Background: Mutations in the TREX1 exonuclease gene cause a spectrum of autoimmune diseases.
Results: The TREX1 Arg-114 residue acts across the stable dimer interface.
Conclusion: TREX1 residues in one protomer contribute to DNA degradation catalyzed in the opposing protomer.
Significance: These data help to explain the heterozygous disease condition.
Keywords: Autoimmune Diseases, DNA Enzymes, Enzyme Mechanisms, Enzyme Mutation, Genetic Diseases, Nucleic Acid Enzymology
Abstract
Mutations in the TREX1 gene cause Aicardi-Goutières syndrome (AGS) and are linked to the autoimmune disease systemic lupus erythematosus. The TREX1 protein is a dimeric 3′ DNA exonuclease that degrades DNA to prevent inappropriate immune activation. One of the most common TREX1 mutations, R114H, causes AGS as a homozygous and compound heterozygous mutation and is found as a heterozygous mutation in systemic lupus erythematosus. The TREX1 proteins containing R114H and the insertion mutations aspartate at position 201 (D201ins) and alanine at position 124 (A124ins), found in compound heterozygous AGS with R114H, were prepared and the DNA degradation activities were tested. The homodimer TREX1R114H/R114H exhibits a 23-fold reduced single-stranded DNA (ssDNA) exonuclease activity relative to TREX1WT. The TREX1D201ins/D201ins and TREX1A124ins/A124ins exhibit more than 10,000-fold reduced ssDNA degradation activities. However, the TREX1R114H/D201ins and TREX1R114H/A124ins compound heterodimers exhibit activities 10-fold greater than the TREX1R114H/R114H homodimer during ssDNA and double-stranded DNA (dsDNA) degradation. These higher levels of activities measured in the TREX1R114H/D201ins and TREX1R114H/A124ins compound heterodimers are attributed to Arg-114 residues of TREX1D201ins and TREX1A124ins positioned at the dimer interface contributing to the active sites of the opposing TREX1R114H protomer. This interpretation is further supported by exonuclease activities measured for TREX1 enzymes containing R114A and R114K mutations. These biochemical data provide direct evidence for TREX1 residues in one protomer contributing to DNA degradation catalyzed in the opposing protomer and help to explain the dimeric TREX1 structure required for full catalytic competency.
Introduction
Nucleases process DNA and RNA to prevent inappropriate immune activation. TREX1 is the most abundant 3′→5′ exonuclease detected in mammalian cells and degrades both ssDNA2 and dsDNA (1–4). Deletion of the TREX1 gene in mice elicits an interferon-dependent autoimmune phenotype characterized by inflammatory myocarditis and a dramatically shortened lifespan (5–7). More than 40 mutations in human TREX1 are linked to a spectrum of autoimmune disorders including Aicardi-Goutières syndrome (AGS) (8, 9), familial chilblain lupus (10, 11), retinal vasculopathy with cerebral leukodystrophy (12), and systemic lupus erythematosus (SLE) (13, 14). These autoimmune disorders show clinical overlap with findings of raised type-I interferon, cerebrospinal fluid lymphocytosis, circulating antibodies to ssDNA and dsDNA, lupus-like rash, and progressive encephalopathy with intracranial calcifications (9, 15–18). Thus, persistent DNA resulting from the dysfunctional TREX1 enzyme activates the immune system leading to autoimmunity.
The TREX1 gene encodes a 314-amino acid protein. The TREX1 N-terminal 242 amino acids contain the catalytic domain and the C-terminal region contains a proposed transmembrane helix that is not found in TREX2. TREX1 and TREX2 have similar homodimeric structures with distinct elements that point to different biological roles (19, 20). Deletion of the TREX2 gene in mice increases susceptibility to induced skin carcinogenesis (21), contrasting the autoimmune phenotype of the TREX1 knock-out mouse. The TREX1 dimer has an 8-amino acid polyproline II helix on the top and bottom of the front face and the TREX2 dimer has a β-hairpin in the same position. The TREX1 enzyme has a flexible region positioned adjacent to the active site that has a sequence distinct from the DNA binding loop adjacent to the TREX2 active site. The TREX1 C-terminal region localizes the enzyme to the endoplasmic reticulum in the perinuclear space of cells (6, 12, 22). TREX1 relocates to the nucleus upon activation of a cell death pathway or by treatment of cells with DNA-damaging agents (7, 22) where it presumably acts to degrade DNA.
