Background: Microarray experiments capture details of cellular responses upon perturbations, including Mycobacterium tuberculosis (Mtb) infection.
Results: Novel bioinformatics approach “Express Path Analysis” identifies Src at the core of the network regulating divergent responses to virulent and avirulent Mtb infection.
Conclusion: Src acts through regulating key antimycobacterial properties, acidification of phagolysosome and autophagy induction.
Significance: Results may help evolve novel host targeted anti-tuberculosis therapy strategy.
Keywords: Macrophages, Microarray, Mycobacterium tuberculosis, Src, Systems Biology, Autophagy, Functional Association Network, Pathway Analysis, Phagosome Maturation
Abstract
Global gene expression profiling has emerged as a major tool in understanding complex response patterns of biological systems to perturbations. However, a lack of unbiased analytical approaches has restricted the utility of complex microarray data to gain novel system level insights. Here we report a strategy, express path analysis (EPA), that helps to establish various pathways differentially recruited to achieve specific cellular responses under contrasting environmental conditions in an unbiased manner. The analysis superimposes differentially regulated genes between contrasting environments onto the network of functional protein associations followed by a series of iterative enrichments and network analysis. To test the utility of the approach, we infected THP1 macrophage cells with a virulent Mycobacterium tuberculosis strain (H37Rv) or the attenuated non-virulent strain H37Ra as contrasting perturbations and generated the temporal global expression profiles. EPA of the results provided details of response-specific and time-dependent host molecular network perturbations. Further analysis identified tyrosine kinase Src as the major regulatory hub discriminating the responses between wild-type and attenuated Mtb infection. We were then able to verify this novel role of Src experimentally and show that Src executes its role through regulating two vital antimicrobial processes of the host cells (i.e. autophagy and acidification of phagolysosome). These results bear significant potential for developing novel anti-tuberculosis therapy. We propose that EPA could prove extremely useful in understanding complex cellular responses for a variety of perturbations, including pathogenic infections.
Introduction
Response of cells to various signaling cues is a dynamic process involving reprogramming of the cellular transcriptional machinery. The transcriptional reprogramming then drives the cellular physiology to an altered state that corresponds to the cellular response in a context-specific manner. Consequently cellular responses to various stimuli have often been characterized by the nature and level of perturbation to the global gene expression profile (1, 2). The analysis of expression data like gene ontology enrichment provides details of various functional classes that are specifically regulated in a given context and helps to postulate hypotheses about how specific cellular response is brought about (3, 4). Unsupervised analyses like k-clustering and hierarchical clustering allow identification of novel response patterns, associations and confirmation of data with prior knowledge (5). Other analytical strategies like Boolean and probability-based Bayesian models help in obtaining mechanistic insights based on microarray data (6, 7). Although useful as such, the Boolean framework suffers severely from its inherent deterministic logic because we understand that biological processes, including gene expression, are stochastic in nature (8). On the contrary, Bayesian models are extremely powerful because fractional controls can also be accommodated, establishing causal relationships among molecules (9). However, the Bayesian framework requires a prior estimate of probability of an event and therefore is not entirely unbiased (7). Clearly, the microarray data are not analyzed in a manner that permits novel unbiased inferences to be drawn.
Recent years have witnessed a surge in network-based approaches for understanding large complex data sets driven by the concept that no biological function is carried out by a molecule in isolation. Rather, they represent dynamic interactions among various molecular regulators to achieve specific function (10). It is also noted that, more than network composition, it is the interaction topology that contains functional information. Therefore, topological changes in the network could be the readjustment through which specific cellular responses are derived (11). Microarray data, somewhat complementarily, capture such topological alterations at the gene expression level. However, integration of microarray data with network analysis has not been used extensively to understand response regulation. We therefore decided to combine microarray results with the functional association network of proteins to gain unbiased insights into regulatory controls of cellular responses.
We chose the highly relevant context of Mycobacterium tuberculosis (Mtb)3 infection of human macrophages to identify the response regulators of the infection process. It is known that cellular responses to a virulent Mtb infection starkly differ from responses to a non-virulent Mtb infection (12–15). They together therefore provide an attractive model to understand specific cellular responses to virulent Mtb infection. In the present study, we monitored the temporal global expression pattern of host macrophages upon Mtb infection. Then, applying the novel analytical strategy named express path analysis (EPA), we identified molecular pathways that were differentially regulated under the virulent and non-virulent infections. The pathways identified were intricately involved with each other, giving rise to molecular networks that captured differential cellular responses. Further network analysis, including measures like centrality parameter, identified tyrosine kinase Src as the key node regulating differential responses to virulent and non-virulent infection. We then performed experiments that established the role of Src kinase in regulating cellular responses to virulent Mtb infection. This is the first report implicating Src as the key regulator of cellular responses in the case of Mtb infection and therefore opens novel avenues for anti-tubercular therapy. The strategy described here therefore has the potential to be exploited under different environmental settings for unearthing novel response regulators.
EXPERIMENTAL PROCEDURES
Cells, Cultures, and Media
The human promonocytic cell line THP1 (American Type Culture Collection) was used in this study. THP1 cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FCS (Hyclone) and were maintained between 2 and 10 × 105 cells/ml at 37 °C in a humidified, 5% CO2 atmosphere. Before infection, cells were seeded in 6-well plates at 2 × 106 cells/well and treated with 20 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma) overnight; cells were allowed to adhere and become differentiated into macrophages at 37 °C in a humidified, 5% CO2 incubator; the cells were washed twice with warm RPMI to remove the PMA. Bacteria were grown as stationary cultures in Middlebrook 7H9 broth (Difco) supplemented with 10% ADC (BD Biosciences), 0.4% glycerol, and 0.05% Tween 80 until the mid-log phase. Antibodies used in the study were procured from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Cell Signaling Technology (Beverly, MA).
Bacterial Suspension Preparation and Infection
Bacterial whole suspension was dispersed by aspiration five times each with a 24- and then a 26-gauge needle, followed by an additional dispersion 15 times through a 30-gauge needle. This was then vortexed until no bacterial clumps were detectable, and the dispersed bacteria were allowed to stand for 5 min. The upper half of the suspension was then used for the experiments. Quantitation of bacteria was done by taking absorbance at a 600-nm wavelength (0.6 OD corresponds to ∼100 × 106 bacteria).
RNA Isolation and Microarray Experiments
Total RNA was isolated from 5 × 106 THP1 cells 6, 12, 48, and 90 h after infection with M. tuberculosis and from uninfected cells as control for the experiment using TRIzol reagent (Invitrogen) in accordance with the manufacturer's instructions. For microarray experiments, total RNA was processed for each RNA sample and hybridized to the Custom Human 4 × 44k Array (Agilent Microarray Design Identifier: 25085) according to standard protocols. Microarray experiments were done at Genotypic Technology (Bangalore, India).
