Background: BST-2/tetherin inhibits virus release by tethering virions to the cell surface.
Results: The putative GPI anchor signal of BST can function as a TM region and can be replaced by heterologous TM segments.
Conclusion: BST-2 contains a second TM region instead of a GPI anchor.
Significance: Understanding the molecular structure of BST-2 is crucial to understanding how the protein inhibits detachment of many enveloped viruses.
Keywords: Plasma Membrane, Protein Chimeras, Protein Conformation, Protein Domains, Protein Structure, Virus Assembly, BST-2, GPI Anchor Modification, Tetherin
Abstract
BST-2/CD317/HM1.24/tetherin is a host factor that inhibits the release of HIV-1 and other enveloped viruses. Structurally, tetherin consists of an N-terminal transmembrane (TM) region, a central coiled coil motif, and a putative C-terminal glycosylphosphatidylinositol (GPI) anchor motif. A current working model proposes that BST-2 inhibits virus release by physically tethering viral particles to the cell surface via its TM motif and GPI anchor. Here we analyzed the functional importance of the C-terminal GPI anchor motif in BST-2. We replaced the GPI anchor motif in BST-2 with the TM regions of several surface markers and found that the TM motifs of CD40 and transferrin receptor, but not that of CD45, could functionally substitute for a GPI anchor in BST-2. Conversely, replacing the TM region of CD4 by the putative GPI anchor signal of human BST-2 resulted in proper membrane targeting and surface expression of the chimeric protein, indicating that the BST-2 GPI anchor signal can function as a bona fide TM region. In fact, attempts to demonstrate GPI anchor modification of human BST-2 by biochemical methods failed. Our results demonstrate that the putative C-terminal GPI anchor motif in human BST-2 fulfills the requirements of a bona fide TM motif, leading us to propose that human BST-2 may in fact contain a second TM segment rather than a GPI anchor.
Introduction
HIV-1 must overcome several host defense mechanisms to establish an efficient infection. Trim-5α, APOBEC3G, and BST-2/tetherin are among those restriction factors that target different stages of viral replication (for reviews, see Refs. 1 and 2). APOBEC3G and BST-2 are targeted and functionally inactivated by dedicated viral proteins. Although Vif is expressed by almost all primate lentiviruses and is the only protein in these viruses capable of controlling APOBEC3G, there is no unique viral gene product targeting BST-2. In fact, three different lentiviral proteins have been implicated in the control of BST-2: HIV-1 uses Vpu, HIV-2 uses its Env glycoprotein, and simian immunodeficiency virus uses Nef. All three proteins are either integral membrane proteins (Vpu and Env) or membrane-associated by means of a myristic acid modification (Nef) and are thought to interfere with BST-2 function via direct physical interaction.
BST-2 was originally identified as a membrane protein in terminally differentiated human B cells of patients with multiple myeloma (3, 4) and later found to be the interferon-inducible host factor responsible for the Vpu-sensitive inhibition of HIV-1 virus release (5, 6). BST-2 is a 30–36-kDa type II transmembrane protein consisting of 180 amino acids (7). The protein is predicted to have an N-terminal transmembrane (TM)2 region and a C-terminal glycosylphosphatidylinositol (GPI) anchor (8). These two domains are separated by ∼120 residues that constitute the ectodomain of the protein and are predicted to form a 16–17-nm long rodlike coiled coil structure (9–12). The BST-2 ectodomain encodes 3 cysteine residues (3, 4, 13, 14), all of which can independently contribute to the formation of cysteine-linked dimers (13, 14). Furthermore, BST-2 is modified by N-linked glycosylation (4, 8, 13); however, the functional importance of BST-2 glycosylation for inhibition of virus release is under debate (13, 14). BST-2 protein associates with lipid rafts at the cell surface and on internal membranes, presumably the trans-Golgi network (8, 15–17), and C-terminal GPI anchor modification has been implicated with raft targeting of BST-2.
A current model suggests that BST-2 tethers mature virions to the cell surface by means of its N-terminal TM region and C-terminal GPI anchor (5). Indeed, immune electron microscopy confirmed that BST-2 could be found on virions tethered to the cell surface (14, 18–20). In addition, Perez-Caballero et al. (14) demonstrated that an artificial tetherin consisting of the N-terminal TM region of transferrin receptor, a coiled coil ectodomain of the cytoplasmic dimeric protein dystrophia myotonica protein kinase, and a GPI anchor signal derived from urokinase plasminogen activator receptor (uPAR) is capable of inhibiting the release of HIV-1 virions tethered to the cell surface. However, much of the evidence for BST-2 containing a GPI anchor in addition to a transmembrane region consists of experimental data performed on rat BST-2, which is only 33% identical to the human protein, and is largely indirect (8, 21). Typical GPI-anchored proteins contain initially two hydrophobic motifs. The N-terminal hydrophobic motif acts as signal peptide that targets the protein to the ER and is removed by N-terminal peptidase. A second hydrophobic segment at the C terminus is part of the GPI anchor signal and is removed upon GPI anchor modification. Thus, the vast majority of mature GPI-anchored proteins lack a TM region (22), making BST-2 one of only a few proteins carrying a TM region in addition to a GPI anchor. In fact, Kupzig et al. (8) reported that, aside from rat BST-2 analyzed in their study, only four other naturally occurring proteins are known to be anchored in the membrane by both a TM region and a GPI anchor.
Experimental verification of GPI anchor modification of proteins containing an additional TM region is technically challenging. The most common assay used for typical GPI-anchored proteins is the release of the proteins from the membrane by phosphatidylinositol-specific phospholipase C (PI-PLC) treatment, which cleaves the protein at the GPI anchor and releases it from the membrane (23). However, proteins containing a TM region will remain membrane-associated under such conditions. Kupzig et al. (8) used a variety of methods to demonstrate GPI anchor modification of rat BST-2. Among those is PI-PLC treatment, which made rat BST-2 susceptible to Triton X-100 extraction (i.e. resulted in loss of raft association). In addition, PI-PLC treatment of BST-2-expressing rat cells resulted in positive staining by an anti-cross-reactive determinant antibody, which can bind to a cross-reactive determinant epitope that is exposed upon PI-PLC treatment (24). Furthermore, treatment of cells with PI-PLC decreased the internalization of BST-2 from the cell surface and lipid raft association, which would implicate a GPI anchor in this process (21). Although all of these experiments are suggestive of a GPI anchor modification, none of them provides direct experimental evidence.
As mentioned above, human BST-2 shares only 33% amino acid identity with the rat protein. However, like the rat protein, human BST-2 is predicted by bioinformatics tools to be GPI anchor-modified. Unfortunately, bioinformatics tools are not foolproof especially because there is no consensus sequence for GPI anchor modification. It is therefore necessary to experimentally verify GPI anchor addition. Given that GPI anchor modification of transmembrane proteins appears to be extremely rare in nature and given the lack of direct experimental evidence for GPI anchor modification of either rat or human BST-2, the goal of the current study was to further investigate GPI anchor modification of human BST-2. We used a variety of biochemical assays, including PI-PLC treatment, aerolysin treatment, and gradual truncation of the putative GPI anchor signal. We were unable to verify GPI anchor modification of human BST-2. Instead, we found strong evidence that the C-terminal putative GPI anchor signal in human BST-2 represents in fact a second TM region. This conclusion is supported by the following observations. (i) The C-terminal putative GPI anchor motif can be transferred to a heterologous protein and function as a TM motif. (ii) C-terminally epitope-tagged BST-2 is functional. Importantly, the C-terminal tag was not subject to proteolytic removal by the GPI modification machinery and localized to the cytoplasmic side of the plasma membrane. (iii) Deletion of 2 C-terminal residues from untagged BST-2 did not affect BST-2 function but can be visualized as a shift in protein mobility, arguing against proteolytic removal of the GPI signal peptide. (iv) Replacing the GPI anchor motif in human BST-2 with the TM segment of heterologous proteins can yield BST-2 capable of inhibiting virus release.
EXPERIMENTAL PROCEDURES
Plasmids
The full-length infectious HIV-1 molecular clone pNL4-3 and the Vpu deletion mutant pNL4-3/Udel have been described (25, 26). Plasmid pcDNA-BST-2 is a vector for the expression of human BST-2 under the control of the cytomegalovirus immediate-early promoter (13). C-terminal deletion mutants of BST-2 (Δ2, Δ3, Δ4, Δ5, Δ10, Δ15, Δ18, and Δ21; see Fig. 7A) were constructed in the backbone of pcDNA-BST-2 N1/N2 (13). BST-2 N1/N2 encodes non-glycosylated but functional BST-2 and was used here to facilitate visualization of differences in the electrophoretic mobility of BST-2 deletion mutants. HA-tagged BST-2 constructs encoding HA epitope tags at the N terminus (BST-2N), in the ectodomain following BST-2 residue 148 (BST-2I), or at the C terminus (BST-2C) were constructed using PCR-based methodologies. Plasmid pcDNA-CD4 encoding CD4 with a shortened 3′-untranslated region was constructed by subcloning a 1.7-kb EcoRI/BamHI fragment from pHIV-CD4 (27) into pcDNA3.1. Plasmid pcDNA-CD4-TM1 was constructed by replacing the CD4 TM region in pcDNA-CD4 (residues Met397–Phe418) with the N-terminal TM segment of BST-2 (Leu22–Ile46) using PCR-based strategies. Similarly, pcDNA-CD4-TM2 was constructed by replacing the CD4 TM region (residues Met397–Phe418) with the C-terminal putative TM region of BST-2 (Ala164–Leu179). pcDNA-CD4-ΔTM was constructed by removing the CD4 TM region (residues Met397–Phe418). To construct pcDNA-BST-2 CD40, pcDNA-BST-2 CD45, and pcDNA-BST-2 transferrin receptor (TfR) chimeras, the GPI anchor signal/putative C-terminal TM region in BST-2 (Ser162–Leu178) was replaced with the TM region sequence of CD40 (ALVVIPIIFGILFAILLVLVFI), CD45 (ALIAFLAFLI IVTSIALLVVLY) (28), or TfR (CSGSICYGTIAVIVFFLIGFMIGYLGYC) (29), respectively (see Fig. 1, A and B). All constructs were verified by sequence analysis.
