU localization from the cytoplasm toward the nucleus at 6 h (Fig. 2B). Interestingly, the accumulation of nCLU within nuclei of exposed cells was temporary, with gradual decreases in total and nuclear nCLU proteins observed by 18 h after IR. Levels of total nCLU protein returned to the pre-irradiated state at 24 h followed by a second peak of nuclear localization at 30 h (Fig. 2). This second increase in nuclear localization of mature nCLU in cells destined to die was previously noted and detected by confocal microscopy (17). The rapid disappearance of nCLU from the cytoplasm with simultaneous accumulation in the nucleus at 6 h after IR treatment led us to a hypothesis that retention of nCLU in the cytoplasm of nonirradiated MCF-7 cells might be, at least partially, mediated by nuclear export processes, possibly by the CRM-1/exportin protein.
FIGURE 2.
nCLU protein is transiently induced and accumulates in the nuclei of IR-treated MCF-7 cells. A, MCF-7 cells were mock-irradiated (0 Gy) or exposed to IR (5 Gy), and whole cell extracts were prepared from half of the cells at various times after IR. Western blot analyses were performed using anti-nCLU (CY-1, upper), or anti-β-actin (lower) antibodies. B, half of the cells from the experiment described in A were subjected to subcellular fractionation. Nuclear and cytoplasmic fractions were separated and analyzed using Western blotting. The CY-1 antibody was used to detect nCLU protein. The purity of each fraction was verified using YY-1 and LDH antibodies for nuclear and cytoplasmic fractions, respectively. Immunoblots shown are representative of experiments performed three or more times.
LMB Enhances nCLU Accumulation after IR
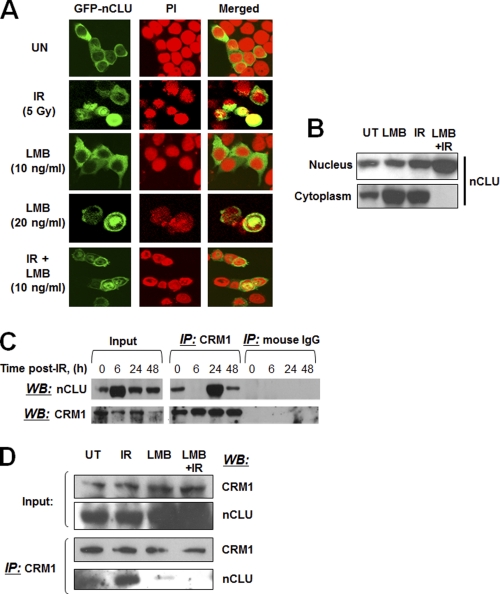
Overexpressed GFP-nCLU accumulated in the nuclei of irradiated MCF-7 cells with the same kinetics and IR dose response as endogenous nCLU protein in irradiated MCF-7 cells (8, 17). Thus, hrGFP-nCLU-transfected MCF-7 cells were used as a model to study the regulatory mechanisms of cytosolic retention of nCLU in MCF-7 cells before and after IR. To test the role of CRM-1 in excluding nCLU from the nucleus, we examined the effects of LMB on the intracellular localization of nCLU before and after IR exposure. Treatment of log phase MCF-7 cells with 20 ng/ml, but not 10 ng/ml LMB (Fig. 3A), resulted in accumulation of hrGFP-nCLU (28 ± 2%) in the nuclei of treated versus control MCF-7 cells (8 ± 1) (Table 1). As observed previously, exposure of MCF-7 cells to IR (5 Gy) significantly enhanced nuclear nCLU levels (17), and adding LMB (10 ng/ml) further increased nCLU levels (18 ± 3% and 41 ± 5%, respectively, Table 1). Interestingly, LMB alone at 20 ng/ml induced significant accumulation of endogenous nCLU in the cytoplasm (Fig. 3B) of exposed cells. This effect was most likely due to the cytotoxicity of LMB resulting in new nCLU protein synthesis. Similar nCLU induction was observed after exposure of cells to a number of other cytotoxic and stress-inducing agents, such as vanadium and heat shock (15, 25). Increases in nuclear levels of nCLU as a result of LMB treatments of IR-exposed MCF-7 cells corresponded to loss of cytoplasmic levels of nCLU (Fig. 3B). Binding of CRM1 to nCLU protein was diminished at 6 h after exposure to IR (Fig. 3C); this time point coincided exactly with the maximum accumulation of nCLU in the nuclei of cells exposed to IR (Fig. 2B). Interestingly, at 24 h after IR treatment the binding of CRM1 to the nCLU protein, as observed by co-immunoprecipitation analyses, increased >2-fold (Fig. 3C). Addition of LMB (10 ng/ml) significantly decreased CRM1-nCLU binding; the observed LMB-mediated decrease in CRM1-nCLU binding was further enhanced (∼3-fold) in LMB-exposed, IR-treated MCF-7 cells (Fig. 3D). These data are consistent with the theory that IR exposure stimulates pnCLU translocation to the nucleus, however, a LMB-sensitive CRM1-mediated process acts to prevent nCLU accumulation and spare apoptotic cell death.
