Background: Dimerization is a prerequisite for protein O-mannosyltransferase activity.
Results: Transferase activity depends on an intact Asp-Glu motif located in a region crucial for acceptor binding/catalysis in both complex partners.
Conclusion: Complex formation leads to the assembly of a composite catalytic center.
Significance: Defining the role of complex formation is crucial for understanding the catalytic mechanism of protein O-mannosyltransferases.
Keywords: Glycoprotein Biosynthesis, Glycosyltransferases, Membrane Proteins, Post-translational Modification, Yeast, PMT, POMT, Mannosylation
Abstract
Protein O-mannosylation is an essential modification in fungi and mammals. It is initiated at the endoplasmic reticulum by a conserved family of dolichyl phosphate mannose-dependent protein O-mannosyltransferases (PMTs). PMTs are integral membrane proteins with two hydrophilic loops (loops 1 and 5) facing the endoplasmic reticulum lumen. Formation of dimeric PMT complexes is crucial for mannosyltransferase activity, but the direct cause is not known to date. In bakers' yeast, O-mannosylation is catalyzed largely by heterodimeric Pmt1p-Pmt2p and homodimeric Pmt4p complexes. To further characterize Pmt1p-Pmt2p complexes, we developed a photoaffinity probe based on the artificial mannosyl acceptor substrate Tyr-Ala-Thr-Ala-Val. The photoreactive probe was preferentially cross-linked to Pmt1p, and deletion of the loop 1 (but not loop 5) region abolished this interaction. Analysis of Pmt1p loop 1 mutants revealed that especially Glu-78 is crucial for binding of the photoreactive probe. Glu-78 belongs to an Asp-Glu motif that is highly conserved among PMTs. We further demonstrate that single amino acid substitutions in this motif completely abolish activity of Pmt4p complexes. In contrast, both acidic residues need to be exchanged to eliminate activity of Pmt1p-Pmt2p complexes. On the basis of our data, we propose that the loop 1 regions of dimeric complexes form part of the catalytic site.
Introduction
Protein O-mannosylation is a conserved modification among fungi, animals, and some bacteria (1). O-Mannosyl glycans have been implicated in a variety of physiological processes such as stability, sorting and localization of proteins, and ligand interactions. In fungi, reduced O-mannosylation affects cell polarity, morphogenesis, cell wall integrity, and endoplasmic reticulum (ER)5 protein quality control, whereas loss of O-mannosylation is lethal (1–3). In higher eukaryotes, impaired O-mannosylation results in severe developmental defects, and in humans, mutations that affect synthesis of O-mannosyl glycans lead to congenital muscular dystrophies with neuronal migration defects (4, 5).
Protein O-mannosylation is initiated in the ER by the transfer of a mannose residue from dolichol monophosphate-activated mannose to hydroxyl groups of serine and threonine residues of nascent polypeptide chains. This transfer reaction is catalyzed by the family of dolichyl phosphate mannose-dependent protein O-mannosyltransferases (PMTs). PMT family members have been characterized throughout the fungal and animal kingdoms and in some bacterial species (6–12).
PMTs have been best characterized in Saccharomyces cerevisiae, where the PMT family comprises at least six members (Pmt1p–Pmt6p) (1). Topological analysis of S. cerevisiae Pmt1p revealed seven transmembrane domains (TMDs) with the N and C termini situated in the cytosol and the ER lumen, respectively (13). Two prominent hydrophilic loops situated between TMD1 and TMD2 (loop 1) and TMD5 and TMD6 (loop 5) are facing the ER lumen. Based on highly similar hydropathy profiles of PMT proteins, this topology seems to be applicable to all PMT family members.
Phylogenetic analyses indicate that the PMT family is subdivided into PMT1, PMT2, and PMT4 subfamilies, whose members include transferases closely related to S. cerevisiae Pmt1p, Pmt2p, and Pmt4p, respectively (5). In bakers' yeast, PMT1 (S. cerevisiae Pmt1p and Pmt5p) and PMT2 (S. cerevisiae Pmt2p and Pmt3p) subfamily members form heterodimeric complexes, whereas Pmt4p, the sole member of the PMT4 family, forms homodimeric complexes. Besides Pmt4p complexes, Pmt1p-Pmt2p complexes account for the major transferase activities in yeast, although alternative complexes can be formed (14).
Based on amino acid sequence similarities, glycosyltransferases (GTs) have been classified into >90 GT families (15). According to their structural features, GT families have been further grouped into the GT-A and GT-B superfamilies (16). A third superfamily was predicted by iterative sequence searches (GT-C) (16). Due to their predicted architecture, GT-C members are large polytopic integral membrane proteins located in the ER or the plasma membrane (16, 17). The vast majority of these enzymes utilize lipid phosphate-activated sugar donors, and glycosyl transfer leads to an inversion of the stereochemistry of the glycosidic bond. PMTs that define the GT39 family represent generic GT-C superfamily members. To date, the very little information available on three-dimensional structures of GT-C transferases is impeding access to the molecular mechanism also of PMTs. Thus, in this study, we used biochemical and genetic methods to elucidate structure-function relationships of PMTs.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
The following S. cerevisiae strains were used for this study: pmt1Δ (MATα, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ901, ura3-52, suc2-Δ9, pmt1Δ::HIS3) (18), pmt2Δ (MATα, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ901, ura3-52, suc2-Δ9, pmt2Δ::LEU2) (18), pmt4Δ (MATα, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ901, ura3-52, suc2-Δ9, pmt4Δ::TRP1) (19), pmt1Δpmt2Δ (MATα, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ901, ura3-52, suc2-Δ9, pmt1Δ::HIS3, pmt2Δ::LEU2) (18), and pmt2Δpmt3Δ (MATa, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ901, ura3-52, suc2-Δ9, pmt2Δ::LEU2, pmt3Δ::HIS3) (7). Yeast strains were grown under standard conditions and transformed according to Ref. 20 with the following plasmids: YEp352 (21), pSB53 (PMT1) (13), pSB56 (PMT1HA) (22), pSB101 (PMT1HAΔ76–124; Δloop 1HA) (22), pVG13 (PMT1HAΔ304–531; Δloop 5HA) (22), pSB112 (PMT1HA-E78A) (22), pSB113 (PMT1HA-D96A) (22), pVG80 (PMT2HA) (14), pJK4-BI (PMT4FLAG) (14), and the plasmids listed below. Standard procedures were used for all DNA manipulations (23). All cloning steps were carried out in Escherichia coli host SURE® 2 (Stratagene). PCR fragments were routinely checked by sequence analysis.6 Amino acid substitutions were constructed by site-directed mutagenesis using recombinant PCR as described previously (24).