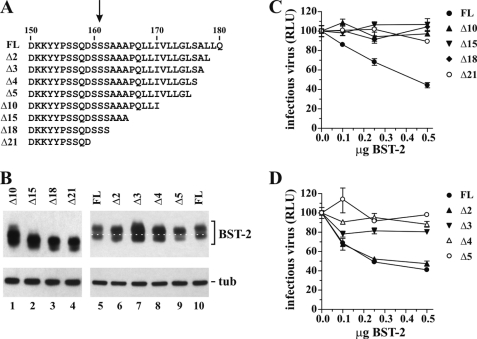
FIGURE 7.
Deletion of C-terminal residues of BST-2 results in shift in size and loss of function. A, schematic depicting the mutations made in BST-2 N1/N2. The arrow points to the predicted GPI anchor signal cleavage site. FL, full-length BST-2 N1/N2. B, cell lysates of 293T cells transfected with 0.5 μg of the indicated BST-2 constructs were subjected to immunoblot analysis with BST-2 antibody. The blot was subsequently reprobed with an antibody to tubulin to control for equal sample loading. The white dotted line connects the lower bands of the full-length (FL) controls and was added to better visualize the mobility shift induced by deletion of 2–5 residues in the mutants. C and D, 293T cells were transfected with 5 μg of NL4-3/Udel and varying amounts of BST-2 DNA as indicated. Virus-containing supernatants were harvested 24 h later and used to infect HeLa TZM-bl cells. Infection of TZM-bl cells was determined 48 h later and was used to calculate virus release as described for Fig. 1C. Values are expressed as mean of three experiments. Error bars, S.E. Infectivity of the virus in the absence of BST-2 was defined as 100%. RLU, relative luciferase units; tub, tubulin.
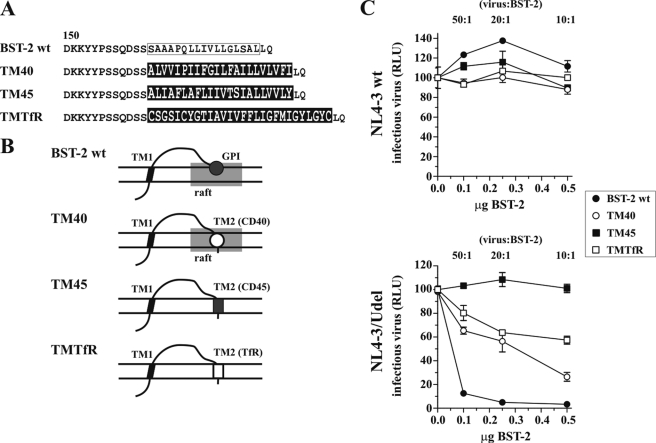
FIGURE 1.
TM regions of CD40 and TfR can functionally replace putative GPI anchor of human BST-2. A, amino acid alignment of BST-2 WT, BST-2 TM40, BST-2 TM45, and BST-2 TMTfR. Shown are the sequences beginning with residue 150 in BST-2. The boxed sequence represents a putative TM domain in BST-2. TM40, TM45, and TMTfR sequences are indicated by white letters on a black background. B, schematic structure of the TM40, TM45, and TMTfR variants compared with BST-2 WT. Shaded rectangles represent lipid rafts. Symbols for TM2 correspond to those in C. C, 293T cells were transfected with 5 μg of NL4-3 WT (top) or NL4-3/Udel (bottom) together with varying amounts of BST-2 WT, TM40, TM45, or TfR chimeric DNAs as indicated. Virus-containing supernatants were collected 24 h after transfection and used for the infection of HeLa TZM-bl indicator cells. Virus-induced luciferase activity was recorded 48 h later and was used as a measure of virus release. Values are expressed as mean of three experiments. Error bars, S.E. The infectivity of NL4-3 or NL4-3/Udel virus produced in the absence of BST-2 (0.0 μg of BST-2) was defined as 100%. RLU, relative luciferase units.
Antisera
Rabbit polyclonal BST-2 antiserum directed against the extracellular portion of BST-2 has been described (13, 30) and is available through the National Institutes of Health AIDS Research and Reference Reagent Program (catalog number 11721). Monoclonal antibodies to tubulin and actin were obtained from Sigma. A monoclonal antibody to CD55 was from Santa Cruz Biotechnology (Santa Cruz, CA). HA-specific mouse monoclonal antibody was from Roche Diagnostics. A rabbit polyclonal antibody reacting with the CD4 ectodomain was produced by immunizing a rabbit with recombinant CD4-IgG fusion protein (generously provided by Genentech Inc.). A polyclonal antibody specific for the CD4 cytoplasmic domain, CD4cyto, was produced by immunizing a rabbit with keyhole limpet hemocyanin-coupled peptide CVRCRHRRRQAERMSQIKRLLSEKKTCQC (31). A mouse monoclonal antibody to the CD4 ectodomain (OKT4) was obtained from eBioscience (San Diego, CA). Serum from an HIV-positive patient was used to detect HIV-1-specific capsid protein. A mouse monoclonal antibody to aerolysin was obtained from Dr. Peter Howard (University of Saskatchewan).
Tissue Culture and Transfections
HeLa and 293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). HeLa TZM-bl cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health (donated by Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc.) and propagated as the HeLa cells. For transfection, cells were grown in 25-cm2 flasks to about 80% confluence. Cells were transfected using TransIT®-LT1 (Mirus, Madison WI) or Lipofectamine PLUSTM (Invitrogen) following the manufacturer's recommendations. A total of 5 μg of plasmid DNA/25-cm2 flask was used. Total amounts of transfected DNA were kept constant in all samples of any given experiment by adding empty vector DNA as appropriate. Cells were harvested 24 h post-transfection.
Immunoblotting
For immunoblot analysis of intracellular proteins, whole cell lysates were prepared as follows. Cells were washed once with PBS, suspended in PBS (400 μl/107 cells), and mixed with an equal volume of sample buffer (4% sodium dodecyl sulfate, 125 mm Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 10% glycerol, 0.002% bromphenol blue). Proteins were solubilized by boiling for 10–15 min at 95 °C with occasional vortexing of the samples to shear cellular DNA. Residual insoluble material was removed by centrifugation (2 min at 15,000 rpm in an Eppendorf minicentrifuge). Cell lysates were subjected to SDS-PAGE, and proteins were transferred to PVDF membranes and incubated with appropriate antibodies as described in the text. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences) and visualized by enhanced chemiluminescence (ECL; Amersham Biosciences).
Fractionation
Cells were washed three times with PBS, then scraped, and moved to an Eppendorf tube. Cells were spun at 400 × g for 5 min at 4 °C. The cell pellet was then resuspended in 500 μl of HB buffer (250 mm sucrose, 3 mm imidazole, pH 7.4, 1 mm EDTA) and centrifuged at 1100 × g for 10 min at 4 °C. The pellet was then resuspended in 500 μl of HB buffer and lysed by passage through a 22-gauge syringe 20 times. Cell lysis was verified by microscopy. The lysate was then centrifuged at 1600 × g for 10 min at 4 °C. The postnuclear supernatant was divided into two aliquots: one for the total fraction and one for membrane analysis. One fraction of postnuclear supernatant was centrifuged at 51,000 rpm in a TLA 100.1 rotor for 1 h at 4 °C. The supernatant was moved to a fresh tube on ice for the soluble fraction. The pellet was resuspended in 100 μl of fresh Na2CO3 (100 mm, pH 11.0) and incubated on ice for 15 min. The suspension was centrifuged at 83,000 rpm in a TLA 100.1 rotor for 10 min at 4 °C. The supernatant was moved to a fresh tube on ice for the peripheral membrane fraction. The pellet was again resuspended in 100 μl of fresh Na2CO3, incubated on ice for 15 min, and centrifuged at 83,000 rpm in a TLA 100.1 rotor for 10 min at 4 °C. The supernatant was added to the peripheral membrane fraction. The final pellet, which contains integral membranes, was resuspended in 200 μl of HB buffer containing 1.0% Triton X-100.
Immunofluorescence and Confocal Microscopy
293T cells were transfected as indicated in the text. Transfected cells were trypsinized, and single cell suspensions were distributed into 12-well plates containing 0.13-mm coverslips. Cells were grown for 15 h at 37 °C in DMEM containing 10% FBS. Cells were fixed in 1% paraformaldehyde (Electron Microscopy Sciences, Hatfield PA) for 30 min at room temperature followed by three washes in PBS. For permeabilization, cells were incubated in 0.1% Triton X-100 in PBS for 4 min followed by two washes in PBS. Cells were blocked with 1% BSA in PBS for 30 min. For antibody staining, coverslips were incubated with appropriate primary antibodies in 1% BSA in PBS for 30 min at room temperature. Cells were washed three times with PBS and incubated with appropriate secondary antibodies for 30 min at room temperature. Cells were then washed three times with PBS and mounted onto microscope slides with glycerol gelatin (Sigma-Aldrich) containing 0.1 m N-propyl gallate (Sigma) to prevent photobleaching. For confocal microscopy, a Zeiss LSM410 inverted laser-scanning microscope equipped with a krypton/argon mixed gas laser was used. Images were acquired with a Plan-Apochromat 63×/1.4 oil immersion objective (Zeiss). Image quality was enhanced during data acquisition using the LSM line average feature (8×). Postacquisition digital image enhancement was performed using the LSM software.