FIGURE 3.
Nuclear accumulation of hrGFP-nCLU increases after IR or LMB. A, log phase MCF-7 cells were transfected with hrGFP-nCLU or hrGFP alone. Twenty-four hours after transfection, cells were treated with IR + LMB. Forty-eight hours after transfection, cells were analyzed for hrGFP-nCLU expression with nuclei counterstained with PI. The photomicrographs shown are representative of studies performed three or more times. See Table 1 for quantification of hrGFP-nCLU nuclear localization. B, LMB enhances IR-induced nuclear accumulation of nCLU. MCF-7 cells were treated with 10 ng/ml LMB and/or 5 Gy. Cells were harvested 24 h after exposure, and the samples were separated into nuclear and cytoplasmic fractions. nCLU was detected by Western blotting using the CY-1 antibody. C, IR-dependent profiling of nCLU binding to CRM1 is shown. Whole cell extracts and immunoprecipitations (IP) were performed using the anti-CRM1 or control normal IgGs. The same CRM1 antibody was used to detect CRM1 in immunoprecipitates, and the CY-1 antibody was used to detect nCLU by Western blotting (WB). Input lanes show steady-state levels of nCLU and CRM1 proteins detected in the same whole cell extracts used for the immunoprecipitations. D, LMB (10 ng/ml) abrogates nCLU-CRM1 binding. IR-, LMB-, or IR-LMB-exposed cells were lysed, and co-immunoprecipitations were done as in C. Input indicates steady-state levels of the nCLU and CRM1 proteins detected by Western blotting in the same whole cell extracts used for the immunoprecipitations.
TABLE 1.
Exposure of cells to LMB enhances accumulation of nCLU in the nucleus and cell death
| IR | Treatment conditiona | Cells with hrGFP− nCLU+ cytoplasm | hrGFPnCLU+ nuclei | Apoptosis |
|---|---|---|---|---|
| % | % | % | ||
| UT | UT | 92 ± 2 | 8 ± 1b | 6 ± 2b |
| LMBc | 73 ± 2 | 28 ± 2b | 10 ± 3b | |
| 10 Gy | UT | 82 ± 3 | 18 ± 3b | 32 ± 6b |
| LMB2 | 59 ± 5 | 41 ± 5b | 89 ± 4b |
a Log phase MCF-7 cells were mock- or IR-exposed (5 Gy), then treated with or without various doses of LMB. UT, untreated.
b p < 0.01 by Student's t tests.
c LMB, a fairly specific inhibitor of CRM1, was added at 10 ng/ml for 24 h.
nCLU-induced Cell Death Is Bax-dependent
We previously defined a site within Ku70 that bound nCLU and showed that nCLU could prevent Ku end binding responses in double strand break repair after IR (13, 17). Separately, we also demonstrated that Ku70 protected against Bax-dependent cell death and defined Ku70-Bax interactions (26). Thus, a role for Bax in nCLU-mediated cell death was examined. Isogenic bax−/−/bak−/−, bax+/+/bak−/−, bax−/−/bak+/+, and wild-type (WT) immortalized MEFs were transfected with either hrGFP or hrGFP-nCLU (18). One day (24 h) after transfection, cells were collected and stained with PI, and cell death responses were calculated as percentage sub-G1/G0 cells using flow cytometry (Fig. 4).
