Background: ZIP13 protein is important for connective tissue development, which has not been characterized in detail.
Results: ZIP13 is an eight-transmembrane protein with a unique hydrophilic region that forms a homo-dimer.
Conclusion: ZIP13 is a homo-dimerized zinc transporter that possesses domains that are not found in other LZT families.
Significance: The data and materials provide useful information and opportunity for further structural and functional analyses of ZIP13.
Keywords: Membrane Proteins, Protein Conformation, Protein Structure, Transport Metals, Zinc, Protein Characterization, SLC39/ZIP, ZIP13, Zinc Transporter
Abstract
The human SLC39A13 gene encodes ZIP13, a member of the LZT (LIV-1 subfamily of ZIP zinc transporters) family. The ZIP13 protein is important for connective tissue development, and its loss of function is causative for the spondylocheiro dysplastic form of Ehlers-Danlos syndrome. However, this protein has not been characterized in detail. Here we report the first detailed biochemical characterization of the human ZIP13 protein using its ectopic expressed and the purified recombinant protein. Protease accessibility, microscopic, and computational analyses demonstrated that ZIP13 contains eight putative transmembrane domains and a unique hydrophilic region and that it resides with both its N and C termini facing the luminal side on the Golgi. Analyses including cross-linking, immunoprecipitation, Blue Native-PAGE, and size-exclusion chromatography experiments indicated that the ZIP13 protein may form a homo-dimer. We also demonstrated that ZIP13 mediates zinc influx, as assessed by monitoring the expression of the metallothionein gene and by detecting the intracellular zinc level with a zinc indicator, FluoZin-3. Our data indicate that ZIP13 is a homo-dimerized zinc transporter that possesses some domains that are not found in other LZT family members. This is the first biochemical characterization of the physiologically important protein ZIP13 and the demonstration of homo-dimerization for a mammalian ZIP zinc transporter family member. This biochemical characterization of the human ZIP13 protein provides important information for further investigations of its structural characteristics and function.
Introduction
There is increasing evidence for the importance of zinc (Zn) in biology (1–5). Zn2+ is an essential trace element, and low Zn2+ causes Zn2+ deficiency syndrome, which is characterized by growth retardation and skin and bone abnormalities (4, 6). Excess Zn2+ causes symptomatic anemia, as a result of Zn2+-induced copper deficiency (7). Not unexpectedly, a disturbance in the normal balance of Zn2+ results in aberrant cellular functions, and such imbalances have been implicated in the etiology of several human diseases (8–10). Computational analysis suggests that 4–10% of the proteins encoded by the human genome possess Zn2+ binding sequences (11). Collectively, these observations indicate that a balance between intracellular and extracellular Zn2+ levels is critical for basic cellular functions involved in growth, proliferation, differentiation, and migration. Thus, the Zn2+ balance must be tightly regulated, and this is accomplished by Zn2+ transport proteins (8).
The Zn2+ level in a cell is controlled through the coordinated actions of the ZIP/SLC39A and ZnT2/SLC30A Zn2+ transporter families (8, 12); the ZIP family members mediate Zn2+ influx into the cytoplasm, and zinc transporters of the ZnT family mediate its efflux to decrease cytoplasmic Zn2+ (13). Several reports on the mechanisms of Zn2+ transport based on cell biological assays have shown that at least two members of the ZIP family, ZIP8 and ZIP14, function as Zn2+/HCO3−-dependent symporters; a HCO3− gradient across the membrane is used to import Zn2+ into the cytoplasm (14–16). On the other hand, ZIPB, a bacterial ZIP protein homolog, showed a channel-like Zn2+ flux in a proteoliposome study (17), indicating that the mechanism of the mammalian ZIP proteins might require further investigation.
The ZIP family can be divided into four subfamilies based on amino acid sequence similarity: ZIPI, ZIPII, GufA, and LZT (8, 12). The LZT family members, which are only found in eukaryotic cells, share the Pro-Ala-Leu (PAL) and His-Glu-X-X-His (HEXXH, where X is any amino acid) motifs (12). The conserved PAL motif is in the N-terminal region and appears to be involved in protein processing (18). The highly conserved, potential metalloprotease HEXXH motif is located within TM5 (12).
ZIP13 belongs to the LZT family (8, 12); it is mainly expressed by and functions in mesenchyme-originating cells (19, 20). By analyzing Zip13 knock-out (KO) mice, we previously demonstrated that ZIP13 is required for connective tissue formation, which is mediated at least in part by the bone morphogenetic protein (BMP)/transforming growth factor-β (TGF-β)-induced nuclear localization of Smad proteins (19). In a connection between mouse research and human disease, we found that a rare inherited human connective tissue disorder, diagnosed as a novel type of Ehlers-Danlos Syndrome (EDS), spondylocheiro dysplastic-EDS, exhibits phenotypes that are highly correlated with those of Zip13-KO mice (19). Recently the SLC39A13 genetic locus was implicated in fasting glucose homeostasis and in type 2 diabetes risk (21). These findings established that ZIP13 has an indispensable role in connective tissue development and its involvement in disorders; however, the structural characterization of the human ZIP13 protein, such as its topology, conformation, and unique motifs, are still unknown.
Here we characterized the human ZIP13 protein by a series of biochemical experiments as well as bioinformatics analyses. These data and materials will be useful for further investigations, in particular, in determining the three-dimensional structure of the ZIP13 protein.
EXPERIMENTAL PROCEDURES
Cell Culture and Construction of Expression Plasmids
293T cells were maintained in RPMI medium supplemented with 10% FBS and antibiotics at 37 °C. To prepare the Zn2+-depleted culture medium, FBS was incubated with 10 g of Chelex-100 (Bio-Rad) per 50 ml of FBS for 1 h at 4 °C and then filtered for sterilization. FLAG-tagged ZIP13 (see Fig. 5, C and D) was generated as previously described (19). To construct V5-tagged ZIP13, the PCR-amplified product was ligated into pcDNA6.2/V5-DEST (Invitrogen) using Gateway Technology according to the manufacturer's instructions. The plasmids were transfected using Lipofectamine 2000 (Invitrogen) following standard procedures. For recombinant protein production with the BaculoDirect system (Invitrogen), a linker composed of Arg-Leu-Leu-Pro-Val-Thr-Thr-Glu-Gly-Pro-Ser-Gly (RLLPVTTEGPSG) and a thrombin recognition site composed of Leu-Val-Pro-Arg-Gly-Ser (LVPRGS) were attached to the C terminus of ZIP13 in pDONR 221 (Invitrogen).
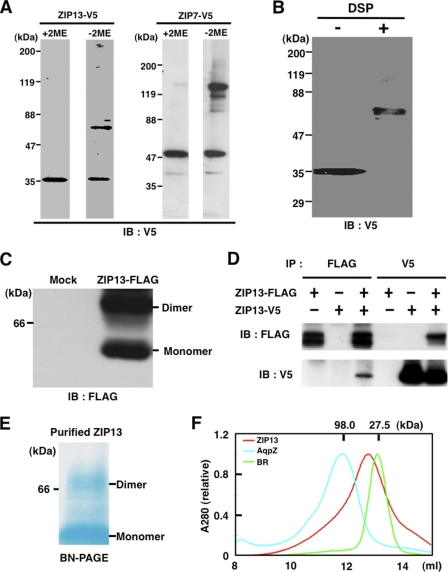
FIGURE 5.
