Background: Cyclin E/Cdk2 is a protein kinase in cell cycle regulation.
Results: MCM3 is a novel substrate of cyclin E/Cdk2, which phosphorylates MCM3 at the site of Thr-722 and regulates its loading onto chromatin.
Conclusion: Excess of MCM3 loading onto DNA will activate the checkpoint pathway.
Significance: We revealed the novel role of MCM3 regulated by cyclin E/Cdk2 in controlling the S phase checkpoint.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, Cyclins, DNA Replication, Protein Phosphorylation, MCM3 Protein, Cyclin E/Cdk2
Abstract
MCM2–7 proteins form a stable heterohexamer with DNA helicase activity functioning in the DNA replication of eukaryotic cells. The MCM2–7 complex is loaded onto chromatin in a cell cycle-dependent manner. The phosphorylation of MCM2–7 proteins contributes to the formation of the MCM2–7 complex. However, the regulation of specific MCM phosphorylation still needs to be elucidated. In this study, we demonstrate that MCM3 is a substrate of cyclin E/Cdk2 and can be phosphorylated by cyclin E/Cdk2 at Thr-722. We find that the MCM3 T722A mutant binds chromatin much less efficiently when compared with wild type MCM3, suggesting that this phosphorylation site is involved in MCM3 loading onto chromatin. Interestingly, overexpression of MCM3, but not MCM3 T722A mutant, inhibits the S phase entry, whereas it does not affect the exit from mitosis. Knockdown of MCM3 does not affect S phase entry and progression, indicating that a small fraction of MCM3 is sufficient for normal S phase completion. These results suggest that excess accumulation of MCM3 protein onto chromatin may inhibit DNA replication. Other studies indicate that excess of MCM3 up-regulates the phosphorylation of CHK1 Ser-345 and CDK2 Thr-14. These data reveal that the phosphorylation of MCM3 contributes to its function in controlling the S phase checkpoint of cell cycle in addition to the regulation of formation of the MCM2–7 complex.
Introduction
During each cell cycle, vast amounts of genetic information must be faithfully duplicated once and only once before cell division. This remarkable accomplishment is ensured by the DNA replication licensing system. Two fundamental stages central to this system are the assembly of pre-replicative complexes (pre-RCs)2 in late M and G1 phases and their subsequent activation at the G1/S boundary (1, 2). A core component of pre-RC is the putative DNA helicase, known as the MCM2–7 complex, which loads onto chromatin in an origin recognition complex-, CDC6-, and CDT1-dependent manner (3). The recruitment of the MCM2–7 complex to an origin signals the completion of pre-RC assembly and renders the competence of the origin to initiate DNA synthesis. Conversion of the MCM2–7 complex into an active helicase is triggered by a concerted activity of S phase kinase cyclin E/Cdk2 and CDC7/DBF4 (4), which ultimately results in recruitment of CDC45 and initiation of DNA replication.
Thus, tight regulation of DNA replication can be divided into two discrete steps, which are closely correlated with oscillations in Cdk activity. That is, the pre-RC complex can only assemble during late M and G1 phases when Cdk activity is low (5–7). Then, increased Cdk activity at the G1/S boundary triggers the activation of MCM2–7 complex helicase activity. In addition to triggering initiation, high levels of Cdk persist throughout S and G2 phases until the cyclin-dependent kinases are inactivated at the end of M phase, ensuring that origins initiate only once per cell cycle. However, the mechanism by which Cdks prevent re-replication and identity of their inhibitory targets are poorly understood. In mammals, some studies have focused on origin recognition complex and CDC6 as targets of negative regulation by Cdks, possibly by means of proteolytic destruction or nuclear export (8–10). However, these results are not mutually consistent, and there is significant controversy regarding precise mechanisms regulating the licensing of replication origins (11, 12). In recent years, there are experimental evidence emerged to suggest that MCM2–7 is not only involved in DNA unwinding but also subject to direct regulation in DNA synthesis and transcription (13–18). Multiple MCM2–7 phosphorylation sites were found to be scattered in each of the protein subunits, which provides a point of potential regulation by kinase signaling pathways. Phosphorylation of Xenopus MCM4 by CDC2 inhibits the activity of MCM2–7 complex and prevents illegitimate DNA replication between late S phase and mitosis (14). Hisao et al. (19) found that phosphorylation of MCM4 by CDC7 kinase facilitates its interaction with CDC45 on the chromatin to initiate DNA replication. Stillman and co-workers (15) demonstrated that the CDC7 kinase can promote S phase by alleviating an inhibitory activity in MCM4. Cortez et al. (17) reported that MCM2 and MCM3 are substrates for ATM and ATR checkpoint kinases, respectively, but the biological consequence remains to be assessed. In a work by Lin et al., MCM3 was found to be phosphorylated at Ser-112 by Cdk1, and this modification regulates the assembly and activity of the MCM2–7 complex (20).
Previously, we identified MCM3 as one of the Cdk2-associated proteins by tandem affinity purification (21). In this report, we confirm that MCM3 is the substrate of cyclin E/Cdk2, which phosphorylates MCM3 at the site of Thr-722 and regulates its loading onto chromatin. Moreover, overexpression of wild type MCM3 but not MCM3 T722A mutant inhibits the S phase entry. Finally, we demonstrate that excess of MCM3 loading onto DNA will activate the phosphorylation of Chk1 and cause the up-regulation of inhibitory phosphorylation of Cdk2.
