Background: New drugs active against persistent Mycobacterium tuberculosis are needed.
Results: The fructose-1,6-bisphosphate aldolase (FBA-tb) is essential for growth of M. tuberculosis, is expressed by replicating and non-replicating bacilli, and displays plasminogen binding activity.
Conclusion: FBA-tb is an essential TB enzyme that might also play a role in host/pathogen interactions.
Significance: FBA-tb shows potential as a novel anti-TB therapeutic target.
Keywords: Bacterial Metabolism, Enzyme Inhibitors, Gluconeogenesis, Glycolysis, Plasminogen, Mycobacterium, Tuberculosis, Aldolase
Abstract
The search for antituberculosis drugs active against persistent bacilli has led to our interest in metallodependent class II fructose-1,6-bisphosphate aldolase (FBA-tb), a key enzyme of gluconeogenesis absent from mammalian cells. Knock-out experiments at the fba-tb locus indicated that this gene is required for the growth of Mycobacterium tuberculosis on gluconeogenetic substrates and in glucose-containing medium. Surface labeling and enzymatic activity measurements revealed that this enzyme was exported to the cell surface of M. tuberculosis and produced under various axenic growth conditions including oxygen depletion and hence by non-replicating bacilli. Importantly, FBA-tb was also produced in vivo in the lungs of infected guinea pigs and mice. FBA-tb bound human plasmin(ogen) and protected FBA-tb-bound plasmin from regulation by α2-antiplasmin, suggestive of an involvement of this enzyme in host/pathogen interactions. The crystal structures of FBA-tb in the native form and in complex with a hydroxamate substrate analog were determined to 2.35- and 1.9-Å resolution, respectively. Whereas inhibitor attachment had no effect on the plasminogen binding activity of FBA-tb, it competed with the natural substrate of the enzyme, fructose 1,6-bisphosphate, and substantiated a previously unknown reaction mechanism associated with metallodependent aldolases involving recruitment of the catalytic zinc ion by the substrate upon active site binding. Altogether, our results highlight the potential of FBA-tb as a novel therapeutic target against both replicating and non-replicating bacilli.
Introduction
The rise in antibiotic-resistant bacterial infections and the lack of drugs capable of efficiently eradicating persistent microorganisms responsible for life-long infections in humans emphasize the need for novel antimicrobial agents with mechanisms of action different from those of presently existing drugs. Tuberculosis (TB)5 in particular is a disease for which novel drugs capable of killing persistent bacilli would be of great benefit (1). Mycobacterium tuberculosis, the etiologic agent of TB, claims about 1.7 million lives annually, and the global number of TB cases is still rising at a rate of 0.6% per year, accompanied by an increase in the number of cases attributable to multidrug-resistant strains (2). It has been estimated that one-third of the world's population is latently infected with the tubercle bacillus. The resulting latent stage of infection is associated with a few bacteria surviving for years in a latent or “semidormant” state with low metabolic activity (3–9). Persistent bacteria appear to be resistant to common chemotherapy and may be reactivated, resulting in active disease.
Although the physiological state of persistent M. tuberculosis bacilli during latent human infection is largely unknown, there is now substantial evidence that M. tuberculosis undergoes important metabolic changes to ensure a constant supply of carbon and energy from alternative sources and pathways (1). Analysis of patient autopsy specimens, transcriptional profiling of bacteria recovered from tuberculous lesions, and studies with various inhibitors and knock-out mutants of M. tuberculosis all indicate that persistent bacilli encounter low oxygen tension, which may contribute in part to their non-replicating state. They are also subject to nutrient deprivation, which obliges them to divert carbon from host-derived fatty acids into gluconeogenesis (4–14). Targeting pathways involved in the adaptation of M. tuberculosis to latent infection may thus represent a promising approach to the eradication of persistent bacilli.
The search for drug therapies against persistent TB has led to our interest in class II fructose-1,6-bisphosphate aldolase (FBA-tb), a key enzyme of glycolysis/gluconeogenesis more abundantly produced and/or secreted by M. tuberculosis grown under low oxygen tension (15–17). FBA-tb is a homotetrameric enzyme dependent on zinc for activity (18), and the crystal structures of three different protein·substrate/product complexes were recently solved (19). Fructose-bisphosphate aldolases (FBAs) (EC 4.1.2.13) are enzymes involved in glycolysis where they reversibly catalyze cleavage of fructose 1,6-bisphosphate (FBP) to yield dihydroxyacetone phosphate and glyceraldehyde 3-phosphate. They are also active in gluconeogenesis and the Calvin cycle where they catalyze the reverse condensation. FBAs are divided into two classes depending on their reaction mechanism. Class I aldolases are present in higher organisms (animals and plants), green algae, and a few prokaryotes. They form a Schiff base intermediate between the carbonyl substrate (FBP or dihydroxyacetone phosphate) and a lysine residue of the active site. In contrast, class II aldolases require a divalent metal ion (usually zinc or cobalt) to polarize the carbonyl group of the substrate (FBP or dihydroxyacetone phosphate) and to stabilize the carbanion intermediate during catalysis. They are mainly found in lower organisms such as bacteria (eubacteria and archeabacteria), fungi, protozoa, and some green algae. Class I and class II enzymes show almost no sequence similarities and are assumed to have arisen from separate evolutionary origins (20). The absence of class II FBAs from mammalian cells and the specificity of their structure and catalytic mechanism thus offer the opportunity to design drugs that selectively inhibit microbial class II enzymes without affecting the gluconeogenetic and glycolytic pathways of the host. This study was undertaken with the goals of assessing the therapeutic potential of FBA-tb and providing biochemical and structural bases for future drug design.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Escherichia coli DH5α, the strain used for cloning experiments, was propagated in LB broth (pH 7.5) (BD Biosciences). M. tuberculosis H37Ra (ATCC 25177) was grown in Middlebrook 7H9 broth (Difco) supplemented with ADC and 0.05% Tween 80 or on solid Middlebrook 7H11 agar supplemented with OADC. Kanamycin, streptomycin, and hygromycin were added to final concentrations of 20, 20, and 50 μg ml−1, respectively. When required, 2% sucrose and anhydrotetracycline were added to the media.
For growth on defined carbon sources, 7H9-tyloxapol (0.025%) broth and 7H11 agar containing 0.5% BSA and 0.085% NaCl were used and supplemented with ADC, 40 mm succinate, or 40 mm acetate. Low oxygen tension experiments were performed according to Wayne and Hayes (21) with appropriate controls to check for oxygen depletion except that Sauton medium (without detergent) instead of Dubos medium was used to allow FBA-tb activity to be measured in the culture filtrate (i.e. in a medium devoid of albumin).
Allelic Replacement at fba-tb Locus of M. tuberculosis
The Ts-sacB method (22) was used to achieve allelic replacement at the fba-tb (MRA_0372) locus of M. tuberculosis H37Ra (100% identical in sequence to Rv0363c of M. tuberculosis H37Rv). The M. tuberculosis gene and flanking regions were PCR-amplified from M. tuberculosis H37Rv genomic DNA, and a disrupted allele, fba-tb::kan, was obtained by inserting the kanamycin resistance cassette from pUC4K (Amersham Biosciences) into the SalI restriction site of fba-tb. fba-tb::kan was then cloned into the XbaI site of pPR27-xylE (22) to obtain pPR27fba-tbKX, the construct used for allelic replacement. pNIP40b-fba-tb, one of the plasmids used for complementation, was obtained by first cloning the PCR-amplified coding sequence of the fba-tb gene into the pVV16 expression plasmid (23) and then transferring the fba-tb gene under control of the phsp60 promoter into the XbaI restriction site of the integrative plasmid pNIP40b (24). pGMCS-10M-P1-fba, the integrative rescue plasmid used in gene silencing experiments, was constructed by cloning the PCR-amplified fba-tb gene into pDO23A (25) by BP recombination (Invitrogen), resulting in pEN23A-fba. After confirmation that fba-tb was cloned without mutations, an LR recombination (Invitrogen) was performed with pEN41A-T10M, pEN12A-P1, and pDE43-MCS (25) to generate pGMCS-10M-P1-fba. pEN41A-T10M and pEN12A-P1 are Gateway entry plasmids containing tetR10 (26) and Pmyc1tetO (27), respectively. pDE43-MCS is a Gateway destination plasmid that includes the integrase and attP site of the mycobacteriophage L5 and a streptomycin resistance gene. BP and LR ClonaseTM mixtures were purchased from Invitrogen and used according to the manufacturer's instructions. Allelic replacement at the fba-tb locus was confirmed by PCR using primers located outside the rescue copy of the gene carried by pNIP40b-fba-tb and pGMCS-10M-P1-fba and outside the disrupted copy of fba-tb used in the allelic exchange experiment. Primer sequences are available upon request.