Express Path Analysis
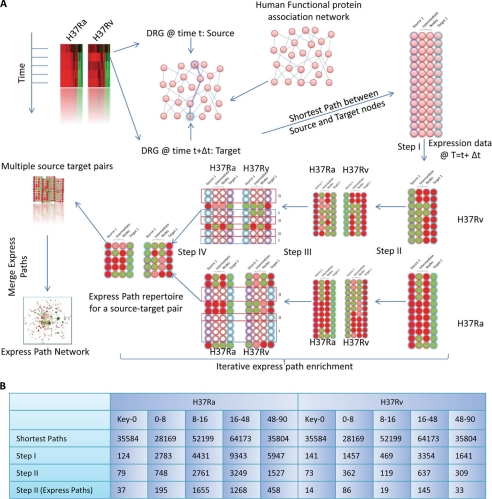
The undirected human interaction database was downloaded from the STRING database. Interactions were parsed and retained only those interactions showing a combined score of >500. Differentially regulated genes (DRGs) at each time point between H37Ra (Ra)- and H37Rv (Rv)-infected cells were identified using standard SQL commands (see “Results”). Key nodes for genes differentially regulated at the 0 h time point were generated using the ExPlain tool of the Transfac database. All possible shortest paths from key regulators to DRG at 0 h and DRS at 0–8, 8–16, 16–48, and 48–90 h were calculated using the built-in function “get.all.shortest.paths” in the R software package. Then, for each path, every step was compared for their expression in Ra- and Rv-infected cells. Paths that showed differential regulation at multiple steps for a given paths were filtered out (see Fig. 2 for a detail description).
FIGURE 2.
EPA, a novel method for analyzing microarray data. The express path analysis strategy is schematically represented in A. The two contrasting environments for this study were THP1 cells infected with Ra and Rv, respectively; however, the approach is applicable to any set of contrasting environments. In the time course microarray experiment, DRGs between the two environments were identified at each of the time points. Then in the human functional protein association network (from the STRING database), the shortest paths were traced between DRGs at one time point to the following time point. The ensemble of shortest paths was then iteratively enriched based on their expression values in the respective environments, eventually yielding a set of shortest paths that are not similarly regulated at any of the nodes between the two conditions. Briefly, once the shortest path ensemble is created, in Step I, we filter out only those shortest paths that are significantly regulated at each of the intermediate steps. Because the expression of intermediates in the Ra- and Rv-infected cells is different, Step I yields a different subset of shortest paths for Ra and Rv. Note the different number of paths for Ra and Rv sets after Step I in the figure (for a given source-target pair). Also important to note here is the fact that the respective paths identified in either Ra or Rv sets were regulated (i.e. down-regulated (green) or up-regulated (red)) at each of the intermediate steps as well. In the next step (Step II), for the subset of pathways in the Ra set, corresponding expression of intermediates in Rv is monitored, and vice versa. If any of the intermediates for a shortest path shows a similar expression pattern in both Ra and Rv, that path is excluded out, consequently enriching differentially regulated paths or express paths. A similar exercise performed on all source-target pairs for a given time window is then merged together to get the shortest path network. The number of shortest paths remaining after every enrichment step is shown in B.
Centrality Analysis of Express Path Networks
Express Paths identified for any given source-target set in both Ra- and Rv-infected sets were then merged together, which yielded the time point-specific express path network. These networks were then imported into Cytoscape for visualization and analyses of centrality parameters. Betweenness centrality was calculated using the “Network Analyzer” function in Cytoscape and was then mapped on to the network as a node attribute.
Confocal Microscopy
Bacteria were stained using the membrane stain PKH67 (Sigma-Aldrich) according to the manufacturer's protocol. PMA-differentiated THP1 cells were seeded at a density of 0.3 × 106 cells/coverslip, onto 12-mm diameter glass coverslips precoated with fibronectin (0.05 μg/μl) in a 24-well tissue culture plate. The cells were infected with stained bacteria at a multiplicity of infection of 10 (10 bacteria/cell). After 4 h of infection, the cells were washed twice with warm RPMI and treated with amikacin (200 μg/ml) for 2 h to remove any remaining extracellular bacteria. The cells were then maintained in complete media containing PP2 inhibitor at a concentration of 50 nm for 48 h. Cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) at 48 h post-bacterial infection. The fixed cells were stained with DAPI (300 nm in 1× PBS). The coverslips were washed thoroughly with PBS and were mounted on slides with Antifade gold reagent (Invitrogen). Four fields were acquired randomly from each set of stained cells with a Nikon EclipseTi-E laser-scanning confocal microscope equipped with a 20×/0.75 numerical aperture plan apochromat differential interference contrast objective lens using the blue diode laser (excitation at 408 nm and emission at 450 nm ) and argon laser (excitation at 488 nm and emission at 515 nm). All images were acquired using the software EZ-C1 3.8, and analyses were performed using the software Image-Pro Plus version 6.0. Magnified images were acquired using a 60×/1.4 numerical aperture oil planapochromat differential interference contrast objective lens. For co-localization studies, 1 h prior to the time point, complete medium containing LysoTracker dye (Molecular Probes, Invitrogen) was added to the cells for staining of acidified lysosomes. Cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 20 min, followed by three washes with 1× PBS. The cells were permeabilized using 0.2% (w/v) Triton X-100 (in 1× PBS) for 20 min. Blocking was performed using 3% (w/v) BSA and 0.5% Tween 20 in 1× PBS for 1 h. This was followed by primary and secondary antibody staining for a duration of 1 h each. The coverslips were washed thoroughly with 1× PBS and were mounted onto glass slides with Antifade reagent (Sigma). For each image, 20 sections covering 10 μm on the z axis at intervals of 0.5 μm were acquired randomly from each set of stained cells with a Nikon EclipseTi-E laser-scanning confocal microscope equipped with a 60×/1.4 numerical aperture plan apochromat differential interference contrast objective. All images were acquired using the software NIS-Elements, and analyses were performed using the software Image-Pro Plus version 6.0.
RESULTS
Infection of THP1 Cells with Ra or Rv Leads to Global Reprogramming of Host Gene Expression
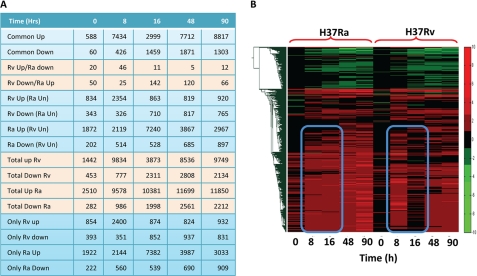
We infected PMA-differentiated THP1 cells (human macrophage-like cell line) with Rv or Ra in separate experiments at a 1:10 multiplicity of infection (see “Experimental Procedures” for details). We first monitored survival or killing kinetics of Ra and Rv inside the THP1 cells (supplemental Fig. S1), and as expected, our experimental conditions too showed killing of Ra and survival of Rv inside the host macrophages. In the separate experiment, cells were harvested at various time intervals like immediately after infection (0 h) and 8, 16, 48, and 90 h postinfection with either Ra or Rv. Host total RNA was isolated from these samples using TRIzol reagent, and their microarray profiling was done at Genotypic Technologies Inc. using the Agilent human genome array, consisting of about 42,465 probe sets together covering around 25,607 genes. In parallel, we also had RNA samples from uninfected THP1 cells as control, to be used as a base value while calculating log2 ratios for changes in individual gene expression. Because a majority of genes were represented by more than one probe set, we selected the non-redundant expression data for the whole genome using an algorithm that selects the most consistent expression value for a given gene (see “Experimental Procedures”). The overall expression pattern of 25,607 genes across five different time points in Ra- or Rv-infected THP1 cells are represented as a heat map in supplemental Fig. S2A. The expression data are presented in supplemental Table S1. A quick review of the expression pattern between Ra- and Rv-infected THP1 cells showed significant differences in the nature of perturbation (on magnitude and kinetics) inflicted on the hosts by the two strains. The number of genes regulated at each of the time points and the overall pattern followed by them varied between Ra and Rv in a unique but overlapping manner. A more detailed comparison of the level and nature of perturbation of Ra and Rv on host macrophages is presented in Fig. 1A. Fig. 1A compares genes that are commonly regulated between Ra- and Rv-infected cells at a given time point and those that are regulated in a contrasting manner. Thus, there are genes that are up-or down-regulated in Ra and Rv both, whereas others are up-regulated in one while down-regulated in the other. Still others are genes that are up- or down-regulated in one case while unaffected in the other (Fig. 1A).