PMT1 Mutants
PMT1HA point mutations were generated using the following mutagenic oligonucleotide pairs: 580/581 (F76A), 582/583 (D77A), 584/585 (H80A), 586/587 (F81A), 588/589 (Y88A), 590/591 (V97A), 592/593 (H98A), 594/595 (P99A), and 596/597 (P100A) in combination with the outer primers vg1 and oligo195. The resulting 1463-bp PCR fragments were subcloned into pGEM®-T-Easy (Promega). A 1323-bp PmlI-KpnI fragment was then excised and subcloned into pSB56 cut with the same enzymes, resulting in the following plasmids: pAS40 (F76A), pAS41 (D77A), pAS42 (H80A), pAS43 (F81A), pAS44 (Y88A), pAS45 (V97A), pAS46 (H98A), pAS47 (P99A), and pAS48 (P100A).
To generate mutant PMT1-D77A/E78A, the mutagenic primer pair 1649/1650 was used in combination with oligo1516 and oligo1651. The resulting 1028-bp PCR fragment was cut with PshAI and Pfl23II and subcloned into pSB53 cut with the same enzymes, resulting in plasmid pScML5 (PMT1-D77A/E78A).
PMT2 Mutants
To generate plasmid pScML6 (PMT2HA), a 2905-bp PstI-SalI fragment containing the PMT2 promoter and coding regions fused to three copies of the hemagglutinin epitope was excised from pVG80 and subcloned into vector pRS424 cut with PstI and XhoI. To obtain mutant PMT2HA-D92A/E93A, the mutagenic primer pair 1652/1653 in combination with the outer primers oligo1518 and oligo1654 were used to generate a 1028-bp PCR fragment as described above. Plasmid pVG80 was used as a template. The PCR product was cut with XhoI and MluI and subcloned into pScML6 cut with the same enzymes, resulting in plasmid pScML7 (PMT2HA-D92A/E93A).
PMT4 Mutants
To generate PMT4 mutants the mutagenic primer pairs 1470/1471 (D80A), 1695/1696 (E81A), 1697/1698 (D80E), 1699/1700 (E81D), and 1701/1702 (D80E/E81D) in combination with the outer primers oligo1469 and oligo1694 were used to produce mutated PCR fragments as described above. For homologous recombination, 100 ng of pJK4-BI (linearized with SphI and ClaI) and 500 ng of PCR product were cotransformed into yeast strain pmt4Δ as described previously (25).
pVG12 (hyPMT1HA)
pSB53 was cut with SphI and NarI, treated with DNA polymerase I (Klenow fragment), and religated to remove a HindIII site from the multiple cloning site. The resulting plasmid (pSB73) was ligated via a HindIII site with a PCR fragment encoding PMT4 loop 5 that had been amplified on genomic DNA using oligo175B and oligo176. Subsequently, the plasmid created (pSB74) was digested with RsrII and XhoI and ligated with a 432-bp RsrII-XhoI fragment isolated from pSB56, fusing six copies of the HA epitope to the PMT1/PMT4 loop 5 hybrid.
Computer Analyses
Multiple sequence alignments of PMT4 family members were prepared using ClustalW2 (26), and rendering was done using BoxShade.
Preparation of Crude Membranes
Yeast cells from exponentially growing cultures were harvested, and cell fractionation was performed as described previously (22).
Preparation of Cts1p
Yeast strains were grown to stationary phase (A600 = 5), and the culture medium was collected by centrifugation at 3000 × g for 5 min at 4 °C. Ice-cold trichloroacetic acid was added to the medium at a final concentration of 14% (v/v), and proteins were allowed to precipitate for 30 min at 4 °C. The protein precipitate was recovered by centrifugation at 20,000 × g for 30 min at 4 °C. Protein pellets were washed with 500 μl of ice-cold acetone and air-dried. Protein pellets corresponding to cells at A600 = 2 of were resuspended in 10 μl of SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
Immunoprecipitation
Protein extracts were prepared as described (22) substituting the deoxycholate buffer with Triton buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.3 mm MgCl2, 10% glycerol, 0.5% Triton X-100, 1 mm PMSF, 1 mm benzamidine, 0.25 mm Nα-p-tosyl-l-lysine chloromethyl ketone, 50 mg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone, 10 mg/ml antipain, 1 mg/ml leupeptin, and 1 mg/ml pepstatin). Immunoprecipitation of HA-tagged proteins was performed using 400 μl of Triton extracts and 15 μl of anti-HA affinity matrix (clone 3F10, Roche Applied Science) for 1–2 h at 4 °C. Immunoprecipitates were washed four times with 1 ml of prechilled Triton buffer and once with 1 ml of Tris-buffered saline.
Photoaffinity Labeling
The photoreactive peptide substrate *Tyr-Ala-Thr-Ala-Val-Lys-biotin (*YATAVK-biotin) was generated by incubating 2 μl of 50 mm biotinylated peptide (NH2-YATAVK(biotin)-COOH (YATAVK-biotin); Thermo Hybaid) with 1 μl of 200 mm N-5-azido-2-nitrobenzoyloxysuccinimide (Pierce) for 1 h in the dark. Immunoprecipitates of Pmt1pHA, Pmt1pHA mutants, and Pmt2pHA were prepared as described above and suspended in 27 μl of buffer (120 mm Tris-HCl (pH 7.5), 7.5 mm MgCl2, and 0.15% Triton X-100) including 0.3 mm HA-peptide (Sigma) to release HA-tagged PMTs from the affinity matrix and the photoreactive probe. Peptides Ac-NH-Tyr-Ala-Thr-Ala-Val-COOH (YATAV), Ac-NH-Tyr-Ala-Ser-Ala-Val-COOH (YASAV), and Ac-NH-Ser-Ser-Ser-Ser-Ser-COOH (SSSSS) (Eurogentec) were added as required (final concentrations between 1.7 and 7.5 mm). To induce cross-linking, the reaction mixture was exposed for 10 min to UV light (312 nm). 5 μl of 5× SDS sample buffer was added, and proteins separated by SDS-PAGE and analyzed by Western blotting.