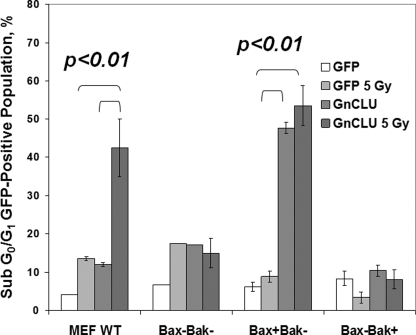
FIGURE 4.
Bax is required for nCLU-induced cell death. Log phase MEFs were transiently transfected with hrGFP-vector or phrGFP-nCLU. Twenty-four hours later, cells were trypsinized, fixed, and stained with PI. The fraction of sub-G1/G0 cells within the total cell population for each treatment condition was determined using flow cytometry. Parallel experiments included hrGFP-vector alone or phrGFP-nCLU-transfected cells exposed to 5 Gy. Each dataset represents three independent transfections performed in triplicate each. Bars represent means ± S.E. for each condition.
Overexpression of hrGFP-nCLU caused a Bax-dependent cell death response in irradiated MEFs (Fig. 4) because all cells lacking bax were resistant to nCLU-induced cell death responses (Fig. 4). IR-exposed wild-type MEF cells or bax+/bak− cells exhibited extensive apoptosis (44 ± 4% or 55 ± 7%, respectively) following hrGFP-nCLU transfection. In contrast, irradiated bax−/−/bak−/− cells were not sensitive (<20% of apoptosis) to nCLU overexpression. These data strongly suggested that nCLU-dependent apoptosis was mediated by Bax.
nCLU-resistant Cell Populations Have Significant Loss of Bax Expression
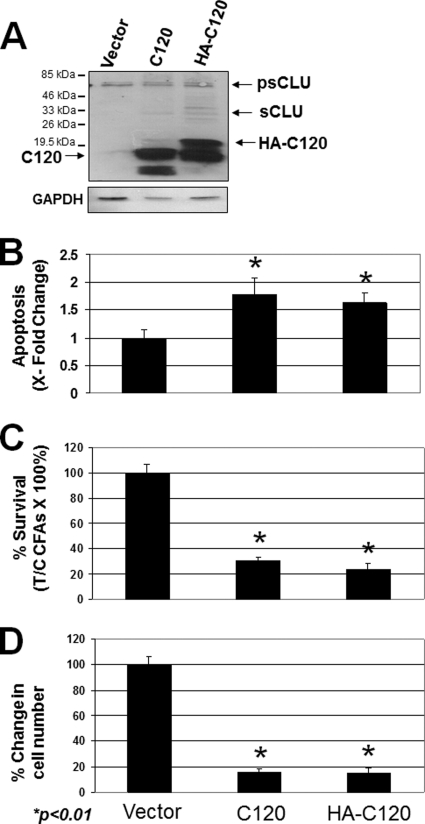
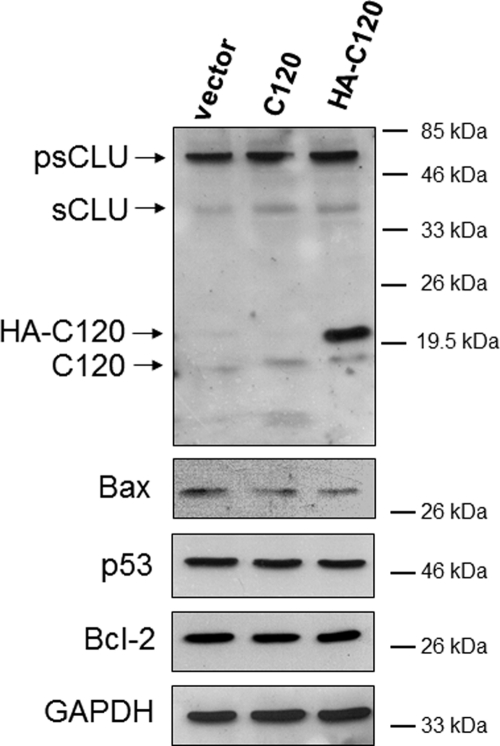
A plasmid expressing the C-terminal polypeptide fragment of nCLU (i.e. C120) was minimally required to induce nCLU-mediated apoptosis functionally (17). To examine further the function of the nCLU isoform, a C-terminal polypeptide of nCLU (C120 fragment with or without an HA tag for protein-protein interaction analyses) was overexpressed in U-2 OS cells. Overexpression of C120 and HA-C120 polypeptides was noted (Fig. 5A). As expected (17), transfection of U-2 OS cells with C120 or HA-C120 caused significant apoptosis (Fig. 5B), loss of viable cells (Fig. 5D) within 24 h, as well as significant loss of long term survival monitored by colony forming ability (Fig. 5C). Indeed, transfection of U-2 OS cells with C120 or HA-C120 compared with vector alone control cells resulted in >75% lethality (Fig. 5D), corresponding to an ∼2-fold increase in apoptosis at 24 h (Fig. 5B). Furthermore, colonies formed by cells overexpressing C120 or HA-C120 were significantly smaller, as confirmed by cell counting (Fig. 5D). Analyses revealed ∼85% fewer cells in C120- or HA-C120-overexpressing colonies compared with mock- or vector-alone-transfected controls. At random, colonies from each transfection were isolated and grown under Geneticin (G418) selection. Interestingly, populations expressing stably HA-C120 or C120 proteins grew with kinetics identical to vector-alone-expressing, G418-resistant cells; doubling times and basal apoptotic levels were not altered in C120, HA-C120, or vector-alone cell populations. Expression analyses of these cell lines revealed a significant down-regulation of basal Bax protein levels (3–5-fold, Fig. 6) in HA-C120- or C120-transfected versus mock or vector-alone cell lines. In contrast, there were no changes in p53 or Bcl-2 protein levels using GAPDH as loading (Fig. 6).
FIGURE 5.
Phenotypic effects of forced C120 or the HA-tagged C120 peptide overexpression in the U-2 OS cells. A, immunoblot analyses of extracts derived after transient transfection of C120-related plasmids in U-2 OS cells results in overexpressed levels of the corresponding C120- and HA-C120 nCLU-related polypeptides. Molecular mass markers are shown for each blot. Protein loading was monitored by GAPDH levels. psCLU denotes the intracellular sCLU precursor isoform; sCLU denotes the α- and β-chains of the heterodimeric mature sCLU isoform. B, U-2 OS cells were transfected with C120 or HA-C120 plasmids as in A, wherein elevated levels of C120 or HA-C120 promote cell death. ×-fold changes in apoptosis were calculated by monitoring sub-G0/G1 cell populations formed in 48 h as described under “Experimental Procedures.” C, cells were treated as described above, and changes in cell number were monitored. D, U-2 OS cells were treated as described above, and survival was monitored by colony forming ability assays. In B–D, results are means ± S.E. from experiments performed three or more times, each in triplicate.
FIGURE 6.
Down-regulation of Bax in C120-resistant U-2 OS cells. Immunoblot analyses of whole cell lysates derived from pools of U-2 OS cells stably transduced with vector alone, C120, or HA-C120 plasmids are shown. Although C120 exerts a potent pro-death activity in U-2 OS cells, a small fraction of cells survived with low expression levels of C120 or HA-tagged C120 polypeptides (see expression of these polypeptides in the top panel). Bax expression was monitored and found to be significantly lower in these resistant populations. In contrast, p53 or Bcl-2 proteins remained unchanged. Molecular mass markers are shown, and equal protein loading was monitored by GAPDH levels. Relative Bax levels were calculated with respect to GAPDH levels as loading controls. Western blots shown are representative of experiments performed three or more times.