Fba-tb Expression and Purification
fba-tb was PCR-amplified from M. tuberculosis H37Rv genomic DNA using primers fba_NdeI_fwd (5′-ggaattccatatgcctatcgcaacgcccgagg-3′) and fba_BamHI_rev (5′-cgcggatcctcagtgggttagggactttccg-3′), and the PCR product was ligated into the NdeI and BamHI restriction sites of the expression vector pET29a (Novagen). FBA-tb production in E. coli BL21(DE3) (Novagen) was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside (MP Biomedicals) and allowing the cells to grow for 16 h at 18 °C. Cells were harvested and resuspended in 50 mm Tris-HCl, pH 8.0 (solution A) containing protease inhibitors (Complete EDTA-free, Roche Applied Science) and disrupted by sonication, and nucleic acids were degraded by the addition of Benzonase (Novagen). The soluble fraction was applied to a Q Sepharose column (GE Healthcare) equilibrated with solution A, and the column was washed with solution A until no absorbance at 280 nm was detected. Elution was performed with a linear gradient of 0–0.5 m NaCl in solution A at 5 ml min−1. Fractions containing FBA-tb were pooled, and a solution of saturated ammonium sulfate was added dropwise until a final concentration of 30% was reached. The solution was incubated at 4 °C for 1 h, and the soluble fraction was loaded onto a phenyl-Sepharose column (GE Healthcare) equilibrated with solution A with 1.5 m ammonium sulfate. The column was washed with solution A containing 1.5 m ammonium sulfate until no absorbance at 280 nm was detected. Elution was performed by a linear gradient of 1.5–0 m ammonium sulfate in solution A. Fractions containing FBA-tb were pooled and concentrated over a Vivaspin 20 membrane (10,000-Da cutoff; Sartorius). The concentrate was then dialyzed against solution A and loaded at 1 ml min−1 onto a Superdex 200 column (GE Healthcare) equilibrated with solution A containing 150 mm NaCl. Fractions were collected, pooled, concentrated to 50 mg ml−1, and stored at −80 °C.
Polyclonal Antibody Production, Immunoblotting, and Flow Cytometry
Polyclonal antibodies against purified FBA-tb were produced in an outbred New Zealand White rabbit (Western Oregon Rabbit Co., Philomath, OR). Subcellular fractions from M. tuberculosis H37Rv grown in glycerol-alanine-salt medium and Mycobacterium leprae purified from armadillo tissues were obtained from the TB Vaccine Testing and Research Materials Contract (NIAID, National Institutes of Health N01-AI-40091) and the Leprosy Research Support Contract (NIAID, National Institutes of Health N01-AI-25469), respectively. The polyclonal anti-FBA-tb antibody and the secondary goat anti-rabbit antibody conjugated to alkaline phosphatase were used at a concentration of 1:3,500 and 1:5,000, respectively, in immunoblotting experiments carried out with the M. tuberculosis and M. leprae extracts. The polyclonal rabbit anti-PimA antibody was used at a concentration of 1:3,000.
For the cell surface localization of FBA-tb by flow cytometry, polyclonal rabbit IgG was purified using the Dynabeads® Protein A and DynaMagTM-2 kits (Invitrogen) according to the manufacturer's recommendations. M. tuberculosis H37Ra bacilli grown in Sauton medium were dispersed by gentle vortexing with 5-mm glass beads for 2 min. Bacteria were resuspended in 1 ml of PBS buffer containing 0.1% BSA, 2% fetal bovine serum (FBS), and 5 μg of anti-FBA-tb IgG; incubated overnight at 4 °C; washed three times with PBS; and incubated for another 2 h in the dark with goat anti-rabbit IgG-Alexa Fluor 647 conjugate (Invitrogen) diluted 1:2,000 in PBS, 0.1% BSA, and 2% FCS. Bacteria were then washed twice in PBS and finally resuspended in 0.4 ml of PBS containing 4% formaldehyde. Untreated bacteria and bacteria incubated with non-immunized (control) rabbit serum, the primary anti-FBA-tb antibody alone, or the secondary antibody alone were used as controls. Samples were analyzed for fluorescence using a CyAn flow cytometer. Under the experimental conditions used, only the FBA-tb protein accessible to the antibodies at the surface of the bacilli but not the intracellular protein should be stained. Cells were detected using forward and log side scatter dot plots, and a gating region was set to exclude cell debris and bacterial aggregates; 20,000 bacterial cells were analyzed.
Auramine-Rhodamine Staining
Lung sections from IFN-γ-KO mice and guinea pigs infected with M. tuberculosis Erdman and M. tuberculosis H37Rv for 18 and 28 days, respectively, were stained with TB fluorescent stain kit T (BD Biosciences) according to the manufacturer's instructions except that counterstaining was performed with hematoxylin QS (Vector Laboratories, Inc., Burlingame, CA) for ∼5 s. The slides were washed with double distilled H2O and mounted using ProLong® Gold antifade reagent (Invitrogen) (28).
Immunohistochemistry
Formalin-fixed, paraffin-embedded lung tissue sections were digested with 30 units ml−1 achromopeptidase and 1 mg ml−1 lysozyme in 10 mm Tris, pH 8.0 for 40 min at 37 °C. Endogenous peroxidases were blocked using a 3% H2O2 solution for 40 min at 25 °C. Slides were then placed in a RetrieverTM 2100 pressure cooker (Pickcell Laboratories, Amsterdam, The Netherlands) at 121 °C for 15 min and then incubated with 1% goat serum for 30 min. The slides were incubated with a 1:500 dilution of the serum containing the rabbit anti-FBA-tb antibody and a mouse anti-GroES antibody (Antibody SA-12, Colorado State University TB Vaccine Testing and Research Materials Contract) along with 1% goat serum for 16 h at 4 °C. The slides were then incubated with a 1:100 dilution of horseradish peroxidase-labeled goat anti-rabbit antibody and also an Alexa Fluor 488-labeled goat anti-rabbit IgG (Invitrogen) in 1% goat serum for 40 min at 25 °C. The signal was amplified using a 1:400 dilution of tyramide-Alexa Fluor 568 in amplification buffer (Invitrogen). Nuclei were stained with DAPI (Invitrogen) at 200 ng ml−1 for 10 min and mounted with ProLong Gold antifade mounting medium (Invitrogen).
Human Plasminogen Binding and Activation Assays
The binding of plasminogen (Plg) to FBA-tb, BSA, and fibrinogen (Sigma) was analyzed by ELISA in Immulon 2HB plates (Thermo Scientific). Wells coated with 2 μg of FBA-tb or control proteins were incubated with different concentrations of Plg in PBS-Tween 20 with 1% BSA for 1 h at 37 °C. Mouse anti-Plg (Pierce) was used as the first antibody, and goat anti-mouse IgG HRP-conjugated antibodies (Thermo Scientific) were used as the secondary antibodies. Peroxidase activity was revealed with 3,3′,5,5′-tetramethylbenzidine substrate solution (Thermo Scientific), and absorbance at 450 nm was measured. A chromogenic assay using Chromozym (Roche Applied Science) was used to monitor the activation of Plg (1 μg) into plasmin at A405 nm in the presence of FBA-tb (2 μg), 0.0875 units (24 ng) of streptokinase (Sigma), and other control proteins (2 μg) with or without added human tissue Plg activator (tPA) (0.025 μg) (Calbiochem). Assays were performed either in solution or in the presence of a fibrin matrix (prepared in the wells from fibrinogen with human thrombin (Sigma)), and assay conditions were as described earlier (29). In studies aimed at analyzing the regulation of plasmin activity by α2-antiplasmin (Innovative Research), FBA-tb (2 μg) and human Plg (1 μg) were first preincubated for 1 h at 37 °C to allow FBA-tb·Plg complexes to form prior to the addition of tPA (0.025 μg) and finally α2-antiplasmin (2 μg) at which point protease activity was monitored over time. All assays were run in duplicates or triplicates, and the results of representative experiments are shown.
FBA-tb Enzymatic Assays
The coupled assay used to measure the enzymatic activity of FBA-tb is based on that of Richards and Rutter (30). The standard reaction mixture contained 50 mm HEPES, pH 7.5, 80 μm NADH (Fluka), 5 units ml−1 triose-phosphate isomerase (Sigma), 2 units ml−1 α-glycerophosphate dehydrogenase (Sigma), 20 μm FBP (Fluka), and 0.2–1.5 μg ml−1 purified enzyme in a total volume of l ml. 15 μg of M. tuberculosis H37Ra cellular extracts or capsular or culture filtrate proteins replaced the purified protein in the assays conducted on whole mycobacterial cultures. Capsular and culture filtrate proteins were prepared as described (31). Cellular extracts were prepared by breaking M. tuberculosis H37Ra cells using a FastPrep® instrument (MP Biomedicals) and centrifuging the lysates at 4,000 rpm for 20 min at 4 °C to remove the unbroken cells. A correction was made for the NADH oxidase and putative class I aldolase activities of the crude extracts by running assays alongside in which FBP was omitted or EDTA (1 mm) was added. All experiments were repeated at least twice, and activity was measured in duplicate on each sample.