The ZIP13 protein forms homo-dimers. A, 293T cells were transfected with an expression plasmid for ZIP13-V5, and the lysates were analyzed by immunoblotting (IB) with (+) or without (−) 2-mercaptoethanol (2ME). The ZIP7-V5 protein is also shown in comparison with its oligomeric state under non-reducing conditions. B, a cross-linking experiment indicated that the ZIP13 protein forms homo-dimers. The membrane fraction from ZIP13-V5-transfected 293T cells was treated with 0.5 mm dithiobis(succinimidyl propionate) (DSP), then subjected to SDS-PAGE and immunoblotting with an anti-V5 antibody. C, the dodecylmaltoside-solubilized membrane fraction from 293T cells expressing ZIP13-FLAG was applied to BN-PAGE followed by immunoblotting with an anti-FLAG antibody. D, 293T cells were co-transfected with expression plasmids for both ZIP13-V5 and FLAG-tagged ZIP13 (ZIP13-FLAG) followed by immunoprecipitation with the indicated antibodies. Immunoblotting was performed with either aniti-V5 or anti-FLAG antibodies. E, the purified recombinant ZIP13 from Sf9 cells was separated by BN-PAGE and visualized by destaining of the protein gel, which was stained by Coomassie Brilliant Blue dye during electrophoresis. F, the purified recombinant ZIP13 protein from Sf9 cells was size-fractionated by Superdex 200 10/300 GL (GE Healthcare) gel filtration. Aquaporin Z (AqpZ, 98.0 kDa) and bacteriorhodopsin (BR; 27.5 kDa) are shown as size markers. AquaporinZ was from E. coli; bacteriorhodopsin was from H. salinarum.
SDS-PAGE, BN-PAGE, and Immunoblotting
For SDS-PAGE, samples were boiled for 5 min in SDS-PAGE sample buffer containing 0.125 m Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% 2-mercaptoethanol, and 0.004% bromphenol blue. Samples of protein (20 μg) were loaded onto 5–20 or 10–20% gradient gels. Native-PAGETM Novex Bis-Tris gels (for BN-PAGE) were purchased from Invitrogen, and the sample preparation and electrophoresis were performed according to the manufacturer's instructions. For immunoblotting analysis, the gel was electroblotted to a PVDF membrane, which was blocked in 5% skim milk and incubated with an anti-FLAG M2 antibody (Sigma) or anti-V5 antibody (Invitrogen). Secondary detection was carried out using a goat anti-mouse antibody (Zymed Laboratories Inc.).
Fluorescence Microscopy
Cells cultured on glass coverslips in 35-mm glass base dishes (Iwaki) were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS containing 1% BSA for 5 min, and then incubated with an anti-V5 antibody (Invitrogen), anti-GM130 antibody (BD Transduction Laboratories), anti-calnexin antibody (Cell Signaling), and DAPI (Molecular Probes). Fluorescence was detected after secondary staining with the Alexa Fluor 488-conjugated F(ab′)2 fragment of goat anti-mouse IgG (Molecular Probes) and the Alexa Fluor 546-conjugated F(ab′)2 fragment of goat anti-rabbit IgG (Molecular Probes). For FluoZin-3 staining, cells were incubated with 100 μm ZnSO4 for 6 h treated with or without 50 μm Zn2+ chelator TPEN (Dojindo) for 30 min and treated with 5 μm FluoZin-3 (Invitrogen) for 30 min.
Immunoprecipitation
Twenty-four hours after being transfected with plasmids, cells were lysed in lysis buffer containing 20 mm HEPES, pH 7.5, 1% Nonidet P-40, 150 mm NaCl, 10% glycerol, and a protease inhibitor mixture (Sigma). The insoluble material was removed by centrifugation at 15,000 × g for 5 min, and the soluble fraction was incubated with an anti-FLAG M2 antibody (Sigma) or an anti-V5 antibody (Invitrogen) for 1 h. The immune complexes were pulled down with protein G-Sepharose (GE Healthcare) and washed with a buffer containing 20 mm HEPES, pH 7.5, 0.05% Nonidet P-40, 150 mm NaCl, 10% glycerol, and a protease inhibitor mixture (Sigma). Samples were eluted with SDS-PAGE sample buffer and analyzed by immunoblotting.
Proteinase Assay
293T cells expressing the V5-tagged ZIP13 protein were homogenized with 10 strokes of a Dounce homogenizer in buffer containing 20 mm HEPES, pH 7.5, 150 mm NaCl, and 10 mm MgCl2 (HEPES buffer), and the cell debris was removed by centrifugation at 5000 × g for 5 min. The membrane fraction was isolated by further ultracentrifugation at 100,000 × g for 30 min and resuspended in HEPES buffer containing 10% glycerol with or without 1% Triton X-100. The proteinase reaction was carried out at 37 °C for 1 h for trypsin (Promega) and for 1 min for proteinase K (Merck). The reactions were quenched by adding SDS-PAGE sample buffer and boiling for 5 min. The expression plasmid for the double-tagged ZIP13 protein containing C-terminal His6 and the V5 epitope (ZIP13-V5-His) was constructed using the BaculoDirect system (Invitrogen). The ZIP13-V5-His protein was expressed in Sf9 cells and purified with Ni-NTA resin, and the tags were removed by thrombin digestion. The purified ZIP13 protein was treated with proteinase K and separated by SDS-PAGE. After electroblotting onto a nitrocellulose membrane, the membrane was stained with Coomassie Brilliant Blue. The stained protein bands were individually excised from the gel for N-terminal amino acid sequencing.
PNGase F Treatment
Samples of the isolated membrane fraction from 293T cells containing 20 μg protein were treated with PNGase F (New England Biolabs) at 37 °C for 1 h according to the manufacturer's instructions. The samples were then analyzed by immunoblotting.
Cross-linking
Samples of the isolated membrane fraction from 293T cells containing 20 μg of protein were treated with 0.5 mm dithiobis(succinimidyl propionate) (DSP) (Sigma) at 4 °C for 2 h. The reaction was quenched by adding 50 mm Tris-HCl, pH 8.0. The samples were analyzed by immunoblotting.
Quantitative Real-time PCR
Total RNA was reverse-transcribed into cDNA by ReverTra Ace (Toyobo) primed with Oligo(dT)12–18 (Invitrogen). The measurement was performed using SYBR Premix Ex Taq (Takara). The primer sequences used for human ZIP13 and MT1A (Takara) are available upon request.
Statistical Analysis
The two-tailed Student's t test was used to analyze the difference between two groups.
RESULTS
Expression and Structural Features of the Human ZIP13 Protein
We first determined the potential translational start codon for the ZIP13 protein. NetStart1.0 predicted two potential start codons in the ZIP13 transcript, Start1 and Start2 (Fig. 1, A and B). The ZIP13 protein generated using the Start1 codon would be 10 amino acids longer than that obtained using Start2 (Fig. 1C). To determine the de facto start codon, we constructed an expression vector for each, transfected it into cells, and assessed the transcript level. As shown in Fig. 1D, the ZIP13 mRNA level driven from the Start1 expression plasmid (pStart1) was undetectable, whereas the transcription from Start2 (pStart2) was clearly detectable, suggesting that Start2 is the start codon used for translation in the ectopic expression system. We, therefore, used the pStart2 expression vector in our experiments.