DISCUSSION
Our data demonstrate that endogenous nCLU protein contributes to IR-induced cytotoxicity at clinically relevant doses (≥2 Gy) of IR. Selective knockdown of endogenous nCLU using siRNA resulted in dramatic resistance to IR-induced lethality. These data strongly suggest that endogenous nCLU plays a significant role in lethality induced by IR. This function of endogenous nCLU is opposite that demonstrated for sCLU, where sCLU protein knockdown significantly enhanced sensitivity to chemotherapy (12) and IR (11, 22) in MCF-7, as well as other cell lines examined. Collectively, these data suggest that CLU gene expression plays a key role in the intracellular decisions determining life and death in irradiated cells. Our data are the first to demonstrate a direct function of nCLU in cell death responses in human and mouse cells.
Accumulation of nCLU in the nuclei of MCF-7 cells following IR induces cell death in an apoptotic process that we demonstrate requires Bax expression. We showed previously that the C terminus of nCLU contained a coiled-coil domain that bound Ku70 and blocked Ku end binding activity (13, 17). The C terminus of nCLU also contained a functional NLS domain that allowed its accumulation in the nuclei of irradiated MCF-7 cells, corresponding to IR-induced cell death (8, 17). The presence of a strong functional C-terminal NLS within the nCLU protein appeared to contradict the apparent, largely cytoplasmic localization of nCLU in nonirradiated control cells, such as in MCF-7 breast cancer cells. Therefore, we investigated the regulation of nCLU distribution in irradiated compared with untreated MCF-7 cells.
Recent findings have shown that the Bax pro-apoptotic protein forms a complex with Ku70 and is released in response to apoptotic stimuli (23). Additionally, recent studies suggest that nCLU contains a putative BH3 motif in its C-terminal coiled-coil domain. Using this domain, nCLU can sequester Bcl-XL and, as a consequence as we have shown, cause release of pro-apoptotic Bax to promote apoptosis. Cell death was accompanied by cytochrome c release and activation of caspase-3 (6). A structural model of the Bcl-XL-nCLU BH3 peptide complex revealed that the binding mode is remarkably similar to those of other Bcl-XL-BH3 peptide complexes (7). Our data showing that nCLU-mediated cell death is Bax-dependent is consistent with these findings.
A number of proteins are shuttled out of the nucleus via the CRM1-dependent regulatory pathway. The CRM1 protein forms a complex with RanGTP and a nuclear export sequence (NES)-containing cargo protein (27, 28). The NES-containing cargo is exported to the cytoplasm, where the RanGTP-CRM1-cargo complex is dissociated upon GTP hydrolysis. LMB binds directly to CRM1 and inhibits its interaction with the NES motif of the cargo protein (29). Although no classical leucine-rich NES in the nCLU protein was noted, this protein may have a functional NES that either contains a nontraditional sequence or is transported out of the nucleus in a complex composed of as yet unidentified NES-containing protein.
As mentioned above, binding of nCLU protein to CRM1 and the overall nuclear export of wild-type nCLU protein were sensitive to IR. The co-immunoprecipitation data clearly suggest a close interaction between nCLU-CRM1, but this protein-protein interaction was abrogated after cells were exposed to IR. The effects of IR on CRM1-nCLU protein-protein association and possible IR-induced modification of nCLU are currently under investigation in our laboratory.