Western Blot Analyses
Proteins were separated by SDS-PAGE and transferred to nitrocellulose. Anti-Pmt1p (27) and anti-Gas1p (kindly provided by L. Popolo) polyclonal antibodies were used at 1:2000 dilution. Anti-Pmt2p (28) and anti-Cts1p (7) polyclonal antibodies were diluted 1:1000. Anti-HA (16B12, Covance) and anti-FLAG (M2, Sigma) monoclonal antibodies were used at 1:8000 dilution. Peroxidase-coupled goat anti-biotin antibody, peroxidase-coupled rabbit anti-mouse IgG antibody, and peroxidase-coupled goat anti-rabbit IgG antibody (all from Sigma) were used at a dilution of 1:5000. Protein-antibody complexes were visualized by enhanced chemiluminescence using the SuperSignal West Pico chemiluminescent System (Pierce).
In Vitro Dolichyl Phosphate Mannose (Dol-P-Man)-dependent PMT Assay
The activity of Pmt1p-Pmt2p was analyzed using the donor substrate Dol-P-[14C]Man or Dol-P-[3H]Man (American Radiolabeled Chemicals), 15–50 μg of membrane protein, and 3.5 mm acceptor substrate Ac-NH-YATAV-CONH2 (YATAV) as described previously (27).
Alternatively, Pmt1p-Pmt2p activity was analyzed using 100 nm Dol-P-[3H]Man, 5–10 μg of membrane protein, 120 μm acceptor substrate biotin-NH-YATAV-CONH2 (Thermo Fisher Scientific), 20 mm Tris-HCl (pH 8), 10 mm EDTA, 0.5% β-octyl thioglucoside (AppliChem), 2 mm β-mercaptoethanol, and 1 mg/ml phosphatidylcholine from egg yolk (Sigma) in a total volume of 20 μl. After incubation at 20 °C for 10 min, the reaction was stopped by the addition of 250 μl of PBS containing 1% Triton X-100 and incubating the reaction mixture at 95 °C for 3 min. The acceptor substrate was precipitated using 40 μl of a slurry of avidin-agarose (Pierce) for 1 h at 4 °C. Precipitates were washed at least five times with 1 ml of PBS containing 1% Triton X-100 and suspended in 200 μl of PBS. Radioactivity adsorbed to the beads was measured using a liquid scintillation counter (Beckman Coulter).
The in vitro activity of Pmt4p was based on the amount of mannose transferred from Dol-P-[3H]Man to an α-dystroglycan-GST fusion protein (29, 30) according to Catrein et al.7
RESULTS
Photoaffinity Labeling of Pmt1p and Pmt2p with an in Vitro Mannose Acceptor Substrate
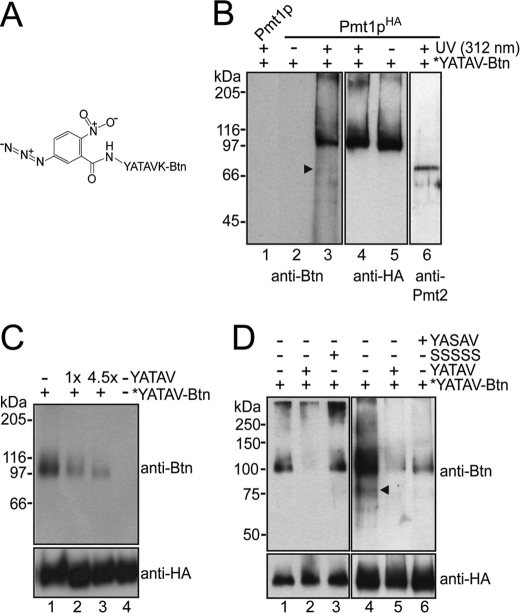
To identify regions in Pmt1p-Pmt2p that are involved in binding of mannose acceptors and/or catalysis, we developed a peptide-based photoaffinity probe. We used the biotinylated peptide YATAVK-biotin, which serves as in vitro mannosyl acceptor substrate of Pmt1p-Pmt2p and is O-mannosylated to a similar extent as the known acceptor peptide YATAV (Table 1). YATAVK-biotin was conjugated via its primary amino group with the heterobifunctional N-hydroxysuccinimide ester and photoactivatable cross-linker N-5-azido-2-nitrobenzoyloxysuccinimide (spacer arm length of 7.7 Å) as described under “Experimental Procedures.” The photoreactive peptide probe *YATAVK-biotin (Fig. 1A) was incubated with Pmt1pHA-Pmt2p complexes that were purified by co-immunoprecipitation. We have shown previously that incorporation of six HA epitopes at the C terminus of Pmt1p (Pmt1pHA; apparent molecular mass of 98 kDa) followed by cell rupture, membrane isolation, solubilization, and then immunoprecipitation under native conditions using an anti-HA monoclonal antibody results in precipitation of Pmt1pHA-Pmt2p complexes (14) that are enzymatically active (data not shown). After photoactivation at 312 nm, samples were resolved on SDS-polyacrylamide gels and analyzed by Western blotting using an anti-biotin monoclonal antibody to specifically detect Pmtp*YATAVK-biotin cross-linking products. Experimental details are described under “Experimental Procedures.” Blots were reprobed with anti-HA antibody and anti-Pmt2p and anti-Pmt1p polyclonal antibodies as needed to control polyacrylamide gel loading and to specifically detect the complex partners Pmt1p and Pmt2p.
TABLE 1.
YATAVK-biotin is a Pmt1p/Pmt2p in vitro mannosyl acceptor substrate
25 μg of membrane proteins from strain pmt1Δ expressing Pmt1pHA was incubated in the in vitro mannosyltransferase assay following the transfer of [14C]mannose from Dol-P-[14C]Man to the indicated peptides (final concentration of 1.8 mm). Values were corrected against the activity detected in a pmt1Δ strain, which is <1% of the activity detected when pSB56 (PMT1HA) was expressed. Results from a representative experiment are shown.
| Peptide | Activity |
|---|---|
| cpm/μg/min | |
| YATAV | 1.76 |
| YATAVK-biotin | 1.40 |
| YASAV | 0.34 |
| SSSSS | 0.08 |
FIGURE 1.