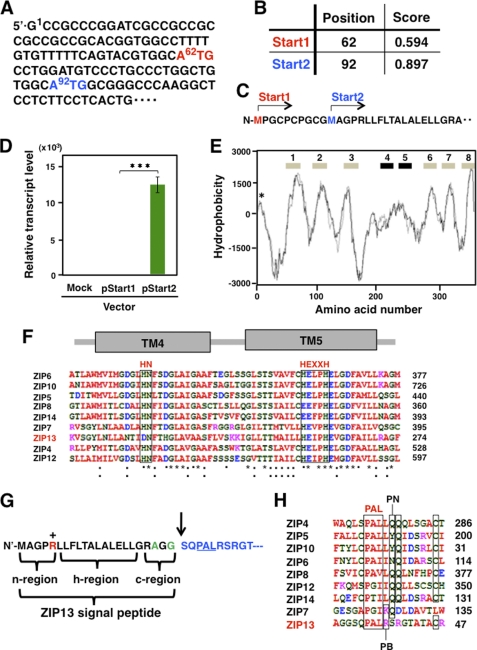
FIGURE 1.
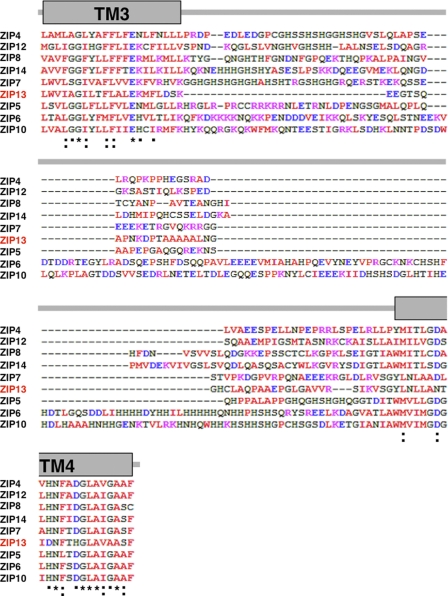
Expression and topology prediction of the ZIP13 protein. A, two predicted translational start codons (62ATG and 92ATG) in the ZIP13 mRNA are shown. B, the nucleotide number for each start codon (Start1 for 62ATG, Start2 for 92ATG) is indicated in the Position column. The Score column shows the probability for the functional start codon calculated by NetStart1.0. C, Start1 and Start2 are marked on the N terminus of the ZIP13 amino acid sequence. D, the ZIP13 mRNA level was determined by quantitative PCR using the RNA from 293T cells transfected with a ZIP13 expression plasmid containing either Start1 (pStart1) or Star2 (pStart2) as the translational start codon. The data were normalized to the Gapdh expression. Data represent the mean ± S.D. of three separate experiments (***, p < 0.001). E, topological features of the human ZIP13 protein are shown. The TM domains (numbers) were predicted on a hydrophobicity plot by the TMpred program. TM4 and TM5 exhibited relatively low hydrophobicity. The location of a signal peptide is marked by an asterisk (*). F, the amino acid alignment of TM4 and TM5 among selected human LZT family members is shown. HN in TM4 and HEXXH in TM5 are putative zinc binding motifs that are highly conserved among the LZT family members. *, identical amino acids in all the aligned sequences; :, conserved substitutions; ·, semi-conserved substitutions. Red, hydrophobic amino acids; blue, acidic amino acids; magenta, basic amino acids; green, hydrophilic amino acids. G, the signal peptide of the ZIP13 protein, with a potential SPC cleavage site between the 22nd and 23rd amino acids, is shown by an arrow. +, positively charged amino acid; green, small amino acids conserved in the ER signal peptide. H, the alignment of the PAL motif among selected human LZT family members is shown. PN, polar and neutral amino acids; PB, polar and basic amino acids.
To further clarify the human ZIP13 structural profile, we performed a hydropathy analysis using the TMpred program, which is considered to be an appropriate algorithm for predicting the TM domains of the LZT family (12). This analysis showed that the ZIP13 protein contains eight membrane-spanning domains (Fig. 1E) and that the hydropathy of the fourth and fifth TM domains (TM4 and TM5) was lower than that of the others (Fig. 1E), probably due to the putative Zn2+ binding HN and HEXXH motifs in TM4 and TM5 (Fig. 1F) (12, 22).
In addition, a small signal peptide-like peak of hydrophobicity was detected close to the initial starting Met residue (Fig. 1E). NetStart1.0 and SignalP revealed a SPC cleavage site between the 22nd and 23rd amino acids from the N terminus, predicting a 22-amino acid long signal peptide (Fig. 1G). The predicted signal peptide could be divided into three regions: an N-terminal region (n-region), a hydrophobic region (h-region), and a C-terminal region (c-region) (Fig. 1G). The N-terminal, hydrophobic, and C-terminal regions, respectively, contained one positively charged residue, a high Leu content, and two small residues (Ala and Gly) ahead of the SPC cleavage site (Fig. 1G). This pattern is identical to that of a typical ER signal peptide (23).
Downstream of the putative SPC-cleavage site, the ZIP13 protein contained a PAL motif, which is a sequence common to all LZT family members (Fig. 1, G and H). Alignment of the LZT family member PAL motif sequences revealed two groups: ZIP13 and ZIP7 have a basic amino acid, i.e. Arg (ZIP13) or Lys (ZIP7), after the PAL motif, whereas in all the other LZT family members the PAL motif is followed by a hydrophobic residue, Leu or Ile (Fig. 1H). In addition, all the LZT members except ZIP13 have a single or tandem Gln residue(s) near the PAL motif, suggesting that the functional redundancy and/or difference among LZT family members may be defined by this motif.
We further examined the membrane topology of human ZIP13 by biochemical experiments. Using the human ZIP13 protein with a V5 sequence at its C terminus (ZIP13-V5), we first assessed whether the ZIP13 protein contains a glycosylation site like other LZT family members (24, 25). The membrane fraction from ZIP13-V5-transfected 293T cells was treated with an N-oligosaccharide glycopeptidase (PNGase F) that digests sugar molecules from N-linked glycoproteins and was then analyzed by SDS-PAGE and immunoblotting. As shown in Fig. 2A, no apparent mobility shift of the ZIP13 protein bands was detected after PNGase F treatment (Fig. 2A, left panel), whereas a shift to lower molecular masses was clearly observed for the ZIP14 protein after PNGase F treatment (Fig. 2A, right panel) (25), indicating that the ZIP13 protein is rarely glycosylated, consistent with the prediction by the NetGlyc software (Fig. 2B, left).
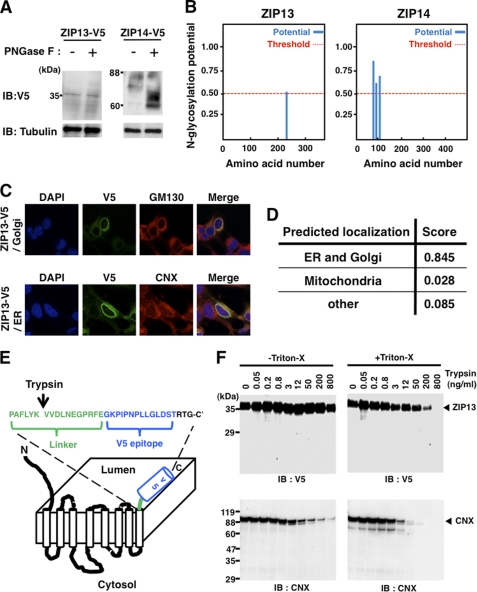
FIGURE 2.