Viral Infectivity
Virus stocks were prepared by transfection of 293T cells with the indicated plasmid DNAs. Virus-containing supernatants were harvested 24 h after transfection. Cellular debris was removed by centrifugation (3 min at 1500 rpm), and clarified supernatants were filtered (0.45 μm) to remove residual cellular debris. 100 μl of viral stock was used to infect 5 × 104 TZM-bl cells in a 24-well plate in a total volume of 1.1 ml. Infection was allowed for 48 h at 37 °C. Medium was removed, and cells were lysed in 300 μl of Promega 1× reporter lysis buffer (Promega Corp., Madison WI) and frozen at −80 °C for a minimum of 30 min. To determine the luciferase activity in the lysates, 5 μl of each lysate was combined with luciferase substrate (Promega Corp.) by automatic injection, and light emission was measured for 10 s at room temperature in a luminometer (Optocomp II, MGM Instruments, Hamden, CT). Each assay was performed at least twice with triplicate infections.
Aerolysin Assay
Aerolysin assays were conducted essentially as described (32). Proaerolysin was obtained from Dr. Peter Howard (University of Saskatchewan). Briefly, HeLa cells were washed with PBS, scraped, and resuspended in 1 ml of PBS. The cells were pelleted, frozen, lysed in 1 ml of lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.5% Triton X-100), and incubated on ice for 20 min. Cell lysates were pelleted at 13,000 × g for 2 min to remove insoluble material. The supernatant was recovered, and the proaerolysin was activated by treatment with trypsin (50 μg/ml) for 10 min at room temperature. The trypsin was inactivated by addition of protease inhibitor (Complete Mini, Roche Applied Science). The lysate was then divided into two equal fractions. Aerolysin (final concentration, 300 ng/ml) was added to one of the fractions. Both fractions were then incubated for 1 h at 4 °C followed by 10 min at 37 °C. Aerolysin-treated and untreated fractions were then divided into two equal aliquots, which were immunoprecipitated with antibody to CD55 or BST-2, respectively, bound to Protein A-Sepharose beads (1 h at 4 °C). Samples were washed twice with wash buffer (50 mm Tris, pH 7.4, 300 mm NaCl, 0.1% Triton X-100). To avoid co-migration of CD55 with immunoglobulin heavy chains, CD55 immunoprecipitates were eluted by heating in buffer lacking 2-mercaptoethanol (10 min at 95 °C). Immunoprecipitated BST-2 was divided into two aliquots: one sample was eluted by heating in buffer lacking 2-mercaptoethanol (10 min at 95 °C) to look for the presence of aerolysin, and the other half was eluted by heating in reducing sample buffer for 10 min at 95 °C to verify BST-2 immunoprecipitation. Samples were separated by SDS-PAGE and subjected to immunoblot analysis as indicated in the text. Antibodies bound to beads were included as a control for IgG.
Protein Structure Prediction
Clone Manager software (v9; Scientific & Educational Software, Cary, NC) was used for hydropathy plot analysis. “MemType-2L” is a web-based server for the prediction of membrane proteins and their types (33). All other tools used for the prediction of GPI anchors or TM regions are listed in Table 1 and can be accessed through the ExPASy Proteomics Server. GPI anchor prediction was done using “big-PI Predictor.” Transmembrane segment predictions were done using “HMMTOP,” “PredictProtein,” “SOSUI” (engine version 1.11), “TMHMM,” “TMpred,” and “TopPred.”
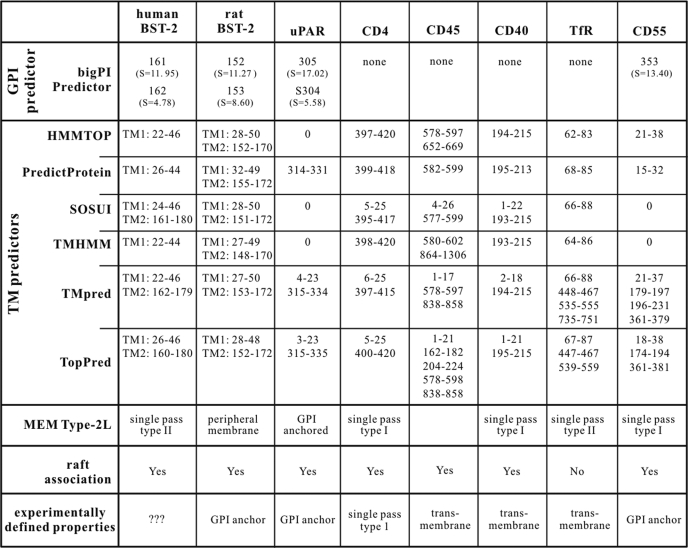
TABLE 1.
In silico analysis of human BST-2 predicts two transmembrane regions
Human and rat BST-2, uPAR, CD4, CD45, CD40, CD55, and TfR were analyzed using the various software programs described. Numbers indicate amino acid positions in the respective protein. “S” values produced by big-PI Predictor are scores assigned by the software to indicate the likelihood of GPI anchor addition. Higher values reflect a higher probability. Information regarding raft association as determined experimentally is included at the bottom.
Membrane Binding Analyses
Raft association of BST-2 was assessed by membrane floatation analysis essentially as described (34). Cells were washed with PBS, pelleted (2000 × g for 2 min), and resuspended in 300 μl of 10 mm Tris-HCl, pH 7.5 supplemented with 4 mm EDTA and CompleteTM protease inhibitor mixture (Roche Diagnostics). After a 10-min incubation on ice, cells were sonicated for 10 s and centrifuged for 3 min at 2000 × g at 4 °C in a microcentrifuge to remove insoluble material and nuclei. The postnuclear supernatants (120 μl) were mixed with 120 μl of TNE lysis buffer (100 mm Tris-HCl, 600 mm NaCl, 16 mm EDTA) supplemented with 0.5% Triton X-100 and an additional 150 mm NaCl and incubated on ice for 20 min. A total of 200 μl of each lysate was mixed with 1 ml of 85.5% sucrose (w/v) in TNE lysis buffer, placed at the bottom of SW55 ultracentrifuge tubes, and overlaid first with 2.5 ml of 65% (w/v) sucrose followed by 1.5 ml of 10% sucrose (w/v) in TNE lysis buffer. The samples were centrifuged at 4 °C in an SW55 rotor for 16 h at 35,000 rpm. Ten equal fractions (550 μl each) were collected from the top, mixed with 4× sample buffer, and heated for 10 min at 95 °C. Samples were analyzed by immunoblotting.
PI-PLC Assay
HeLa cells were transfected with 5 μg of pNL4-3/Udel. After 24 h, cells were washed to remove virus-containing supernatants, detached by scraping, and washed again to remove residual virus trapped in the monolayer. Cells were then divided into four equal aliquots and suspended in 100 μl of PI-PLC buffer (10 mm Tris, pH 8.0, 10 mm EDTA, 10% glycerol). Two samples each were incubated for 15 min at 37 °C in the presence or absence, respectively, of PI-PLC (5 units/ml; Sigma). One treated sample and one untreated sample each were then vortexed for 30 s at room temperature. All four samples were then spun at 5000 rpm for 30 s to pellet cells, and the virus-containing supernatants were removed and filtered through 0.45-μm cellulose acetate spin filters (Corning Costar Corp., Cambridge, MA) to remove residual cells and cellular debris. The supernatants were recovered and divided in half. One-half was mixed with 4× sample buffer for immunoblot analysis of CD55; the other half of each sample was pelleted by ultracentrifugation (90 min at 35,000 rpm at 4 °C) through 10 ml of 20% sucrose to concentrate released virions and remove soluble viral proteins that may have been released from lysed cells during the procedure. The virus pellet was suspended in sample buffer and subjected to immunoblot analysis to detect virus-associated Gag protein.
FACS Analysis
Cells were washed twice with ice-cold 20 mm EDTA in PBS followed by two washes in ice-cold 1% BSA in PBS. Cells were treated for 10 min with 50 μg of mouse IgG (Millipore, Temecula, CA) to block nonspecific binding sites. Cells were incubated with Alexa Fluor 647-conjugated anti-human CD317 antibody (BioLegend, San Diego, CA) for 30 min at 4 °C in the dark. Cells were then washed twice with ice-cold 1% BSA in PBS and suspended in 1% paraformaldehyde in PBS. Finally, cells were analyzed on a FACSCalibur (BD Biosciences Immunocytometry Systems). Data analysis was performed using Flow Jo (Tree Star, San Carlos, CA). For gating of transfected cells, pEGFP-N1 (Clontech) was cotransfected.
RESULTS
In Silico Analysis of BST-2: Does C-terminal Hydrophobic Region Constitute GPI Anchor Signal or Second TM Motif?
Initial biochemical studies on BST-2 were done on rat protein, which was shown to be a type II TM protein with an N-terminal TM region and a C-terminal GPI anchor (8). Human and rat BST-2 share only 33% amino acid identity, but they have similar hydropathy profiles. Both proteins contain an N-terminal hydrophobic segment representing the TM region, and they have a second hydrophobic region at the very C terminus assumed to be part of the GPI anchor signal. However, GPI anchor modification of human BST-2 has to our knowledge never been experimentally verified. Nonetheless, both the N-terminal TM region and the hydrophobic putative C-terminal GPI anchor signal are critical for inhibiting HIV-1 virion release (5). In fact, Perez-Caballero et al. (14) demonstrated that an artificial tetherin consisting of the N-terminal TM motif of transferrin receptor, a coiled coil ectodomain of the cytoplasmic dimeric protein dystrophia myotonica protein kinase, and a GPI anchor signal derived from uPAR is capable of inhibiting the release of HIV-1 virions tethered to the cell surface.