Crystallization and Data Collection
FBA-tb crystals were grown by vapor diffusion from a 1:1 mixture of protein solution (10 mg ml−1 initial protein concentration made up in 25 mm Tris HCl, pH 7.0) and precipitant buffer (20% PEG 8000, 1.8 m Li2SO4, and 50 mm Tris/HOAc, pH 7.5), and 4-μl drops were equilibrated at 23 °C against 1-ml reservoirs of precipitant solution. Crystals grew in 2 weeks. Enzymatic assays revealed complete inhibition of FBA-tb under conditions of the crystallization buffer (1.8 m Li2SO4 and 50 mm Tris/HOAc, pH 7.5). Residual activity at saturating substrate concentration was <1% of maximal activity. Inhibition was corroborated by monitoring hydrazone formation at 240 nm in a chemical assay based on reaction of hydrazine with the nascent aldehyde (32).
FBA-tb crystals were soaked in TD3 inhibitor buffer (mother liquor plus 10 mm TD3 compound) for 10 min. The structure of TD3 is identical to that of compound 1 shown in Ref. 33. Prior to data collection, crystals were cryoprotected by transfer through a cryobuffer solution (compound TD3 plus 10% glycerol) and immediately flash frozen in a stream of gaseous N2 cooled to 100 K. Diffraction data were collected from single crystals at beamline X29 of the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY) with an Area Detector Systems Corp. Quantum 315r detector to 1.9-Å resolution. All data sets were processed with XDS (34) and SCALA from the CCP4 suite (35), and the results are summarized in Table 2. A control data set without TD3 was also collected to 2.35-Å resolution under identical experimental conditions.
TABLE 2.
Data collection and refinement statistics
All values in parentheses are given for the highest resolution shell. r.m.s., root mean square.
| FBA-tb in complex with compound TD3 | Native FBA-tb | |
|---|---|---|
| Data collection | ||
| Resolution (Å) | 47.9–1.9 (2.0–1.9) | 46.5–2.35 (2.5–2.35) |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Unique reflections/multiplicity | 114,281/3.3 (16,596/3.4) | 61,704/3.5 (8,877/3.4) |
| Completeness (%) | 99.5 (99.7) | 99.3 (98.8) |
| Average I/σ(I) | 10 (2.1) | 3.8 (1.9) |
| Rpima | 0.08 (0.50) | 0.11 (0.33) |
| Space group | C2 | C2 |
| Unit cell parameters | ||
| a, b, c (Å) | 335.4, 43.0, 102.6 | 335.9, 43.2, 103.0 |
| α, β, γ (°) | 90, 99.4, 90 | 90, 99.6, 90 |
| Refinement | ||
| Number of reflection used | 107,083 | 56,699 |
| Number of atoms | ||
| Protein | 9,848 | 9,384 |
| Water | 866 | 804 |
| Hetero | 48 | 42 |
| Rcryst (%)b | 16.9 | 21.1 |
| Rfree (%)c | 21.4 | 26.1 |
| Root mean square deviation | ||
| Bond length (Å) | 0.008 | 0.006 |
| Bond angle (°) | 1.02 | 0.98 |
| Average B-factor/r.m.s. (Å2) | 17.7/4.9 | 23.6/5.6 |
| Ramachandran analysisd (%) | ||
| Favored regions | 98.8 | 98.0 |
| Allowed regions | 1.2 | 2.0 |
| Luzzati error in coordinates (Å) | 0.23 | 0.32 |
a Rpim = Σhkl √(1/N − 1)Σi|Ii(hkl) − ¯Ii(hkl)|/Σhkl ΣiIi(hkl) with i running over the number of independent observations of reflection hkl.
b Rcryst = Σhkl‖Fo(hkl)| − |Fc(hkl)‖/Σhkl|Fo(hkl)|.
c Rfree = ΣhklT‖Fo(hkl) − |Fc(hkl)‖/ΣhklT|Fo(hkl)| where T is a test data set randomly selected from the observed reflections prior to refinement. The test data set was not used throughout refinement and contained 7.5% of the total unique reflections.
d Analyzed by MolProbity.
Structure Solution and Refinement
FBA-tb structures with and without TD3 were solved by molecular replacement with the program AutoMR (Phenix suite) (36) using native FBA-tb in complex with FBP (19) (Protein Data Bank code 3elf) as the search model. The structure in complex with compound TD3 belongs to space group C2 (dimer of homodimers in the asymmetric unit). The best solution was used as the starting point for refinement of the liganded structure and was solved by iterative rounds of refinement (simulated annealing and minimization) with Phenix and model building using Coot (37). The MolProbity server (38) and the Coot validating tools were used to optimize the structures during the refinement. Water molecules were automatically added by Phenix in the initial rounds and manually near the end of the refinement. Loop regions (residues 165–180) in each subunit were associated with regions of weak electron density. In the final round of refinement, the occupancy of TD3 was adjusted to take into account competition by sulfate ions for the TD3 phosphate oxyanion binding sites. Refinement of the unbound native structure used sulfate ions positioned coincident with phosphate oxyanion binding sites of TD3. The loop region corresponding to residues 211–221 and adjacent to residues 165–180 was associated with weak electron density in all subunits of the native FBA-tb and was not modeled into the structure.
Ligand modeling was based on interpretation of electron density shapes of 2Fo − Fc and Fo − Fc annealed omit maps and using the Phenix.elbow command for topology and parameter generation. Binding by compound TD3 was readily discernable and associated with clearly defined electron densities in the active site in two subunits of the same homodimer. In the remaining homodimer, active site electron densities showed no evidence for detectable binding by TD3. Occupancies of the catalytic zinc ion closely matched that of TD3 and were set equal to TD3 occupancy in the final round of refinement. Difference electron density (Fo − Fc) annealed omit maps calculated in the final round of refinement confirmed identical binding of TD3 in protomers. Final model statistics calculated with Phenix, MolProbity, and SFCHECK (39) are shown in Table 2. The coordinates and structure factors of native FBA-tb (Protein Data Bank code 4a21) and in complex with TD3 (Protein Data Bank code 4a22) have been deposited with the Protein Data Bank. The final structure of the native FBA-tb and FBA-tb in complex with compound TD3 was refined to Rcryst (Rfree) values of 0.211 (0.261) and 0.191 (0.229). The corresponding Luzzati atomic coordinate errors were estimated at 0.32 and 0.23 Å, respectively. Ramachandran analysis with the MolProbity server placed at least 98.0% of non-glycine and non-proline residues of the two structures in the most favorable region with the remainder found in allowed regions, attesting to good model geometry in the structures.
Structure Comparisons
Superimpositions were performed with the super command in PyMOL with use of Cα atom coordinates of identical regions of amino acid sequences or by LSQ fit in Coot. Root mean square deviations are reported based on superimposition of equivalent Cα atoms in FBA-tb structures and for positional comparisons of compounds based on superimposition of equivalent non-hydrogen atoms.
RESULTS AND DISCUSSION
M. tuberculosis Aldolase Is an Essential Glycolytic and Gluconeogenetic Enzyme of M. tuberculosis
The requirement of FBA-tb for M. tuberculosis growth was examined genetically by knocking out the chromosomal copy of fba-tb in the presence or absence of a rescue copy of this gene carried by an integrative plasmid. Attempts to inactivate fba-tb in M. tuberculosis H37Ra and M. tuberculosis H37Rv in the absence of a rescue copy of this gene yielded no candidate mutant at the last selection step of the procedure, suggesting that, unlike the situation in Neisseria meningitidis (40) but similar to those in E. coli, Streptomyces, and Candida albicans (41–43), the class II FBA gene of M. tuberculosis is required for growth even under optimal laboratory growth conditions where both glucose and oleic acid are present in the culture medium. As proposed in the case of E. coli (44), it is likely that this requirement is related to the role of FBA-tb in preventing the toxic effect of FBP accumulation in the cells. FBP is indeed an allosteric effector of several enzymes in central carbon metabolism (pyruvate kinase, phosphoenolpyruvate carboxylase, ADP-glucose pyrophosphorylase, glucose-6-phosphate dehydrogenase, and 6-phosphogluconate dehydrogenase) whose deregulation is likely to affect cell growth (44–46). Inhibition of glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase by high levels of FBP in the absence of FBA-tb for instance would prevent flux through the oxidative branch of the pentose phosphate pathway (46) and inhibit reductive biosyntheses as well as induce oxidative stress.