Cross-linking of Pmt1p with a photoreactive mannosyl acceptor peptide. A, structure of the photoreactive probe *YATAVK-biotin (Btn). B–D, Pmt1pHA-Pmt2p complexes isolated from the yeast strains pmt1Δ/pSB56 (Pmt1pHA) and pmt1Δ/pSB53 (Pmt1p) were labeled using the photoreactive probe *YATAVK-biotin as described under “Experimental Procedures.” Photoactivation of the affinity probe was induced by UV light at 312 nm. Samples were resolved on 8% SDS-polyacrylamide gels. Western blots were sequentially probed with anti-biotin and anti-HA antibodies. Arrowheads highlight labeling products with the apparent molecular mass of Pmt2p. B, *YATAVK-biotin predominantly reacted with Pmt1pHA (lanes 3 and 4). In control reactions without Pmt1p-Pmt2p (lane 1) and in which photoactivation was omitted (lanes 2 and 5), no labeling was observed. A sample identical to that shown in lanes 3 and 4 was analyzed using anti-Pmt1p (not shown) and anti-Pmt2p (lane 6) polyclonal antibodies. C, cross-linking reactions were performed in the absence (lane 1) or presence of 1- and 4.5-fold molar excesses (lanes 2 and 3) of the mannosyl acceptor peptide YATAV. D, cross-linking reactions were performed in the absence (lanes 1 and 4) or presence of a 4.5-fold molar excess of YATAV (lanes 2 and 5), YASAV (lane 6), and SSSSS (lane 3).
As shown in Fig. 1, a predominant cross-linking product could be detected that represents Pmt1pHA (Fig. 1B, lanes 3 and 4). In control reactions in which photoactivation was omitted (Fig. 1B, lanes 2 and 5) or in which heterobifunctional cross-linkers with different spacer arm lengths were used (up to 19.9 Å) (data not shown), no cross-linking products were obtained. To further control the specificity of the labeling, cross-linking reactions were performed in the presence of varyingly effective in vitro mannosyl acceptor peptides that should compete with *YATAVK-biotin for Pmt1pHA binding (Table 1). As expected, increasing amounts of the peptide YATAV efficiently hampered *YATAVK-biotin labeling (Fig. 1C). Compared with YATAV, this effect was less pronounced when the poorer mannosyl acceptor peptide YASAV was used (Fig. 1D, right panel, and Table 1). Accordingly, the pentapeptide SSSSS, which is not mannosylated by Pmt1p-Pmt2p, did not interfere with *YATAVK-biotin binding (Fig. 1D, left panel, and Table 1), further proving the specificity of the photoaffinity labeling.
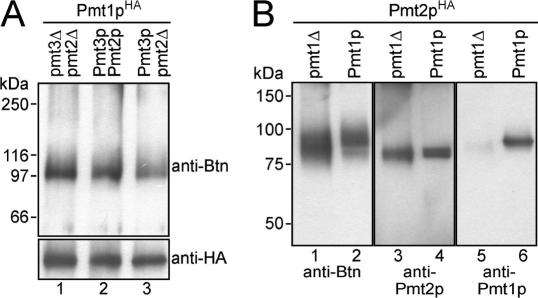
Our data show that *YATAVK-biotin was preferentially cross-linked to Pmt1p. Minor signals became evident only when exposure times of the blots were significantly increased (Fig. 1, B and D, arrowhead), suggesting labeling of Pmt2p. To further follow this issue, we analyzed Pmt1pHA in the absence of known complex partners. Pmt1p interacts with Pmt2p and, mainly when Pmt2p is absent, also with Pmt3p (14). Thus, we expressed Pmt1pHA in the absence of Pmt2p (pmt2Δ) as well as Pmt2p and Pmt3p (pmt2Δpmt3Δ) and performed photoaffinity labeling as described above. As shown in Fig. 2A, Pmt1pHA still interacted with the peptide substrate. Furthermore, we immunoprecipitated Pmt2pHA (apparent molecular mass of ∼88 kDa) that was expressed in a pmt2Δ and a pmt1Δpmt2Δ mutant background. As shown previously (14), immunoprecipitation under native conditions using an anti-HA monoclonal antibody resulted in precipitation of Pmt1p-Pmt2pHA complexes (Fig. 2B, lanes 4 and 6). Photoaffinity labeling of Pmt1p-Pmt2pHA revealed that the photoreactive probe reacted predominantly with Pmt1p (Fig. 2B, lanes 2, 4, and 6). Pmt2pHA was significantly labeled only when Pmt1p was absent (Fig. 2B, lane 1).
FIGURE 2.
*YATAVK-biotin preferentially reacts with Pmt1p. A, Pmt1pHA interacts with the photoreactive probe in the absence of Pmt2p. Pmt1pHA was isolated in the presence of Pmt2p and Pmt3p (strain pmt1Δ/pSB56; lane 1), in the absence of Pmt2p (strain pmt1Δpmt2Δ/pSB56; lane 2), and in the absence of both Pmt2p and Pmt3p (strain pmt2Δpmt3Δ/pSB56; lane 3). Cross-linking reactions and analyses of the samples were performed as described in the legend to Fig. 1. B, *YATAVK-biotin preferentially reacts with Pmt1p. Pmt2pHA was immunoprecipitated from strains pmt1Δpmt2Δ/pVG80 (pmt1Δ; lanes 1, 3, and 5) and pmt2Δ/pVG80 (Pmt1p; lanes 2, 4, and 6) and labeled with the photoreactive probe as described under “Experimental Procedures.” Samples were resolved on 8% SDS-polyacrylamide gels, and Western blots were sequentially probed with anti-biotin (Btn; lanes 1 and 2) and anti-Pmt2p (lanes 3 and 4) antibodies. Blots were stripped and reprobed with anti-Pmt1p antibodies (lanes 5 and 6).
In summary, our data demonstrate that both Pmt1p and Pmt2p have the capacity to bind the peptide substrate. However, under the experimental conditions used, cross-linking of the *YATAVK-biotin probe to Pmt1p is favored.
Pmt1p Loop 1, Especially Glu-78, Is Crucial for Cross-linking of the Photoreactive Peptide
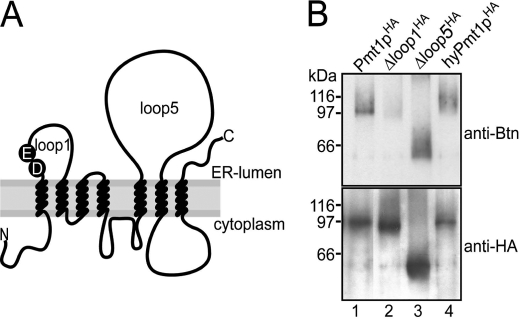
To further narrow down regions involved in peptide binding, we analyzed Pmt1pHA mutants in which the major ER-oriented loop regions, loop 1 (amino acids 71–135) and loop 5 (amino acids 294–586), were deleted (Fig. 3A). Following the transfer of mannose from Dol-P-Man to the pentapeptide YATAV, we demonstrated previously that, compared with Pmt1pHA, the in vitro activity of mutants Δloop 1HA (Δ76–124) and Δloop 5HA (Δ304–531) is diminished to 0.7 and 7%, respectively. In vivo, both mutants resemble a pmt1Δ phenotype (22). Although mutant Δloop 1HA was stably expressed and efficiently immunoprecipitated, cross-linking to the *YATAVK-biotin probe was lost (Fig. 3B, lane 2, and supplemental Fig. 1A). In contrast, deletion of loop 5 did not affect binding of the photoreactive probe (Fig. 3B, lane 3). The same was true for a Pmt1pHA hybrid protein (∼15% residual in vitro activity) (data not shown) in which the Pmt1p loop 5 had been replaced with the Pmt4p loop 5 region (Fig. 3B, lane 4).