We used web-based analytical tools as detailed under “Experimental Procedures” for the prediction of GPI anchor signals and TM regions in human and rat BST-2 as well as in a series of reference proteins, including CD4, CD40, CD45, CD55, TfR, and uPAR (Table 1). big-PI Predictor uses a complex algorithm to assign score values where values of 2 or higher are considered predictive of GPI anchor modification (37, 38). As expected, big-PI Predictor found a strong prediction for GPI anchor modification at position 305 in uPAR (S = 17.02) and at position 353 in CD55 (S = 13.40), both of which have been experimentally verified to carry GPI anchor modifications (35, 36). GPI anchor signals were also predicted with slightly lower score values for human (S = 11.95) and rat BST-2 (S = 11.27) at residues 161 and 152, respectively. Alternative GPI anchor addition sites with low score values were predicted at adjacent positions (Table 1). As expected, no GPI anchor signals were predicted for CD4, CD40, CD45, and TfR.
In addition to big-PI Predictor, we used six different programs for predicting transmembrane motifs. All six tools correctly predicted an N-terminal TM region in human and rat BST-2 albeit with slight fluctuations regarding the specific positioning of the TM motif within the protein (Table 1, TM1). In addition, three of the programs predicted a second TM motif at the C terminus of human BST-2 mapping to the same area where big-PI Predictor predicted a GPI anchor modification (Table 1, TM2). Remarkably, all six programs predicted a second TM region at the C terminus of rat BST-2 (Table 1, TM2). Although this is not surprising given that both GPI anchor signals and TM motifs tend to be hydrophobic in nature (for a review, see Ref. 39), it raises questions about the reliability of GPI anchor versus TM motif predictions and emphasizes the need for experimental validation. All predictors properly predicted a TM region near the C terminus of CD4 again with slight variations in the specific positioning within the protein. Several TM predictors additionally suggested a second TM region at the N terminus of CD4; the sequence identified here represents the hydrophobic signal peptide that targets CD4 into the membrane but is clipped from the mature protein. Of note, both uPAR and CD55 were predicted by at least three programs to contain TM motifs in the region known to function as GPI anchor signals. Predictors properly calculated the TM regions of CD40, CD45, and TfR; however, as with CD4, some predictors suggested additional TM motifs, including the N-terminal signal peptides (Table 1). Predictions of protein type using MemType-2L yielded mixed results. uPAR was correctly identified as a GPI-anchored protein, and CD4 was correctly identified as a single pass transmembrane protein, but the GPI anchor-modified CD55 was incorrectly predicted to be a single pass type I transmembrane protein (Table 1). Human and rat BST-2 proteins were also not identified by MemType-2L as GPI-anchored proteins but were predicted to be a single pass type II transmembrane or peripheral membrane protein, respectively. Taken together, in silico analysis of human and rat BST-2 by a variety of web-based analytical tools in parallel with proteins known to be GPI anchor-modified or known to not have a GPI anchor produced inconsistent results, thus highlighting the difficulties of accurately predicting protein modifications solely based on amino acid sequences.
C-terminal GPI Anchor in BST-2 Can Be Replaced by Heterologous TM Region
Because of the inconsistent results from our in silico analysis, we designed a study with the aim (a) to experimentally address whether a C-terminal GPI anchor in BST-2 can be replaced by a second TM motif to yield functional protein with the ability to tether virions to the cell surface and (b) to determine whether the C-terminal putative GPI anchor signal in human BST-2 is in fact a GPI anchor or a second TM region.
Previously, BST-2 was shown to be associated with lipid rafts (8, 16); in fact, raft association of BST-2 was attributed to the presence of a C-terminal GPI anchor (16). We constructed three BST-2 chimeras carrying the TM regions of CD40, CD45, and TfR, respectively, in place of the putative BST-2 GPI anchor signal. CD40 has a TM motif known to target the protein to lipid rafts, whereas the TM region of the raft protein CD45 is not involved in lipid raft targeting (28). Finally, the TM region of TfR was chosen because TfR is a well studied non-raft-associated protein (40). An alignment of the C-terminal residues of BST-2 WT, BST-2-CD40 (TM40), BST-2-CD45 (TM45), and BST-2-TfR (TMTfR) downstream of residue 150 is shown in Fig. 1A. The schematic structures of the proteins are outlined in Fig. 1B.
We tested the ability of the TM40, TM45, and TfR chimeras to inhibit virus particle release. HIV-1 pNL4-3 WT or vpu-defective pNL4-3/Udel plasmids were transfected into 293T cells lacking endogenous BST-2 expression either alone or in combination with BST-2 WT, TM40, TM45, or TMTfR at virus:BST-2 ratios of 50:1, 20:1, and 10:1. Virus-containing supernatants were harvested 24 h later and used for the infection of TZM-bl indicator cells. Relative virus titer was determined by measuring the virus-induced expression of luciferase in the TZM-bl cells 48 h later. The signal produced by NL4-3 or NL4-3/Udel virus in the absence of BST-2 was each defined as 100%. As expected, BST-2 WT inhibited the release of NL4-3/Udel virions in a dose-dependent manner (Fig. 1C, bottom panel), whereas the release of NL4-3 WT was not inhibited (Fig. 1C, top panel). BST-2 TM45 containing the TM region of CD45 inhibited neither NL4-3 WT nor NL4-3/Udel virus release (Fig. 1C). In contrast, BST-2 TM40 carrying the raft-targeting TM motif of CD40 at its C terminus behaved similarly to BST-2 WT and did not inhibit NL4-3 WT but did inhibit the release of NL4-3/Udel although not to the same extent as BST-2 WT. The difference in inhibition might be due to reduced tethering activity or could be caused by differences in protein expression. Interestingly, BST-2 TMTfR also inhibited virus release albeit slightly less efficiently than the TM40 chimera (Fig. 1C). These results demonstrate that the C-terminal putative GPI anchor in BST-2 is not absolutely required for tethering activity but can be replaced by TM regions from heterologous transmembrane proteins. Of note, replacement of the putative GPI anchor sequence in BST-2 by heterologous TM regions, including the non-raft marker TfR, did not abolish raft association of the resulting BST-2 chimera (supplemental Fig. 1A), suggesting that sequences in addition to (or instead of) the putative GPI anchor signal are required for raft targeting of the chimera. All BST-2 chimeras, including the inactive BST-2 TM45 chimera, were expressed at the cell surface (supplemental Fig. 1B). The lack of correlation between biological function (i.e. inhibition of virus release) and raft association of our BST-2 chimeras suggests that membrane lipid raft association of BST-2 may be necessary but is not sufficient for inhibition of virus particle release.
C-terminal Hydrophobic Region in BST-2 Can Function as Second Transmembrane Motif
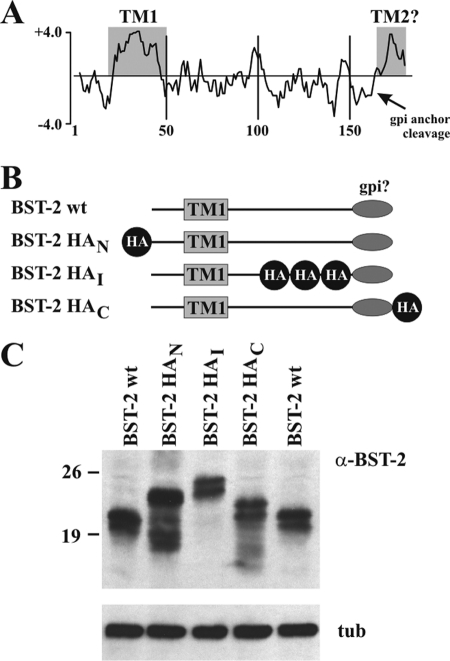
Numerous studies on BST-2 use epitope-tagged protein. In those studies, the epitope tag is either placed at the N terminus of the protein (e.g. Refs. 41 and 42) or near the C terminus in the BST-2 ectodomain (e.g. Refs. 14 and 43). C-terminal epitope tags are not commonly used to study BST-2 function because of the anticipated GPI anchor modification. A C-terminal tag would either be removed during GPI anchor modification, thus preventing identification of the protein by antibody-based assays, or result in inactive protein if the protein fails to be properly GPI anchor-modified. We previously produced an antibody to the ectodomain of BST-2 that allows us to identify the protein irrespective of the presence or absence of an epitope tag (30). We took advantage of this fact to test the above predictions regarding C-terminal epitope tagging of BST-2. To do so, we constructed a series of BST-2 variants containing an HA epitope tag either at the N terminus (BST-2 HAN), in the ectodomain N-terminal to the predicted GPI cleavage site (BST-2 HAI), or at the extreme C terminus (BST-2 HAC) as schematically shown in Fig. 2B. We reasoned that if BST-2 were in fact modified by a C-terminal GPI anchor then BST-2 HAC should not be recognized by an HA-specific antibody due to the removal of the C-terminal peptide (see Fig. 2A). Expression of the proteins was verified by immunoblotting and compared with untagged BST-2 (Fig. 2C). All HA-tagged proteins exhibited a slower mobility than untagged BST-2 consistent with the presence of a 14-residue single HA tag in BST-2 HAN and BST-2 HAC and a 32-residue triple HA tag in BST-2 HAI.
FIGURE 2.
Construction and analysis of epitope-tagged BST-2. A, hydropathy plot of human BST-2. The profile was calculated using Clone Manager software. Shown is a Kyte-Doolittle plot. Hydrophobic residues are indicated as positive values. The known transmembrane region (TM1) as well as a possible second transmembrane region near the C terminus (TM2) is indicated by shaded areas. The arrow points to the predicted GPI anchor cleavage site. B, schematic outline of HA-tagged BST-2 constructs. The HA epitope was added to the N terminus (BST-2 HAN), the ectodomain (BST-2 HAI), or the C terminus (BST-2 HAC) of human BST-2. The number of symbols reflects the number of HA epitopes present in the constructs. C, 293T cells were transfected with 0.5 μg each of HA-tagged BST-2 or 0.1 μg of WT BST-2. Whole cell lysates were prepared after 24 h and subjected to immunoblot analysis using BST-2-specific antibody. Untagged BST-2 was loaded on both sides for reference. tub, tubulin.