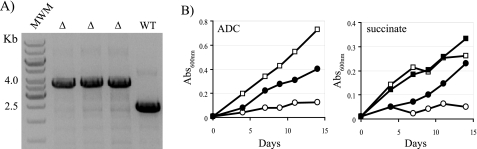
Allelic exchange experiments were then conducted in a merodiploid strain, H37Ra/pNIP40b-fba-tb, carrying an integrated rescue copy of fba-tb. The choice of using M. tuberculosis H37Ra is justified by the fact that the H37Ra strain is considered a class II organism yet requires FBA-tb for growth like its virulent counterpart, M. tuberculosis H37Rv, with which it shares the exact same glycolytic/gluconeogenetic and pentose phosphate pathway enzymes (100% identical in their primary sequence) (47). Mutants in which the wild-type chromosomal copy of the gene was inactivated were easily isolated using this merodiploid strain, confirming the requirement of fba-tb for growth of M. tuberculosis on 7H11-OADC-sucrose plates (Fig. 1A).
FIGURE 1.
fba-tb is required for M. tuberculosis growth. A, evidence for allelic replacement at the fba-tb locus of M. tuberculosis H37Ra in the presence of a rescue copy of the gene integrated in the chromosome. Allelic exchange mutants were rescued with fba-tb expressed from the hsp60 promoter in pNIP40b-fba-tb. Allelic replacement at the fba-tb chromosomal locus was confirmed by PCR (see “Experimental Procedures”). WT, wild-type M. tuberculosis H37Ra. The wild-type 2.4-kb amplification signal is replaced by a 3.6-kb fragment in the allelic exchange mutants (△) due to the insertion of a 1.2-kb kanamycin resistance cassette. MWM, molecular weight marker. B, growth of M. tuberculosis H37Ra wild-type (squares) and M. tuberculosis H37RaΔfba/pGMCS-10M-P1-fba (circles) in the presence of glucose (under the form of ADC supplement) or succinate as carbon sources. Bacteria were inoculated at an initial A600 nm of 0.01 and cultured at 37 °C with constant stirring in 7H9-tyloxapol broth with 40 mm succinate or ADC supplement in the presence (filled symbols) or absence (open symbols) of 100 ng ml−1 anhydrotetracycline. Abs, absorbance.
To assess the requirement of FBA-tb under gluconeogenetic conditions, which might be experienced by tubercle bacilli during the persistence stage of the infection (14), a conditional M. tuberculosis H37Ra mutant was constructed in which the chromosomal copy of fba-tb was knocked out and replaced by a wild-type copy of this gene expressed in cis from a tetracycline-inducible promoter. The successful construction of the conditional mutant, hereafter referred to as H37RaΔfba/pGMCS-10M-P1-fba, was confirmed by PCR. Gene silencing experiments in 7H9 medium containing various carbon sources mapping upstream or downstream of FBP in the glycolytic pathway revealed that the growth of H37RaΔfba/pGMCS-10M-P1-fba in glucose- and succinate-containing media was strictly dependent upon the induction of fba-tb expression with anhydrotetracycline (Fig. 1B). FBA-tb is thus an essential enzyme under both glycolytic and gluconeogenetic conditions.
M. tuberculosis, like E. coli, has been reported to display both class I and class II FBA activities in vitro (15, 16). In the presence of a class I enzyme with a redundant role in glycolysis/gluconeogenesis, the reason for the essentiality of the class II aldolase of M. tuberculosis is thus unclear. The fact that we were not able to detect any significant class I aldolase activity under any of the growth conditions tested in this study (see further section) raised doubts as to its existence. Also, failure to categorically identify a class I aldolase gene in M. tuberculosis despite the availability of a growing number of genome sequences of this bacterium further supported its absence from M. tuberculosis. Indeed, BLAST searches for orthologs of class I FBA genes from prokaryotic (E. coli and Thermococcus gammatolerans) and eukaryotic (human FBA isozymes A, B, and C) sources in the genome of M. tuberculosis H37Rv/H37Ra all yielded negative results. A class I enzyme whose N-terminal sequence matches that of protein MSMEG_3507 from Mycobacterium smegmatis mc2155 was purified to near homogeneity from a M. smegmatis isolate (48) and found to share extensive sequence similarities with other prokaryotic class I enzymes. However, an analysis of the distribution of this gene within the Mycobacterium genus failed to identify any orthologs beyond fast growing non-tuberculous Mycobacterium species and the Mycobacterium avium complex.
Production of Active Form of FBA-tb and Evidence for Its Surface Exposure in M. tuberculosis
With the aim of studying the expression and subcellular localization of FBA-tb in M. tuberculosis grown under different in vitro and in vivo conditions, the FBA-tb protein was produced and purified from E. coli, and polyclonal anti-FBA-tb antibodies were raised in rabbit.
FBA-tb is a homotetrameric 344-amino acid protein constituting a 144-kDa molecule (18). An untagged form of FBA-tb was produced because N-terminal tags have been found to affect dramatically the solubility and activity of FBA-tb produced in E. coli probably by disrupting its quaternary structure (18, 49). Moreover, we found a C-terminal hexahistidine-tagged form of FBA-tb to be inactive when expressed in M. smegmatis.6 The untagged protein was produced as a soluble form in E. coli BL21(DE3) and purified to near homogeneity (see supplemental Fig. S1A). The purified enzyme showed activity in the coupled aldolase assay of Richards and Rutter (30), indicating that the enzyme was properly folded and its active site was competent (supplemental Fig. S1B).
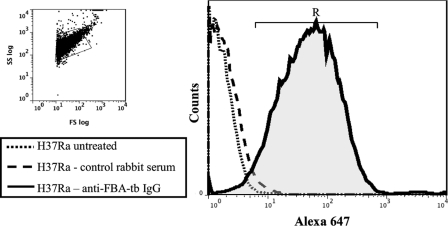
Polyclonal anti-FBA-tb antibodies were used to assess the expression and subcellular localization of this protein in M. tuberculosis. Immunoblots confirmed the specificity of the antibodies for FBA-tb and revealed the presence of the protein in the culture filtrates, cell wall, membrane, and cytosol of M. tuberculosis H37Rv (supplemental Fig. S2). FBA-tb thus appears to translocate across the different compartments of the bacterial cell. The presence of antibody-accessible FBA-tb at the cell surface of the bacterium was further confirmed by direct enzyme activity measurements and by probing intact M. tuberculosis H37Ra bacilli with anti-FBA-tb antibodies followed by flow cytometry analysis (Table 1 and Fig. 2). Untreated bacteria and bacteria treated with control rabbit serum followed by anti-rabbit IgG-Alexa Fluor 647 produced weak fluorescence signals (1.4 and 18.5 mean fluorescence intensities, respectively), whereas cells treated with anti-FBA-tb followed by anti-rabbit IgG-Alexa Fluor 647 conjugate demonstrated a clear shift in fluorescence signal (104 mean fluorescence intensity), confirming the cell surface localization of FBA-tb (Fig. 2). 95% of the M. tuberculosis H37Ra bacilli grown as an undisturbed surface pellicle in Sauton medium were found in the R region, suggesting that the majority of the bacterial population had FBA-tb present on the cell surface. The finding of FBA-tb at the cell surface and in culture filtrates, although surprising in view of the apparent lack of secretion signals in the protein and its glycolytic/gluconeogenetic function, is consistent with the reported surface exposure of class II FBP aldolases from several other bacterial and fungal pathogens (40, 50–54) and earlier proteomics observations on M. tuberculosis (17, 55, 56).
TABLE 1.
Total and subcellular distribution of class II fructose-1,6-bisphosphate aldolase activity of M. tuberculosis bacilli grown under various axenic conditions
NA, not applicable; CF, culture filtrate. The time points (T1–T4) at which the Sauton-tyloxapol cultures were collected are shown in supplemental Fig. S3.
| Growth condition | Total FBA-tb activity | Subcellular fraction | Distribution of the FBA-tb activity in the total culture | Specific FBA-tb activity in each subcellular fraction |
|---|---|---|---|---|
| pmol FBP/min/μg protein | % | pmol FBP/min/μg protein | ||
| 7H11-OADC | 13 | Cells | 100 | NA |
| 7H11-ADC | 17 | Cells | 100 | NA |
| 7H11-acetate | 15 | Cells | 100 | NA |
| Sauton-O2 depletion | 11.3 ± 1.8 | Cells | 95 | 11 ± 1.6 |
| CF | 5 | 16 ± 5.4 | ||
| Sauton-oxygenated | 10.7 ± 0.7 | Cells | 96 | 11 ± 0.6 |
| CF | 4 | 4 ± 1.8 | ||
| Sauton-tyloxapol | ||||
| Early log phase (T1) | 12.9 ± 2.0 | Cells | 99 | 13 ± 2.1 |
| CF | 1 | 4 ± 0 | ||
| Midlog phase (T2) | 13.9 ± 3.9 | Cells | 99 | 14 ± 4.0 |
| CF | 1 | 4 ± 0.12 | ||
| Late log phase (T3) | 13.9 ± 0.3 | Cells | 98 | 14 ± 0.3 |
| CF | 2 | 5 ± 1.7 | ||
| Stationary phase (T4) | 11.9 ± 1.4 | Cells | 96 | 12 ± 1.4 |
| CF | 4 | 8 ± 0.6 | ||
| Sauton-surface pellicle | 19.4 ± 0.3 | Cells | 92 | 19 ± 0 |
| Capsule | 7 | 26 ± 4.3 | ||
| CF | 1 | 13 ± 0.12 | ||
FIGURE 2.