FIGURE 1.
Global gene expression changes in macrophage cells upon infection with Ra and Rv. Gene expression profiles of THP1 cells infected with Ra or Rv were generated at 0, 8, 16, 48, and 90 h after infection. Among the significantly regulated genes (genes showing more than 2-fold increase or decrease from the uninfected control), we compared those that were commonly regulated or differentially regulated between Ra and Rv infection (A). The number of genes showing common regulation (Common Up/Down), contrasting regulation (Rv up/Ra down and Rv Down/Ra Up), and differential regulation Rv up (Ra unaffected (Un)), Rv down (Ra unaffected), and vice versa is listed. A clustergram of the expression profile of genes in both Ra and Rv is shown in B. Commonly and distinctly regulated clusters are clearly visible in the clustergram. The blue box highlights the genes that are differentially regulated.
The time-dependent changes in gene expression profile can be used to categorize genes with similar and distinct effects on their expression. A clustergram analysis clusters genes showing similar behavior across time points into separate categories (16, 17). The clustergram analysis of the Ra- and Rv-infected THP1 cell expression profile showed very distinct patterns (supplemental Fig. S2B). Thus, whereas the clustergram of Ra-infected cells yielded three large broad clusters, that of Rv-infected cells resulted in seven distinct clusters (supplemental Fig. S2B). An immediate inference explaining this difference could be that, as expected, Rv infection inflicts larger perturbation on cellular machinery as compared with infection with Ra. Because avirulent infection fails to manipulate the host system, unlike their virulent counterparts, which influence almost every possible cellular processes, a less diverse expression pattern in the case of avirulent infection seems to be a true representation of global perturbations upon Mtb infection (supplemental Fig. S2B). In order to compare the patterns across both of the categories, we also performed a clustergram analysis with Ra- and Rv-infected cell expression data sets together (Fig. 1B). A quick look at Fig. 1B suggests that the overall patterns of perturbations are very similar between both Ra- and Rv-infected cells. Rv infection, however, interferes with a major cluster of up-regulated genes in a time-dependent manner (Fig. 1B). Specifically, genes up-regulated at 16, 48, and 96 h post-Ra infection are contrastingly regulated in Rv-infected cells. In order to confirm this with the previously described cellular responses upon Mtb infection, we performed gene ontology analysis of the microarray results. Then through a combination of k-cluster analysis followed by functional class enrichment and principle component analysis, we were able to establish two interesting aspects of cellular responses to virulent or non-virulent infections (supplemental Fig. S3). The major functional classes of genes showing discrete regulation between Ra- and Rv-infected cells were immune responses and gene regulation. Second, clusters distinguishing expression pattern between Ra- and Rv-infected cells also distinguished functional class enrichment (supplemental Fig. S3). These results together highlighted the validity of microarray results, ensuring that specific cellular responses were captured through the experiments.
Results so far indicate that the gene expression experiment followed by clustering and gene ontology enrichment identifies specific patterns in the nature of perturbation inflicted by the virulent and avirulent Mtb strains. However, beyond identifying these interesting patterns, these analyses did not divulge much, in terms of regulatory events, mechanistic controls, etc. Gaining mechanistic information from microarray data in the eukaryotes is a complex problem due to the multitiered regulatory events of gene expression (18). Nonetheless, approaches like Bayesian analysis have been used to establish cause-effect relationships and pathway reconstruction in eukaryotic systems (7). Another approach utilizes transcription factor binding site analysis to establish regulatory loops in the expression pattern of genes and their upstream transcription factors (19, 20). A major limitation here is overreliance on specific transcription factor (TF) binding sites upstream from genes. In many cases, TF recruitment is a multifactorial process requiring cooperativity between different molecules, the majority of them regulated through post-translational modification. Here we devised a novel strategy for analyzing the microarray data set to identify cellular responses specific to virulent Mtb infection in an unbiased manner.
EPA Identifies Potential Contrastingly Regulated Paths between Ra- and Rv-infected Cells
In order to perform an unbiased pathway analysis, we first downloaded the STRING database of functional protein-protein association for humans. The database is collective set of 15,404 nodes (genes) and 305,745 edges (interactions). For each of the interactions listed, the database provides a cumulative score that can be used to filter out high confidence interacting partners. For high confidence interaction data, we retained only those interactions from the database that showed a cumulative score of more than 0.5, and the resulting network was used as the base network for all future analysis (Fig. 2A). In parallel, we also identified genes in the expression data at each of the time points that showed differential regulation (DRGs) between Ra- and Rv-infected cells (Fig. 1A, Rv Up/Ra down and Rv Down/Ra Up). Guided by the assumption of cause and effect relationship between genes regulated at earlier and later time points, respectively, we traced the shortest paths between DRGs at a given time point as source nodes and DRGs at the following time point as target nodes. The shortest paths were extracted from the protein functional association network. The shortest path is the one that connects two nodes through minimum number of intermediates in a network (21). Arguably, two nodes can be connected through multiple shortest paths in a network, and therefore we generated an ensemble of all possible shortest paths between each pair of the source and the target. This exercise yielded a large ensemble of time point-specific shortest paths (Fig. 2B). In some cases, the number of total possible shortest paths was >65,000. We also analyzed regulation at the 0 h time point, through identifying key upstream regulatory nodes using the Explain feature of the Transfac database. Key nodes are identified through the Transfac knowledge base, which incorporates signaling events leading to activation/regulation of downstream transcription factors. We identified key nodes for DRGs at the 0 h time point. Then, to identify pathways, the shortest paths were traced between each of the key nodes as source and DRGs as targets with the base network in the background. Because the source and targets were same for both Ra and Rv set, the initial number of shortest paths was the same in both cases. In the next step (Fig. 2A, Step I), we retained only those paths where all of the constituent nodes were significantly regulated (up- or down-regulated). Obviously, this exercise was done separately on the Ra and Rv ensemble, because the nature of regulation under the two conditions is different. This resulted in a marked reduction in total number of shortest paths in both cases across all of the time points (Fig. 2B, Step I). Then, to identify divergent paths of regulation, we started retaining only those paths that showed trend reversal between Ra- and Rv-infected cells at the penultimate steps (node immediately upstream from the target node and also the step before that, Steps II and III in Fig. 2A) iteratively. This exercise yielded a relatively smaller set of shortest paths that were differentially regulated at three consecutive steps beginning from the target node. Essentially, the exercise enriched the shortest path ensemble with paths that were distinctly regulated between Ra- and Rv-infected cells (Fig. 2A, Step IV). These deferentially regulated paths and their relative expression in both conditions are listed in supplemental Table S2. With the accepted limitation of looking only at gene expression patterns, these differentially regulated shortest paths depict the flux of information transfer when a host macrophage encounters virulent or avirulent mycobacteria.