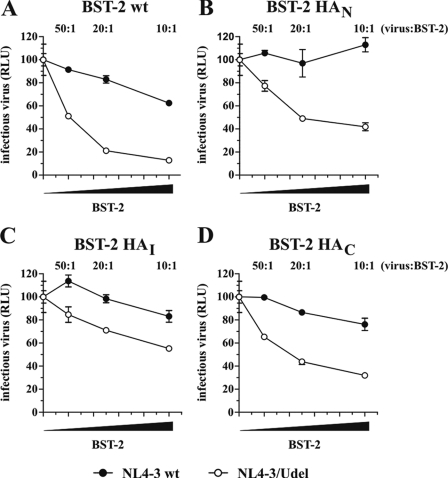
We next tested the functional properties of the HA-tagged BST-2 variants in a virus release assay. For that purpose, NL4-3 WT or NL4-3/Udel was transfected into 293T cells either alone or in combination with BST-2 WT or HA-tagged BST-2 variants at virus:BST-2 ratios of 50:1, 20:1, and 10:1. Relative virus titer was determined as in Fig. 1 by measuring the virus-induced expression of luciferase in TZM-bl cells. As expected, expression of BST-2 WT resulted in a dose-dependent inhibition of NL4-3/Udel virus release (Fig. 3A, open circles), whereas the release of NL4-3 WT was less affected (Fig. 3A, closed circles). Similarly, BST-2 HAN and BST-2 HAI inhibited the release of Udel virus in a dose-dependent manner (Fig. 3, B and C). Although inhibition by BST-2 HAN and BST-2 HAI appeared to be not quite as efficient when compared with untagged BST-2, these results nonetheless indicate that the presence of N-terminal and internal HA tags did not drastically affect the functional properties of BST-2. Importantly, BST-2 HAC also inhibited HIV-1 virus release in a Vpu-sensitive manner and with efficiency similar to that of BST-2 HAN and BST-2 HAI, indicating that the presence of a C-terminal HA tag did not affect the tethering activity of BST-2 (Fig. 3D).
FIGURE 3.
C-terminally epitope-tagged BST-2 is functional. 293T cells were transfected with 5 μg each of NL4-3 WT or NL4-3/Udel together with increasing amounts (0, 0.1 μg (50:1), 0.25 μg (20:1), or 0.5 μg (10:1)) of BST-2 WT (A), BST-2 HAN (B), BST-2 HAI (C), or BST-2 HAC (D). Virus-containing supernatants were collected 24 h later and used to infect HeLa TZM-bl indicator cells. Analysis of the data was performed as in Fig. 1C. Error bars, S.E. The signal produced by NL4-3 or NL4-3/Udel virus in the absence of BST-2 was each defined as 100%. RLU, relative luciferase units.
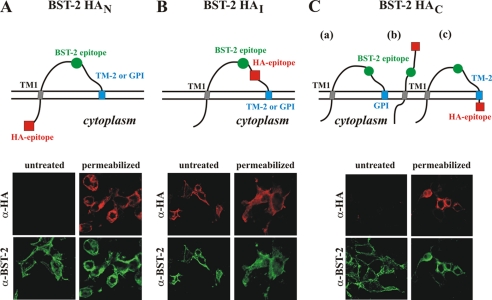
We next studied expression and topology of the HA-tagged BST-2 variants at the surface of transfected 293T cells using confocal microscopy. 293T cells were transfected with the indicated BST-2 constructs and subjected to indirect immunofluorescence either without (Fig. 4, untreated) or subsequent to permeabilization with Triton X-100 (Fig. 4, permeabilized). Samples were double labeled using BST-2-specific rabbit polyclonal antibody and an HA-specific mouse monoclonal antibody. The antibodies specifically recognized BST-2 because mock-transfected cells and untransfected cells present on the coverslips were negative (data not shown). The location of the epitopes recognized by the antibodies is schematically shown at the top of Fig. 4. The orientation of the proteins in the membrane was assessed by comparing BST-2- and HA-specific fluorescence (Fig. 4, bottom panels). As expected, all proteins were recognized by the BST-2-specific polyclonal antibody irrespective of whether the cells had been permeabilized or not. This indicates that all proteins were properly synthesized and expressed at the cell surface. Visualization of BST-2 HAN with the HA-specific antibody was dependent on cell permeabilization (Fig. 4A). This indicates that the N terminus of BST-2 localizes to the cytoplasmic side of the plasma membrane and confirms BST-2 as a type II transmembrane protein. Consistent with this result, BST-2 HAI in which both antibody epitopes are located in the ectodomain was identified by both the BST-2 and HA antibodies regardless of permeabilization of the cells (Fig. 4B). As far as BST-2 HAC is concerned, three different scenarios can be envisioned (Fig. 4C). First, if the protein is modified by a GPI anchor, then the HA tag will be removed, and no HA-specific signal should be detected even in permeabilized cells (Fig. 4C, a). Second, addition of the HA tag prevents the protein from being GPI anchor-modified. In this scenario, the C terminus of BST-2 should not be anchored in the membrane but should extend into the extracellular space, and the HA epitope should be accessible with and without cell permeabilization (Fig. 4C, b). Third, instead of a GPI anchor, BST-2 contains a second TM region. In this case, the HA epitope should remain attached to the protein but only be visible upon cell permeabilization (Fig. 4C, c). Indeed, we found that BST-2 HAC was not visible with the HA antibody in the absence of cell permeabilization but became clearly accessible to the HA antibody upon cell permeabilization. Similar results were obtained with a BST-2 variant containing a C-terminal Myc tag (data not shown). These results indicate that BST-2 HAC is a membrane protein with two TM segments as depicted in model c. This implies that the C-terminal hydrophobic region in BST-2 at least in conjunction with a C-terminal epitope tag acts as a second TM region and is sufficient for tethering activity. It is also conceivable that a small portion of the protein loses its epitope tag as part of a GPI anchor modification to produce the biological activity observed in Fig. 3. This seems unlikely, however, given that the vast majority of BST-2 HAC exhibited a slower mobility in the gel than untagged BST-2 (see Fig. 2C).
FIGURE 4.
Epitope-tagged BST-2 contains a second transmembrane region. A–C, schematics depicting possible orientations of the BST-2 variants and the location of antibody epitopes. The epitope recognized by the BST-2 antibody is symbolized by a green circle. The location of the HA epitope tag is symbolized by a red square. The horizontal double line represents the plasma membrane. The cytoplasmic side of the membrane is below the double line. a, b, and c represent possible scenarios discussed under “Results.” 293T cells were transfected with 0.5 μg each of the indicated BST-2 constructs. 24 h after transfection, cells were fixed with paraformaldehyde (1% for 30 min at room temperature) and either permeabilized or left untreated. Cells were dually stained with a rabbit polyclonal antibody to BST-2 (green channel; bottom panels) or a mouse monoclonal antibody to HA (red channel; top panels). Stained cells were examined by confocal fluorescence microscopy using a Zeiss LSM410. Images were acquired simultaneously and collected in separate image channels.
PI-PLC Treatment Fails to Release Tethered Virions
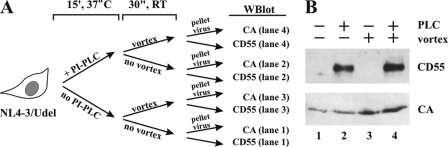
A popular model of BST-2 function predicts the tethering of otherwise fully detached virions to the plasma membrane by means of its membrane-spanning N-terminal TM region and its C-terminal GPI anchor (5). PI-PLC is known to cleave GPI anchors (44). Therefore, we hypothesized that treatment of cells producing Vpu-deficient virus with PI-PLC should result in the release of virions. To test this hypothesis, HeLa cells were transfected with pNL4-3/Udel. Cells were collected 24 h later and subjected to a PI-PLC treatment protocol as described under “Experimental Procedures” and outlined in Fig. 5A. As expected, PI-PLC treatment released CD55, a known GPI anchor protein, into the culture supernatants (Fig. 5B, lanes 2 and 4). Subjecting cells to vortexing did not result in the release of CD55 (Fig. 5B, lane 3), indicating that this procedure did not cause nonspecific cell lysis. Of note, PI-PLC treatment did not induce the release of virions above the level recovered by incubation in PLC buffer without enzyme (Fig. 5B, compare lanes 1 and 2). Importantly, however, the 3–4-fold increase in viral capsid protein recovered after vortexing of untreated as well as PI-PLC-treated cells (Fig. 5B, lanes 3 and 4) indicates that the inability of PI-PLC to release virions is not due to the absence of tethered particles at the cell surface. These results confirm a previous study by Fitzpatrick et al. (18), who also failed to demonstrate PI-PLC-induced virus release. Thus, the failure of PI-PLC treatment to induce the release of tethered virions from the surface of cells further argues against a model involving GPI anchor-dependent tethering of virions by BST-2.
FIGURE 5.
PI-PLC treatment does not release tethered virions. A, the experimental flow chart is schematically shown. B, HeLa cells were transfected with 5.0 μg of NL4-3/Udel, washed, and divided into four equal aliquots. Cells were suspended in 100 μl of PLC buffer (10 mm Tris, pH 8.0, 10 mm EDTA, 10% glycerol) and incubated in the absence (−) or presence (+) of 0.5 unit of PI-PLC for 15 min (15′) at 37 °C. After PI-PLC treatment, one set of samples was subjected to 30 s (30″) of vortexing, and supernatants were processed for immunoblot analysis as detailed under “Experimental Procedures.” CA, capsid protein; WBlot, Western blot.