ZIP13 is a Golgi-localized protein whose C terminus faces the luminal side. A, glycosidase treatment rarely altered the migration of the ZIP13 protein. The isolated membrane fraction from ZIP13-V5-transfected cells was treated with PNGase F before immunoblotting (IB) with an anti-V5 antibody. V5-tagged ZIP14 (ZIP14-V5) was used as a control N-glycosylated LZT family member. B, shown is the prediction by NetGlyc that ZIP13 protein has little N-glycosylation potential, which does not greatly exceed the threshold level. The ZIP14 protein shows three sites of high glycosylation potential. C, human ZIP13 protein is mainly localized to the Golgi. ZIP13-V5 protein was expressed in 293T cells and visualized with an anti-V5 antibody. GM130 (upper panel) and calnexin (CNX, lower panel) were stained as marker proteins for the Golgi and ER, respectively. D, prediction by TargetP that ZIP13 localization may be associated with the secretary pathway through the ER and Golgi is shown. E, shown is a schematic topology model for the ZIP13 protein. The arrow indicates an artificial cleavage site for Trypsin in the linker sequence. F, a trypsin accessibility assay indicated that the C terminus of the ZIP13 protein faces the luminal side. The isolated membrane from ZIP13-V5-transfected cells was incubated with or without Triton X-100 and then with trypsin at the indicated concentration followed by immunoblotting using the indicated antibodies.
The endogenous mouse ZIP13 protein is reported to localize to the Golgi in cells of mesenchymal origin (19). To clarify whether this is also the case for human ZIP13, we established 293T cells expressing human ZIP13-V5 and analyzed them by immunocytochemistry. Using an anti-V5 antibody, we detected the major signal of the ZIP13-V5 protein in the perinuclear space (Fig. 2C). It overlapped well with the Golgi marker GM130 (Fig. 2C, upper panel) and partially with the ER marker Calnexin (Fig. 2C, lower panel), suggesting that the human ZIP13 protein is mainly located at the Golgi, as predicted by the TargetP program (Fig. 2D).
To further define ZIP13 membrane topology (Fig. 1E), we next determined the direction of its C terminus. The isolated membrane fraction from ZIP13-V5-transfected 293T cells was used for protease accessibility experiments. The samples were treated with trypsin, then separated by SDS-PAGE followed by immunoblotting with an anti-V5 antibody to detect the C terminus of the ZIP13 protein. If the C terminus of the ZIP13 protein were located on the luminal side (Fig. 2E), a trypsin recognition site at its C-terminal linker region would be readily exposed to trypsin in the presence of Triton X-100. On the other hand, if the C-terminal region were located on the cytoplasmic side, the cleavage of the C-terminal V5 tag by trypsin could occur independent of Triton X-100 treatment. As shown in Fig. 2F, in the presence of Triton X-100 the cleavage of the C-terminal V5 tag of the ZIP13 protein was clearly trypsin dose-dependent (Fig. 2F, right upper panel) but not in the absence of Triton X-100 (Fig. 2F, left upper panel). Furthermore, no obvious small V5 tag-containing fragments of ZIP13 appeared with trypsin treatment, suggesting that the C-terminal region of the human ZIP13 protein is located on the luminal side.
Calnexin was used as a positive control for the trypsin accessibility experiment (Fig. 2F, lower panels). Calnexin consists of a large (410 amino acid) N-terminal luminal domain, a single TM domain, and a short (90-amino acid) C-terminal cytosolic domain. The large N-terminal luminal domain of Calnexin would be exposed to trypsin by Triton X-100 treatment; thus, a stronger digestion pattern appeared in the Triton X-100-treated than untreated samples labeled with an anti-calnexin antibody specific for a sequence near Ala-51 (Fig. 2F, right lower panel). Together, our results show that the basic structural and membrane topology of human ZIP13 is that of a Golgi-localized, eight-TM-domain protein with little glycosylation and a luminal C terminus.
Characterization of the Hydrophilic Regions of the ZIP13 Protein
All LZT family members have eight TM domains, but the sequences and lengths of the hydrophilic regions vary (12). To investigate these regions on the ZIP13 protein, we performed proteolysis experiments with a serine protease, proteinase K. The isolated membrane fraction from 293T cells expressing the ZIP13-V5 protein was treated with proteinase K in the presence or absence of Triton X-100 followed by immunoblotting with an anti-V5 antibody to locate the cleavage sites. Three major fragments, including ZIP13 itself, appeared after proteinase K treatment both with and without Triton X-100 (Fig. 3A, left), implying that at least one main digestion site was on the hydrophilic region(s) of the ZIP13 protein that faces the cytoplasm. Based on the apparent molecular weight of the cleaved fragments (fragment 1 (F1) and fragment 2 (F2) were about 23 and 12 kDa, respectively) and on the observation that Triton X-100 was not needed for the proteinase K accessibility, we speculated that the cleavage site was located within the longest hydrophilic loop between TM3 and TM4 facing the cytoplasm, which we named (Int-L2) (Fig. 3B). The other cleavage site, which released F2, was predicted to be between TM5 and TM6 (Int-L3) (Fig. 3B), based on the estimated molecular weight (Fig. 3A). We next performed the same digestion experiments using recombinant ZIP13 protein purified from Sf9 cells (Fig. 3C) and observed a similar digestion pattern to that described above. Sequencing of the N-terminal peptide of F1 revealed that it was generated by a cleavage between Leu-197 and Ala-198 in Int-L2 (data not shown), which was further confirmed by a MALDI-TOF/MS spectrum obtained using purified F1 and ZIP13 protein from Sf9 cells (Fig. 3D). The observed molecular weight of ZIP13 (37,379 daltons) was smaller than the expected weight of full-length ZIP13 (39,124 daltons) (Fig. 3D), most likely due to deletion of the signal peptide, as the molecular weight of the predicted signal peptide (Fig. 1G) is 2,241 daltons. The detected molecular mass of ZIP13 (37379) was slightly greater than that calculated for the signal peptide-deleted ZIP13 (36,883 daltons). This might be the result of an unidentified post-translational modification or some other unknown cause.
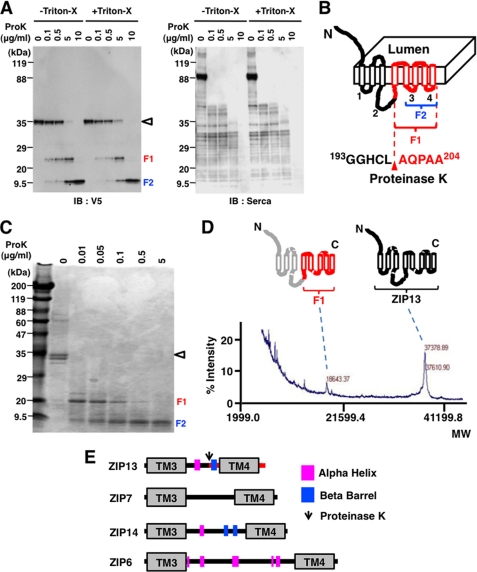
FIGURE 3.