MATERIALS AND METHODS
Plasmids and Antibodies
MCM3 cDNA was amplified by RT-PCR from human embryonic kidney cDNA library and cloned into pENTR-6 (modified form of pENTR-4, Invitrogen) and then into pDEST-FLAG by Gateway Recombination Cloning technology (Invitrogen). The truncated MCM3 mutants (MCM3 1–194, 194–490, and 490–808) were cloned into pCMV FLAG. The mammalian expression vectors for cyclin E and Cdk2 were used as described previously (21). The cDNAs of cyclin D and Cdk4 were cloned into pCMV Myc. Full-length MCM3 and Cdk2 were cloned into pET41b and pET28c (Novagen), respectively.
The polyclonal antibodies against human MCM3 and MCM5 were generated by immunizing the rabbits with GST-MCM3 (490–807 aa) or GST-MCM5 (644–734 aa). FLAG antibody (M2) is purchased from Sigma. Myc (9E10), His (H-3), MCM7 (141–2), CDC45 (H-300), proliferating cell nuclear antigen (PC-10), lamin B (M-20), Cdk2 (D-12), CDC6 (180.2), and β-actin (1–19) antibodies are purchased from Santa Cruz Biotechnology. Phospho-CHK1 (Ser-345) antibody (2341S), and phospho-threonine-proline antibody (9391) is purchased from Cell Signaling. Cdk2 (phospho-Thr-14) antibody (EP2234Y) is purchased from Abcam.
Mass Spectrometry
pDEST FLAG-MCM3 and pCMV Myc-cyclin E/Cdk2 were transfected into 293T cells. The cell lysates were harvested and immunoprecipitated with anti-FLAG antibody. The enriched FLAG-MCM3 was eluted and digested with trypsin and then subjected to LC-MS/MS analysis to identify the phosphopeptides using a Thermo Fisher LTQ linear ion trap mass spectrometer. All MS/MS spectra were processed using the SEQUEST-based Bioworks 3.3.1 (Thermo Fisher, Inc).
Co-immunoprecipitation and GST Pulldown
FLAG-tagged MCM3 or its mutants with Myc-tagged cyclin E and Cdk2 or Myc-tagged cyclin D and Cdk4 were co-expressed in 293T cells by transient transfection. The cell lysates were harvested and immunoprecipitated with FLAG or Myc antibodies and subjected to immunoblotting with indicated antibodies. For the GST pulldown assay, 0.5 μg of GST-MCM3 or GST as a negative control were incubated with the cell lysates from 293T cells overexpressing Myc-tagged cyclin E and Cdk2, or with His-Cdk2 purified from BL21. The glutathione beads were then added and incubated for 1 h. The bound proteins were eluted with sample loading buffer and analyzed by immunoblotting with anti-Myc or Cdk2 antibodies.
Cell Fractionation
Cells harvested from 10-cm dishes were washed with PBS and extracted with CSK buffer (10 mm PIPES, 100 mm NaCl, 300 mm sucrose, 1 mm MgCl2, 1 mm ATP, 0.5% Triton X-100, protease inhibitor tablet (Roche Diagnostics) for 20 min on ice and then subjected to centrifugation at 1000 × g for 5 min. The supernatant was collected as the CSK soluble fraction. The pellet was washed once with CSK buffer and dissolved in SDS loading buffer as the CSK insoluble fraction.
Cell Culture and Synchronization
HEK 293T cells were cultured in DMEM containing 10% fetal calf serum. T-RExTM-HeLa (Invitrogen) cell lines were maintained in DMEM containing 10% fetal calf serum plus 5 μg/ml of blasticidin. The cells were synchronized at G1/S phase by double thymidine treatment as described in a previous report (21). To synchronize the cells to M phase, the cells were treated with thymidine for 16 h and released for 6 h and then treated with nocodazole (100 ng/ml) for 6 h. The mitotic cells were collected by shaking off.
In Vitro Kinase Assay
GST fused MCM3, MCM3 S112A, MCM3 T464A, MCM3 S611A, MCM3 T722A truncated forms of MCM3, cyclin E, and Cdk2 proteins were all expressed in the BL21 strain of Escherichia coli and then purified by standard procedures (21). 1 μg of GST-MCM3 proteins with 1 μg of GST-cyclin E and Cdk2 were incubated in kinase buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 0.02% BSA, 0.04 mm ATP) in the presence of 0.5 μCi of [γ-32P]ATP for 30 min at 30 °C. Samples were resolved by 10% SDS-PAGE and autoradiographed to x-ray film.
RNAi Treatment
The knockdown of MCM3 was achieved by transfection of HeLa cells with two rounds of 100 nm siRNA. Human MCM3 siRNA target sequence is GCATTGTCACTAAATGTTCTCTAGT. Control sequence is GCAGTCACTCAATGTTCTATTTAGT.
Flow Cytometry
For DNA content analysis, cells were fixed in ice-cold 70% ethanol, washed with PBS-1% BSA, and then incubated with PBS-1% BSA containing 20 μg/ml propidium iodide and 100 μg/ml RNase A. The percentage of cells in each phase of the cell cycle was estimated with ModFit. All samples were analyzed on a FACSCalibur cytometer (BD Biosicences).
Generation of Tet-On Stable Cell Lines
FLAG-tagged MCM3, MCM3 T722A were cloned into the NotI-XhoI sites of pcDNATM/TO (Invitrogen). The plasmids and empty vector were transfected into T-RExTM-HeLa cells (Invitrogen), respectively. 48 h after transfection, the cells were selected with 5 μg/ml of blasticidin and 250 μg/ml of zeocin for 3 weeks. The individual clones were picked, and MCM3 expression was analyzed by immunoblotting after tetracycline treatment.