The Shortest Path Network Identifies Key Regulators of the Response Divergence
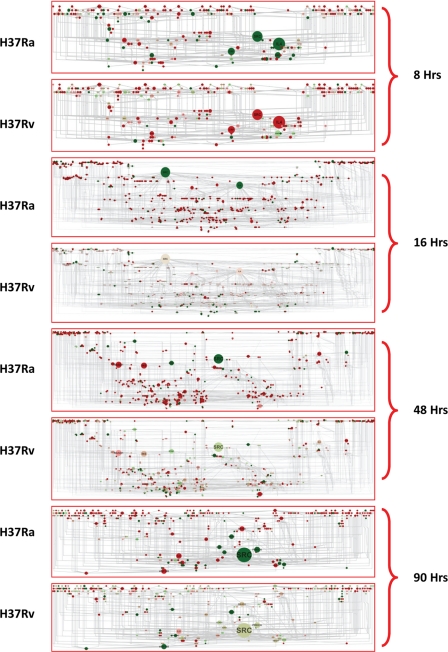
A detailed analysis of the distinctly regulated shortest path ensemble at each time point revealed that many interactions were shared across multiple different paths. Hence, there was the possibility of identifying certain interactions that were central to the overall regulatory events taking place at a particular time point simply based on the frequency of their occurrences in multiple shortest paths. To identify those interactions and thereby the participating nodes, we merged the contrasting shortest path sets from both the Ra and Rv set for a given time point, yielding a time point-specific shortest path network (Fig. 3). Thus, we had shortest path networks for key nodes to 0, 0–8, 8–16, 16–48, and 48–90 h, respectively. The key node to the 0 h network has a relatively small number of molecules; however, it did follow a pattern that reflects differences between Ra- and Rv-infected cells (supplemental Fig. S4). In the shortest path networks, we incorporated two more features in order to clearly visualize the regulated nodes and their importance. First we performed the graph theoretic analysis to calculate the centrality measures of each of the nodes in the networks. One of the most informative centrality parameters is betweenness centrality, signifying the importance of a given node in connecting various modules of a given network (22). A node with higher betweenness value assumes a more important role in the functioning of the network (22). We then mapped the betweenness value of the nodes onto the network and visualized the node size corresponding to their relative betweenness value. Thus, in Fig. 3, nodes with a large diameter have a very high betweenness value. Second, we also incorporated the gene expression value into each of the networks. Up- and down-regulated genes are represented as red and green nodes, respectively (Fig. 3). Therefore, Fig. 3 cumulatively shows the shortest path network at a given time point, the relative importance of the constituent nodes in the functioning of the network, and how they are regulated in Ra- and Rv-infected cells. A quick look at the molecules that showed a significantly high betweenness value shows IL-4, CRP, and APOA1 at 8 h; IL-4, IL-8, and MAPK3 at 16 h; RASD1, INS, EDN1, STAT1, MYOD1, IL-8, and IL-1A at 48 h; and IL-8, RASD1, PRKCA, SMAD4, CCL12, and WNT10B at 90 h (Fig. 3). These molecules were also contrastingly regulated between Ra- and Rv-infected cells. Interestingly, consistent down-regulation of IL-4 and up-regulation of IL-8 in Ra-infected cells augers well with the known functions of these molecules in contrastingly executing the inflammatory responses (23). Although expression-secretion of IL-4 by macrophages is surprising, traditionally known to be secreted by T cells, a few studies do report the expression of IL-4 by alveolar macrophages (24). Similarly, IL-8 is known inflammatory cytokine and is often involved in acute responses; hence, its up-regulation in avirulent infection corresponds well with the clearing of the avirulent infection (23). Other molecules, like STAT1, APOA1, PRKCA, SMADs, RASD1, etc., also are potent regulators of inflammatory responses, and each of these molecules could play a crucial role in establishing and/or executing the divergent responses between avirulent and virulent infections. However, the most outstanding pattern in the express path networks was displayed by the tyrosine kinase family member Src (Fig. 3). It not only showed the highest betweenness value across each network, but it was consistently down-regulated in Ra-infected cells across the time points, whereas in Rv-infected cells, it was either up-regulated (8 h) or close to basal expression levels (16, 48, and 90 h). Src is a tyrosine kinase known to be involved in regulating many physiological processes, including receptor-dependent signaling, cell cycle regulation, and survival (25). Moreover, Src is always among the initial set of molecules that get recruited-activated for multiple cellular stimuli (25). It seemed therefore very logical to assume that differential regulation of Src could serve as the central regulator of divergent host responses. We then characterized the nature of perturbation at Src in detail to understand its role more clearly in regulating cellular response against infection. At least in the case of Mtb infection of macrophages, the role of Src has not been identified so far. One likely explanation for as yet unidentified role of this molecule could be the extreme redundancy displayed by the Src family of kinases. There are at least nine different kinases belonging to the Src family, and many of them overlap in the respective range of functions they execute (26).
FIGURE 3.
Express path network identifies key regulatory nodes. The express paths identified through our analysis (Fig. 2, A and B) were then all merged together, from both Ra and Rv data sets for any given source-target pair time point. This resulted in a network of interaction due to the highly overlapping nature of express paths identified. Express path networks for each of the source-target pairs, along with the betweenness centrality visualization and relative expression differences between Ra and Rv, are represented. Large sized nodes will have a high betweenness value (i.e. more express paths utilize that particular molecule in order to reach the target molecules). Expression data from Ra- and Rv-infected cells were also incorporated into the network, red representing up-regulated genes and green showing down-regulated ones. Note the identical betweenness coefficients for the nodes between Ra- and Rv-infected cells and the largely distinct expression pattern between them.
Src Is the Key Regulator of Cellular Responses to M. tuberculosis Infection
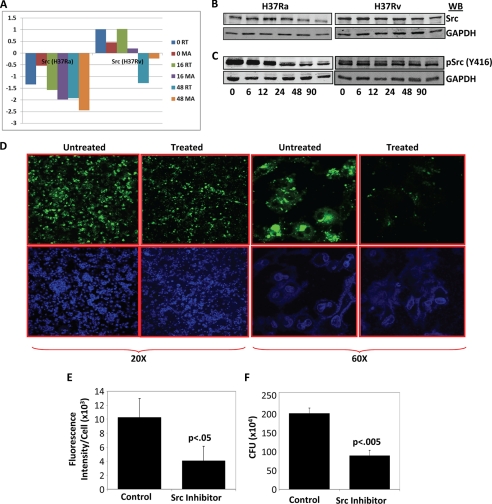
Identification of Src kinase as one of the most important regulatory nodes across all of the observed time points as well as the opposing nature of perturbation on its expression between virulent and avirulent infection throughout the course of infection possibly suggest a key role played by this molecule in deciding cellular responses against Ra or Rv infections. Src expression was significantly down-regulated upon infection with Ra; however, in the case of Rv infection, its expression was either up-regulated or remained unchanged as compared with the uninfected control (supplemental Table S1). The differential expression of Src was further validated by quantitative real-time PCR analysis. A comparison of the real-time versus microarray profile of Src is represented in Fig. 4A. A clear distinction is evident in terms of Src expression between Ra- and Rv-infected cells; however, the mechanisms that lead to such differential regulation of Src in Ra- and Rv-infected cells were not immediately clear. We therefore performed a detailed promoter analysis for Src (∼3 kb upstream of the transcription start site) using the Biobase Biological Database (Transfac) (27, 28). The analysis identified several TFs that could potentially bind to upstream regions of Src and regulate its expression. The details of TFs and their potential binding locations upstream from Src TSS are shown in supplemental Table S3. The microarray data then provided a unique opportunity, allowing us to look for respective regulation of these TFs between Ra- and Rv-infected THP1 cells at different time points. Expression of these TFs at different time points under the two conditions are summarized in supplemental Table S4 along with the number of corresponding transcription factor binding sites identified for a given TF. Surprisingly, most TFs showed almost identical expression profiles in the case of Ra or Rv infection; however, some of them were outstanding due to differential regulation between the two cases (supplemental Fig. S5). Thus, whereas Jun, FOXA2, and ESR1 showed increased expression in Ra-infected cells, TFs like CEBPB, FOS, FOXA3, and SMC1B were selectively up-regulated in Rv-infected cells. On a similar note, ZNF263 and ESRRA were selectively down-regulated in Ra- and Rv-infected cells, respectively. These results, although far from any conclusion regarding mode of differential expression of Src in Ra- and Rv-infected cells, do indicate that some specific events precede the decreased expression of Src in Ra-infected cells and maintained expression in Rv-infected cells. Regulation of eukaryotic gene expression is a highly complex