Characterization of the hydrophilic regions of the ZIP13 protein. A, a protease accessibility assay indicated the presence of an internal flanking sequence between TM4 and TM5. The isolated membrane fraction from 293T cells expressing ZIP13-V5 was incubated with or without Triton X-100 before treatment with proteinase K (ProK) followed by immunoblotting (IB) with an anti-V5 antibody. Endogenous sarcoplasmic reticulum Ca2+-ATPase protein was used as a control for proteinase K digestion. ◁, Zip13; F1, Fragment 1; F2, Fragment 2. B, shown is a schematic model of the hydrophilic flanking region of the ZIP13 protein. The red arrowhead indicates a proteinase K cleavage site that was identified by Edman sequencing. The numbers on the cytosolic side (1–4) indicate intracellular loops. C, shown is a protease accessibility assay using purified ZIP13 protein. Purified Zip13 protein from Sf9 cells was treated with proteinase K and visualized by Coomassie Blue staining. ◁, Zip13; F1, Fragment 1; F2, Fragment 2. D, the molecular weight of the F1 fragment was determined from the MALDI-TOF/MS spectrum. The purified recombinant ZIP13 protein and F1 fragment were determined to have molecular weights of 37,378.89 and 18,643.37 daltons, respectively. E, the secondary structure of the intracellular loop2 of LZT family members was determined using the New-joint method (PDB-REPRDB). Magenta box, α helix; blue box, β barrel.
Int-L2 has neither signatures nor highly conserved motifs (Fig. 3E), and the predicted secondary structures of the corresponding region are extremely variable among LZT family members (Figs. 3E and 4). These results indicate that the ZIP13 protein possesses a hydrophilic Int-L2 that is unique from that of other LZT family members.
FIGURE 4.
Alignment of the intracellular loop2 sequence of human LZT family members. *, amino acids that are identical in all the aligned sequences; :, conserved substitutions; ·, semi-conserved substitutions; red, hydrophobic amino acids; blue, acidic amino acids; magenta, basic amino acids; green, hydrophilic amino acids.
The ZIP13 Protein Forms Homo-dimers
Next, we asked whether the ZIP13 protein forms an oligomer, as some ZIP and ZnT family members are reported to form homo- or hetero-oligomers, and in some cases oligomerization is required for their function (26–28). SDS-PAGE under reducing conditions followed by immunoblotting analysis showed a clear single band that corresponded to the apparent molecular mass of the ZIP13 protein monomer (see Fig. 5A, left panel, +2-mercaptoethanol (+2ME)). On the other hand, an additional band at the dimer position appeared under non-reducing conditions (Fig. 5A, left panel, −2ME). A similar pattern was also observed for the ZIP7 protein (Fig. 5A, right panel), which is the closest ZIP family member to ZIP13 protein (8, 12). These results indicated that the dimerized ZIP13 protein might contain a disulfide bond(s) or possess an oxidation-sensitive site between the monomers. To clarify the oligomeric state of the ZIP13 protein, we performed cross-linking experiments using the cross-linking agent dithiobis(succinimidyl propionate) (DSP) (29). The cross-linked ZIP13-V5 protein migrated more slowly, with the apparent molecular mass of the homo-dimer (Fig. 5B). BN-PAGE of non-cross-linked samples also detected the ZIP13 monomer and dimer (Fig. 5C). To determine whether the interaction was homophilic, we performed immunoprecipitation experiments using whole cell lysates prepared from 293T cells that were co-transfected with expression plasmids encoding V5- and/or FLAG-tagged ZIP13 (ZIP13-FLAG). As shown in Fig. 5D, the ZIP13-V5 and ZIP13-FLAG proteins were detected in immune complexes precipitated with either the anti-FLAG or anti-V5 antibodies, indicating that the complexes included homo-dimers.
To further confirm the oligomeric state of the ZIP13 protein, purified ZIP13 protein from Sf9 cells was subjected to both BN-PAGE and size-exclusion chromatography. The BN-PAGE analysis detected the ZIP13 dimer (Fig. 5E) as shown in 293T cell lysates (Fig. 5C). It is possible that some complexes are dissociated or denatured during the electrophoresis in this system. The observed ZIP13 monomer (Fig. 5, C and E) might have been the result of denaturation and/or oxidation during electrophoresis. The size-exclusion chromatography demonstrated that the purified ZIP13 was eluted between the size of bacteriorhodopsin from Halobacterium salinarum (27.5 kDa, the approximate size of the monomer) and aquaporin Z from Escherichia coli (98 kDa, the tetramer size) with high homogeneity (Fig. 5F). Because of the binding of lipids and detergent to membrane proteins, it is generally difficult to estimate the precise molecular mass of a membrane protein using size-exclusion chromatography, but this method is useful for judging its homogeneity. Taken together, these results suggest that the purified ZIP13 protein also forms a dimer.
The ZIP13 Protein Controls Intracellular Zn2+ Homeostasis
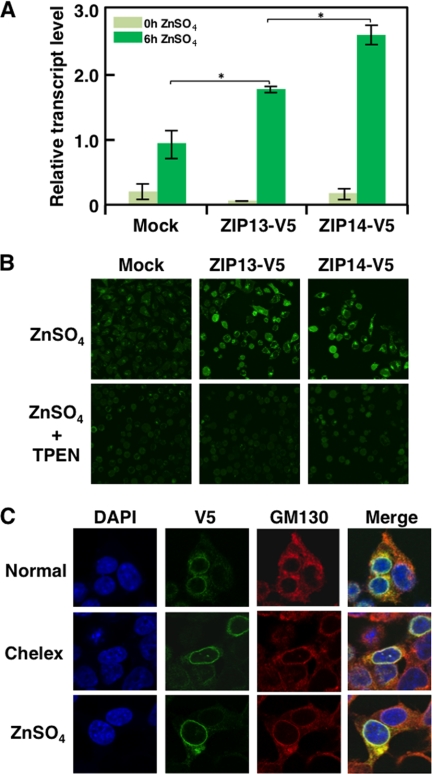
Disruption of the ZIP13 protein causes the dysregulation of intracellular Zn2+ homeostasis (19). However, the effect of ZIP13 protein expression on the regulation of the intracellular Zn2+ level or whether ZIP13-mediated intracellular Zn2+ homeostasis is affected by Zn2+ itself have been unknown. To address whether ectopically expressed ZIP13 protein alters the intracellular Zn2+ homeostasis, we measured the mRNA level of MT1A in ZIP13-V5-transfected 293T cells. The MT1A mRNA level, which is up-regulated by the Zn2+ sensor MTF1, is often measured as a reflection of the intracellular Zn2+ level (18, 30, 31). After Zn2+ addition, the MT1A mRNA level was significantly higher in the ZIP13-V5-transfected 293T cells than in the mock-transfected cells (Fig. 6A), suggesting that the ectopically expressed ZIP13 protein affected the intracellular Zn2+ homeostasis and that extracellular Zn2+ might stimulate ZIP13 function even though ZIP13 is located at the Golgi. To visualize the intracellular Zn2+ accumulation, the ZIP13-V5-transfected cells were stained with a Zn2+ indicator, FluoZin-3 (Fig. 6B). Fluorescent intensity of FluoZin-3 in ZIP13-V5-transfected cells was elevated by Zn2+ treatment, and it was reduced in the basal level by the Zn2+ chelator TPEN treatment. This result showed that the ZIP13-V5-transfected cells accumulated Zn2+ more efficiently than the control cells after Zn2+ treatment (Fig. 6B). ZIP14-V5-transfected cells were used as a positive control for Zn2+ accumulation. To examine whether Zn2+ regulates the cellular localization and/or expression of ZIP13 protein, because extracellular Zn2+ alters the cellular localization of some Zn2+ transporters (16, 18), we performed Zn2+ addition or depletion using ZIP13-V5-transfected 293T cells and fluorescence microscopy. The results did not reveal significant changes in either the protein level or localization of ZIP13 in the presence of Zn2+ (Fig. 6C). These data together suggest that ZIP13 is a persistently Golgi-localized Zn2+ transporter that controls intracellular Zn2+ homeostasis.