BST-2 Fails to Bind Aerolysin
As described above, PI-PLC is known to cleave GPI anchors. Indeed, PI-PLC treatment rendered rat BST-2 sensitive to Triton X-100 treatment of unfixed cells in immunocytochemical analyses, consistent with the loss of raft association of BST-2 (8). We attempted to verify GPI anchor modification of human BST-2 using two strategies. First, we treated HeLa cell extracts with PI-PLC to identify a possible shift in the mobility of endogenous BST-2 as reported for other proteins (45, 46). Second, we subjected PI-PLC-treated HeLa cell extracts to floatation gradient analysis to identify a potential change in the raft association of BST-2. However, both assays failed, and we saw neither a shift in mobility nor a change in raft association of BST-2 (data not shown). Although these results argue against a GPI anchor modification of human BST-2, they represent negative data. It is possible that PI-PLC was simply ineffective in cleaving a GPI anchor on human BST-2 despite the efficient release of CD55 in Fig. 5. We also contemplated assessing cross-reactive determinant recognition of PI-PLC-treated human BST-2; however, our attempts to obtain anti-cross-reactive determinant antibody were unsuccessful.
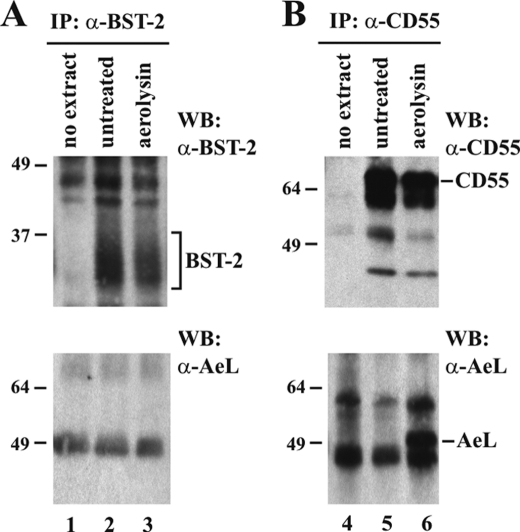
Another way of identifying GPI anchor modifications involves the binding of aerolysin. Aerolysin is a toxin produced by Aeromonas hydrophilia that binds with high affinity to GPI moieties and can then be detected by an aerolysin-specific antibody (47, 48). Aerolysin is secreted as an inactive precursor, proaerolysin, that is proteolytically cleaved to yield the active form of aerolysin (49). We obtained proaerolysin and activated the toxin by trypsin cleavage as described under “Experimental Procedures.” HeLa cell extracts were then exposed to activated aerolysin. The sample was then divided into equal aliquots and immunoprecipitated with an antibody to BST-2 (Fig. 6A) or CD55, a known cellular GPI anchor protein (Fig. 6B). Immunoprecipitates were subjected to immunoblot analysis using CD55- or BST-2-specific antiserum to ensure successful precipitation of the target proteins (Fig. 6, A and B, top panels, lanes 2 and 3). Parallel samples of immunoprecipitated proteins were probed with antibody to aerolysin to detect GPI-bound aerolysin (Fig. 6, A and B, bottom panels). Immunoprecipitation of CD55 enriched the protein in both untreated and aerolysin-treated samples (Fig. 6B, top panel, lanes 5 and 6), and probing the precipitates with an aerolysin-specific antibody revealed that, as predicted, aerolysin indeed bound to CD55 (Fig. 6B, bottom panel, lane 6). Like CD55, BST-2 was efficiently precipitated from the HeLa extracts (Fig. 6A, top panel, lanes 2 and 3). However, probing the BST-2 precipitates with an aerolysin-specific antibody failed to reveal a specific signal (Fig. 6A, bottom panel, compare lane 3 with lanes 1 and 2). Thus, whereas we were able to confirm aerolysin binding to CD55, confirming its status as a GPI-anchored protein and validating the experimental approach, we failed to detect evidence for GPI anchor modification of human BST-2 using the aerolysin binding assay.
FIGURE 6.
Lack of aerolysin binding suggests that endogenous BST-2 does not contain GPI anchor. HeLa cells were used to analyze aerolysin binding to endogenous BST-2. Cells were either left untreated (lanes 2 and 5) or were treated with aerolysin as described under “Experimental Procedures” (lanes 3 and 6). A, half of each untreated or aerolysin-treated cell extract was immunoprecipitated with BST-2-specific antibody (lanes 2 and 3). Antibody-bound beads not exposed to cell extract were included as a negative control (lane 1). Immunoprecipitates were probed with an antibody to BST-2 (top panel) or aerolysin (bottom panel). B, CD55 is a known GPI anchor protein and was used as a positive control. The second half of each aerolysin-treated or untreated cell lysate was immunoprecipitated with antibody to CD55 (lanes 5 and 6). A mock immunoprecipitation (IP) (CD55 antibodies without cell extract) was included as a negative control (lane 4). The CD55 immunoprecipitates were then either probed with antibody to CD55 (top panel) or antibody to aerolysin (bottom panel). The positions of BST-2, CD55, and aerolysin (AeL) are indicated. WB, Western blot.
C-terminal Residues of BST-2 Are Important for Tethering Activity
GPI anchor modification of human BST-2 is predicted to replace the 21 C-terminal residues of BST-2 with a GPI moiety. To further support our hypothesis that BST-2 contains a second TM region, we constructed C-terminal truncations of BST-2. Because BST-2 is glycosylated and normally runs as multiple bands (see Fig. 2C), we utilized a previously characterized construct, BST-2 N1/N2, lacking both signals for N-linked glycosylation (13). Of note, BST-2 N1/N2 is expressed at the cell surface, inhibits virus release in the absence of Vpu, and thus has the functional properties of BST-2 WT (13). Interestingly, transiently expressed BST-2 N1/N2 ran as two distinct bands (see Fig. 7B). The reason is unclear; however, it was conceivable that one of the bands represents unprocessed BST-2 (a significant portion of transiently expressed BST-2 is retained in the ER (13)), whereas the other band represents GPI anchor-modified BST-2. If this were the case, we reasoned that removal of C-terminal residues in BST-2 should eliminate the doublet (if GPI anchor modification is affected) and/or shift the mobility of one of the bands (the one representing unmodified BST-2).
We created a series of C-terminal deletion mutants of BST-2 as illustrated in Fig. 7A. In an initial series, we deleted 10, 15, 18, or 21 C-terminal residues in BST-2, which is apparent by the increased mobility of the resulting proteins (Fig. 6B, lanes 1–4). Of note, deletion of 21 residues corresponds to the removal of the entire putative GPI anchor signal (Fig. 7A, arrow). Importantly, the change in mobility caused by the deletions equally affected both protein bands, and the relative distance between the protein doublets remained constant in all mutants (Fig. 7B). Thus, the protein doublet observed upon transient expression of BST-2 N1/N2 does not represent GPI-modified and unmodified BST-2, respectively, and the nature of the protein doublet therefore remains unclear. The biological function of the BST-2 deletion mutants was assessed by cotransfecting NL4-3/Udel into 293T cells together with increasing amounts of full-length BST-2 or BST-2 mutants and measuring virus production in a standard luciferase assay as described for Fig. 1. As can be seen in Fig. 7C, all four truncation mutants were biologically inactive and failed to inhibit HIV-1 particle release in contrast to the full-length BST-2 N1/N2 variant (Fig. 7C, FL).
The C-terminal hydrophobic region in human BST-2, representing either a GPI anchor signal or a second TM region, encompasses residues 163–179 (see Fig. 2A). Thus, truncation of 10 or more residues might have prevented the protein from being processed for GPI anchor modification or from forming a second TM segment. To test whether smaller deletions from the C terminus of BST-2 would be tolerated more readily, we created a series of smaller truncation mutants lacking 2, 3, 4, or 5 residues from the C terminus of BST-2. Again, we could see a slight increase in electrophoretic mobility with each deletion using for reference the dotted line that connects the two flanking full-length proteins (Fig. 7B, lanes 5–10). Functional analysis of these mutants revealed that deletion of 2 residues from the C terminus was tolerated and did not impair inhibition of virus release by BST-2, whereas deletion of 3 or more residues led to functional impairment (Fig. 7D). The fact that deletion of 2 residues did not affect BST-2 function but was visible as a slight shift in electrophoretic mobility (Fig. 7B, compare lanes 5 and 6) is inconsistent with the removal of the C-terminal 21 residues for GPI anchor modification but supports the hypothesis that the C-terminal hydrophobic region in BST-2 in fact represents a second TM segment.