phenomenon involving multivariate cross-talk among several transcriptional regulators that include trans-activators as well as repressors (29). Moreover, many of the TFs are regulated by post-transcriptional modification for their respective action (30). Clearly, transcriptional control of Src, particularly in the context of Mtb-infected cells requires a focused study to understand the regulatory events that are crucial for the observed pattern of differential Src expression in virulent and avirulent infections. In addition, there is little evidence in the literature to indicate how differential expression of Src could impact the differential host responses under virulent and avirulent Mtb infection conditions. Nonetheless, Src is reported to regulate fusion of phagosome with lysosome, although not directly in the case of Mtb infection (31). Src could potentially do so by regulating the activity levels of some known executors in phagocytosis like PI3K, Syk, Akt, etc. Activation of Src is dependent on various factors, phosphorylation at tyrosine 416 being the most important of them (32). The activation level of Src is then regulated through either dephosphorylation by phosphatases like SHP1 and SHP2 or degradation through ubiquitin ligation (32). Therefore, to infer the functional significance of our observations at the transcript level of Src, it was important to monitor its protein levels and phosphorylation states in Ra- or Rv-infected cells. Surprisingly, the Src protein profile showed remarkable similarity to the corresponding mRNA profiles (Fig. 4B and supplemental Fig. S6A). Upon infection with Ra, Src protein level declined after 6 h and kept decreasing throughout the course of infection. In case of Rv infection, however, there was an initial increase in protein level that started decreasing only at later time points (48 h onward). This was an intriguing observation because even at the protein level, Src was differentially regulated between Ra- and Rv-infected cells. We also monitored Tyr416 phosphorylation of Src using phospho-specific antibodies (Fig. 4C and supplemental Fig. S6B). A comparison between Ra- and Rv-infected cells for the level of phospho-Tyr416-Src again showed a distinct pattern in their kinetics. In the case of Ra infection, Src was dephosphorylated immediately after infection and continued to decrease further over the course of infection (Fig. 4C and supplemental Fig. S6B). Interestingly, Rv-infected cells showed a marked increase in Src phosphorylation, and some decrease was visible only 24 h onward (Fig. 4C and supplemental Fig. S6B). Thus, clearly differential regulation of Src between Ra- and Rv-infected cells was not restricted to the transcript level; rather, a larger mechanism seem to be working, which influenced Src phosphorylation, protein levels, and transcript levels. At this stage, what could not be established was whether Src was becoming fast dephosphorylated or degraded or both in the case of Ra infection. In a separate experiment, we monitored the stability of Src in cyclohexamide-treated THP1 cells under either Ra or Rv infection. The overall degradation rate of Src in Ra- and Rv-infected cells did not show any significant differences (supplemental Fig. S6C). This suggests an interesting mechanism of Src regulation in the case of Mtb infection. Because the rate of Src degradation is proportional to its activation rate, in both Ra- and Rv-infected cells, Src activation takes place as a normal cellular response process. However, because Ra infection of cells also leads to down-regulation of Src expression, the de novo synthesis of Src fails to match the degradation rate. Unlike Ra, in the case of Rv infection, the rate of activation-degradation is unaltered; through some mechanism, the expression level of the Src transcript is retained at a basal or higher level, thereby keeping the protein concentration constant. The decision between survival and killing of the infecting pathogen seemed to be decided based on these differences.
FIGURE 4.
Src is a key regulator of differential cellular response between Ra- and Rv-infected cells. The differential gene expression pattern of Src in Ra- and Rv-infected cells was validated through real-time PCR at 0, 16, and 48 h after infection. The bar plot in A shows Src expression at different time points in microarray (MA) or real-time (RT) experiments. B, Src protein levels at various time points after infection compared between Ra- and Rv-infected cells. In Ra-infected cells, 6 h after infection, there was sudden decrease in Src protein content, whereas in the case of Rv infection, there was no such significant change until 90 h after infection. Results are representative of three separate experiments. C, relative phosphorylation of Src at tyrosine 416, an indicator of its activation. Src seems to be dephosphorylated/degraded very fast in the case of Ra infection. D, results of the experiment where PKH67-labeled Rv (green fluorescent)-infected cells were treated with either vector or Src inhibitor PP2. Cells were monitored under the laser confocal microscope at ×20 and ×60 magnification. One of the several fields from three different experiments is shown here. There is a clear difference in green fluorescence between treated and untreated cells, indicative of clearance of Rv from the cells that were treated with the Src inhibitor. The overall fluorescent intensities were quantitated in terms of per unit DAPI (stained for the nuclei), and the values are shown here as a bar graph (E). In a parallel experiment, cells were lysed at 90 h after infection and plated for cfu. Treatment with PP2 resulted in a similar decrease in cfu compared with mock-treated (F) (data from four separate experiments shown as mean ± S.D. (error bars)). Respective p values for the t test are indicated.
In order to establish that differential regulation at the level of Src indeed could lead to differential cellular responses to Mtb infection, we performed an experiment whereby cells were infected with PKH67 (green-fluorescing)-labeled Ra or Rv, and Src activity was inhibited using the specific Src inhibitor PP2. We observed the overall Mtb load of the cell both qualitatively (at ×20 and ×60 magnification) and quantitatively at various time points after infection through a laser-scanning confocal microscope (Fig. 4D). There was a clear distinction in Mtb load between cells treated with the Src inhibitor and those that were untreated. We counted the overall intensity of green fluorescence (for Mtb) per unit of cells (counted as blue DAPI dots) and observed more than 60% reduction in the Mtb load in the cells that were treated with Src inhibitor as compared with the control (Fig. 4E). In a parallel experiment, we also plated the cell lysates at 90 h after infection and estimated bacterial load using the cfu technique. The cfu technique also showed a nearly 70% decrease in the Mtb load upon Src inhibitor treatment (Fig. 4F). Importantly, treatment with PP2 of cells infected with Ra did not show any significant difference from the control (supplemental Fig. S7, A and B). Because Ra infection itself down-regulates and destabilizes Src signal, further inhibition of Src did not show any significant effect on host responses.
Src Regulates Cellular Responses to Mtb Infection by Regulating Autophagy and Phagosome Maturation
Having established clear role of Src in regulating intracellular survival of Mtb, we also explored for possible mechanisms through which Src could execute these responses. Mtb is known to modulate the bactericidal activity of host phagocytes by blocking maturation of the phagosome, the compartment where Mtb is believed to reside for prolonged periods. Among the ultimate steps in the phagosome-mediated killing of pathogens are fusion of phagosome with lysosomal compartment followed by acidification of lysosomes that in turn activates various hydrolytic enzymes of lysosomes (33). Mtb, through its manipulative strategies, can neutralize/block the acidification of lysosomes and hence helps its survival (34). Another important mechanism, implicated recently in regulating intracellular Mtb survival, is autophagy (35–37). Autophagy is a standard cellular degradative process mostly associated with stress responses, including nutritional and other stress (38). Several recent studies indicate a role of autophagy in regulating intracellular bacterial and viral pathogen survival, including Mtb, dengue, etc. (39, 40). We therefore decided to explore whether Src could regulate these two processes.