FIGURE 6.
Expression of the ZIP13 protein increases the intracellular Zn2+ level. A, the ectopic expression of ZIP13 enhanced the MT1A mRNA level in cells. 293T cells expressing either ZIP13-V5 or ZIP14-V5 (as a control (25, 41)) were treated with 100 μm ZnSO4 for 6 h followed by quantification of the MT1A mRNA level by quantitative real-time PCR. Data are representative of three experiments (*, p < 0.05). B, the ectopic expression of ZIP13 increased the labile/free Zn2+ level in cells. HeLa cells transfected with expression plasmids for ZIP13-V5 or ZIP14-V5 were treated with 100 μm ZnSO4 for 6 h with or without 50 μm Zn2+ chelator TPEN for 30 min followed by Zn2+ visualization using FluoZin-3. C, neither the addition nor the depletion of Zn2+ changed the intracellular localization of the ZIP13 protein. 293T cells expressing ZIP13-V5 were treated with either Chelex or 30 μm ZnSO4 for 6 h followed by visualization using an anti-V5 antibody and anti-GM130 antibody. DAPI was used to stain nuclei.
DISCUSSION
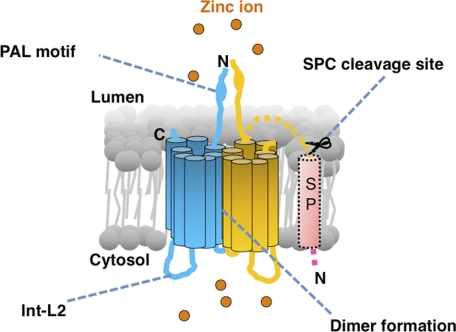
In this study we molecularly characterized the human ZIP13 protein and revealed that 1) it contains eight putative TM domains, with both the N and C termini facing the luminal side of the Golgi, 2) it possesses a unique intracellular loop, 3) it forms a homo-dimer, and 4) it mediates Zn2+ influx (Fig. 7).
FIGURE 7.
Schematic model of the human ZIP13 protein. The ZIP13 protein forms a homo-dimer and possesses luminal termini, a unique flexible intracellular loop (Int-L2), and a PAL motif and controls intracellular Zn2+ homeostasis. The signal peptide (SP) is thought to be removed after insertion of the protein into the ER membrane. The orange spheres represent Zn2+ ions.
The membrane topology model of the ZIP13 protein defined by the results of protease accessibility experiments and by bioinformatics suggested that ZIP13 contains eight putative TM domains, with both termini facing the lumen of the Golgi (Fig. 2E). This is consistent with the proposed topology models for other LZT family members, which also have eight TM domains with both the N and C termini facing either the extracellular or the luminal side, based on alignment analysis and antibody accessibility experiments (12, 18). We also defined the N-terminal signal peptide in the ZIP13 sequence (Fig. 1G), which is identical to a traditional ER signal peptide (23). Based on previous studies (23, 32, 33), the positively charged Arg in the N-terminal region of the ZIP13 signal peptide may function to anchor the ZIP13 polypeptide on the ER membrane, enabling it to pass through the ER membrane using its hydrophobic region, and the signal peptide may then be removed by SPC, creating the luminal N terminus of ZIP13. The precise roles of the N-terminal and hydrophobic regions in the ZIP13 N-terminal signal peptide still need to be verified.
The PAL motif in the N-terminal region upstream of TM1 is a common feature of LZT family members (8, 12) and has been proposed to be the proteolytic processing site of several LZT family members in response to the extracellular Zn2+ level (18). LZT family members also have conserved sequences consisting of one hydrophobic and two polar residues following the PAL motif (PAL(L/I)(Q/Y/N)Q), except for ZIP7 and ZIP13 (Fig. 1H). In particular, the PAL motif of ZIP13 is followed not by hydrophobic, but by basic residues (PALRSR). Although the precise function(s) of the PAL motif remains to be established, it and its surrounding amino acids may confer different functions on different LZT family members.
Of particular interest from the comparative study of the primary structure of ZIP13 is the intracellular loop termed Int-L2 between TM3 and TM4 (Fig. 4). Each LZT family member possesses a unique sequence in Int-L2 (Fig. 4) (12). The corresponding region of ZIP13 is relatively short (Fig. 4), and it is predicted to be almost unstructured (Fig. 3E). Although the role(s) of this region in LZT family members remain to be determined, one possibility is that it participates in the degradation of the LZT protein. For example, Zrt1, a ZIP family member in yeast undergoes endocytosis under high Zn2+ conditions accompanied by the ubiquitination of Lys-195 in Int-L2, suggesting that this modification serves as a signal for Zrt1 recruitment to early endosomes via clathrin-coated pits, leading to its down-regulation (34); therefore, the Int-L2 in Zrt1 plays an important role in its own Zn2+-responsive post-translational inactivation. Similarly, the ZIP4 protein has a His-rich cluster in Int-L2, and this sequence is involved in ZIP4 ubiquitination and degradation in response to added Zn2+ (35). We examined whether a similar scenario occurs with ZIP13, but no clear alteration in its protein level or localization was observed with Zn2+ addition or depletion (Fig. 6C). Collectively, these observations suggest that the Int-L2 in ZIP13 may have distinct roles from those of the corresponding region of other LZT family members for Zn2+ transport and/or controlling its downstream signals. The amino acid sequence of Int-L2 has little similarity among the LZT family members, including ZIP13 (Fig. 4). Deletion mutation studies and identification of associating molecules should help to reveal its potential roles.
We showed that the ZIP13 protein forms a homo-dimer (Fig. 5). This is similar to the oligomerization state of the bacterial ZIP homolog ZIPB (17), and we believe our report is the first to demonstrate the homo-dimeric state of a mammalian ZIP protein. Metal transporters show various oligomerization states. For example, the copper transporter Ctr1 contains three TM domains and forms a trimer (36), and this oligomerization is theoretically needed for its transport function. In the case of ZIP13, although each ZIP13 protein has multiple TM domains, whether the pocket/pore for Zn2+ flux is formed by the monomer alone is not yet known. The bacterial cation diffusion facilitator YiiP forms a dimer, and its homo-dimerization is important for its ability to sense and transport Zn2+ (27). In addition to homo-oligomerization, the role of the hetero-oligomerization of transporters has been discussed (26, 27, 37, 38). The SLC30A/ZnT (zinc transporter) family members ZnT5 and ZnT6 form a hetero-dimer, and this state is important for the Zn2+ efflux that leads to alkaline phosphatase activation (37, 39). The band at the dimer position appeared under non-reducing conditions (Fig. 5A, left panel, −2-mercaptoethanol (−2ME)), suggesting that the dimerized ZIP13 protein may possess an oxidation-sensitive site between the monomers. The role and the mechanisms of the ZIP13 homo-dimer and whether ZIP13 forms functional hetero-oligomers with other LZT family members are still open questions. Because the similar results was obtained in the case of ZIP7 protein (Fig. 5A), which is the closest to ZIP13 protein (8), these two ZIP family members may share the mechanisms and roles of their oligomerization.