nCLU appears to be one of a number of proteins whose nuclear export is altered by IR or other cytotoxic agents that cause DNA double strand breaks. IR inhibits CRM1-dependent nuclear export of cyclin B1 and activation-induced cytosine deaminase (30, 31). Similar to nCLU, IR-induced accumulation of cyclin B1 in the nucleus is associated with apoptosis (31). In addition to IR, nuclear export of activation-induced cytosine deaminase is inhibited by bleomycin and H2O2, but not UV (30), suggesting that only agents that cause DNA double strand breaks abrogate CRM1-mediated export. In contrast, CRM1-dependent nuclear export of BRCA1 was enhanced after IR (32). This suggests that CRM1 remains fully active after IR and that inhibition of nuclear export may be due to changes in cargo proteins and not to the export machinery itself. Changes in cargo proteins may include covalent modifications and/or conformational changes that make NES motifs inaccessible to CRM1. The conformational changes or covalent modifications in nCLU that lead to its dissociation from CRM1 remain to be determined. Our prior data suggested that the N- and C-terminal coiled-coil regions of nCLU could bind each other (8), making self-folding/unfolding a potential mechanism for exposing its NLS.
We hypothesize that nCLU activation and accumulation into the nuclei of IR-treated MCF-7 cells may sequester Ku70, possibly in conjunction with stress-induced acetylation of Ku70 (33). Thus, nCLU in MCF-7 cells may compete with and affect Ku70-Bax interaction interactions. Data in Fig. 5 are consistent with a role of nCLU in Bax-mediated cell death responses.
Our laboratory is investigating the details of Bax-dependent cell death induced by nCLU after IR. The recent discovery that nCLU binds Bcl-XL via a novel BH3 domain (6, 7) directly implicates nCLU in the regulation of Bax-mediated cell death.
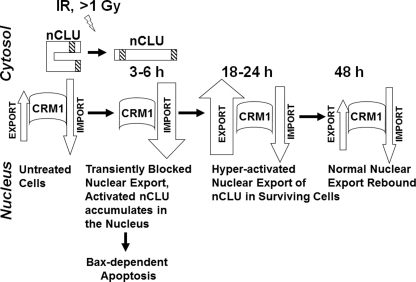
Based on our data with LMB, we conclude that a basal level of active nCLU is continually exported from the nucleus to the cytoplasm in untreated cells. Cells appear to maintain a controlled level of nCLU in both cellular compartments. The presence of nCLU in the nucleus of cells under control conditions is consistent with our prior results (13, 17) and strongly suggests a normal basal function for nCLU in the nucleus. Exposure of cells to IR causes activation of pnCLU (∼49 kDa) to mature ∼55-kDa nCLU, resulting in its accumulation in the nuclei of exposed cells. This accumulation was greatly enhanced by inhibiting CRM1-mediated export (Figs. 2 and 3) as modeled in Fig. 7.
FIGURE 7.
Model for nuclear import/export of the nCLU cell death protein in control and IR-exposed cells. We propose that in untreated cells, a majority of nCLU is in an inactive conformation as pnCLU, a form most likely folded onto itself to hide its functional NLS. Under normal growth conditions, a low level of expressed and active mature ∼55-kDa nCLU binds CRM1. Most likely an intermediate carrier protein is involved, and this complex is actively exported out of the nucleus. This mechanism of export maintains critical balanced levels of nCLU and prevents cell death. Exposure to IR (≥1 Gy) causes activation of nCLU via a currently unknown mechanism. The net result is the inability of CRM1 to bind activated nCLU after IR, resulting in accumulation of nCLU in the nuclei of cells, which enhances cell death and inhibits double strand break DNA repair. By this mechanism, the nuclear import/export balance of activated nCLU is shifted toward import. nCLU activation, in turn, disrupts Ku70-Bax complexes, resulting in Bax release. This mechanism appears to explain the resistance to nCLU-mediated cell death in bax-deficient cells. Activated nCLU also binds BH3 domain proteins, sequestering pro-survival proteins such as Bcl-Xl.