Flow cytometry analysis of axenically grown M. tuberculosis bacilli for FBA-tb surface exposure. M. tuberculosis H37Ra bacilli grown as a surface pellicle in Sauton medium were gently dispersed with glass beads and submitted to flow cytometry analysis for FBA-tb surface localization as described under “Experimental Procedures.” Bacteria were either untreated, treated with control rabbit serum followed by anti-rabbit IgG-Alexa Fluor 647, or treated with anti-FBA-tb antibodies followed by anti-rabbit IgG-Alexa Fluor 647. The histogram area in “R” represents the population of fluorescently labeled bacilli. SS, side scatter; FS, forward scatter.
FBA-tb Is Expressed by Replicating and Non-replicating Bacilli during Adaptation to Stationary Phase, Low Oxygen Tension, and Changes in Carbon Sources
Because of its involvement in gluconeogenesis and reported induction and/or oversecretion under low oxygen tension (15–17), we next sought to assess the potential regulation of fba-tb in M. tuberculosis grown under various axenic conditions including some thought to mimic the physical environment encountered by the bacilli during persistence in vivo. Enzyme assays were performed on cellular extracts, surface extracts, and culture filtrates of M. tuberculosis grown in the presence of glucose or fatty acids as carbon sources, under high or low oxygen tension, and at various stages of growth (Table 1). Culture filtrates and capsular fractions were checked for cell lysis by immunoblot with antibodies directed against PimA (a cytosolic GDP-Man-utilizing mannosyltransferase) (23) (supplemental Fig. S3).
Results confirmed the production of an active FBA-tb enzyme under all conditions tested including low oxygen tension (21). Interestingly, no significant up- or down-regulation of the overall class II aldolase activity of the cultures was found whatever the growth conditions tested (Table 1). Although surprising in light of what had been reported earlier for other glyoxylate cycle and gluconeogenetic genes (6, 12, 13, 57), our results are consistent with those of some 30 transcriptomics studies performed on M. tuberculosis bacilli grown under various stress conditions (e.g. inside macrophages and in the presence of SDS, drugs, NO, low oxygen tension, low iron, low nutrient, and various mutant backgrounds). Overall, the percentage of class II aldolase activity found in the culture filtrates never exceeded 4% of the total FBA-tb activity of the culture, and that found in the capsular surface-exposed material of surface pellicle-grown bacteria represented less than 7% of the total activity.
FBA-tb Is Expressed during Host Infection
To assess fba-tb expression during host infection, immunohistochemistry experiments were undertaken using our polyclonal anti-FBA-tb antibody and M. tuberculosis-infected lung tissues from IFN-γ-KO mice and guinea pigs. The location of auramine-rhodamine-positive bacilli in the lungs of mice and guinea pigs over time is detailed in Ref. 28. Representative micrographs of these experiments are presented in Fig. 3. Results unambiguously pointed to the strong expression of FBA-tb by M. tuberculosis bacilli inside the granulomatous lung tissues of IFN-γ-KO mice (Fig. 3, A and B). Expression of FBA-tb was also clearly detectable in the necrotic core of primary lung granulomas from infected guinea pigs (Fig. 3, C–E). Although the precise physiological state under which the bacilli depicted in Fig. 3, C–E, exist cannot be ascertained, it is noteworthy that persistent bacilli have been shown to reside within such lesions (1, 58). As expected, few bacilli were detected by acid-fast auramine-rhodamine staining in necrotic cores of guinea pigs (Fig. 3C). However, within these lesions, FBA-tb co-localized with the GroES protein (Rv3418c in M. tuberculosis H37Rv), confirming the association of the protein with M. tuberculosis bacilli (Fig. 3, D and E). Moreover, no signal was observed for the control experiments in which lung sections were treated with the secondary antibody in the absence of the primary antibody. Interestingly, FBA from M. leprae (ML0286c), which shares 87% amino acid identity with FBA-tb, was also detected in subcellular fractions of the leprosy bacillus purified from chronically infected armadillo tissues (supplemental Fig. S2). Thus, consistent with its apparent constitutive expression in both replicating and non-replicating bacilli in vitro (see previous section) and requirement for growth under glycolytic and gluconeogenetic conditions, FBA-tb is also actively produced during host infection.
FIGURE 3.
Production of FBA-tb by M. tuberculosis bacilli in vivo. A and B, lung granulomas from M. tuberculosis Erdman-infected IFN-γ-KO mice (×40). A, fluorescent auramine-rhodamine stain of M. tuberculosis bacilli. B, FBA-tb (red) was detected by immunohistochemistry using polyclonal anti-FBA-tb antibodies and tyramide amplification signal. C–E, lung granulomas from M. tuberculosis H37Rv-infected guinea pigs (×20). C, fluorescent auramine-rhodamine stain of M. tuberculosis bacilli. Fluorescent (D) and confocal (E) micrographs of co-localizing FBA-tb (red) and GroES (green) proteins that were detected by immunohistochemistry within the necrotic core of guinea pig lung tissues are shown. White arrows denote co-localization.
FBA-tb Binds to Human Plasminogen
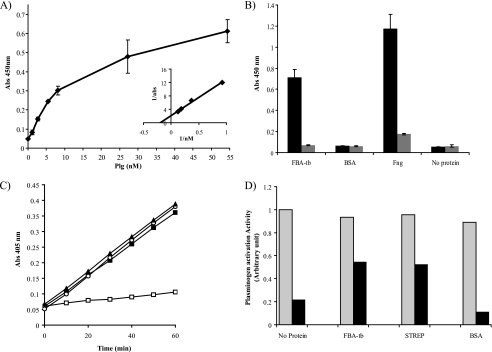
Proteomics analysis had identified FBA-tb among M. tuberculosis H37Rv candidate culture filtrate proteins with human Plg binding capabilities (29). ELISAs with the recombinant native FBA-tb protein confirmed that the protein bound human Plg with an apparent Kd of 6.7 ± 3 nm (Fig. 4A). Binding was inhibited by the lysine analog ϵ-aminocaproic acid (Fig. 4B) but was not affected by the addition of a large excess of a competitive inhibitor of FBA-tb (TD3; IC50 ∼ 4 nm; see Ref. 33 and below) to the reaction mixture (Kd of 6.8 nm in the presence of 0.7 μm TD3). Thus, binding is predominantly mediated by the lysine binding sites of Plg and lysine residues within the FBA-tb protein and is not dependent on the catalytic activity of FBA-tb. In a solution assay (data not shown) or in the presence of fibrin matrices (Fig. 4C), FBA-tb-bound Plg was activated to plasmin by human tPA, but FBA-tb by itself did not activate Plg. In contrast, streptokinase, a bacterial protein known to activate Plg, displayed potent Plg activating activity in the absence of tPA in the reaction mixture (Fig. 4C). Importantly, the ability of FBA-tb-bound plasmin to respond to regulation by the host serpin α2-antiplasmin in the fibrin matrix assay was significantly decreased compared with plasmin incubated either with no additional protein or in the presence of a control protein that does not bind plasmin(ogen) such as BSA (Fig. 4, B–D). Whereas α2-antiplasmin inhibited plasmin activity by 78–88% in the fibrin matrix wells containing Plg + tPA or Plg + tPA + BSA, the percentage of inhibition in the presence of Plg + tPA + FBA-tb was only 42%. Under the conditions of this assay, the decreased response of FBA-tb-bound plasmin to α2-antiplasmin regulation was similar to that observed for streptokinase·plasmin complexes (45.4% inhibition of plasmin activity) (Fig. 4D). Our findings, which support an involvement of the Lys binding sites of the kringle domains of plasmin(ogen) in the attachment of this host molecule to FBA-tb (Fig. 4B), and the fact that the same domains are known to mediate α2-antiplasmin/plasmin interactions (59) suggest that the inhibition of plasmin regulation by α2-antiplasmin in the presence of FBA-tb results from a competitive mechanism between the two proteins for the same binding sites on plasmin.
FIGURE 4.