RESULTS
MCM3 Interacts with Cyclin E/Cdk2
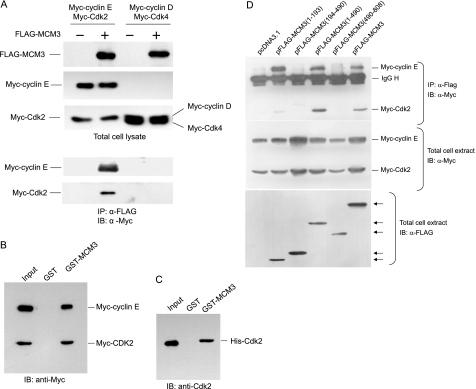
We previously have identified a number of novel Cdk2-associated proteins by tandem affinity purification (21). One of these is the MCM3 protein, a subunit of the MCM2–7 complex known as replicative DNA helicase in eukaryotes. To confirm whether MCM3 is a bona fide Cdk2-interacting partner, we further analyzed the association between MCM3 and Cdk2. FLAG-tagged MCM3 and Myc-tagged cyclin E/Cdk2 constructs were co-transfected into 293T cells. The cell lysates were subjected to immunoprecipitate with FLAG antibody and immunoblotted with Myc antibody. As shown in Fig. 1A, Myc-tagged cyclin E/Cdk2 were co-immunoprecipitated with FLAG-MCM3. GST pulldown assay with GST-MCM3 also showed MCM3 could interact with cyclin E and Cdk2 from cell lysates (Fig. 1B). To verify the direct interaction between MCM3 and Cdk2, GST-MCM3 and His-Cdk2 were used for the GST pulldown assay. The data showed that MCM3 interacts with Cdk2 in vitro (Fig. 1C).
FIGURE 1.
MCM3 interacts with cyclin E/Cdk2. A, co-immunoprecipitation assay. 293T cells were transfected Myc-tagged cyclin E and Cdk2 or Myc-tagged cyclin D and Cdk4 with pcDNA3.1 or FLAG-tagged MCM3. Cell lysates were harvested for immunoprecipitation with FLAG antibody followed by immunoblotting with Myc antibody. B, 293T cells were transfected with Myc-tagged cyclin E and Myc-tagged Cdk2 plasmids. The cell lysates were harvested and subject to GST pulldown with GST or GST-MCM3 fusion proteins and then immunoblotted (IB) with Myc antibody. C, GST protein or GST-MCM3 immobilized on glutathione beads was incubated with purified His-Cdk2. The associated protein was eluted with SDS loading buffer and immunoblotted with His antibody. D, 293T cells were co-transfected with Myc-tagged cyclin E and Myc-tagged Cdk2 with pcDNA3.1 or FLAG-tagged MCM3 (1–193 aa), MCM3 (194–490 aa), MCM3 (1–490 aa), MCM3 (490–807 aa), and full-length MCM3. Cell lysates were subjected to immunoprecipitation with FLAG antibody followed by immunoblotting with Myc antibody. Arrows indicate the bands of FLAG-MCM3 and its truncated forms.
There are eight RXL motifs in MCM3, which are potentially important for its interaction with cyclin E/Cdk2. To map which region of MCM3 is critical for its binding with cyclin E/Cdk2, we generated a set of truncated MCM3 mutants for co-immunoprecipitation assay. The truncated FLAG-tagged MCM3 and Myc-tagged cyclin E/Cdk2 were co-expressed in 293T cells. The cell lysates were immunoprecipitated with FLAG antibody and immunoblotted with Myc antibody. As shown in Fig. 1D, MCM3 (1–194 aa) with four of RXL motifs and MCM3 (1–490 aa) can interact with cyclin E/Cdk2, whereas MCM3 (194–490 aa) and MCM3 (490–808 aa) failed to bind with cyclin E/Cdk2. These data demonstrate that the N-terminal region of MCM3 is essential for its interaction with cyclin E/Cdk2.
MCM3 Is Phosphorylated by Cyclin E/Cdk2
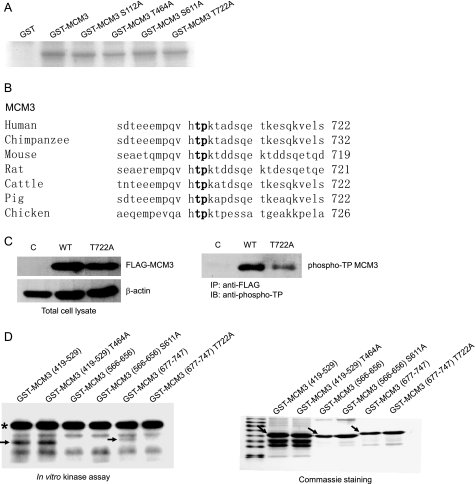
To examine whether MCM3 can be phosphorylated by cyclin E/Cdk2, GST-MCM3 and MCM3 mutants were prepared for in vitro kinase assay. As shown in Fig. 2A, MCM3 is phosphorylated by cyclin E/Cdk2 and the mutation at all four potential phosphorylation sites of MCM3 reduced its phosphorylation. These four sites at MCM3 do not strictly match the consensus sequence (S/T)PX(K/R) but fit the relaxed consensus sequence (S/T)P (Ser-112, Thr-464, Ser-611, Thr-722). To identify the phosphorylation site of MCM3 by cyclin E/Cdk2, FLAG-MCM3 and Myc-cyclin E/Cdk2 were co-expressed in 293T cells. Affinity-purified MCM3 was digested with trypsin and subjected to mass spectrometry analysis. The data showed that Ser-711 and Thr-722 at MCM3 were phosphorylated (supplemental Table S1). By comparing the peptide sequence, including Ser-711 and Thr-722 among different species, we found that Thr-722 but not Ser-711 is conserved from chicken to human (Fig. 2B). To further confirm that MCM3 is phosphorylated at Thr-722 in vivo, HeLa cells were transfected with FLAG-MCM3 and FLAG-MCM3 T722A mutant. Cell lysates were immunoprecipitated with FLAG antibody and immunoblotted with phospho-threonine proline antibody. As shown in Fig. 2C, the phosphorylation of MCM3 T722A was reduced significantly compared with that of wild type MCM3, indicating that Thr-722 of MCM3 is phosphorylated in vivo.