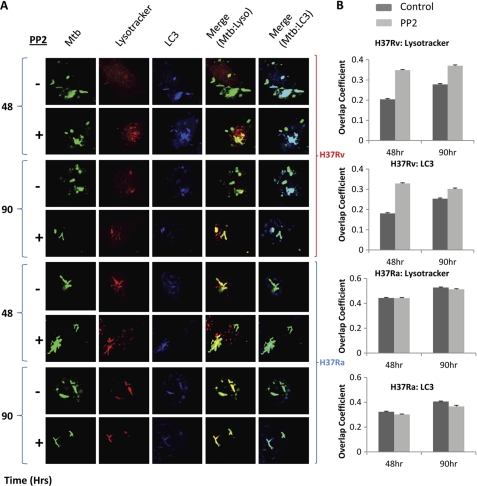
PMA-differentiated THP1 cells were infected with fluorescent dye PKH67-labeled Ra or Rv. We then checked at 48 and 90 h after infection for localization of Mtb in the acidified lysosomal compartment and autophagosomal compartment using confocal microscopy, in the presence or absence of Src inhibitor PP2. Fig. 5 summarizes the results from these experiments. Surprisingly, Src seemed to regulate both phagolysosome acidification and autophagosome formation. At both 48 and 96 h postinfection, localization of Rv to the acidified lysosomal compartment and autophagosome increased significantly where the Src inhibitor PP2 was included in the culture (Fig. 5A). However, an even more exciting observation was that PP2 treatment had virtually no effect on Mtb localization to acidified lysosome and autophagosome in the case of Ra infection. It is important to note here that these results correlate well with the results in Fig. 4, where we observe that, in contrast to Rv, PP2 treatment has no effect on intracellular survival of Ra. Clearly, these results indicate that virulent Mtb infection actively blocks the apparently natural tendency of the host cells to down-regulate Src expression and activity. Because natural host response remains unabated in the case of avirulent infection, further inhibition of the activity does not elicit any response in these cells.
FIGURE 5.
Src regulates phagolysosome acidification and Mtb targeting to the autophagosome. Localization of Mtb to the phagolysosome and autophagosome was monitored through confocal microscopy. A, PMA-treated cells were infected with PKH67-labeled Ra or Rv (green). Infected cells were maintained in the absence (−) or presence (+) of Src inhibitor PP2. At 48 and 90 h postinfection, cells were labeled with Lysotracker (marker for acidified lysosome; red) and the autophagy marker LC3 (blue). Cells were then fixed and examined under the laser-scanning confocal microscope at ×60 magnification. The effect of Src inhibition on the co-localization of Mtb with Lysotracker (Merge (Mtb:Lyso), yellow) or LC3 (Merge Mtb:LC3, cyan) is distinctly visible here. Rv-infected cells showed a marked increase in the yellow (with lysosome) and cyan (with autophagosome) colors, whereas Ra-infected cells did not show any significant effect upon Src inhibition. The co-localization coefficient for red-green (yellow) and blue-green (cyan) were calculated using the software Image-pro version 6.0. Data for the overlap coefficient (y axis) from more than 10 fields across two different experiments consisting of above 50 cells each are represented as bar plots in B (error bars represent S.E.).
DISCUSSION
In the context of cellular perturbation, gene expression profiling has proved immensely helpful to understanding the course of cellular responses (4, 41–43). Not surprisingly, therefore, the differential gene expression profiles of Mtb-infected macrophages upon Ra or Rv infection have been reported in several studies (44–46). One common observation from most large scale expression studies is differential regulation of genes belonging to functional classes like immune regulation and apoptosis between pathogenic and non-pathogenic infection (46). In addition, functional classes as diverse as adhesion, signal transduction, transcription, actin polymerization, and growth regulation are also shown to be distinctly regulated (46). These studies, although they have proved beyond doubt the distinct transcriptional reprogramming of host macrophages upon infection, mostly remain observational in nature. Therefore, apart from listing the sets of genes being modulated and possible implications in pro- or anti-pathogen responses, few conclusions had been drawn from them. Moreover, many of these studies relied on a select set of genes specifically known for their immune regulation properties, thereby biasing the study against regulators that particularly have not been characterized as immune regulators (47, 48). Expectedly, it is known that immune regulator genes like CXCL1, HIF1α, IL12B, ITGA5, TNF, etc. are repressed upon infection by Mtb (2). These results, although informative, fail to provide mechanistic clues about diverse regulatory events that determine the cellular response.
Here, by using a novel approach, EPA, to analyze whole genome gene expression data, we tried to identify molecules that were responsible for establishing the differential response against contrasting environments. EPA involved identification of molecules that are contrastingly regulated at each time point. We reasoned that differential expression at any given time would impinge on the differential regulation at later time points. With this assumption, we traced the shortest path between genes that were contrastingly regulated at one time point to the genes that were contrastingly regulated at the following time point. Because there could be multiple shortest paths between a pair of source and target genes, we started enriching the shortest paths, where each of the intermediates were also differentially regulated. We called these paths express paths because we were identifying the shortest paths of regulation through the gene expression data. As is evident in Fig. 2B, of several possible paths, only a select few are regulated differentially between Ra- and Rv-infected cells. Combining these paths to generate a time point-specific shortest path network yielded some very interesting information. The network identified, through analysis of network parameters like the betweenness centrality measure, molecules that were more frequently present in the differentially regulated shortest paths. Although there were several molecules with higher frequency of occurrence, Src was outstanding because it was present in each of the time point-specific shortest path networks, had a very high betweenness coefficient, and throughout showed decreased expression in Ra-infected cells. Our subsequent analysis showed that in Ra-infected cells, Src protein levels and activity both decreased sharply after infection unlike in Rv-infected cells, where they retained the basal level throughout except at a very late stage of infection. More importantly, inhibiting Src activity in Rv-infected cells led to increased killing of Mtb, although it did not have any significant effect on cells infected with Ra (supplemental Fig. S7, A and B). Our observation clearly established a hitherto unexplored link between Mtb survival and Src activity. The role of Src kinases in sensing environmental changes and subsequent signal transduction is well documented. They are actively recruited downstream to the majority of cell surface receptors, including BCR, TCR, growth factors, and adhesion and cytokine/chemokine receptors (25). Incidentally, Src functions as the key signaling molecule in growth, proliferation, cell cycle regulation, and survival (25, 49). An immediate implication therefore of decreased Src levels in the case of Ra infection could be to help macrophages initiate the apoptotic pathway. Induction of apoptosis upon infection is a known antimicrobial activity of macrophages. However, in a separate experiment, we checked for induction of apoptosis in cells that were infected with Ra or Rv in the presence or absence of Src inhibitor. We could not observe any significant difference as a result of Src inhibition on apoptosis of THP1 cells. Thus, the Src-dependent survival strategy of virulent mycobacteria does not relate directly with the blocking of apoptotic responses. It also suggests that signaling events downstream from Src play a more diverse role in the case of Mtb infection. An interesting observation in this context is the reported phosphorylation of the Helicobacter pyroli virulence factor CagA by Src kinase (50). A similar target discovery in the case of Mtb infection that could be modulated by Src could open an entirely new field of investigation in tuberculosis research.