Human Plg binding properties of FBA-tb: impact on Plg activation and regulation of plasmin by α2-antiplasmin. A, binding of human Plg to FBA-tb measured by ELISA over a range of Plg concentrations (see “Experimental Procedures”). The inset shows a double reciprocal plot of 1/bound (1/A450 nm) against 1/[Plg] used to determine the Kd. Each point was measured in triplicate, and the mean values and S.D. (error bars) are shown. B, the binding of human Plg (5 μg) to FBA-tb, fibrinogen (Fng) (positive control), BSA (negative control), or no protein was determined by ELISA in the presence (gray bars) and absence (black bars) of ϵ-aminocaproic acid (50 mm). 2 μg of FBA-tb, fibrinogen, or BSA coated each well. C, the activation of Plg into plasmin in the presence or absence of tPA was measured in the presence of fibrin matrices using Chromozym as the substrate (see “Experimental Procedures”). The complete reaction mixture contained FBA-tb (2 μg), Plg (1 μg), and tPA (0.025 μg) (filled squares). In some assays, tPA was omitted from the reaction mixture (open squares), no FBA-tb or streptokinase was present (open circles), or streptokinase (0.0875 units; 24 ng) replaced FBA-tb (filled triangles). No tPA was present in the wells containing streptokinase. Each point was measured in triplicate, and the mean values and S.D. (error bars) are shown. The values obtained in the FBA-tb assays lacking Plg were similar to those obtained when tPA was omitted. D, regulation of plasmin activity by α2-antiplasmin. Plasmin activity was measured in the presence of fibrin matrices, Plg, and tPA as described in C and in the absence (gray bars) or presence (black bars) of α2-antiplasmin (2 μg). FBA-tb (2 μg) or BSA (2 μg) was added to some wells at the same time as Plg (see “Experimental Procedures”). Wells containing streptokinase (STREP) (0.0875 units; 24 ng) did not contain tPA. Protease activity was calculated from the slopes of kinetic assays performed in duplicate for each condition (see C), and all values were expressed relative to the reaction rate measured for the “no protein/no α2-antiplasmin” control well arbitrarily set to 1. No protease activity was detected in the presence of ϵ-aminocaproic acid (75 mm) in the reaction mixture. Abs, absorbance.
Despite lacking identifiable secretion signals, a number of glycolytic enzymes including FBP aldolases, glyceraldehyde-3-phosphate dehydrogenases, and enolases have been found to exhibit non-glycolytic functions at the cell surface of several bacterial pathogens contributing to tissue invasiveness and dissemination (40, 60). In the context of TB infection, a deregulation of plasmin-dependent pathways could have a major impact on the inflammatory response and induction of host metalloproteinases with consequences on granuloma formation (61), lung tissue destruction (62), and bacterial dissemination. Experiments using cellular models of infection and in vivo studies involving conditional fba-tb gene silencing in M. tuberculosis H37Rv (63) are in progress to directly test these hypotheses and assess the essential character of FBA-tb throughout the different stages of the infection.
Structure Determination of Apo Form of FBA-tb and of FBA-tb·Inhibitor Complex Reveals Previously Unknown Reaction Mechanism for Class II Aldolases
The recombinant form of the protein crystallized by Pegan et al. (19) carries a C-terminal His tag, which we found to inhibit the activity of recombinant FBA-tb when expressed in a mycobacterial host.6 Moreover, the crystallization conditions of the C-terminal His tag enzyme corresponded to acidic pH that may affect interaction between phosphorylated substrate/products and the active site residues. These considerations prompted us to search for another crystal structure of the native active protein at a more physiological pH and without N-terminal or C-terminal tags that incidentally may be more amenable to the future design and optimization of inhibitors.
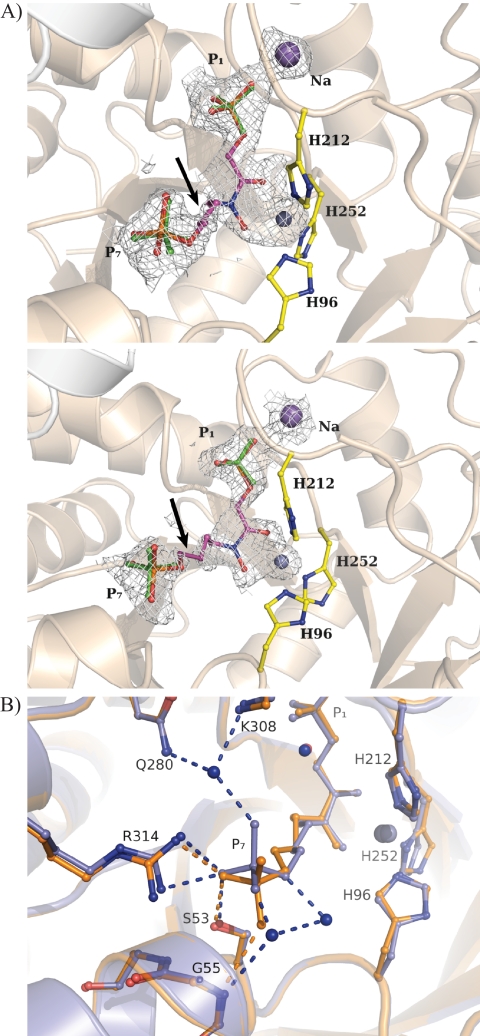
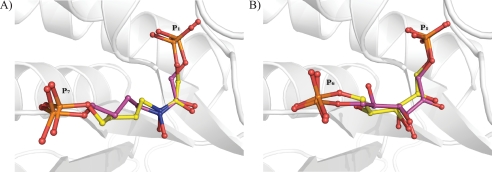
The structural analysis of the native enzyme indicated a subunit polypeptide fold identical to the repeating (βα)8 motif found in numerous other class II aldolases (Table 2) (19, 64–66). The crystallographic structure of FBA-tb bound to TD3, a hydroxamate analog of sedoheptulose 1,7-bisphosphate and potent inhibitor of the enzyme (33), is shown in Fig. 5A. The hydroxamate moiety of the inhibitor interacts with the catalytic zinc ion, which results in a loosely coordinated trigonal bipyramid geometry around the metal ion. This geometry is identical to that previously reported for the zinc ion and TD3 in the class II aldolase of Helicobacter pylori (33, 64). The absence of substituents at C5 and C6 in TD3 capable of interacting with active site residues makes for a backbone with considerable flexibility and correlates with weak electron density at the C5 and C6 atoms. Otherwise, the binding locus of the inhibitor completely overlaps with that of the bound FBP substrate reported previously (19), consistent with TD3 being a competitive inhibitor (supplemental Fig. S4). A subtle difference in TD3 binding between the two subunits of the same FBA-tb homodimer lies in the binding by the P7 oxyanion and solvation of the oxyanion in the high occupancy subunit, which was not observed in the low occupancy subunit (Fig. 5B).
FIGURE 5.
Binding by substrate analog TD3 in FBA-tb subunits. A, difference electron density (Fo − Fc) simulated annealed omit map showing fit to the electron density map by inhibitor TD3 bound in the active site of FBA-tb. The subunit corresponding to high occupancy binding (0.6) by TD3 is shown in the top panel, whereas that of low occupancy binding (0.3) is shown in the middle panel. P1 and P7 phosphates of TD3 are identified. Residual electron density at the P1 and P7 phosphates binding loci was refined as sulfate ions that are shown in green. Side chains of the histidine residues chelating the catalytic zinc ion are also depicted. The figure was drawn and the superimposition was prepared using the program PyMOL. The catalytic zinc ion is shown as a gray sphere in all figures. TD3 binding further activates sodium ion binding with the P1 oxyanion. The arrows point to the C5-C6 bond in TD3 that corresponds to the weakest region of electron density in the difference map for TD3 in both subunits. The electron density of the difference omit, which is shown outlining the active binding event, was set to the 2σ level instead of 4σ to reflect the partial occupancy by TD3 and metal ion in the active site. B, superimposition of FBA-tb subunits binding the TD3 inhibitor. The TD3 binding locus is identical in the two subunits. The inhibitor bound in the high occupancy subunit is shown in blue, and it is shown in orange for the low occupancy subunit. In the subunit corresponding to high TD3 occupancy (0.6), the P7 oxyanion participates in three well defined hydrogen bonding interactions with water molecules that are not present in the subunit exhibiting lower TD3 occupancy (0.3). The presence of solvating water molecules in the high occupancy subunit and their absence in the other subunits were corroborated by electron density omit maps (data not shown). The same binding geometry corresponding to the high occupancy TD3 site was also observed for zinc ion chelating FBP in FBA-tb (19). The extensive superimposition of FBP and TD3 molecules in the FBA-tb active site shown in supplemental Fig. S4 reinforces the role of TD3 as a competitive inhibitor. Binding by TD3, however, does not induce any additional conformational changes with respect to active site binding by FBP.