FIGURE 3.
Pmt1p loop 1 is crucial for cross-linking of the photoreactive peptide. A, schematic representation of the ER membrane topology of yeast PMTs. The conserved DE motif is indicated. B, wild-type and mutant versions of Pmt1pHA were isolated from strain pmt1Δ transformed with pSB56 (Pmt1pHA; lane 1), pSB101 (Δloop 1HA; lane 2), pVG13 (Δloop 5HA; lane 3), and pVG12 (hyPmt1pHA; lane 4). After photoaffinity labeling, samples were resolved on 8% SDS-polyacrylamide gels. Western blots were sequentially probed with anti-biotin (Btn) and anti-HA antibodies.
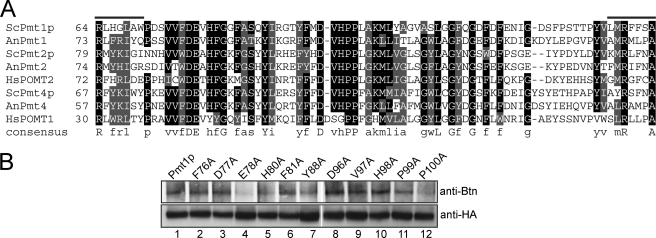
Our data suggest that the loop 1 region contains specific amino acids that are involved in peptide substrate binding and/or catalysis. The loop 1 domain is highly conserved among eukaryotic as well as prokaryotic PMTs. By protein sequence alignments with ClustalW, we identified peptide motifs that are conserved in loop 1 of all PMTs analyzed (Fig. 4A and supplemental Fig. 2). To test the role of the conserved amino acids, we used site-directed mutagenesis and replaced them individually with alanine. The mutant Pmt1pHA proteins were expressed and characterized in a pmt1Δ background. Mutations did not alter protein stability, and only mutants Y88A and P100A moderately decreased complex formation with Pmt2p (supplemental Fig. 3). Surprisingly, only mutation E78A resulted in reduction of in vitro mannosyltransferase activity by >50% compared with Pmt1pHA (Table 2). Accordingly, photoaffinity labeling of this mutant protein was decreased (Fig. 4B). In summary, our data show that Glu-78 is important for acceptor binding and/or catalysis.
FIGURE 4.
Photocross-linking of Pmt1p loop 1 point mutants. A, alignment of S. cerevisiae Pmt1p loop 1 with other PMT family members. Protein sequences are from S. cerevisiae (Sc), Aspergillus nidulans (An), and Homo sapiens (Hs). Loop 1 of S. cerevisiae Pmt1p from Arg-64 to Ala-79 is aligned with members of the three PMT subfamilies. Residues showing at least 80% identity (black) or similarity (gray) are shaded. B, immunoprecipitations with anti-HA antibody were performed on extracts isolated from the yeast strain pmt1Δ expressing Pmt1pHA and individual point mutants thereof. Photoaffinity labeling and analysis of the samples were performed as described in the legend to Fig. 1. Btn, biotin.
TABLE 2.
In vitro mannosyltransferase activity of Pmt1p loop 1 mutant proteins
20 μg of membrane proteins from strain pmt1Δ expressing wild-type and individual mutant Pmt1p proteins was incubated in the in vitro mannosyltransferase assay following the transfer of [3H]mannose from Dol-P-[3H]Man to the mannosyl acceptor peptide biotin-NH-YATAV-CONH2. Values are corrected against the activity detected in a pmt1Δ strain. Average values of two biological replicates with at least four technical replicates each are shown.
| Relative activity | |
|---|---|
| % | |
| Pmt1pHA | 100 |
| F76A | 78.79 ± 4.36 |
| D77A | 71.95 ± 5.39 |
| E78Aa | 46.68 ± 11.97 |
| H80A | 83.03 ± 13.27 |
| F81A | 71.71 ± 2.82 |
| Y88A | 73.79 ± 5.06 |
| D96Aa | 63.21 ± 4.97 |
| V97A | 80.23 ± 8.97 |
| H98A | 62.77 ± 8.52 |
| P99A | 61.77 ± 9.72 |
| P100A | 59.67 ± 8.97 |
| D77A/E78A | 0.19 ± 1.53 |
a Mutants are described in Ref. 22.
The Loop 1 Asp-Glu Motif Is Essential for Activity of Pmt1p-Pmt2p Complexes
In agreement with our findings, a recent study suggested that the loop 1 Asp-Glu (DE) motif of the Mycobacterium tuberculosis PMT homolog is crucial for mannosyltransferase activity (11). But, in contrast to the Pmt1p mutations D77A (∼72% residual activity) and E78A (∼47% residual activity) (Table 2), the corresponding single mutations in the bacterial PMT homolog resulted in complete loss of enzymatic activity. We decided to go further into these differences and generated a Pmt1p double mutant in which both Asp-77 and Glu-78 were replaced with alanine (Pmt1p DE → AA). Pmt1p DE → AA was first characterized in a pmt1Δ background. Change of the DE motif did not alter the stability of the mutant protein and its interaction with Pmt2p (supplemental Fig. 3B). To access in vivo activity, we analyzed the glycosylation status of the specific Pmt1p-Pmt2p substrate chitinase (Cts1p) (31), which is highly O-mannosylated. As shown in Fig. 5 (lanes 1 and 2), Cts1p isolated from strain pmt1Δ was less glycosylated compared with pmt1Δ expressing Pmt1pHA. No significant differences in the glycosylation of Cts1p could be detected between strains expressing Pmt1pHA or mutants D77A and E78A (Fig. 5, lanes 3 and 4), whereas Pmt1p DE → AA failed to restore Cts1p to its normal glycosylation levels (lane 5). Furthermore, compared with the single mutants, the double mutant Pmt1p DE → AA showed basically no in vitro mannosyltransferase activity (Table 2). We further created a Pmt2pHA DE → AA mutant protein with Asp-92 and Glu-93 replaced with alanine (Fig. 4A) and measured the in vitro mannosyltransferase activity of the wild-type and mutant proteins in a pmt1Δpmt2Δ mutant background. Both the Pmt1p DE → AA-Pmt2pHA and Pmt1p-Pmt2pHA DE → AA complexes were inactive (Table 3). The residual in vitro activity detected (∼3% compared with Pmt1p-Pmt2pHA) is most likely due to alternative Pmt1p-Pmt3p or Pmt2pHA-Pmt5p complexes (14). When mutant proteins Pmt1pHA DE → AA and Pmt2pHA DE → AA were coexpressed, mannosyltransferase activity was completely abolished (Table 3).