FIGURE 2.
MCM3 is phosphorylated by cyclin E/Cdk2 at Thr-722. A, in vitro kinase assay. 1 μg of GST, GST-MCM3, GST-MCM3 S112A, GST-MCM3 T464A, GST-MCM3 S611A, and GST-MCM3 T722A were incubated with GST-cyclin E and GST-Cdk2 in the presence of [γ-32P]ATP. Phosphorylation of MCM3 was assessed by SDS-PAGE followed by autoradiography of the gel. B, comparison of amino acid sequences of human and other species on MCM3 peptide around Thr-722. The number at the end of each sequence represented the position of threonine in boldface. C, MCM3 is phosphorylated at Thr-722 in vivo. Tet-on inducible cell lines (vector, MCM3, or MCM3 T722A) were treated with tetracycline for 24 h. Cell lysates were immunoprecipitated with FLAG antibody, followed by immunoblotting (IB) with phospho-threonine-proline antibody. D, GST-MCM3 (419–529 aa), GST-MCM3 (566–656 aa), GST-MCM3 (677–747 aa), and their corresponding phosphorylation site mutations (either Ser or Thr to Ala substitutions) were prepared and subject to in vitro kinase assay as described in B. The arrows indicated the band of phosphorylated MCM3, and the asterisk pointed out the phosphorylated cyclin E (left panel). The purified GST-MCM3 in Coomassie-stained gel is indicated by arrows as well (right panel).
To further confirm phosphorylation site of MCM3 by cyclin E/Cdk2, truncated GST-MCM3 (566–656, 677–747) and their mutants were prepared for in vitro kinase assay. As shown in Fig. 2D, wild type MCM3 (677–747) but not MCM3 (566–656) can be phosphorylated by cyclin E/Cdk2, whereas the phosphorylation of MCM3 (677–747) T722A mutant was abolished entirely. These data indicated that cyclin E/Cdk2 phosphorylates the carboxyl-terminal region of MCM3 exclusively at Thr-722.
Phosphorylation of MCM3 Affects Its Loading onto Chromatin
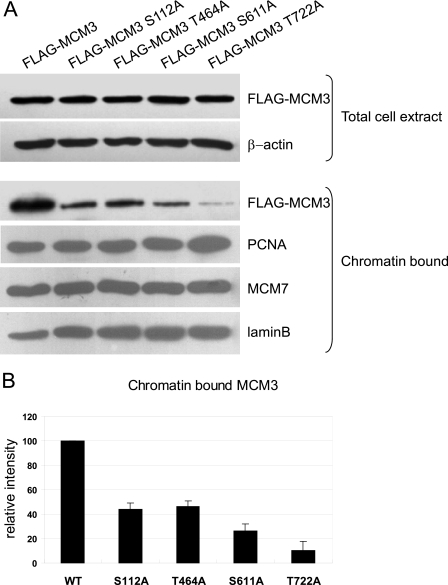
The MCM complex is loaded onto chromatin during late M and G1 phase before DNA replication initiation. 6-dimethylaminopurine, an inhibitor of serine/threonine kinases, can block the loading of Xenopus MCM3 to chromatin, which implies that phosphorylation of MCM3 is necessary for its association with chromatin. We then asked whether phosphorylation of MCM3 affects its loading onto chromatin. Wild type and phosphorylation site-mutated MCM3 were transiently introduced into 293T cells, respectively. The amount of MCM3 in total cell lysates and CSK-insoluble fraction was analyzed by immunoblotting. As shown in Fig. 3A, the mutations at all phosphorylation sites in MCM3 cause the reduction of chromatin loading. The quantification data (Fig. 3B) indicated that the MCM3 T722A mutant almost entirely lost its ability to bind to chromatin. These results suggested that the phosphorylation of MCM3 regulates its chromatin loading.
FIGURE 3.
Phosphorylation of MCM3 affects its loading onto chromatin. A, 293T cells were transfected with indicated plasmids. Total cell lysates and chromatin-bound fraction were prepared and subjected to immunoblot with indicated antibodies. β-Actin and lamin B were used as the loading controls. B, quantitcation of chromatin-bound MCM3 and its mutants. The wide-type MCM3 was arbitrarily set to 100. Data are represented as mean ± S.D. (n = 3).