C-terminal Hydrophobic Region of BST-2 Can Act as Transmembrane Motif in Heterologous Protein
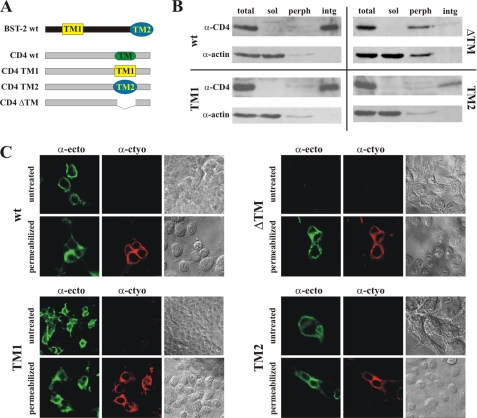
If the C-terminal hydrophobic segment of BST-2 acts as a second TM region, then this region of BST-2 should function as a TM region in heterologous proteins. To test this hypothesis, we replaced the TM region of CD4 with either the N-terminal TM region of BST-2 (TM1) or the predicted C-terminal TM region (TM2) as depicted in Fig. 8A. As a control, we also created a construct lacking the CD4 TM region (ΔTM) to verify that this region is necessary and sufficient for membrane association of CD4. Proper membrane targeting and integration of the resulting chimera was assessed biochemically in transiently transfected 293T cells using a cell fractionation assay as described under “Experimental Procedures.” Postnuclear supernatants were either analyzed directly (Fig. 8B, total) or following further fractionation by high speed ultracentrifugation into soluble (Fig. 8B, sol) and membrane fractions. The membrane fraction was extracted with sodium carbonate to further discriminate between peripheral membrane proteins (Fig. 8B, perph) and integral membrane proteins (Fig. 8B, intg). The samples were then subjected to immunoblot analysis using CD4 WT and actin as controls. As expected, CD4 WT was located primarily in the integral membrane fraction, whereas actin is a soluble protein (Fig. 8B, wt). Of note, CD4-TM1 and CD4-TM2 behaved like CD4 WT and were enriched in the integral membrane protein fraction (Fig. 8B, TM1 and TM2). In contrast, deletion of the CD4 TM motif changed the properties of the resulting protein, which no longer was purified in the peripheral membrane protein fraction but was sensitive to sodium carbonate extraction (Fig. 8B, ΔTM). Thus, the CD4 TM region is critical for stable integration into membranes. We therefore conclude that the C-terminal hydrophobic region of BST-2 is able to function as a TM motif when transferred to a heterologous protein, allowing for proper insertion into the membrane.
FIGURE 8.
Second transmembrane segment of BST-2 can function as transmembrane motif in heterologous protein. A, schematic depicting the CD4 chimera used in the analysis. TM1 and TM2 represent the N-terminal genuine TM motif and the C-terminal putative TM motif of human BST-2, respectively. The ΔTM construct lacks the TM motif of CD4. B, 293T cells were transfected with 5 μg each of the indicated CD4 chimeras and fractionated into a postnuclear supernatant, which was separated into a cytosolic soluble fraction (sol) and a membrane fraction. The membrane fraction was further fractionated into integral (intg) and peripheral (perph) fractions as described under “Experimental Procedures.” Whole cell extracts (total) were included as positive controls. The fractions were separated by SDS-PAGE and subjected to immunoblotting with antibodies to CD4 and actin. C, 293T cells were transfected with 5 μg of the indicated CD4 constructs. 24 h after transfection, cells were fixed with paraformaldehyde (1% for 30 min at room temperature) and either permeabilized or left untreated. Cells were then dually labeled with a rabbit polyclonal antibody to the cytoplasmic domain of CD4 (red channel; α-cyto) or a monoclonal antibody to the ectodomain of CD4 (green channel; α-ecto). Stained cells were examined by confocal fluorescence microscopy as described for Fig. 4. Bright field images of the sections analyzed are shown on the right.
Surface expression and membrane topology of the CD4-TM1 and CD4-TM2 chimeras were assessed by indirect immunofluorescence of unpermeabilized and permeabilized transfected 293T cells as described for Fig. 4. We used two antibodies to CD4: one selectively recognizes the cytoplasmic domain, which should be visible only upon permeabilization, and one reacts with the CD4 ectodomain, which should be accessible to staining irrespective of permeabilization. As expected, CD4, CD4-TM1, and CD4-TM2 were identified by the CD4 ectodomain antibody in non-permeabilized cells, indicating that the proteins were expressed at the cell surface (Fig. 8C, untreated). In contrast, CD4-ΔTM was not detected on non-permeabilized cells, indicating that this protein is not expressed at the cell surface (Fig. 8C, ΔTM, untreated). Surface expression of CD4-TM1 and CD4-TM2 was also confirmed by FACS using a directly conjugated commercial antibody to the CD4 ectodomain (data not shown). As expected, CD4 WT reacted with the cytoplasmic domain antibody only upon permeabilization (Fig. 8C, wt, α-cyto). Similarly, recognition of CD4-TM1 and CD4-TM2 by the antibody to the CD4 cytoplasmic domain also required permeabilization of the cells. Finally, CD4-ΔTM was recognized by both antibodies upon permeabilization, demonstrating that the lack of surface labeling in non-permeabilized cells was not due to lack of protein expression or antibody recognition (Fig. 8C, ΔTM). Thus, both the N-terminal TM region of BST-2 and the C-terminal hydrophobic region function as TM motifs when transferred into CD4 and allow for proper membrane orientation and cell surface targeting of CD4. As such, the C-terminal hydrophobic region of human BST-2 fulfills the criteria expected of a bona fide transmembrane motif.
DISCUSSION
In analogy to the rat protein, human BST-2 is commonly presumed to carry a C-terminal GPI anchor. GPI anchor addition to proteins occurs in the lumen of the endoplasmic reticulum and is catalyzed by GPI transamidase (for a review, see Ref. 50). This process involves the removal of a C-terminal GPI signal sequence. Based on in silico analysis, signal cleavage is predicted to occur on Ser-161 or Ser-162 in human BST-2 (see Table 1 and Fig. 7A, arrow). Thus, we expected epitope tags added to the C terminus of human BST-2 to be either clipped off as part of the GPI anchor addition or to result in a biologically inactive protein lacking a GPI anchor and anchored in the membrane solely by its N-terminal TM motif. Surprisingly, neither prediction turned out to be true. In fact, C-terminally tagged human BST-2 retained its tag and was biologically active. Moreover, both the N-terminal domain of BST-2 and the C-terminal epitope tag were located on the cytoplasmic side of the plasma membrane, indicating that C-terminally tagged human BST-2 contained two genuine TM regions. These observations inspired us to reinvestigate whether human BST-2 was indeed modified by GPI anchor addition as predicted from in silico analysis and in analogy to the rat protein or whether untagged and endogenous human BST-2 contains two genuine TM segments.
Our results failed to provide experimental evidence for C-terminal GPI anchor modification of human BST-2. Instead, we propose that the C-terminal hydrophobic region in human BST-2 serves as a genuine second TM motif. This conclusion is supported by our observation that replacement of the CD4 TM region by the BST-2 C terminus resulted in protein that was properly oriented in the membrane and expressed at the cell surface, suggesting that the presumed GPI anchor signal of human BST-2 has the propensity to act as a TM region when transferred to a heterologous protein. Furthermore, small deletions introduced at the C terminus resulted in identifiable shifts in electrophoretic mobility of human BST-2, suggesting that the C-terminal putative GPI anchor signal peptide in BST-2 is not removed. Importantly, replacement of the presumed GPI anchor signal in human BST-2 by the TM segments of CD40 and TfR resulted in biologically active protein capable of inhibiting the release of HIV-1 virions. This result is of critical importance as it provides proof of concept that a TM region can in principle replace a putative GPI anchor to yield BST-2 with the ability to inhibit HIV-1 virus release.
Several previous studies have indicated that the C-terminal region of BST-2 was important for lipid raft association (8, 16, 21). Our results are consistent with those observations. However, instead of GPI anchor-driven raft targeting, we propose that human BST-2 is targeted to rafts by a C-terminal region upstream of the proposed TM motif. Reports that a known GPI anchor can substitute for this region are not surprising because GPI proteins are known to associate with lipid rafts (14). On the other hand, there is precedent in the literature for TM regions capable of targeting the protein to lipid rafts (28). In fact, using one of these TM segments encoded by CD40, we obtained a BST-2 variant that was capable of inhibiting virus release and that was properly targeted to rafts. However, we were also able to target BST-2 variants to rafts using TM regions of TfR and CD45, which do not possess raft targeting properties, suggesting that some other region of BST-2 is important for raft association. In addition, raft association is not sufficient for function as BST-2 containing the TM region of CD45 associated with lipid rafts but was not biologically active.
Previous work examining whether BST-2 contained a GPI anchor was primarily performed on rat BST-2. However, results from the rat studies cannot be automatically extrapolated to the human protein. In fact, there are several known differences in the behavior of rat and human BST-2 proteins. For instance, although rat BST-2 lacking the GPI anchor signal was mislocalized to the ER (8), human BST-2 lacking the putative GPI anchor signal was efficiently transported to the plasma membrane (5, 14). Furthermore, treatment with PI-PLC decreased internalization of rat BST-2 (21) but increased internalization of human BST-2 (16). Both studies found that treatment of cells with PI-PLC resulted in reduced affinity of BST-2 to detergent-resistant membranes. However, these experiments do not address whether PI-PLC treatment directly targets BST-2 or induces an indirect effect by cleaving other raft-associated cellular protein(s). In fact, it is possible that another GPI-anchored lipid raft protein is involved in targeting BST-2 to lipid rafts. This would explain why C-terminal truncations in BST-2 result in loss of function but have only partial effects on the raft association of BST-2. Our data confirm a recent study that failed to observe enhanced virus release following PI-PLC treatment of cells (18). It cannot be ruled out of course that PI-PLC fails to cleave BST-2 when it is associated with tethered virions due to conformation constraints or steric hindrance.
The strongest experimental evidence that human BST-2 is modified by a GPI anchor comes from studies using a CHO cell line lacking phosphatidylinositol glycan class L (PIGL), an ER-resident enzyme required for the addition of GPI anchors (14). When WT BST-2 was expressed in this cell line, the protein was retained in the ER. In contrast, BST-2 lacking the C-terminal putative GPI anchor signal properly localized to the cell surface (14). Although this suggests that the C-terminal region of BST-2 is important for the protein to reach the cell surface, it does not rule out the possibility that the effect is indirect and mediated by another GPI-containing protein interacting with the C-terminal region of BST-2 and involved in BST-2 trafficking. Therefore, whereas the data by Perez-Caballero et al. (14) strongly imply the involvement of GPI-anchored protein(s) in the trafficking of BST-2, they do not provide direct experimental evidence that BST-2 itself carries a GPI anchor modification.