FIGURE 5.
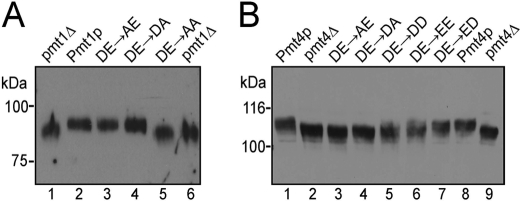
In vivo mannosyltransferase activity of DE motif mutants. A, chitinase was precipitated from culture medium as described under “Experimental Procedures.” PMT1HA and the indicated point mutants were individually expressed in strain pmt1Δ. B, Gas1p was analyzed in microsomal membrane preparations. PMT4FLAG and the indicated point mutants were individually expressed in strain pmt4Δ. Proteins were resolved on 6% SDS-polyacrylamide gels and analyzed by Western blotting using anti-Cts1p (A) and anti-Gas1p (B) antibodies.
TABLE 3.
In vitro mannosyltransferase activity of loop 1 DE motif mutants
25 μg of membrane proteins from strain pmt1Δpmt2Δ expressing wild-type and individual mutant Pmt1p and Pmt2p proteins was incubated in the in vitro mannosyltransferase assay following the transfer of [3H]mannose from Dol-P-[3H]Man to the mannosyl acceptor peptide NH-YATAV-CONH2. Values are corrected against the activity detected in a pmt1Δpmt2Δ strain. Average values of two biological replicates with three technical replicates each are shown.
| Relative activity | |
|---|---|
| % | |
| Pmt1p, Pmt2p | 100 |
| Pmt1p, pmt2Δ | 3.02 ± 2.28 |
| pmt1Δ, Pmt2p | 5.93 ± 3.64 |
| Pmt1p, Pmt2p-D92A/E93A | 3.63 ± 1.69 |
| Pmt1p-D77A/E78A, Pmt2p | 3.59 ± 0.48 |
| Pmt1p-D77A/E78A, Pmt2p-D92A/E93A | 0.24 ± 0.97 |
Taken together, our results demonstrate that the DE motif is important for the enzymatic activity of yeast Pmt1p-Pmt2p complexes. But why is there a discrepancy between eukaryotic Pmt1p-Pmt2p and the bacterial PMT homolog where mutation of one individual residue of the DE motif already abolishes mannosyltransferase reaction?
Characterization of the Loop 1 DE Motif of Homodimeric Pmt4p Complexes
For various GTs, it has been suggested that acidic amino acid residues contribute to the substrate binding/catalytic domain (17). Thus, two individual loop 1 regions could contribute to such a catalytic site (Fig. 4A). This is one possibility to explain why complex formation is obligatory for PMT activity (1, 30), although complex formation of bacterial PMTs has not been analyzed yet. It might also explain why mutation of a single amino acid of the loop 1 DE motif is less deleterious for heteromeric complexes. To track this possibility, we characterized the loop 1 DE motif of Pmt4p, which forms homomeric complexes (14).
For this purpose, we generated the Pmt4p mutant proteins D80A (DE → AE), D80E (DE → EE), E81A (DE → DA), E81D (DE → DD), and D80E/E81D (DE → ED) and characterized them in a pmt4Δ background. Individual changes in the DE motif did not alter the stability of the mutant proteins (supplemental Fig. 3). The glycosylation status of the β-1,3-glucanosyltransferase Gas1 (Gas1p) (31), which is O-mannosylated specifically by Pmt4p, was analyzed to access the in vivo activity of the mutant proteins. As shown in Fig. 5B, Gas1p isolated from strain pmt4Δ was hypoglycosylated compared with pmt4Δ expressing Pmt4p (compare lanes 1 and 8 and lanes 2 and 9). Both mutant proteins Pmt4p DE → AE and Pmt4p DE → DA failed to restore Gas1p glycosylation (Fig. 5B, lanes 3 and 4). In contrast, Gas1p glycosylation was partially re-established when the acidic character of the DE motif (DD or EE) was maintained (Fig. 5B, lanes 5 and 6). No significant differences in the glycosylation of Gas1p could be detected between the strains expressing Pmt4p DE → ED and wild-type Pmt4p (Fig. 5B, lane 7).
To further quantify Pmt4p activity in more detail, we established an assay to monitor Pmt4p-specific mannosyltransferase activity in vitro. As shown in Table 4, exchange of each Asp-80 and Glu-81 with alanine resulted in a drop in Pmt4p activity of >96%. With the negative character of the exchanged amino acid conserved (DE → DD and DE → EE), enzymatic activity is less severely affected (∼50% compared with wild-type Pmt4p activity), whereas reversing the DE motif (DE → ED) does not change activity (Table 4).
TABLE 4.
In vitro mannosyltransferase activity of Pmt4p loop 1 mutant proteins
25 μg of membrane proteins from strain pmt4Δ expressing wild-type and individual mutant Pmt4p proteins was incubated in the in vitro mannosyltransferase assay following the transfer of [3H]mannose from Dol-P-[3H]Man to α-dystroglycan-GST. Average values of two biological replicates with three technical replicates each are shown.
| Relative activity | |
|---|---|
| % | |
| Pmt4pFLAG | 100 |
| Pmt4p-D80A | 3.21 ± 0.94 |
| Pmt4p-E81A | 1.43 ± 0.52 |
| Pmt4p-D80E | 52.98 ± 3.83 |
| Pmt4p-E81D | 46.59 ± 3.75 |
| Pmt4p-D80E/E81D | 116.54 ± 12.20 |
In agreement with our assumption, our data demonstrate that a non-conservative exchange of one amino acid of the DE motif causes a dramatic decrease in Pmt4p activity. In addition, our results show that not only the charge but also the nature of the acidic amino acids of the DE motif is important for mannosyltransferase activity.