The binding site of TD3 was further examined using automated in silico docking as described in supplemental Data S5. The docking calculations led to extensive overlap between predicted and observed binding sites for TD3 (root mean square deviation, 0.67 Å between identical TD3 atoms) (Fig. 6A). To benchmark the predictive capability of the docking software, the binding site predicted for FBP was compared with the FBP binding site reported for FBA-tb (19) (Fig. 6B) and showed similar extensive overlap (root mean square deviation, 0.77 Å between identical FBP atoms). The high quality of the in silico predictions substantiates the crystallographically determined binding site for TD3 in the liganded FBA-tb crystal structure. Further comparison of the structures of the ligand-bound complexes with that of the native enzyme indicated similar folding of the polypeptide sequence in each subunit of the enzyme regardless of ligand occupancy in the active site and no additional conformational changes upon either TD3 or FBP binding (root mean square deviations, 0.383 and 0.324 Å, respectively).
FIGURE 6.
Automated docking using AutoDock 4 to predict binding sites for substrate and inhibitor corresponding to cluster of lowest interaction energy in FBA-tb. A, comparison of the TD3 binding site as predicted by AutoDock 4 (yellow) and observed in the crystal structure of the high occupancy subunit of FBA-tb (magenta). B, comparison of the FBP binding site as predicted by AutoDock 4 (yellow) and observed in the crystal structure of a C-terminal His-tagged variant of FBA-tb (Protein Data Bank code 3elf) (magenta).
A distinguishing feature of the FBA-tb·TD3 complex was the substoichiometric binding of TD3 and Zn2+ to the enzyme. Subunit occupancies of TD3 and Zn2+ indicated limited binding in one homodimer and undetectable levels in the remaining homodimer. Relative levels of TD3 and Zn2+ occupancy amounted to an approximate 1:1 stoichiometric ratio in each of the bound subunits. In the active site of the native FBA-tb structure, electron density corresponding to the catalytic Zn2+ ion was not detected. This unexpected finding is in contrast to the previous report of the substoichiometric presence of zinc ion in a native form of FBA-tb (0.5 zinc ion per protomer) purified from E. coli (18) and is most likely a consequence of competitive zinc ion sequestration by sulfate ions present in the crystallization buffer (67). A substoichiometric zinc content per subunit in class II FBP aldolases has recently been documented (68) and is as low as 0.16 zinc ion per protomer in the case of FBA from Pseudomonas aeruginosa. Active site binding by the TD3 ligand thus entails a previously unknown reaction mechanism associated with class II aldolases involving apparent stoichiometric recruitment of the catalytic zinc ion by TD3 (a consequence of the chelating properties of the hydroxamate moiety in this compound). Further supporting this reaction mechanism, the same FBP-zinc ion stoichiometry was observed in the structure of the FBA-tb·FBP complex (19), suggesting that ligand-activated Zn2+ binding may be a general recruitment mechanism to maximize binding and catalytic activity. Such ligand-activated Zn2+ binding has been reported for metallo-β-lactamases where the apo form is the prevailing state under physiological conditions in the absence of substrates (69). Substrate availability apparently induces a spontaneous self-activation due to a decrease of the dissociation constants, resulting in the formation of fully active enzyme. Serendipitously, the crystallization conditions used in this study that rely on molar concentrations of sulfate ion in the crystallization buffer responsible for competitive zinc ion sequestration (67) have provided the first structural corroboration for this observation. For the same reason, the crystallization conditions probably mitigated maximal TD3 activated zinc binding in the crystal lattice, resulting in only 0.9 active sites being occupied of four possible sites.
In addition to competitive sequestration of Zn2+, sulfate ions at molar concentration also compete for the phosphate binding sites, thereby inhibiting active site binding, consistent with the complete loss of enzymatic activity observed in the crystallization buffer. The absence of catalytic Zn2+ and TD3 binding in one homodimer may thus reflect tighter active site binding by sulfate ions due to differential lattice packing between the homodimers. Enhanced stabilization of the sulfate ion by merely 1 kcal mol−1 in this homodimer would effectively diminish the observed Zn2+- and TD3-bound population to an occupancy of <0.1, which would not be detectable by structural analysis.
Conclusions
Because of their central involvement in glycolysis, gluconeogenesis, and ATP synthesis under low oxygen tension, fructose-1,6-bisphosphate aldolases represent attractive targets for the development of novel drugs. Accordingly, recent studies have exploited the fact that glycolysis serves as the major source of ATP in some human parasitic protozoans to explore the potential of class I and class II FBAs as therapeutic targets in Giardia lamblia (class II FBA) and in Trypanosoma brucei, Leishmania mexicana, and Plasmodium falciparum (class I FBAs) (66, 70). Despite their widespread distribution in bacteria (including most major bacterial pathogens) and absence from mammalian cells, prokaryotic class II FBAs have been much less studied. The results of our study clearly highlight the potential of the class II fructose-1,6-bisphosphate aldolase of M. tuberculosis as a therapeutic target both from the perspective of its apparent constitutive expression and requirement for growth and for its potential involvement in the immunopathology of the disease. The lack of effect of a potent substrate analog of FBA-tb on the binding of this protein to Plg alludes to two spatially distinct binding loci that may be useful for target-based drug design. Complementary to our efforts to precisely define the role(s) of FBA-tb during host infection, work is in progress to identify drug-like inhibitors of this promising target.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Michael R. McNeil and Patrick J. Brennan (Mycobacteria Research Laboratories, Department of Microbiology, Immunology, and Pathology, Colorado State University) for helpful discussions and Drs. Karolin Luger and Dr. Mark van der Woerd (Department of Biochemistry and Molecular Biology, Colorado State University) for providing access to their protein purification facility and for help with data collection and processing. Some of this work was carried out at the National Synchrotron Light Source, Brookhaven National Laboratory, which is supported by the United States Department of Energy, Division of Materials Sciences and Division of Chemical Sciences under Contract DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grants AI078126 from the NIAID and NS066438 from the NINDS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Data S5.
The atomic coordinates and structure factors (codes 4a21 and 4a22) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
P. M. Gest and M. Jackson, unpublished results.
- TB
- tuberculosis
- FBP
- fructose 1,6-bisphosphate
- FBA
- fructose-bisphosphate aldolase
- FBA-tb
- M. tuberculosis fructose-1,6-bisphosphate aldolase
- Plg
- plasminogen
- tPA
- tissue Plg activator
- OADC
- oleic acid-albumin-dextrose-catalase
- ADC
- albumin-dextrose-catalase
- TD3
- N-(4-hydroxybutyl)-glycolohydroxamic acid bisphosphate.