Overexpression of MCM3 Blocks S Phase Entry
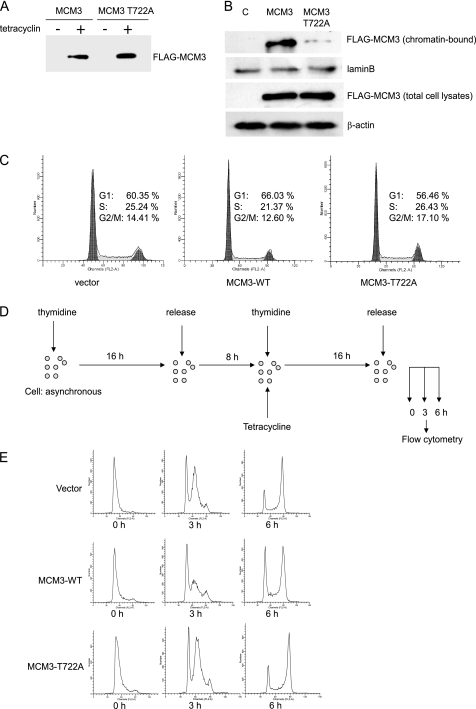
To unravel the biological role of MCM3, the inducible cell lines were generated by transfecting FLAG-MCM3 and FLAG-MCM3 T722A into T-RExTM-HeLa cells. The data from immunoblotting with FLAG antibody showed that both MCM3 and MCM3 T722A were expressed upon tetracycline treatment, whereas there was no expression without tetracycline (Fig. 4A). The pattern of MCM3 and MCM3 T722A associated with chromatin in HeLa cells was similar as that in 293T cells (Fig. 4B). Immunoblotting with anti-MCM3 antibody, which recognizes both endogenous and FLAG-MCM3 indicated that there is a 4–5-fold increase of induced MCM3 at 16 h after the addition of tetracycline (data not shown).
FIGURE 4.
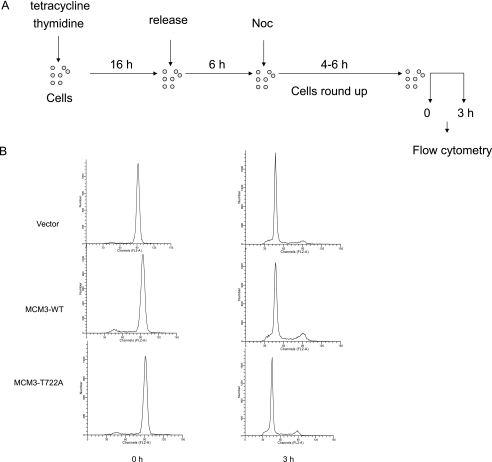
Overexpression of MCM3 blocks S phase entry. A, Tet-on inducible HeLa cells (MCM3 and MCM3 T722A) were treated with or without tetracycline for 24 h. Cell extracts were prepared and subjected to immunoblot with anti-FLAG antibody. B, Tet-on inducible HeLa cells for control, FLAG-MCM3, and FLAG-MCM3 T722A were treated with tetracycline. The chromatin-bound fraction and total cell lysates were prepared for immunoblotting with indicated antibodies. C, Tet-on inducible HeLa cells for control, FLAG-MCM3, and FLAG-MCM3 T722A were treated with tetracycline and subject to flow cytometry analysis. D, schematic view of experimental design. Tet-on inducible cells were treated with 2 mm thymidine for 16 h and released in fresh medium for 8 h and then treated again with 2 mm thymidine and tetracycline for 16 h. Cells then were released for 3 and 6 h and subjected to flow cytometry analysis. E, cell cycle profiles were analyzed by flow cytometry analysis.
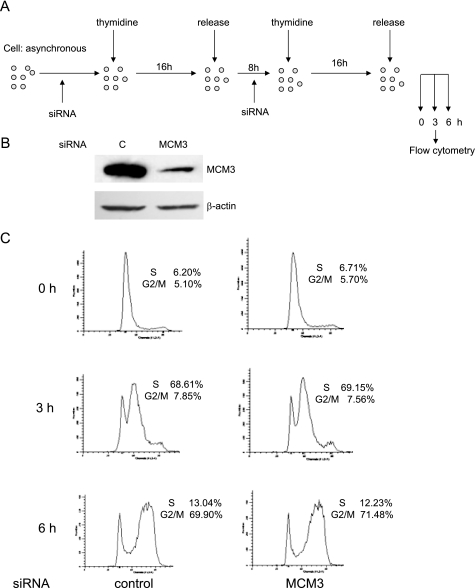
First, we examined the cell cycle profile by flow cytometry in asynchronous MCM3-induced cells. Compared with control and MCM3 T722A-overexpressed cells, MCM3-overexpressed cells had higher G1 phase and lower S phase (Fig. 4C). To analyze whether excess MCM3 affects cell cycle progression, cells were synchronized at the G1/S boundary by double thymidine treatment and released for 3 and 6 h, respectively. Then, they were collected and analyzed by flow cytometry (Fig. 4C). As shown in Fig. 4E, all three cell lines were synchronized at the G1/S boundary. At 3 h after release, >60% of control and MCM3 T722A overexpressed cells entered S phase, whereas only 45% of MCM3-overexpressed cells entered S phase. At 6 h after release, there was still >27% of MCM3-overexpressed cells in G1 phase, whereas the majority of control and MCM3 T722A-overexpressed cells entered G2/M phase. These results suggested that overexpression of wild type MCM3 but not the MCM3 T722A mutant inhibits S phase entry or S phase progression. To analyze whether overexpression of wild type MCM3 affected M to G1 phase progression, cells were synchronized at M phase by thymidine treatment followed by nocodazole treatment and then released for 3 h (Fig. 5A). Flow cytometry analysis revealed that there is no difference between control, MCM3, and MCM3 T722A-overexpressed cells (Fig. 5B), which indicates that overexpression of wild type MCM3 does not block M to G1 phase progression. Then, we took the RNA interference approach to analyze whether knockdown of MCM3 will affect the S phase entry (Fig. 6A). The immunoblotting data showed the MCM3 in RNAi-treated cells was reduced to ∼20% compared with control cells (Fig. 6B). The flow cytometry data indicated that knockdown of MCM3 did not block S phase entry (Fig. 6C). Similar results were obtained in cells treated with two additional SiRNAs (data not shown). Taken together, these results implied that certain amount of MCM3 is sufficient for S phase entry, whereas excess of MCM3 may exert a negative effect in S phase progression in cell cycle.