Finally, we showed that the ectopic expression of ZIP13 in mammalian cells changed the intracellular Zn2+ level after Zn2+ addition (Fig. 6, A and B). These findings support the intriguing possibility that Zn2+-sensing molecules on the plasma membrane may transmit unknown signals to stimulate ZIP13 Zn2+ transporting activity (40). Another operational possibility is that ZIP13 passively takes up excess intracellular Zn2+ into the Golgi, which affects the Zn2+ uptake from the extracellular region into the intracellular area, thereby increasing the rate of Zn2+ influx through the plasma membrane. Such a scenario may be supported by a recent report showing that the bacterial ZIP homolog ZIPB facilitates passive Zn2+ uptake driven by a Zn2+ concentration gradient (17). Further research that includes a crystallographic analysis to determine the structure of the purified ZIP13 protein and a zinc transport analysis that makes use of liposome-based and/or patch clamp-based approaches will reveal the detailed mechanisms by which the ZIP13 protein regulates cellular Zn2+ levels, including its dimerization.
In the present work, we obtained biochemical, microscopic, and software-based data that together suggested that the ZIP13 protein is localized to the Golgi independent of the Zn2+ level, possesses a unique intracellular loop, forms homo-dimers, and is indeed involved in Zn2+ homeostasis. These data and materials provide useful information and unique opportunity for further structural and functional analyses of ZIP13 at the molecular level and, in particular, studies aiming to reveal the three-dimensional structure of the ZIP13 protein and pathogenic mechanisms involving ZIP13.
Acknowledgments
We are grateful to Masami Kawamura, Ayumi Ito, and Mayumi Hara for excellent technical assistance. We also thank Mizuki Shimura, Reiko Kimura, and Ryoko Masuda for secretarial assistance.
This work was supported by KAKENHI and the Japan Science and Technology Agency-Core Research for Evolutional Science and Technology program and the Naito Foundation (to T. F.).
- ZnT
- zinc transporter
- TM
- transmembrane
- BN
- blue native
- LZT
- LIV-1 subfamily of ZIP zinc transporters
- SPC
- signal peptidase complex
- ER
- endoplasmic reticulum
- PNGase F
- N-oligosaccharide glycopeptidase
- Int-L
- intracellular loop
- Sf9
- Spodoptera frugiperda 9
- MT1A
- metallothionein 1A
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TPEN
- N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine.
REFERENCES
- 1. Fukada T., Yamasaki S., Nishida K., Murakami M., Hirano T. (2011) J. Biol. Inorg. Chem. 16, 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hirano T., Murakami M., Fukada T., Nishida K., Yamasaki S., Suzuki T. (2008) Adv. Immunol. 97, 149–176 [DOI] [PubMed] [Google Scholar]
- 3. MacDonald R. S. (2000) J. Nutr. 130, 1500S–1508S [DOI] [PubMed] [Google Scholar]
- 4. Prasad A. S. (1995) Nutrition 11, 93–99 [PubMed] [Google Scholar]
- 5. Yamasaki S., Sakata-Sogawa K., Hasegawa A., Suzuki T., Kabu K., Sato E., Kurosaki T., Yamashita S., Tokunaga M., Nishida K., Hirano T. (2007) J. Cell Biol. 177, 637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamaguchi M. (2010) Mol. Cell. Biochem. 338, 241–254 [DOI] [PubMed] [Google Scholar]
- 7. Pawa S., Khalifa A. J., Ehrinpreis M. N., Schiffer C. A., Siddiqui F. A. (2008) Am J. Med. Sci. 336, 430–433 [DOI] [PubMed] [Google Scholar]
- 8. Fukada T., Kambe T. (2011) Metallomics 3, 662–674 [DOI] [PubMed] [Google Scholar]
- 9. Nishida K., Hasegawa A., Nakae S., Oboki K., Saito H., Yamasaki S., Hirano T. (2009) J. Exp. Med. 206, 1351–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foster M., Petocz P., Samman S. (2010) Atherosclerosis 210, 344–352 [DOI] [PubMed] [Google Scholar]
- 11. Andreini C., Banci L., Bertini I., Rosato A. (2006) J. Proteome Res. 5, 196–201 [DOI] [PubMed] [Google Scholar]
- 12. Taylor K. M., Nicholson R. I. (2003) Biochim. Biophys. Acta 1611, 16–30 [DOI] [PubMed] [Google Scholar]
- 13. Kambe T., Yamaguchi-Iwai Y., Sasaki R., Nagao M. (2004) Cell. Mol. Life Sci. 61, 49–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He L., Girijashanker K., Dalton T. P., Reed J., Li H., Soleimani M., Nebert D. W. (2006) Mol. Pharmacol. 70, 171–180 [DOI] [PubMed] [Google Scholar]
- 15. Girijashanker K., He L., Soleimani M., Reed J. M., Li H., Liu Z., Wang B., Dalton T. P., Nebert D. W. (2008) Mol. Pharmacol. 73, 1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Z., Li H., Soleimani M., Girijashanker K., Reed J. M., He L., Dalton T. P., Nebert D. W. (2008) Biochem. Biophys. Res. Commun. 365, 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin W., Chai J., Love J., Fu D. (2010) J. Biol. Chem. 285, 39013–39020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kambe T., Andrews G. K. (2009) Mol. Cell. Biol. 29, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukada T., Civic N., Furuichi T., Shimoda S., Mishima K., Higashiyama H., Idaira Y., Asada Y., Kitamura H., Yamasaki S., Hojyo S., Nakayama M., Ohara O., Koseki H., Dos Santos H. G., Bonafe L., Ha-Vinh R., Zankl A., Unger S., Kraenzlin M. E., Beckmann J. S., Saito I., Rivolta C., Ikegawa S., Superti-Furga A., Hirano T. (2008) PLoS One 3, e3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fukada T., Asada Y., Mishima K., Shimoda S., Saito I. (2011) J. Oral Biosci. 53, 1–12 [Google Scholar]