Finally, BST-2 orthologues from several species of owl monkey have been described that differ in their ability to inhibit HIV-1 virus release. Interestingly, a single amino acid change in the predicted GPI anchor signal peptide was found to be responsible for the different phenotypes (51). The amino acid variation does not affect GPI anchor predictions based on in silico analyses (data not shown). However, the observed functional impact of the amino acid variation is difficult to explain if the region is removed as part of a GPI anchor modification. Thus, a second TM region in owl monkey BST-2 instead of a GPI anchor would be more consistent with the observed differential effects on virus release.
The most direct evidence for determining whether or not endogenous BST-2 is modified by a GPI anchor could come from mass spectrophotometry of the protein. However, attempts to purify endogenous BST-2 to sufficient concentration and purity have thus far been unsuccessful in part due to the inherent heterogeneity of the protein caused by the presence of two N-linked carbohydrate chains. In addition, transient overexpression results in the production of predominantly immature forms of BST-2 (13), the vast majority of which is clearly not GPI anchor-modified based on the results of our current study and is therefore not suited for mass spectrometry. Our study provides compelling evidence against a GPI anchor modification of human BST-2 and strongly supports a model in which BST-2 is anchored in the membrane(s) by two genuine TM motifs. Thus, the overall model in which BST-2 prevents virus release by tethering otherwise fully detached virions to the plasma membrane of the virus-producing cell does not need to be revised based on these data.
Supplementary Material
Acknowledgments
We thank Drs. Eri Miyagi, Robert C. Walker, Jr., Sarah Welbourn, and Takeshi Yoshida for helpful suggestions and critical comments on the manuscript. We further thank Dr. Alicia Buckler-White and Ronald Plishka for conducting nucleotide sequence analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant AI000669 from the Intramural Research Program of NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- TM
- transmembrane
- GPI
- glycosylphosphatidylinositol
- uPAR
- urokinase plasminogen activator receptor
- ER
- endoplasmic reticulum
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- TfR
- transferrin receptor.
REFERENCES
- 1. Andrew A., Strebel K. (2011) J. Interferon Cytokine Res. 31, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrew A., Strebel K. (2010) Mol. Aspects Med. 31, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goto T., Kennel S. J., Abe M., Takishita M., Kosaka M., Solomon A., Saito S. (1994) Blood 84, 1922–1930 [PubMed] [Google Scholar]
- 4. Ohtomo T., Sugamata Y., Ozaki Y., Ono K., Yoshimura Y., Kawai S., Koishihara Y., Ozaki S., Kosaka M., Hirano T., Tsuchiya M. (1999) Biochem. Biophys. Res. Commun. 258, 583–591 [DOI] [PubMed] [Google Scholar]
- 5. Neil S. J., Zang T., Bieniasz P. D. (2008) Nature 451, 425–430 [DOI] [PubMed] [Google Scholar]
- 6. Van Damme N., Goff D., Katsura C., Jorgenson R. L., Mitchell R., Johnson M. C., Stephens E. B., Guatelli J. (2008) Cell Host Microbe 3, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishikawa J., Kaisho T., Tomizawa H., Lee B. O., Kobune Y., Inazawa J., Oritani K., Itoh M., Ochi T., Ishihara K., Hirano T. (1995) Genomics 26, 527–534 [DOI] [PubMed] [Google Scholar]
- 8. Kupzig S., Korolchuk V., Rollason R., Sugden A., Wilde A., Banting G. (2003) Traffic 4, 694–709 [DOI] [PubMed] [Google Scholar]
- 9. Hinz A., Miguet N., Natrajan G., Usami Y., Yamanaka H., Renesto P., Hartlieb B., McCarthy A. A., Simorre J. P., Göttlinger H., Weissenhorn W. (2010) Cell Host Microbe 7, 314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schubert H. L., Zhai Q., Sandrin V., Eckert D. M., Garcia-Maya M., Saul L., Sundquist W. I., Steiner R. A., Hill C. P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17951–17956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang H., Wang J., Jia X., McNatt M. W., Zang T., Pan B., Meng W., Wang H. W., Bieniasz P. D., Xiong Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18428–18432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swiecki M., Scheaffer S. M., Allaire M., Fremont D. H., Colonna M., Brett T. J. (2011) J. Biol. Chem. 286, 2987–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrew A. J., Miyagi E., Kao S., Strebel K. (2009) Retrovirology 6, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perez-Caballero D., Zang T., Ebrahimi A., McNatt M. W., Gregory D. A., Johnson M. C., Bieniasz P. D. (2009) Cell 139, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubé M., Roy B. B., Guiot-Guillain P., Mercier J., Binette J., Leung G., Cohen E. A. (2009) J. Virol. 83, 4574–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masuyama N., Kuronita T., Tanaka R., Muto T., Hirota Y., Takigawa A., Fujita H., Aso Y., Amano J., Tanaka Y. (2009) J. Biol. Chem. 284, 15927–15941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hauser H., Lopez L. A., Yang S. J., Oldenburg J. E., Exline C. M., Guatelli J. C., Cannon P. M. (2010) Retrovirology 7, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fitzpatrick K., Skasko M., Deerinck T. J., Crum J., Ellisman M. H., Guatelli J. (2010) PLoS Pathog. 6, e1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Habermann A., Krijnse-Locker J., Oberwinkler H., Eckhardt M., Homann S., Andrew A., Strebel K., Kräusslich H. G. (2010) J. Virol. 84, 4646–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammonds J., Wang J. J., Yi H., Spearman P. (2010) PLoS Pathog. 6, e1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rollason R., Korolchuk V., Hamilton C., Schu P., Banting G. (2007) J. Cell Sci. 120, 3850–3858 [DOI] [PubMed] [Google Scholar]
- 22. Brown D., Waneck G. L. (1992) J. Am. Soc. Nephrol. 3, 895–906 [DOI] [PubMed] [Google Scholar]
- 23. Doering T. L., Englund P. T., Hart G. W. (2001) Curr. Protoc. Mol. Biol. Chapter 17, Unit 17.8 [DOI] [PubMed] [Google Scholar]
- 24. Cardoso de Almeida M. L., Turner M. J. (1983) Nature 302, 349–352 [DOI] [PubMed] [Google Scholar]
- 25. Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klimkait T., Strebel K., Hoggan M. D., Martin M. A., Orenstein J. M. (1990) J. Virol. 64, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willey R. L., Maldarelli F., Martin M. A., Strebel K. (1992) J. Virol. 66, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bock J., Gulbins E. (2003) FEBS Lett. 534, 169–174 [DOI] [PubMed] [Google Scholar]
- 29. Zerial M., Melancon P., Schneider C., Garoff H. (1986) EMBO J. 5, 1543–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyagi E., Andrew A. J., Kao S., Strebel K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bour S., Perrin C., Strebel K. (1999) J. Biol. Chem. 274, 33800–33806 [DOI] [PubMed] [Google Scholar]
- 32. Iacovache I., Paumard P., Scheib H., Lesieur C., Sakai N., Matile S., Parker M. W., van der Goot F. G. (2006) EMBO J. 25, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chou K. C., Shen H. B. (2007) Biochem. Biophys. Res. Commun. 360, 339–345 [DOI] [PubMed] [Google Scholar]
- 34. Ono A., Freed E. O. (1999) J. Virol. 73, 4136–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ploug M., Rønne E., Behrendt N., Jensen A. L., Blasi F., Danø K. (1991) J. Biol. Chem. 266, 1926–1933 [PubMed] [Google Scholar]
- 36. Medof M. E., Walter E. I., Roberts W. L., Haas R., Rosenberry T. L. (1986) Biochemistry 25, 6740–6747 [DOI] [PubMed] [Google Scholar]
- 37. Eisenhaber B., Schneider G., Wildpaner M., Eisenhaber F. (2004) J. Mol. Biol. 337, 243–253 [DOI] [PubMed] [Google Scholar]
- 38. Eisenhaber B., Bork P., Eisenhaber F. (1999) J. Mol. Biol. 292, 741–758 [DOI] [PubMed] [Google Scholar]
- 39. Udenfriend S., Kodukula K. (1995) Annu. Rev. Biochem. 64, 563–591 [DOI] [PubMed] [Google Scholar]
- 40. Ono A. (2010) Biol. Cell 102, 335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mangeat B., Gers-Huber G., Lehmann M., Zufferey M., Luban J., Piguet V. (2009) PLoS Pathog. 5, e1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goffinet C., Homann S., Ambiel I., Tibroni N., Rupp D., Keppler O. T., Fackler O. T. (2010) J. Virol. 84, 4089–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gupta R. K., Hué S., Schaller T., Verschoor E., Pillay D., Towers G. J. (2009) PLoS Pathog. 5, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rhee S. G., Suh P. G., Ryu S. H., Lee S. Y. (1989) Science 244, 546–550 [DOI] [PubMed] [Google Scholar]
- 45. Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. (1987) Cell 51, 229–240 [DOI] [PubMed] [Google Scholar]
- 46. Walmsley A. R., Zeng F., Hooper N. M. (2001) EMBO J. 20, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buckley J. T., Halasa L. N., Lund K. D., MacIntyre S. (1981) Can. J. Biochem. 59, 430–435 [DOI] [PubMed] [Google Scholar]
- 48. Fukushima K., Ikehara Y., Kanai M., Kochibe N., Kuroki M., Yamashita K. (2003) J. Biol. Chem. 278, 36296–36303 [DOI] [PubMed] [Google Scholar]
- 49. Howard S. P., Buckley J. T. (1985) J. Bacteriol. 163, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orlean P., Menon A. K. (2007) J. Lipid Res. 48, 993–1011 [DOI] [PubMed] [Google Scholar]
- 51. Wong S. K., Connole M., Sullivan J. S., Choe H., Carville A., Farzan M. (2009) J. Virol. 83, 8771–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.