FIGURE 5.
Overexpression of MCM3 does not affect M phase exit. A, schematic view of experimental design. Tet-on inducible cells (control, MCM3, and MCM3 T722A) were arrested at G1/S phase by thymidine treatment for 16 h and then released for 6 h followed by treatment with nocodazole for 6 h. Cells were released for 3 h and subjected to flow cytometry analysis. B, cell cycle profiles at 0 and 3 h after release from nocodazole treatment were analyzed by flow cytometry.
FIGURE 6.
Knockdown of MCM3 does not affect S phase entry. A, schematic view of experimental design. HeLa cells were treated with thymidine and transfected with two rounds of siRNA against MCM3 at the time indicated. B, HeLa cell were treated as described in A. Cells were harvested at 0 h (before final release into S phase) after two rounds of siRNA 1 transfection, and cell lysates were subjected to immunoblot with MCM3 antibody. β-Actin was used as a loading control. C, S phase dynamics after MCM3 depletion. HeLa cells released at 0, 3, and 6 h after double thymine treatment and MCM3 depletion were harvested and subjected to flow cytometry analysis.
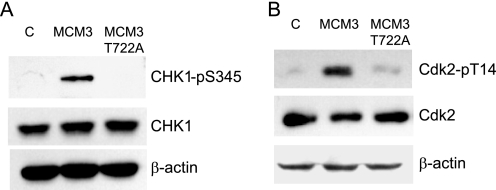
Excess of MCM3 Up-regulates Phosphorylation of CHK1 Ser-345 and Inhibitory Phosphorylation of Cdk2
It has been reported that ATM phosphorylates MCM3 and regulate its chromatin loading (23). Because the main differences in MCM3 and MCM3 T722A mutant are their ability to associate with chromatin and influence on S phase entry, we asked whether excess of MCM3 could trigger the ATM/ATR checkpoint pathway. First, we examined the CHK1 phosphorylation at Ser-345, which is a marker of the activated ATM/ATR checkpoint pathway. As shown in Fig. 7A, the phosphorylation of CHK1 Ser-345 was increased in MCM3-overexpressed cells compared with that in control and MCM3 T722A-overexpressed cells. It is known that CHK1 activation will cause the ubiquitin-mediated degradation of CDC25A, so we analyzed protein level of CDC25A in control, MCM3, or MCM3 T722A-overexpreesed cells. There is no significant difference observed among these cells (data not shown). It is known that activation of CHK1 causes the inhibitory phosphorylation of Cdk and block the cell cycle progression (24). Therefore, we examined the phosphorylation status of Cdk2 Thr-14 as cells were synchronized at G1/S phase by double thymidine block. The result showed that the phosphorylation of Cdk2 Thr-14 was much higher in MCM3-overexpressed cells than that in control and MCM3 T722A cells (Fig. 7B). Taken together, our data indicated that excess accumulation of MCM3 in chromatin will cause the activation of CHK1 and inhibition of Cdk2 and eventually block S phase entry.
FIGURE 7.
Overexpression of MCM3 up-regulates the phosphorylation of Chk1 Ser-345 (A) and Cdk2 Thr-14 (B). Tet-on inducible cells (control, MCM3, and MCM3 T722A) were treated with tetracycline and synchronized at the G1/S phase. Total cell lysates were harvested and subjected to immunoblot with the indicated antibodies.