The TREX1 dimeric structure is maintained by an extensive interface that involves 1650 Å2 of buried surface area from each monomer, equivalent to 15% of the monomer surface (20). The hydrogen-bonding network between protomers includes backbone contacts of the β3 strands and six pairs of side chain contacts. The antiparallel α4 helices contribute hydrophobic packing interactions between the protomers. The dimeric structure of TREX1 is unusual for 3′→5′ exonucleases and contributes to the dominant autoimmune phenotype in some TREX1 mutations (23). The metal ion-binding residues Asp-18, Glu-20, Asp-130, and Asp-200 positions at the TREX1 active site, and the D18N, D200N, and D200H heterozygous mutations have been identified in dominant familial chilblain lupus and AGS (10, 11, 18, 24, 25). The TREX1 enzymes containing dominant D18N, D200N, and D200H mutations are defective in dsDNA degradation activity, and these TREX1 mutants inhibit the dsDNA degradation activity of the TREX1WT enzyme explaining the dominant phenotypes of these TREX1 mutant alleles (23, 26).
One of the most common TREX1 mutations found in autoimmune disease is the single nucleotide Gua→Ade missense mutation at position 341 causing the R114H substitution. The TREX1 R114H is found as homozygous and compound heterozygous in more than 60 AGS-associated disease genotypes (8, 9, 17) and R114H is mostly heterozygous in SLE with a single homozygous R114H SLE patient identified (13, 14). The TREX1 Arg-114 residue is about 15 Å removed from the active site positioned at the dimer interface where it hydrogen bonds with two backbone carbonyl oxygens of the opposing protomer. The TREX1R114H/R114H homodimer exhibits severely reduced enzymatic activity on ss- and dsDNA compared with TREX1WT (20, 23).
Compound heterozygote individuals carrying the TREX1 R114H and D201ins alleles or the TREX1 R114H and A124ins alleles have been identified in AGS (8). The TREX1 D201ins mutation is caused by a three-nucleotide GAT insertion mutation between nucleotides 600 and 601 resulting in the insertion of an extra aspartate at amino acid position 201. The TREX1 A124ins mutation is caused by a three-nucleotide GGC insertion mutation between nucleotides 368 and 369 resulting in the insertion of an extra alanine at position 124.
The mechanism by which R114H causes TREX1 catalytic dysfunction was determined. The considerable distance of Arg-114 from the TREX1 catalytic center and its location at the dimer interface raised the possibility that this residue affects activity in the opposing protomer of the TREX1 dimer. The TREX1R114H/D201ins and TREX1R114H/A124ins compound heterodimers were prepared and the remarkable stability of the TREX1 dimer was demonstrated. The activities of TREX1 R114H-containing homodimers, heterodimers, and compound heterodimers were compared with TREX1WT using quantitative ss- and dsDNA degradation assays. The activities measured in these TREX1 R114H mutants demonstrate that Arg-114 from one TREX1 protomer participates in DNA degradation occurring in the opposing protomer. This conclusion is further substantiated in TREX1 mutant homodimers, heterodimers, and compound heterodimers containing R114A and R114K. These data help to explain the stable TREX1 dimeric structure necessary for biological function and support the observed autoimmune phenotypes.
EXPERIMENTAL PROCEDURES
Materials
The synthetic 30-mer oligonucleotide 5′-ATACGACGGTGACAGTGTTGTCAGACAGGT-3′ with 5′ fluorescein was from Eurofins. Plasmid 1 (9.4 kb) was nicked with the Nt.BbvCI restriction endonuclease as previously described (23).
Enzyme Preparation
The human TREX1WT and mutant homo- and heterodimer enzymes were prepared as previously described (20, 23). Briefly, mutant TREX1-containing plasmids were produced using a PCR site-directed mutagenesis strategy (27) and confirmed by DNA sequencing. The TREX1WT and mutant homodimer enzymes (amino acids 1–242) were produced in bacteria as N-terminal maltose-binding protein (MBP) fusions with a PreScission Protease recognition sequence between the MBP and TREX1. The MBP-TREX1 fusion protein was bound to an amylose resin (New England Biolabs), washed, and the TREX1 was separated from MBP with PreScission Protease (GE Biosciences). The TREX1 was collected and purified to homogeneity using phosphocellulose chromatography.
To generate TREX1 heterodimers, one TREX1-containing plasmid was engineered to express MBP-TREX1 and a second to express His-NusA-TREX1. The two plasmids were co-expressed in the same bacterial cell to generate a mixture of TREX1 homodimers containing only the MBP, only the His-NusA affinity tags, and TREX1 heterodimers containing both affinity tags. The TREX1 heterodimers were separated from the homodimers by sequential chromatography using nickel-nitrilotriacetic acid (Qiagen) and amylose resins (New England Biolabs). The TREX1 heterodimers were purified by phosphocellulose chromatography.