DISCUSSION
Among the well over 90 GT families annotated in the CAZy Database, two general folds (GT-A and GT-B) have been observed among all structures of nucleotide sugar-dependent GTs (17). Additionally, a third fold of GTs termed GT-C was recently predicted, but the GT-C fold is merely speculative, and only limited structural information is available. The predicted architecture of the GT-C fold is that of a large hydrophobic integral membrane protein having between 7 and 13 transmembrane helices (16).
PMTs (GT39) have been suggested to adopt a GT-C fold. PMTs are polytopic transmembrane proteins bearing seven TMDs. Two major hydrophilic regions (loops 1 and 5) reside on the luminal side of the ER membrane (Fig. 3A) (13). Deletion of either loop region of S. cerevisiae Pmt1p results in highly reduced transferase activity (22); therefore, the catalytic site could not be clearly assigned to one of the loop regions. Here, we used a photoreactive peptide substrate (*YATAVK-biotin) (Fig. 1A) to further address this issue. Photoaffinity labeling was observed for wild-type Pmt1p and the Δloop 5 mutant protein but was strongly reduced for a Pmt1p loop 1 deletion mutant (Fig. 3B). Our data indicate that the loop 1 region constitutes or is part of the acceptor binding and/or catalytic site.
A highly conserved DE motif is present in the loop 1 domains of PMT proteins ranging from bacteria to humans. It is also found in PMT homologous sequences of archaea that we identified by protein BLAST analyses using Pmt1p from S. cerevisiae (Fig. 4A and supplemental Fig. 2). Here, we have demonstrated that the DE motif is crucial for PMT activity. Interestingly, single mutations in the DE motif of S. cerevisiae Pmt1p resulted in only a moderate inhibition of enzymatic activity (Table 2) (22), whereas in bacteria, single mutations of the conserved acidic residues abolish transferase activity (11). This discrepancy might be explained by complex formation of O-mannosyltransferases, which is obligatory for enzymatic activity in eukaryotes (22, 30), although PMT complexes have not been studied in bacteria. To date, it is unclear in which way the physical interaction of two Pmt molecules contributes to their function. Two possible scenarios are conceivable. (i) Each monomer possesses an intrinsic transferase activity, but interaction with the complex partner synergistically stimulates mannosyl transfer. (ii) Only upon complex formation is a composite catalytic center formed, whereas the monomers are not able to catalyze sugar transfer.
Mutation of the entire DE motif (DE → AA) in either S. cerevisiae Pmt1p or Pmt2p resulted in repression of in vitro transferase activity by >95%. The extremely slight residual activity (∼4%) (Table 3) is most likely due to less abundant Pmt1p-Pmt3p and Pmt2p-Pmt5p complexes (14). These findings support the latter assumption. Furthermore, participation of both loop 1 regions within the complex leads to the presence of four acidic amino acids derived from the two DE motifs. Removal of one of these negatively charged amino acids affects enzymatic activity. Deletion of two negative charges in either Pmt1p or Pmt2p results in complete loss of complex function. For homodimeric Pmt4p complexes, the same effect is observed when one charge is eliminated in the DE motif of the loop 1 region (D80A or E81A) (Table 4). These data suggest that all four acidic amino acids are needed to grant full enzymatic activity. Conservative exchange of either Asp-80 or Glu-81 in Pmt4p to some extent diminishes mannosyl transfer, whereas upon inversion of the motif (DE → ED), Pmt4p activity is retained (Table 4). In summary, our findings strongly support that, in PMT complexes, two loop 1 regions contribute to formation of the catalytic site.
The catalytic site of GT-C superfamily members was suggested to reside in a luminally oriented loop region of the enzyme containing conserved variations of a DxD motif corresponding to ExD, DxE, DDx, or DEx residues (16, 32). This motif is located close to the C terminus of the first TMD and is often followed by a small patch of hydrophobic amino acids (16). Mutational analyses of the acidic amino acids revealed their importance for the function of GT-C family members (33, 34). Very recently, the three-dimensional structure of a GT-C family member has been solved and is shedding light on the role of acidic motifs. Campylobacter lari PglB is a homolog of the STT3 subunit of the eukaryotic oligosaccharyltransferase involved in the N-glycosylation of Asn-X-Ser/Thr sequons (35). The structure revealed that PglB features two major domains, an N-terminal transmembrane region and a C-terminal periplasmic region. The transmembrane region consists of 13 TMDs connected by short loops, with the exception of two long external loops situated between TMD1 and TMD2 and between TMD9 and TMD10 (35). The transmembrane segment constitutes the substrate-binding and catalytic sites. The catalytic pocket features four acidic amino acids, including Asp-154 and Asp-156, which derive from a conserved DxD motif, and Asp-56 and Glu-319, which are also highly conserved in STT3 homologs. With regard to the enzyme function, Asp-56, Asp-154, and Glu-319 seem to coordinate a divalent cation that, on the one hand, properly positions Asp-56 and Glu-319 to interact with the substrate Asn and, on the other hand, electrochemically stabilizes the lipid phosphate leaving group. Mutational analyses of Asp-56 and Glu-319 led to >90% reduction of enzymatic activity, whereas exchange of Asp-154 resulted in >50% inhibition, confirming their crucial function for sugar transfer (35). STT3 has been suggested to be distantly related to PMTs,8 indicating that similarities in either the architecture of the enzyme or the catalytic mechanism might be present.
Like PglB/STT3, PMTs are inverting GTs, changing the stereochemistry at the anomeric center of the carbohydrate (35, 36). At least three acidic amino acids are often involved in the reaction mechanism of sugar-transferring enzymes. Inverting GTs employ a direct Sn2-like reaction mechanism, thereby, an active side chain serves as a base catalyst that deprotonates the nucleophile of an acceptor, facilitating direct Sn2-like displacement of the leaving group (37). Asp and Glu often function as such a general base in GTs. However, the moderate decrease in enzymatic activity of Pmt1p mutants D77A and E78A does not support a role as direct catalyst for one of the residues of the DE motif. It has to be noted that, within the loop 5 region of eukaryotic PMTs, at least three acidic amino acids are present that are conserved also (22). However, alanine substitutions of these residues did not severely affect Pmt1p-Pmt2p activity in S. cerevisiae (data not shown). Because bacterial PMTs, which likely employ a similar reaction mechanism, feature a truncated loop 5 lacking these acidic residues, their involvement in the transfer reaction is unlikely.