DISCUSSION
Using tandem affinity purification and in vitro kinase assay, we identified MCM3, a subunit of the putative replicative DNA helicase, is a substrate of cyclin E/Cdk2. It is phosphorylated by cyclin E/Cdk2 at Thr-722. Our data indicated that phosphorylation of MCM3 at Thr-722 promotes loading of MCM3 onto chromatin. MCM complex at replication origins is activated by the concerted actions of cyclin-dependent kinases and the CDC7 protein kinase (25), which leads to initiation of DNA synthesis. These data allow us to propose a positive role of Cdk2-dependent MCM3 phosphorylation in origin firing and movement of the replication forks. To reveal the biological consequence of Thr-722 phosphorylation of MCM3, we generated Tet-on inducible HeLa cell lines expressing MCM3 and MCM3 T722A. Surprisingly, our data show that overexpression of MCM3 elicits a dramatic G1 phase arrest and S phase delays instead of accelerating S phase progression. However, substitution of Thr-722 to alanine completely alleviated this cell cycle defect. Meanwhile, we found that MCM3 T464A mutant exhibited a similar phenotype as MCM3-WT (data not shown). These findings strongly suggest that Cdk2 mediated phosphorylation of MCM3 at Thr-722 might exert an inhibitory effect on DNA replication. A number of studies have shown that Cdk2 kinase activity is indispensable in S phase initiation (6, 26, 27). Consistent with this, we found that HeLa cells synchronized in G1 phase cannot progress into S phase by roscovitine, an inhibitor of Cdk2, implying that Cdk2-dependent phosphorylation events are essential in pre-RC activation (data not shown). However, cyclin-dependent kinases also play a negative role in regulating DNA replication. This dual role of Cdks is likely to be conducted in targeting different substrates. The MCM2–7 complex consists of six related but different subunits and plays a central role in the initiation and elongation of DNA replication (28), thus rendering it a good candidate for regulation by Cdks at multiple spots. Ishimi et al. (13) reported that the DNA helicase activity of the MCM4–6-7 complex is negatively regulated by Cdk2 phosphorylation. Ishimi et al. (13) also suggested that MCM4 was involved in DNA replication checkpoint. Phosphorylation of MCM4 by cyclin A/Cdk2 after activation of DNA damage checkpoint inhibits DNA replication through the inactivation of the MCM complex. Also, Ishimi et al. (13) indicated that phosphorylation of MCM4 occurred even in the absence of hydroxyurea, implying this phosphorylation event might exist in normal cell cycle progression. Our data suggest that phosphorylation of MCM3 by cyclin E/Cdk2 may also be involved in DNA replication checkpoint control. We did observe the up-regulation of phosphorylation of Chk1 Ser-345, which is the downstream target of ATM/ATR in MCM3-overexpressed cells. Because overexpression of MCM3 T722A did not result in obvious Chk1 activation, it is likely that Chk1 activation may be dependent on MCM3 phosphorylation at Thr-722. However, the detailed mechanisms still need to be elucidated.
It has been reported that MCM3 is a direct substrate of ATM in response to ionizing radiation (23). We examined the MCM3 T722 phosphorylation in cells treated with caffeine, an inhibitor of ATR/ATM. There was no difference of the MCM3 phosphorylation at Thr-722 in control and caffeine-treated cells (data not shown), indicating that Thr-722 of MCM3 is not a common phosphorylation site of Cdk2 and ATR but is specific to Cdk2. However, human Thr-722 is in close proximity to two ATM-dependent phosphorylation sites of MCM3, Ser-725 and Ser-732 (23). Based on the results here, it is tempting to speculate that Thr-722, Ser-725, and Ser-732 phosphorylation by different kinases may execute similar biological outputs.
In our experiments, we show that knockdown of MCM3 to a very low level by RNA interference still allowed DNA replication in both asynchronous cells (data not shown) and cells released from G1/S phase (Fig. 6, B and C). This result is consistent with a previous report by Ibarra et al. (3). All of these findings come to the same conclusion that only a small number of MCM2–7 complexes are needed to drive normal fork progression. However, there is a large excess of MCM complexes loaded at origins and distributed along the chromatin. It is called the “MCM paradox” (29, 30). More evidence implies that excess MCM proteins might not be important for DNA replication but involved in checkpoint function and genome stability. An important work by Cortez et al. (17) showed that decreasing the amount of MCM7 protein to a level that did not significantly affect DNA replication led to an intra-S phase checkpoint defect, implying that MCM is involved in other function rather than origin firing. Ibarra et al. (3) proposed a model in which the distribution of MCM proteins sets the limits for total origin availability. In normal conditions, some MCM proteins are chosen as the effective DNA helicase, whereas the others serve as backups. After firing one of the origins, a signal will be sent from the forks to suppress the activity of nearby origins. Recent studies in different systems reveal that this inhibitory signal is mediated by checkpoint kinases (32–35), even in the absence of DNA damage. These cryptic origins can be activated through inhibition of Chk1 (36, 37), thus providing an important backup function during replicative stress. In mammalian cells, Chk1 participates in the intra-S phase checkpoint that regulates late origins in an unperturbed S phase. It has been reported that ATR is activated in response to ssDNA (26, 31, 34, 38), a common intermediate of both normal replication and damaged DNA metabolism. During DNA replication, single-stranded DNA will emerge as the MCM complex melts the template. Then ssDNA-binding protein replication protein A would bind ssDNA and recruit ATR-ATRIP complex, which ultimately activates Chk1 and down-regulates unfired origins. The strength of the checkpoint signal generated by ATR seems to correlate with the amount of ssDNA generated by DNA damage or normal DNA replication. All of these results suggested that ATR/ATRIP coupled to MCM proteins may monitor and regulate the extent of ssDNA-replication protein A intermediates generated during normal DNA replication or after DNA damage (22). Indeed, we found that Chk1 phosphorylation was up-regulated in MCM3-overexpressed cells, which suggested that excess chromatin-bound MCM3 may stabilize the ssDNA transiently and amplify the checkpoint signal by activating ATR-Chk1 pathway. We propose that MCM3 may play a role in keeping the checkpoint signal in an appropriate way to ensure the proper DNA replication in normal condition and hold DNA replication in the presence of genotoxic stress. However, the detailed mechanism of MCM3 in keeping the threshold of checkpoint signal merits further study.
Increasingly more evidence supports the idea that MCM subunits play important roles in DNA replication checkpoint not only as DNA damaged but also during normal DNA replication. Furthermore, our data point to the possibility that the MCM complex can function upstream in checkpoint signaling pathways. The MCM proteins are attractive candidates for regulation by checkpoints, thus providing a platform for multiple regulatory inputs to regulate DNA replication. Further studies will be needed to delineate the more detailed mechanism underlying MCM proteins and activation of DNA replication checkpoint.