REFERENCES
- 1. Barry C. E., 3rd, Boshoff H. I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R. J., Young D. (2009) Nat. Rev. Microbiol. 7, 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization (ed) (2009) WHO Report 2009—Global Tuberculosis Control. Epidemiology, Strategy, Financing, World Health Organization, Geneva [Google Scholar]
- 3. Parrish N. M., Dick J. D., Bishai W. R. (1998) Trends Microbiol. 6, 107–112 [DOI] [PubMed] [Google Scholar]
- 4. Wayne L. G., Lin K. Y. (1982) Infect. Immun. 37, 1042–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manabe Y. C., Bishai W. R. (2000) Nat. Med. 6, 1327–1329 [DOI] [PubMed] [Google Scholar]
- 6. McKinney J. D., Höner zu Bentrup K., Muñoz-Elías E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. (2000) Nature 406, 735–738 [DOI] [PubMed] [Google Scholar]
- 7. Höner zu Bentrup K., Russell D. G. (2001) Trends Microbiol. 9, 597–605 [DOI] [PubMed] [Google Scholar]
- 8. Wayne L. G., Sohaskey C. D. (2001) Annu. Rev. Microbiol. 55, 139–163 [DOI] [PubMed] [Google Scholar]
- 9. Stewart G. R., Robertson B. D., Young D. B. (2003) Nat. Rev. 1, 97–105 [DOI] [PubMed] [Google Scholar]
- 10. Monack D. M., Mueller A., Falkow S. (2004) Nat. Rev. Microbiol. 2, 747–765 [DOI] [PubMed] [Google Scholar]
- 11. Muñoz-Elías E. J., McKinney J. D. (2005) Nat. Med. 11, 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Timm J., Post F. A., Bekker L. G., Walther G. B., Wainwright H. C., Manganelli R., Chan W. T., Tsenova L., Gold B., Smith I., Kaplan G., McKinney J. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14321–14326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bai N. J., Pai M. R., Murthy P. S., Venkitasubramanian T. A. (1974) FEBS Lett. 45, 68–70 [DOI] [PubMed] [Google Scholar]
- 16. Bai N. J., Pai M. R., Murthy P. S., Venkitasubramanian T. A. (1982) Methods Enzymol. 90, 241–250 [DOI] [PubMed] [Google Scholar]
- 17. Rosenkrands I., Slayden R. A., Crawford J., Aagaard C., Barry C. E., 3rd, Andersen P. (2002) J. Bacteriol. 184, 3485–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramsaywak P. C., Labbé G., Siemann S., Dmitrienko G. I., Guillemette J. G. (2004) Protein Expr. Purif. 37, 220–228 [DOI] [PubMed] [Google Scholar]
- 19. Pegan S. D., Rukseree K., Franzblau S. G., Mesecar A. D. (2009) J. Mol. Biol. 386, 1038–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsh J. J., Lebherz H. G. (1992) Trends Biochem. Sci. 17, 110–113 [DOI] [PubMed] [Google Scholar]
- 21. Wayne L. G., Hayes L. G. (1996) Infect. Immun. 64, 2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jackson M., Camacho L. R., Gicquel B., Guilhot C. (2001) in Mycobacterium tuberculosis Protocols (Parish T., Stocker N. G. eds) pp. 59–75, Humana Press, Totowa, NJ [Google Scholar]
- 23. Korduláková J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B., Jackson M. (2002) J. Biol. Chem. 277, 31335–31344 [DOI] [PubMed] [Google Scholar]
- 24. Méderlé I., Bourguin I., Ensergueix D., Badell E., Moniz-Peireira J., Gicquel B., Winter N. (2002) Infect. Immun. 70, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blumenthal A., Trujillo C., Ehrt S., Schnappinger D. (2010) Plos One 5, e15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klotzsche M., Ehrt S., Schnappinger D. (2009) Nucleic Acids Res. 37, 1778–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., Schnappinger D. (2005) Nucleic Acids Res. 33, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ryan G. J., Hoff D. R., Driver E. R., Voskuil M. I., Gonzalez-Juarrero M., Basaraba R. J., Crick D. C., Spencer J. S., Lenaerts A. J. (2010) PLoS One 5, e11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xolalpa W., Vallecillo A. J., Lara M., Mendoza-Hernandez G., Comini M., Spallek R., Singh M., Espitia C. (2007) Proteomics 7, 3332–3341 [DOI] [PubMed] [Google Scholar]
- 30. Richards O. C., Rutter W. J. (1961) J. Biol. Chem. 236, 3177–3184 [PubMed] [Google Scholar]
- 31. Ortalo-Magné A., Dupont M. A., Lemassu A., Andersen A. B., Gounon P., Daffé M. (1995) Microbiology 141, 1609–1620 [DOI] [PubMed] [Google Scholar]
- 32. Jagannathan V., Singh K., Damodaran M. (1956) Biochem. J. 63, 94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Daher R., Coinçon M., Fonvielle M., Gest P. M., Guerin M. E., Jackson M., Sygusch J., Therisod M. (2010) J. Med. Chem. 53, 7836–7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 35. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 36. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 37. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 38. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 39. Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 40. Tunio S. A., Oldfield N. J., Berry A., Ala'Aldeen D. A., Wooldridge K. G., Turner D. P. (2010) Mol. Microbiol. 76, 605–615 [DOI] [PubMed] [Google Scholar]
- 41. Wehmeier U. F. (2001) FEMS Microbiol. Lett. 197, 53–58 [DOI] [PubMed] [Google Scholar]
- 42. Gerdes S. Y., Scholle M. D., Campbell J. W., Balázsi G., Ravasz E., Daugherty M. D., Somera A. L., Kyrpides N. C., Anderson I., Gelfand M. S., Bhattacharya A., Kapatral V., D'Souza M., Baev M. V., Grechkin Y., Mseeh F., Fonstein M. Y., Overbeek R., Barabási A. L., Oltvai Z. N., Osterman A. L. (2003) J. Bacteriol. 185, 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodaki A., Young T., Brown A. J. (2006) Eukaryot. Cell 5, 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singer M., Walter W. A., Cali B. M., Rouviere P., Liebke H. H., Gourse R. L., Gross C. A. (1991) J. Bacteriol. 173, 6249–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hueck C. J., Hillen W. (1995) Mol. Microbiol. 15, 395–401 [DOI] [PubMed] [Google Scholar]
- 46. Moritz B., Striegel K., De Graaf A. A., Sahm H. (2000) Eur. J. Biochem. 267, 3442–3452 [DOI] [PubMed] [Google Scholar]
- 47. Zheng H., Lu L., Wang B., Pu S., Zhang X., Zhu G., Shi W., Zhang L., Wang H., Wang S., Zhao G., Zhang Y. (2008) PLoS One 3, e2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marsh J. J., Wilson K. J., Lebherz H. G. (1989) Plant Physiol. 91, 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rellos P., Sygusch J., Cox T. M. (2000) J. Biol. Chem. 275, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 50. Ling E., Feldman G., Portnoi M., Dagan R., Overweg K., Mulholland F., Chalifa-Caspi V., Wells J., Mizrachi-Nebenzahl Y. (2004) Clin. Exp. Immunol. 138, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marsollier L., Brodin P., Jackson M., Korduláková J., Tafelmeyer P., Carbonnelle E., Aubry J., Milon G., Legras P., André J. P., Leroy C., Cottin J., Guillou M. L., Reysset G., Cole S. T. (2007) PLoS Pathog. 3, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crowe J. D., Sievwright I. K., Auld G. C., Moore N. R., Gow N. A., Booth N. A. (2003) Mol. Microbiol. 47, 1637–1651 [DOI] [PubMed] [Google Scholar]
- 53. Blau K., Portnoi M., Shagan M., Kaganovich A., Rom S., Kafka D., Chalifa Caspi V., Porgador A., Givon-Lavi N., Gershoni J. M., Dagan R., Mizrachi Nebenzahl Y. (2007) J. Infect. Dis. 195, 1828–1837 [DOI] [PubMed] [Google Scholar]
- 54. Wu Z., Zhang W., Lu C. (2008) FEMS Immunol. Med. Microbiol. 53, 52–59 [DOI] [PubMed] [Google Scholar]
- 55. Jungblut P. R., Schaible U. E., Mollenkopf H. J., Zimny-Arndt U., Raupach B., Mattow J., Halada P., Lamer S., Hagens K., Kaufmann S. H. (1999) Mol. Microbiol. 33, 1103–1117 [DOI] [PubMed] [Google Scholar]
- 56. Målen H., Berven F. S., Fladmark K. E., Wiker H. G. (2007) Proteomics 7, 1702–1718 [DOI] [PubMed] [Google Scholar]
- 57. Shi L., Sohaskey C. D., Pfeiffer C., Datta P., Parks M., McFadden J., North R. J., Gennaro M. L. (2010) Mol. Microbiol. 78, 1199–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lenaerts A. J., Hoff D., Aly S., Ehlers S., Andries K., Cantarero L., Orme I. M., Basaraba R. J. (2007) Antimicrob. Agents Chemother. 51, 3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gerber S. S., Lejon S., Locher M., Schaller J. (2010) Cell. Mol. Life Sci. 67, 1505–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boyle M. D., Lottenberg R. (1997) Thromb. Haemost. 77, 1–10 [PubMed] [Google Scholar]
- 61. Taylor J. L., Hattle J. M., Dreitz S. A., Troudt J. M., Izzo L. S., Basaraba R. J., Orme I. M., Matrisian L. M., Izzo A. A. (2006) Infect. Immun. 74, 6135–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Elkington P. T., D'Armiento J. M., Friedland J. S. (2011) Sci. Transl. Med. 3, 71ps6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gandotra S., Schnappinger D., Monteleone M., Hillen W., Ehrt S. (2007) Nat. Med. 13, 1515–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fonvielle M., Coinçon M., Daher R., Desbenoit N., Kosieradzka K., Barilone N., Gicquel B., Sygusch J., Jackson M., Therisod M. (2008) Chemistry 14, 8521–8529 [DOI] [PubMed] [Google Scholar]
- 65. Blom N. S., Tétreault S., Coulombe R., Sygusch J. (1996) Nat. Struct. Biol. 3, 856–862 [DOI] [PubMed] [Google Scholar]
- 66. Galkin A., Kulakova L., Melamud E., Li L., Wu C., Mariano P., Dunaway-Mariano D., Nash T. E., Herzberg O. (2007) J. Biol. Chem. 282, 4859–4867 [DOI] [PubMed] [Google Scholar]
- 67. Owen B. B., Curry R. W. (1938) J. Am. Chem. Soc. 60, 3074–3078 [Google Scholar]
- 68. Labbé G., de Groot S., Rasmusson T., Milojevic G., Dmitrienko G. I., Guillemette J. G. (2011) Protein Expr. Purif.. in press [DOI] [PubMed] [Google Scholar]
- 69. Wommer S., Rival S., Heinz U., Galleni M., Frere J. M., Franceschini N., Amicosante G., Rasmussen B., Bauer R., Adolph H. W. (2002) J. Biol. Chem. 277, 24142–24147 [DOI] [PubMed] [Google Scholar]
- 70. Dax C., Duffieux F., Chabot N., Coincon M., Sygusch J., Michels P. A., Blonski C. (2006) J. Med. Chem. 49, 1499–1502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.