Acidic amino acids can also be involved in facilitating the departure of the phosphate leaving group by the coordination of divalent cations (Mn2+ or Mg2+) that electrostatically stabilize the developing negative charge (17, 35). Previous work has shown that PMT activity is significantly stimulated by the presence of divalent Mg2+ cations (30, 38, 39). However, chelating agents such as EDTA do not inhibit in vitro mannosyltransferase activity, suggesting either that PMTs have a stronger affinity for divalent cations than many other GTs or that assay conditions used hamper the accessibility of the chelator (30, 38, 39). At this time, it cannot be ruled out that the DE motif is involved in the coordination of Mg2+ ions within PMT complexes, but this issue needs to be further investigated.
The exact function of the DE motif cannot be clearly defined yet. Hence, structural analysis of PMT complexes will be necessary in the future to elucidate the molecular mechanisms underlying the mannosyl transfer reaction.
Supplementary Material
Acknowledgments
We are grateful to W. Tanner and L. Popolo for generously providing yeast strains and antibodies. We thank A. Metschies and C. Müller for excellent technical assistance, E. Ragni for stimulating discussions, and I. Catrein for critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB638.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Oligonucleotide sequences are available upon request.
I. Catrein, T. Jank, M. Schmid, and S. Strahl, manuscript in preparation.
A. Bateman, unpublished data.
- ER
- endoplasmic reticulum
- PMT
- protein O-mannosyltransferase
- TMD
- transmembrane domain
- GT
- glycosyltransferase
- Dol-P-Man
- dolichyl phosphate mannose.
REFERENCES
- 1. Lommel M., Strahl S. (2009) Glycobiology 19, 816–828 [DOI] [PubMed] [Google Scholar]
- 2. Arroyo J., Hutzler J., Bermejo C., Ragni E., García-Cantalejo J., Botías P., Piberger H., Schott A., Sanz A. B., Strahl S. (2011) Mol. Microbiol. 79, 1529–1546 [DOI] [PubMed] [Google Scholar]
- 3. Goder V., Melero A. (2011) J. Cell Sci. 124, 144–153 [DOI] [PubMed] [Google Scholar]
- 4. Endo T. (2005) Acta Myol. 24, 64–69 [PubMed] [Google Scholar]
- 5. Willer T., Valero M. C., Tanner W., Cruces J., Strahl S. (2003) Curr. Opin. Struct. Biol. 13, 621–630 [DOI] [PubMed] [Google Scholar]
- 6. Strahl-Bolsinger S., Immervoll T., Deutzmann R., Tanner W. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8164–8168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gentzsch M., Tanner W. (1996) EMBO J. 15, 5752–5759 [PMC free article] [PubMed] [Google Scholar]
- 8. Martín-Blanco E., García-Bellido A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6048–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jurado L. A., Coloma A., Cruces J. (1999) Genomics 58, 171–180 [DOI] [PubMed] [Google Scholar]
- 10. Willer T., Amselgruber W., Deutzmann R., Strahl S. (2002) Glycobiology 12, 771–783 [DOI] [PubMed] [Google Scholar]
- 11. VanderVen B. C., Harder J. D., Crick D. C., Belisle J. T. (2005) Science 309, 941–943 [DOI] [PubMed] [Google Scholar]
- 12. Mahne M., Tauch A., Pühler A., Kalinowski J. (2006) FEMS Microbiol. Lett. 259, 226–233 [DOI] [PubMed] [Google Scholar]
- 13. Strahl-Bolsinger S., Scheinost A. (1999) J. Biol. Chem. 274, 9068–9075 [DOI] [PubMed] [Google Scholar]
- 14. Girrbach V., Strahl S. (2003) J. Biol. Chem. 278, 12554–12562 [DOI] [PubMed] [Google Scholar]
- 15. Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 16. Liu J., Mushegian A. (2003) Protein Sci. 12, 1418–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 18. Lussier M., Gentzsch M., Sdicu A. M., Bussey H., Tanner W. (1995) J. Biol. Chem. 270, 2770–2775 [DOI] [PubMed] [Google Scholar]
- 19. Immervoll T., Gentzsch M., Tanner W. (1995) Yeast 11, 1345–1351 [DOI] [PubMed] [Google Scholar]
- 20. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. (1986) Yeast 2, 163–167 [DOI] [PubMed] [Google Scholar]
- 22. Girrbach V., Zeller T., Priesmeier M., Strahl-Bolsinger S. (2000) J. Biol. Chem. 275, 19288–19296 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Higuchi R. (1990) in PCR Protocols: A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. eds) pp. 177–183, Academic Press Inc., San Diego, CA [Google Scholar]
- 25. Ma H., Kunes S., Schatz P. J., Botstein D. (1987) Gene 58, 201–216 [DOI] [PubMed] [Google Scholar]
- 26. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 27. Strahl-Bolsinger S., Tanner W. (1991) Eur. J. Biochem. 196, 185–190 [DOI] [PubMed] [Google Scholar]
- 28. Gentzsch M., Immervoll T., Tanner W. (1995) FEBS Lett. 377, 128–130 [DOI] [PubMed] [Google Scholar]
- 29. Lommel M., Willer T., Cruces J., Strahl S. (2010) Methods Enzymol. 479, 323–342 [DOI] [PubMed] [Google Scholar]
- 30. Manya H., Chiba A., Yoshida A., Wang X., Chiba Y., Jigami Y., Margolis R. U., Endo T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gentzsch M., Tanner W. (1997) Glycobiology 7, 481–486 [DOI] [PubMed] [Google Scholar]
- 32. Oriol R., Martinez-Duncker I., Chantret I., Mollicone R., Codogno P. (2002) Mol. Biol. Evol. 19, 1451–1463 [DOI] [PubMed] [Google Scholar]
- 33. Kang J. Y., Hong Y., Ashida H., Shishioh N., Murakami Y., Morita Y. S., Maeda Y., Kinoshita T. (2005) J. Biol. Chem. 280, 9489–9497 [DOI] [PubMed] [Google Scholar]
- 34. Maeda Y., Watanabe R., Harris C. L., Hong Y., Ohishi K., Kinoshita K., Kinoshita T. (2001) EMBO J. 20, 250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011) Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 36. Lehle L., Strahl S., Tanner W. (2006) Angew. Chem. Int. Ed. Engl. 45, 6802–6818 [DOI] [PubMed] [Google Scholar]
- 37. Lairson L. L., Withers S. G. (2004) Chem. Commun. 2243–2248 [DOI] [PubMed] [Google Scholar]
- 38. Sharma C. B., D'Souza C., Elbein A. D. (1991) Glycobiology 1, 367–373 [DOI] [PubMed] [Google Scholar]
- 39. Hendershot L. L., Aeed P. A., Kézdy F. J., Elhammer A. P. (2002) Anal. Biochem. 307, 273–279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.