Several affinity-tagged TREX1 fusion proteins were prepared to perform dimer stability studies. The His-NusA-TREX1WT homodimer was purified by nickel affinity and MonoQ chromatography. The His-MBP-TREX1WT and His-MBP-TREX1R114H homodimers were purified by nickel affinity chromatography and collected in the flow-through from a MonoQ column. The mixture of His-NusA-TREX1WT homodimers with His-NusA-TREX1WT/His-MBP-TREX1WT heterodimers was generated by coexpression of the two affinity-tagged TREX1 fusion protein constructs and subsequent purification by nickel affinity and MonoQ chromatography. All protein concentrations were determined by A280 using molar extinction coefficients for the TREX1 protomer, ϵ = 23,950 m−1 cm−1; His-MBP-TREX1, ϵ = 90,300 m−1 cm−1; His-NusA-TREX1, ϵ = 54,890 m−1 cm−1; and the His-NusA-TREX1, His-NusA-TREX1/His-MBP-TREX1 mixture, ϵ = 63,743 m−1 cm−1.
Dimer Stability Assays
The TREX1 dimer stability assays (500 μl) contained 50 mm Tris (pH 7.5), 50 mm NaCl, 1 mm EDTA, 10% glycerol, and the indicated amounts of TREX1WT or variant and His-MBP-TREX1WT or His-MBP-TREX1R114H. The reactions were incubated as described in the legend of Fig. 2. Amylose resin (30 μl) was added to the reaction and incubated with rotation for 60 min at 4 °C. The resin was washed with 1 ml of buffer A (20 mm Tris, pH 7.5, 200 mm NaCl, 1 mm EDTA) three times and the bound proteins were eluted with 200 μl of buffer A containing 20 mm maltose. Samples of the eluted proteins were analyzed by SDS-PAGE and gels were stained with Coomassie Blue (Bio-Rad).
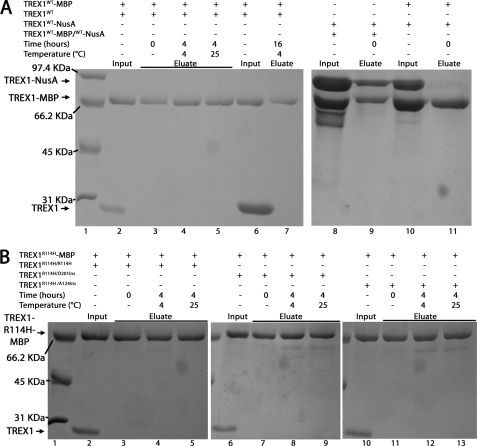
FIGURE 2.
Stability of the TREX1 dimer. A, mixtures of TREX1WT (2.5 μm, lanes 2–5, or 20 μm, lanes 6 and 7) and His-MBP-TREX1WT (2.5 μm, lanes 2–7) were incubated as indicated. Also, a mixture of His-NusA-TREX1WT homodimers with His-NusA-TREX1WT/His-MBP-TREX1WT heterodimers (2.5 μm, lanes 8 and 9) and a mixture of His-MBP-TREX1 homodimers (2.5 μm, lanes 10 and 11) and His-NusA-TREX1 homodimers (2.5 μm, lanes 10 and 11) were prepared. B, mixtures of TREX1R114H/R114H (2.5 μm, lanes 2–5), TREX1R114H/D201ins (2.5 μm, lanes 6–9), or TREX1R114H/A124ins (2.5 μm, lanes 10–13) and His-MBP-TREX1R114H (2.5 μm, lanes 2–13) were incubated as indicated. The MBP-tagged protein was recovered as described under “Experimental Procedures.” The input samples (A, lanes 2, 6, 8, and 10; and B, lanes 2, 6, and 10) and eluate samples (A, lanes 3–5, 7, 9, and 11; and B, lanes 3–5, 7–9, and 11–13) were electrophoresed on 12% SDS-PAGE gels and stained with Coomassie Blue. Lane 1 in A and B contain the indicated molecular weight standards.
Exonuclease Assays
The exonuclease assays contained 20 mm Tris (pH 7.5), 5 mm MgCl2, 2 mm dithiothreitol, 100 μg/ml of bovine serum albumin, 50 nm fluorescein-labeled 30-mer oligonucleotide (ssDNA assay) or 10 μg/ml of nicked plasmid DNA (dsDNA assay), and TREX1 protein as indicated in the legends of Figs. 3–6. Reactions were incubated at 25 °C for 15 min or as indicated, quenched by the addition of 3 volumes of cold ethanol, and dried in vacuo. For ssDNA assays, the reaction products were resuspended in 4 μl of formamide and separated on 23% denaturing polyacrylamide gels. Fluorescently labeled bands were visualized and quantified using a Storm PhosphorImager (GE Healthcare). The fraction of oligomer at each position was multiplied by the number of dNMPs excised from the 30-mer and by the total femtomoles of 30-mer in the starting reaction to determine the activities for TREX1WT and variants (fmol dNMP/s/fmol Enzyme) (19, 20, 23, 26).