Recent clinical data suggest that aggressive tumors of the esophagus, breast, and colon have elevated sCLU with concomitant reduced nCLU protein levels in the nuclei of neoplastic cells (1, 2). In contrast, the few apoptotic cells present in associated normal tissues contained low levels of sCLU, with elevated nCLU protein levels in the nuclei of dying cells. The data presented here, as well as in our prior papers (8, 17), suggest that aggressive colon, esophagus, as well as breast tumors may have altered ratios of sCLU to nCLU protein levels, leading to a pro-survival microenvironment. Collectively, our data strongly suggest that the ratio of sCLU to nCLU expression is a major determinant of aggressiveness of specific cancers. More specifically, the data presented here suggest that tumor cells may have an enhanced ability to exclude nCLU from their nuclei, decrease Bax expression, and/or lose the ability to generate truncated nCLU mRNA and corresponding pro-death nCLU protein (8). We are currently examining tumor compared with associated normal tissue for genetic alterations of the CLU gene that may explain this apparent loss of a pro-death response while simultaneously expressing the pro-survival sCLU protein.
Acknowledgments
We are grateful to the Boothman and Matsuyama laboratory members for helpful discussions.
This work was supported by Department of Energy Grant DE-FG02-09ER64789 (to D. A. B.). This is paper CSCN070 from the University of Texas Southwestern program in cell stress and cancer nanomedicine.
- CLU
- clusterin
- hr
- human recombinant
- IR
- ionizing radiation
- LMB
- leptomycin B
- MEF
- mouse embryonic fibroblast
- nCLU
- nuclear CLU
- NES
- nuclear export sequence
- NLS
- nuclear localization sequence
- PI
- propidium iodide
- pnCLU
- pre-nuclear CLU
- psCLU
- pre-secretory CLU
- sCLU
- secretory CLU
- Scr
- scrambled.
REFERENCES
- 1. Pucci S., Bonanno E., Pichiorri F., Angeloni C., Spagnoli L. G. (2004) Oncogene 23, 2298–2304 [DOI] [PubMed] [Google Scholar]
- 2. He H. Z., Song Z. M., Wang K., Teng L. H., Liu F., Mao Y. S., Lu N., Zhang S. Z., Wu M., Zhao X. H. (2004) World J. Gastroenterol. 10, 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Redondo M., Villar E., Torres-Muñoz J., Tellez T., Morell M., Petito C. K. (2000) Am. J. Pathol. 157, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Debure L., Vayssiere J. L., Rincheval V., Loison F., Le Drean Y., Michel D. (2003) J. Cell Sci. 116, 3109–3121 [DOI] [PubMed] [Google Scholar]
- 5. July L. V., Beraldi E., So A., Fazli L., Evans K., English J. C., Gleave M. E. (2004) Mol. Cancer Ther. 3, 223–232 [PubMed] [Google Scholar]
- 6. Kim N., Yoo J. C., Han J. Y., Hwang E. M., Kim Y. S., Jeong E. Y., Sun C. H., Yi G. S., Roh G. S., Kim H. J., Kang S. S., Cho G. J., Park J. Y., Choi W. S. (May 12, 2011) J. Cell Physiol. 10.1022/jcp.22836 [DOI] [PubMed] [Google Scholar]
- 7. Lee D. H., Ha J. H., Kim Y., Bae K. H., Park J. Y., Choi W. S., Yoon H. S., Park S. G., Park B. C., Yi G. S., Chi S. W. (2011) Biochem. Biophys. Res. Commun. 408, 541–547 [DOI] [PubMed] [Google Scholar]
- 8. Leskov K. S., Klokov D. Y., Li J., Kinsella T. J., Boothman D. A. (2003) J. Biol. Chem. 278, 11590–11600 [DOI] [PubMed] [Google Scholar]
- 9. O'Sullivan J., Whyte L., Drake J., Tenniswood M. (2003) Cell Death Differ. 10, 914–927 [DOI] [PubMed] [Google Scholar]
- 10. Reddy K. B., Jin G., Karode M. C., Harmony J. A., Howe P. H. (1996) Biochemistry 35, 6157–6163 [DOI] [PubMed] [Google Scholar]