γ Within host cells, many targets of the Src PI3K pathway, MAPK pathway, and phospholipase Cγ pathway are particularly important (26, 51). Some of these pathways lead to activation of downstream events like Ca2+ response, Akt activity, calmodulin kinase II activation, etc. that in turn regulate phagolysosome fusion and autophagy induction in addition to altering the expression of several downstream genes (34). In order to obtain mechanistic insights into the novel role attributed to Src and its regulation in the case of Mtb infection, we looked both at upstream regulation and the downstream effects of Src modulation. A detailed promoter analysis of Src revealed binding sites for several TFs, and interestingly, many of those TFs were also differentially regulated between Ra- and Rv-infected cells. This would presumably happen through differential signal processing in Ra- or Rv-infected cells, leading to differential expression/recruitment of the TFs to the Src upstream region. Transcriptional regulation of Src under these conditions therefore seems to be a highly complex phenomenon that warrants a separate study. The study is, however, expected to be highly promising in terms of identifying initial cellular responses specific to virulent infections. We further established that differential expression and activity of Src could have significant bearing on host cell physiology, especially those associated with the natural microbicidal activities. In subsequent experiments, therefore, we show that inhibiting the activity of Src leads to enhanced acidification of virulent Mtb containing phagolysosome. Src inhibition also leads to significantly higher targeting of Rv to the autophagosome, as observed by its co-localization with the autophagosome marker LC3. Interestingly, these effects were specific to Rv infection because Src inhibition had no visible effect on these two physiological events in cells infected with Ra. The results therefore indicate that infection of host macrophage with avirulent Mtb elicits natural response in the host cells that includes decreased expression of Src and consequently higher phagosome maturation rate and autophagy. When infecting Mtb is virulent (Rv in this case), through its manipulative strategies, it actively subverts the down-regulation of Src expression with a concomitant block in phagolysosome acidification and autophagosome targeting. That largely explains why Src inhibition acts selectively in the case of Rv infection and not for Ra. Also, the fact that many of the TFs that could be recruited to regulate Src expression can essentially be regulated by Src through post-translational modifications, thereby setting up a self-regulatory loop (54, 55), highlights the complex nature of regulatory events postinfection. To reconcile these, a simple model is proposed in Fig. 6 that delineates how differential Src regulation could be brought about and therefore how it leads to differential cellular responses in Ra- or Rv-infected cells. At the molecular level, regulation of these physiological processes, however, remains open to investigation. One possibility is that Src could exert its regulatory influence through downstream regulation of Akt, especially because it has been previously shown that an Akt-centric network plays an important role in intracellular survival of M. tuberculosis (52). Alternatively, Src could also act through regulating mTOR via the Akt pathway. Because activated mTOR blocks autophagy response, Rv, by maintaining Src activity, escapes the autophagy-mediated killing. In contrast, decreased Src levels in Ra-infected cells will have an inhibitory influence on mTOR, leading to activation of macroautophagy and killing. The role of autophagy in intracellular mycobacterial survival has been reported in several studies (35, 36, 53). However, involvement of other signaling pathways in Src-mediated control of antimycobacterial autophagy and phagolysosome acidification could not be ruled out at this stage. In either case, very early in the infection, differential regulation of Src could play a crucial role in order to establish the cellular response mode (see supplemental Fig. S6, A and B). The current study therefore certainly establishes a platform for a detailed study dedicated to signaling events upon infection of host cells with Ra or Rv and differences in signaling events downstream from Src. Further, understanding the exact mechanisms of kinetics of TF recruitment to the Src upstream regulatory region could provide further details about the origin, establishment, and perpetuation of differential cellular response in the case of virulent and avirulent Mtb infection.
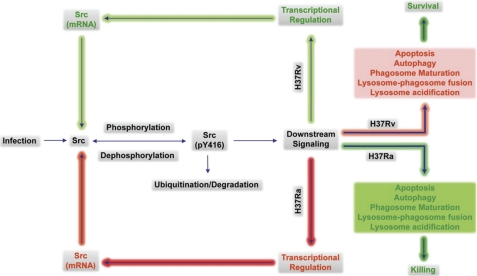
FIGURE 6.
A framework for Src-centric origin and maintenance of differential host response in Ra- and Rv-infected cells. Src appears to be at the center of regulating the diverse cellular responses in the case of Mtb infection. Src phosphorylation is triggered by cellular perturbation, including Mtb infection. The standard mode of controlling Src signaling is through a combination of both dephosphorylation and ubiquitin-mediated degradation. Signaling events downstream from Src vary between Ra- cells, leading to differential TF recruitment and activation patterns, resulting in decreased expression of the Src molecule in Ra-infected cells. Decreased expression of Src interferes with the normal turnover of the Src protein in infected cells, concomitantly bringing the Src molecule levels down in Ra-infected cells. In addition to somewhat regulating its own transcription, Src also appears to regulate the cellular microbicidal processes like phagosome maturation and the induction of autophagy in the case of Mtb infection. This essentially means that, through the complex regulatory mechanism, Src not only ensures its differential regulation; it also defines the downstream events affecting the cellular responses to virulent or avirulent infections. Clearly, an Src-centric self-regulatory loop seems to be playing a role as a central regulator in determining cellular responses against Mtb infection. Processes that are differentially blocked or activated between Ra and Rv are shown in red or green, respectively.
In conclusion, through the present study by supplementing a gene expression profile with cues from global functional associations among the cellular components, we successfully exploited the topological determinants for their role in establishing specific host response. The study identifies regulation of a key signaling molecule, Src, as the central character in determining cellular responses upon infection with Mtb. Our findings conclude that signaling events upstream from transcriptional regulation of Src and downstream from activated Src (pY416) would hold the key to establishing the mechanistic wherewithal of the entire process. Because Src inhibition could not completely eliminate the infection, other parallel mechanisms of regulations are not ruled out. Nonetheless, we believe such understanding would help us to design novel therapeutic strategies for treating tuberculosis in combination with already existing ones. In addition, the novel approach discussed here can potentially be utilized for several other conditions where the regulator of specific cellular responses needs to be identified.
Supplementary Material
This work was supported by an IYBA Grant (DB01/DK/10/363) and a Centre of Excellence grant from the Department of Biotechnology, Government of India, and an Indian Council of Medical Research Grant (to D. K.). This work was also supported in part by a Wellcome-DBT Alliance Grant WTA01/10/355 and an NIH developmental supplement grant P30AI027767 entitled “Creative and Novel Ideas in HIV Research” (to A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S7.