Supplementary Material
Acknowledgments
We thank Dr. Y Li (Institute of Psychology, Chinese Academy of Sciences) for kind help with flow cytometry. We are grateful to G. Nalepa (Indiana University) for critical reading of the manuscript.
This work was supported by National Natural Science Foundation of China Grant 30871238, The Ministry of Science and Technology of China Grants 2009ZX10004-101, 2008ZX10002-009, 2011CB504705, 2012CB519003, and Chinese Academy of Sciences Innovation Projects KSCX2-EW-J-6.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- pre-RC
- pre-replication complex
- aa
- amino acids
- MCM
- minichromosome maintenance
- Cdk2
- cyclin-dependent kinase 2
- ATM
- ataxia telangiectasia-mutated
- ATR
- ATM and Rad3 related.
REFERENCES
- 1. Sclafani R. A., Holzen T. M. (2007) Annu. Rev. Genet. 41, 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun J., Kong D. (2010) ACTA BIOCHIM. BIOPHYS. SIN. 42, 433–439 [DOI] [PubMed] [Google Scholar]
- 3. Ibarra A., Schwob E., Méndez J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tanaka T., Knapp D., Nasmyth K. (1997) Cell 90, 649–660 [DOI] [PubMed] [Google Scholar]
- 5. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 6. Diffley J. F. X. (2004) Curr. Biol. 14, R778–786 [DOI] [PubMed] [Google Scholar]
- 7. Blow J. J., Dutta A. (2005) Nat. Rev. Mol. Cell Biol. 6, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elsasser S., Chi Y., Yang P., Campbell J. L. (1999) Mol. Biol. Cell 10, 3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jallepalli P. V., Brown G. W., Muzi-Falconi M., Tien D., Kelly T. J. (1997) Genes Dev. 11, 2767–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drury L. S., Perkins G., Diffley J. F. (2000) Curr. Biol. 10, 231–240 [DOI] [PubMed] [Google Scholar]
- 11. Early A., Drury L. S., Diffley J. F. (2004) Philos. Trans. R. Soc. B Biol. Sci. 359, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yim H., Erikson R. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19742–19747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishimi Y., Komamura-Kohno Y., Kwon H. J., Yamada K., Nakanishi M. (2003) J. Biol. Chem. 278, 24644–24650 [DOI] [PubMed] [Google Scholar]
- 14. Hendrickson M., Madine M., Dalton S., Gautier J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12223–12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheu Y. J., Stillman B. (2010) Nature 463, 113-U127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takei Y., Swietlik M., Tanoue A., Tsujimoto G., Kouzarides T., Laskey R. (2001) Embo. Rep. 2, 119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cortez D., Glick G., Elledge S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snyder M., Huang X. Y., Zhang J. J. (2009) J. Biol. Chem. 284, 13466–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J. M., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., Arai K. (2006) J. Biol. Chem. 281, 39249–39261 [DOI] [PubMed] [Google Scholar]
- 20. Lin D. I., Aggarwal P., Diehl J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8079–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deng M., Li F., Ballif B. A., Li S., Chen X., Guo L., Ye X. (2009) J. Biol. Chem. 284, 7875–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shechter D., Gautier J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10845–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi Y., Dodson G. E., Mukhopadhyay P. S., Shanware N. P., Trinh A. T., Tibbetts R. S. (2007) J. Biol. Chem. 282, 9236–9243 [DOI] [PubMed] [Google Scholar]
- 24. Laezza C., Pisanti S., Crescenzi E., Bifulco M. (2006) FEBS Lett. 580, 6076–6082 [DOI] [PubMed] [Google Scholar]
- 25. Walter J. C. (2000) J. Biol. Chem. 275, 39773–39778 [DOI] [PubMed] [Google Scholar]
- 26. Namiki Y., Zou L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stillman B. (1996) Science 274, 1659–1664 [DOI] [PubMed] [Google Scholar]
- 28. Bochman M. L., Schwacha A. (2009) Microbiol. Mol. Biol. Rev. 73, 652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edwards M. C., Tutter A. V., Cvetic C., Gilbert C. H., Prokhorova T. A., Walter J. C. (2002) J. Biol. Chem. 277, 33049–33057 [DOI] [PubMed] [Google Scholar]
- 30. Hyrien O., Marheineke K., Goldar A. (2003) Bioessays 25, 116–125 [DOI] [PubMed] [Google Scholar]
- 31. Liu E., Lee A. Y., Chiba T., Olson E., Sun P., Wu X. (2007) J. Cell Biol. 179, 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marheineke K., Hyrien O. (2004) J. Biol. Chem. 279, 28071–28081 [DOI] [PubMed] [Google Scholar]
- 33. Meister P., Taddei A., Ponti A., Baldacci G., Gasser S. M. (2007) EMBO J. 26, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shechter D., Costanzo V., Gautier J. (2004) Nat. Cell Biol. 6, 648–655 [DOI] [PubMed] [Google Scholar]
- 35. Feijoo C., Hall-Jackson C., Wu R., Jenkins D., Leitch J., Gilbert D. M., Smythe C. (2001) J. Cell Biol. 154, 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maya-Mendoza A., Petermann E., Gillespie D. A., Caldecott K. W., Jackson D. A. (2007) EMBO J. 26, 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Syljuåsen R. G., Sørensen C. S., Hansen L. T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. (2005) Mol. Cell Biol. 25, 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi J. H., Lindsey-Boltz L. A., Kemp M., Mason A. C., Wold M. S., Sancar A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.