- 11. Sutton D., Kim S., Shuai X., Leskov K., Marques J. T., Williams B. R., Boothman D. A., Gao J. (2006) International J. Nanomed. 1, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trougakos I. P., So A., Jansen B., Gleave M. E., Gonos E. S. (2004) Cancer Res. 64, 1834–1842 [DOI] [PubMed] [Google Scholar]
- 13. Yang C. R., Yeh S., Leskov K., Odegaard E., Hsu H. L., Chang C., Kinsella T. J., Chen D. J., Boothman D. A. (1999) Nucleic Acids Res. 27, 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H., Kim J. K., Edwards C. A., Xu Z., Taichman R., Wang C. Y. (2005) Nat. Cell Biol. 7, 909–915 [DOI] [PubMed] [Google Scholar]
- 15. Caccamo A. E., Scaltriti M., Caporali A., D'Arca D., Scorcioni F., Candiano G., Mangiola M., Bettuzzi S. (2003) Ann. N.Y. Acad. Sci. 1010, 514–519 [DOI] [PubMed] [Google Scholar]
- 16. Boothman D. A., Trask D. K., Pardee A. B. (1989) Cancer Res. 49, 605–612 [PubMed] [Google Scholar]
- 17. Yang C. R., Leskov K., Hosley-Eberlein K., Criswell T., Pink J. J., Kinsella T. J., Boothman D. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5907–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zong W. X., Ditsworth D., Bauer D. E., Wang Z. Q., Thompson C. B. (2004) Genes Dev. 18, 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poon S., Easterbrook-Smith S. B., Rybchyn M. S., Carver J. A., Wilson M. R. (2000) Biochemistry 39, 15953–15960 [DOI] [PubMed] [Google Scholar]
- 20. Wyatt A., Yerbury J., Poon S., Dabbs R., Wilson M. (2009) Adv. Cancer Res. 104, 89–114 [DOI] [PubMed] [Google Scholar]
- 21. Wyatt A. R., Yerbury J. J., Wilson M. R. (2009) J. Biol. Chem. 284, 21920–21927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Criswell T., Beman M., Araki S., Leskov K., Cataldo E., Mayo L. D., Boothman D. A. (2005) J. Biol. Chem. 280, 14212–14221 [DOI] [PubMed] [Google Scholar]
- 23. Trougakos I. P., Lourda M., Antonelou M. H., Kletsas D., Gorgoulis V. G., Papassideri I. S., Zou Y., Margaritis L. H., Boothman D. A., Gonos E. S. (2009) Clin. Cancer Res. 15, 48–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hatakeyama S., Kitagawa M., Nakayama K., Shirane M., Matsumoto M., Hattori K., Higashi H., Nakano H., Okumura K., Onoé K., Good R. A., Nakayama K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3859–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Markopoulou S., Kontargiris E., Batsi C., Tzavaras T., Trougakos I., Boothman D. A., Gonos E. S., Kolettas E. (2009) FEBS J. 276, 3784–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gomez J. A., Gama V., Yoshida T., Sun W., Hayes P., Leskov K., Boothman D., Matsuyama S. (2007) Biochem. Soc. Trans. 35, 797–801 [DOI] [PubMed] [Google Scholar]
- 27. Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 28. Ossareh-Nazari B., Bachelerie F., Dargemont C. (1997) Science 278, 141–144 [DOI] [PubMed] [Google Scholar]
- 29. Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brar S. S., Watson M., Diaz M. (2004) J. Biol. Chem. 279, 26395–26401 [DOI] [PubMed] [Google Scholar]
- 31. Porter L. A., Cukier I. H., Lee J. M. (2003) Blood 101, 1928–1933 [DOI] [PubMed] [Google Scholar]
- 32. Feng Z., Kachnic L., Zhang J., Powell S. N., Xia F. (2004) J. Biol. Chem. 279, 28574–28584 [DOI] [PubMed] [Google Scholar]
- 33. Cohen H. Y., Lavu S., Bitterman K. J., Hekking B., Imahiyerobo T. A., Miller C., Frye R., Ploegh H., Kessler B. M., Sinclair D. A. (2004) Mol. Cell 13, 627–638 [DOI] [PubMed] [Google Scholar]