- 21. Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., Lindgren C. M., Mägi R., Morris A. P., Randall J., Johnson T., Elliott P., Rybin D., Thorleifsson G., Steinthorsdottir V., Henneman P., Grallert H., Dehghan A., Hottenga J. J., Franklin C. S., Navarro P., Song K., Goel A., Perry J. R., Egan J. M., Lajunen T., Grarup N., Sparsø T., Doney A., Voight B. F., Stringham H. M., Li M., Kanoni S., Shrader P., Cavalcanti-Proença C., Kumari M., Qi L., Timpson N. J., Gieger C., Zabena C., Rocheleau G., Ingelsson E., An P., O'Connell J., Luan J., Elliott A., McCarroll S. A., Payne F., Roccasecca R. M., Pattou F., Sethupathy P., Ardlie K., Ariyurek Y., Balkau B., Barter P., Beilby J. P., Ben-Shlomo Y., Benediktsson R., Bennett A. J., Bergmann S., Bochud M., Boerwinkle E., Bonnefond A., Bonnycastle L. L., Borch-Johnsen K., Böttcher Y., Brunner E., Bumpstead S. J., Charpentier G., Chen Y. D., Chines P., Clarke R., Coin L. J., Cooper M. N., Cornelis M., Crawford G., Crisponi L., Day I. N., de Geus E. J., Delplanque J., Dina C., Erdos M. R., Fedson A. C., Fischer-Rosinsky A., Forouhi N. G., Fox C. S., Frants R., Franzosi M. G., Galan P., Goodarzi M. O., Graessler J., Groves C. J., Grundy S., Gwilliam R., Gyllensten U., Hadjadj S., Hallmans G., Hammond N., Han X., Hartikainen A. L., Hassanali N., Hayward C., Heath S. C., Hercberg S., Herder C., Hicks A. A., Hillman D. R., Hingorani A. D., Hofman A., Hui J., Hung J., Isomaa B., Johnson P. R., Jørgensen T., Jula A., Kaakinen M., Kaprio J., Kesaniemi Y. A., Kivimaki M., Knight B., Koskinen S., Kovacs P., Kyvik K. O., Lathrop G. M., Lawlor D. A., Le Bacquer O., Lecoeur C., Li Y., Lyssenko V., Mahley R., Mangino M., Manning A. K., Martínez-Larrad M. T., McAteer J. B., McCulloch L. J., McPherson R., Meisinger C., Melzer D., Meyre D., Mitchell B. D., Morken M. A., Mukherjee S., Naitza S., Narisu N., Neville M. J., Oostra B. A., Orrù M., Pakyz R., Palmer C. N., Paolisso G., Pattaro C., Pearson D., Peden J. F., Pedersen N. L., Perola M., Pfeiffer A. F., Pichler I., Polasek O., Posthuma D., Potter S. C., Pouta A., Province M. A., Psaty B. M., Rathmann W., Rayner N. W., Rice K., Ripatti S., Rivadeneira F., Roden M., Rolandsson O., Sandbaek A., Sandhu M., Sanna S., Sayer A. A., Scheet P., Scott L. J., Seedorf U., Sharp S. J., Shields B., Sigurethsson G., Sijbrands E. J., Silveira A., Simpson L., Singleton A., Smith N. L., Sovio U., Swift A., Syddall H., Syvänen A. C., Tanaka T., Thorand B., Tichet J., Tönjes A., Tuomi T., Uitterlinden A. G., van Dijk K. W., van Hoek M., Varma D., Visvikis-Siest S., Vitart V., Vogelzangs N., Waeber G., Wagner P. J., Walley A., Walters G. B., Ward K. L., Watkins H., Weedon M. N., Wild S. H., Willemsen G., Witteman J. C., Yarnell J. W., Zeggini E., Zelenika D., Zethelius B., Zhai G., Zhao J. H., Zillikens M. C., Borecki I. B., Loos R. J., Meneton P., Magnusson P. K., Nathan D. M., Williams G. H., Hattersley A. T., Silander K., Salomaa V., Smith G. D., Bornstein S. R., Schwarz P., Spranger J., Karpe F., Shuldiner A. R., Cooper C., Dedoussis G. V., Serrano-Ríos M., Morris A. D., Lind L., Palmer L. J., Hu F. B., Franks P. W., Ebrahim S., Marmot M., Kao W. H., Pankow J. S., Sampson M. J., Kuusisto J., Laakso M., Hansen T., Pedersen O., Pramstaller P. P., Wichmann H. E., Illig T., Rudan I., Wright A. F., Stumvoll M., Campbell H., Wilson J. F., Bergman R. N., Buchanan T. A., Collins F. S., Mohlke K. L., Tuomilehto J., Valle T. T., Altshuler D., Rotter J. I., Siscovick D. S., Penninx B. W., Boomsma D. I., Deloukas P., Spector T. D., Frayling T. M., Ferrucci L., Kong A., Thorsteinsdottir U., Stefansson K., van Duijn C. M., Aulchenko Y. S., Cao A., Scuteri A., Schlessinger D., Uda M., Ruokonen A., Jarvelin M. R., Waterworth D. M., Vollenweider P., Peltonen L., Mooser V., Abecasis G. R., Wareham N. J., Sladek R., Froguel P., Watanabe R. M., Meigs J. B., Groop L., Boehnke M., McCarthy M. I., Florez J. C., Barroso I. (2010) Nat. Genet. 42, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor K. M., Morgan H. E., Johnson A., Nicholson R. I. (2004) Biochem. J. 377, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paetzel M., Karla A., Strynadka N. C., Dalbey R. E. (2002) Chem. Rev. 102, 4549–4580 [DOI] [PubMed] [Google Scholar]
- 24. Hogstrand C., Kille P., Nicholson R. I., Taylor K. M. (2009) Trends Mol. Med. 15, 101–111 [DOI] [PubMed] [Google Scholar]
- 25. Hojyo S., Fukada T., Shimoda S., Ohashi W., Bin B. H., Koseki H., Hirano T. (2011) PLoS One 6, e18059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukunaka A., Suzuki T., Kurokawa Y., Yamazaki T., Fujiwara N., Ishihara K., Migaki H., Okumura K., Masuda S., Yamaguchi-Iwai Y., Nagao M., Kambe T. (2009) J. Biol. Chem. 284, 30798–30806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu M., Chai J., Fu D. (2009) Nat. Struct. Mol. Biol. 16, 1063–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor K. M., Vichova P., Jordan N., Hiscox S., Hendley R., Nicholson R. I. (2008) Endocrinology 149, 4912–4920 [DOI] [PubMed] [Google Scholar]
- 29. Stout J. G., Kirley T. L. (1996) Biochemistry 35, 8289–8298 [DOI] [PubMed] [Google Scholar]
- 30. Lopez V., Foolad F., Kelleher S. L. (2011) Cancer Lett. 304, 41–51 [DOI] [PubMed] [Google Scholar]
- 31. Bi Y., Palmiter R. D., Wood K. M., Ma Q. (2004) Biochem. J. 380, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 33. Zimmermann R., Eyrisch S., Ahmad M., Helms V. (2011) Biochim. Biophys. Acta 1808, 912–924 [DOI] [PubMed] [Google Scholar]
- 34. Gitan R. S.., Eide D. J., (2000) Biochem. J. 346, 329–336 [PMC free article] [PubMed] [Google Scholar]
- 35. Mao X., Kim B. E., Wang F., Eide D. J., Petris M. J. (2007) J. Biol. Chem. 282, 6992–7000 [DOI] [PubMed] [Google Scholar]
- 36. Aller S. G., Unger V. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suzuki T., Ishihara K., Migaki H., Ishihara K., Nagao M., Yamaguchi-Iwai Y., Kambe T. (2005) J. Biol. Chem. 280, 30956–30962 [DOI] [PubMed] [Google Scholar]
- 38. Nose Y., Rees E. M., Thiele D. J. (2006) Trends Biochem. Sci 31, 604–607 [DOI] [PubMed] [Google Scholar]
- 39. Fukunaka A., Kurokawa Y., Teranishi F., Sekler I., Oda K., Ackland M. L., Faundez V., Hiromura M., Masuda S., Nagao M., Enomoto S., Kambe T. (2011) J. Biol. Chem. 286, 16363–16373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hershfinkel M., Silverman W. F., Sekler I. (2007) Mol. Med. 13, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taylor K. M., Morgan H. E., Johnson A., Nicholson R. I. (2005) FEBS Lett. 579, 427–432 [DOI] [PubMed] [Google Scholar]