- Mtb
- M. tuberculosis
- EPA
- express path analysis
- DRG
- differentially regulated gene
- PMA
- phorbol 12-myristate 13-acetate
- Ra
- H37Ra
- Rv
- H37Rv
- TF
- transcription factor
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1. Iyer V. R., Eisen M. B., Ross D. T., Schuler G., Moore T., Lee J. C., Trent J. M., Staudt L. M., Hudson J., Jr., Boguski M. S., Lashkari D., Shalon D., Botstein D., Brown P. O. (1999) Science 283, 83–87 [DOI] [PubMed] [Google Scholar]
- 2. Jenner R. G., Young R. A. (2005) Nat. Rev. Microbiol. 3, 281–294 [DOI] [PubMed] [Google Scholar]
- 3. Alexa A., Rahnenführer J., Lengauer T. (2006) Bioinformatics 22, 1600–1607 [DOI] [PubMed] [Google Scholar]
- 4. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarke R., Ressom H. W., Wang A., Xuan J., Liu M. C., Gehan E. A., Wang Y. (2008) Nat. Rev. Cancer 8, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahoo D., Dill D. L., Gentles A. J., Tibshirani R., Plevritis S. K. (2008) Genome Biol. 9, R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang J. T., Carvalho C., Mori S., Bild A. H., Gatza M. L., Wang Q., Lucas J. E., Potti A., Febbo P. G., West M., Nevins J. R. (2009) Mol. Cell 34, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shmulevich I., Dougherty E. R., Kim S., Zhang W. (2002) Bioinformatics 18, 261–274 [DOI] [PubMed] [Google Scholar]
- 9. Friedman N., Linial M., Nachman I., Pe'er D. (2000) J. Comput. Biol. 7, 601–620 [DOI] [PubMed] [Google Scholar]
- 10. Barabási A. L., Oltvai Z. N. (2004) Nat. Rev. Genet. 5, 101–113 [DOI] [PubMed] [Google Scholar]
- 11. Bar-Yam Y., Epstein I. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4341–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Behr M., Schurr E., Gros P. (2010) Cell 140, 615–618 [DOI] [PubMed] [Google Scholar]
- 13. Brosch R., Philipp W. J., Stavropoulos E., Colston M. J., Cole S. T., Gordon S. V. (1999) Infect. Immun. 67, 5768–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinchey J., Lee S., Jeon B. Y., Basaraba R. J., Venkataswamy M. M., Chen B., Chan J., Braunstein M., Orme I. M., Derrick S. C., Morris S. L., Jacobs W. R., Jr., Porcelli S. A. (2007) J. Clin. Invest. 117, 2279–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porcelli S. A., Jacobs W. R., Jr. (2008) Nat. Immunol. 9, 1101–1102 [DOI] [PubMed] [Google Scholar]
- 16. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramoni M. F., Sebastiani P., Kohane I. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9121–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu D., Olman V., Wang L., Xu Y. (2003) Nucleic Acids Res. 31, 5582–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim H. D., Shay T., O'Shea E. K., Regev A. (2009) Science 325, 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramsey S. A., Klemm S. L., Zak D. E., Kennedy K. A., Thorsson V., Li B., Gilchrist M., Gold E. S., Johnson C. D., Litvak V., Navarro G., Roach J. C., Rosenberger C. M., Rust A. G., Yudkovsky N., Aderem A., Shmulevich I. (2008) PLoS Comput. Biol. 4, e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Managbanag J. R., Witten T. M., Bonchev D., Fox L. A., Tsuchiya M., Kennedy B. K., Kaeberlein M. (2008) PLoS One 3, e3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dunn R., Dudbridge F., Sanderson C. M. (2005) BMC Bioinformatics 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinarello C. A. (2000) Chest 118, 503–508 [DOI] [PubMed] [Google Scholar]
- 24. Pouliot P., Turmel V., Gélinas E., Laviolette M., Bissonnette E. Y. (2005) Clin. Exp. Allergy 35, 804–810 [DOI] [PubMed] [Google Scholar]
- 25. Schlessinger J. (2000) Cell 100, 293–296 [DOI] [PubMed] [Google Scholar]
- 26. Parsons S. J., Parsons J. T. (2004) Oncogene 23, 7906–7909 [DOI] [PubMed] [Google Scholar]
- 27. Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., Kloos D. U., Land S., Lewicki-Potapov B., Michael H., Münch R., Reuter I., Rotert S., Saxel H., Scheer M., Thiele S., Wingender E. (2003) Nucleic Acids Res. 31, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhäuser R., Prüss M., Schacherer F., Thiele S., Urbach S. (2001) Nucleic Acids Res. 29, 281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marr M. T., 2nd, Isogai Y., Wright K. J., Tjian R. (2006) Genes Dev. 20, 1458–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tootle T. L., Rebay I. (2005) BioEssays 27, 285–298 [DOI] [PubMed] [Google Scholar]
- 31. Majeed M., Caveggion E., Lowell C. A., Berton G. (2001) J. Leukoc. Biol. 70, 801–811 [PubMed] [Google Scholar]
- 32. Harris K. F., Shoji I., Cooper E. M., Kumar S., Oda H., Howley P. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenberger C. M., Finlay B. B. (2003) Nat. Rev. Mol. Cell Biol. 4, 385–396 [DOI] [PubMed] [Google Scholar]
- 34. Koul A., Herget T., Klebl B., Ullrich A. (2004) Nat. Rev. Microbiol. 2, 189–202 [DOI] [PubMed] [Google Scholar]
- 35. Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 36. Singh S. B., Davis A. S., Taylor G. A., Deretic V. (2006) Science 313, 1438–1441 [DOI] [PubMed] [Google Scholar]
- 37. Harris J., Master S. S., De Haro S. A., Delgado M., Roberts E. A., Hope J. C., Keane J., Deretic V. (2009) Vet. Immunol. Immunopathol. 128, 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martinet W., Agostinis P., Vanhoecke B., Dewaele M., De Meyer G. R. (2009) Clin. Sci. 116, 697–712 [DOI] [PubMed] [Google Scholar]
- 40. Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coombes B. K., Mahony J. B. (2001) Infect. Immun. 69, 1420–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giacomini E., Iona E., Ferroni L., Miettinen M., Fattorini L., Orefici G., Julkunen I., Coccia E. M. (2001) J. Immunol. 166, 7033–7041 [DOI] [PubMed] [Google Scholar]
- 43. George M. D., Sankaran S., Reay E., Gelli A. C., Dandekar S. (2003) Virology 312, 84–94 [DOI] [PubMed] [Google Scholar]
- 44. Silver R. F., Walrath J., Lee H., Jacobson B. A., Horton H., Bowman M. R., Nocka K., Sypek J. P. (2009) Am. J. Respir. Cell Mol. Biol. 40, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keller C., Lauber J., Blumenthal A., Buer J., Ehlers S. (2004) Tuberculosis 84, 144–158 [DOI] [PubMed] [Google Scholar]
- 46. McGarvey J. A., Wagner D., Bermudez L. E. (2004) Clin. Exp. Immunol. 136, 490–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Volpe E., Cappelli G., Grassi M., Martino A., Serafino A., Colizzi V., Sanarico N., Mariani F. (2006) Immunology 118, 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ragno S., Romano M., Howell S., Pappin D. J., Jenner P. J., Colston M. J. (2001) Immunology 104, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zamoyska R., Basson A., Filby A., Legname G., Lovatt M., Seddon B. (2003) Immunol. Rev. 191, 107–118 [DOI] [PubMed] [Google Scholar]
- 50. Stein M., Bagnoli F., Halenbeck R., Rappuoli R., Fantl W. J., Covacci A. (2002) Mol. Microbiol. 43, 971–980 [DOI] [PubMed] [Google Scholar]
- 51. Singh D. K., Kumar D., Siddiqui Z., Basu S. K., Kumar V., Rao K. V. (2005) Cell 121, 281–293 [DOI] [PubMed] [Google Scholar]
- 52. Kuijl C., Savage N. D., Marsman M., Tuin A. W., Janssen L., Egan D. A., Ketema M., van den Nieuwendijk R., van den Eeden S. J., Geluk A., Poot A., van der Marel G., Beijersbergen R. L., Overkleeft H., Ottenhoff T. H., Neefjes J. (2007) Nature 450, 725–730 [DOI] [PubMed] [Google Scholar]
- 53. Kumar D., Nath L., Kamal M. A., Varshney A., Jain A., Singh S., Rao K. V. (2010) Cell 140, 731–743 [DOI] [PubMed] [Google Scholar]
- 54. Karin M., Smeal T. (1992) Trends Biochem. Sci. 17, 418–422 [DOI] [PubMed] [Google Scholar]
- 55. Ritchie S. A., Pasha M. K., Batten D. J., Sharma R. K., Olson D. J., Ross A. R., Bonham K. (2003) Nucleic Acids Res. 31, 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.