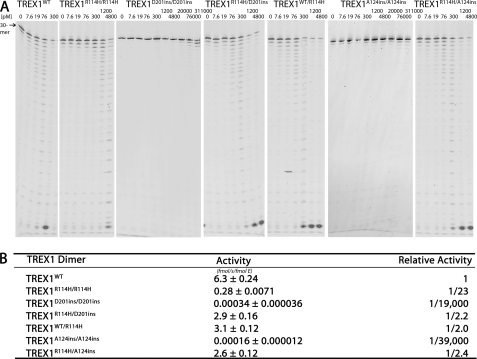
FIGURE 3.
The ssDNA exonuclease activities of TREX1WT, R114H, D201ins, and A124ins variants. A, standard exonuclease reactions were prepared with a fluorescein-labeled 30-mer oligonucleotide and dilutions of the recombinant TREX1WT, TREX1R114H/R114H, TREX1D201ins/D201ins, TREX1A124ins/A124ins, TREX1WT/R114H, TREX1R114H/D201ins, and TREX1R114H/A124ins were prepared at 10 times the final concentrations. Samples (3 μl) containing the TREX1 enzymes were added to reactions to yield the final indicated concentrations and reactions were incubated 15 min at 25 °C. The reaction products were subjected to electrophoresis on 23% urea-polyacrylamide gels and quantified as described under “Experimental Procedures.” B, to precisely quantify results, the TREX1WT and variant enzymes were assayed in triplicate at three different concentrations as described above. TREX1WT was assayed at 8, 15, and 20 pm, TREX1WT/R114H, TREX1R114H/D201ins, and TREX1R114H/A124ins were assayed at 30, 45, and 60 pm, TREX1R114H/R114H was assayed at 150, 200, and 300 pm, and TREX1D201ins/D201ins and TREX1A124ins/A124ins were assayed at 1.5, 2.3, and 3 μm. Plots of activity versus enzyme concentrations were used to confirm the linearity of the assay and generate the enzyme activity values. The average activities and S.E. were determined by regression analysis using SigmaPlot 8.02 (SPSS Science, Inc.). The relative activity was calculated as: relative activity = 100 × ((femtomole of dNMP released/s/fmol of mutant enzyme)/(fmol of dNMP released/s/fmol of WT enzyme)). The position of migration of the 30-mer is indicated.
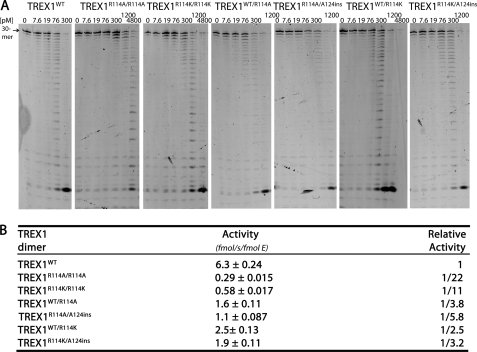
FIGURE 4.
The ssDNA exonuclease activities of TREX1WT, R114A, and R114K variants. A, standard exonuclease reactions were prepared with a fluorescein-labeled 30-mer oligonucleotide and dilutions of the recombinant TREX1WT, TREX1R114A/R114A, TREX1R114K/R114K, TREX1WT/R114A, TREX1WT/R114K, TREX1A124ins/R114A, and TREX1A124ins/R114K were prepared at 10 times the final concentrations. Samples (3 μl) containing the TREX1 enzymes were added to reactions to yield the final indicated concentrations and reactions were incubated 15 min at 25 °C. The reaction products were subjected to electrophoresis on 23% urea-polyacrylamide gels and quantified as described under “Experimental Procedures.” B, to precisely quantify results, the TREX1WT and variant enzymes were assayed in triplicate at three different concentrations as described above. TREX1WT was assayed at 8, 15, and 20 pm, TREX1WT/R114A, TREX1WT/R114K, TREX1R114A/A124ins, and TREX1R114K/A124ins were assayed at 30, 45, and 60 pm, and TREX1R114A/R114A and TREX1R114K/R114K were assayed at 100, 150, and 200 pm. Plots of activity versus enzyme concentrations were used to confirm the linearity of the assay and generate the enzyme activity values. The average activities and S.E. were determined by regression analysis using SigmaPlot 8.02 (SPSS Science, Inc.). The relative activity was calculated as: relative activity = 100 × ((femtomole of dNMP released/s/fmol of mutant enzyme)/(fmol of dNMP released/s/fmol of WT enzyme)). The position of migration of the 30-mer is indicated.
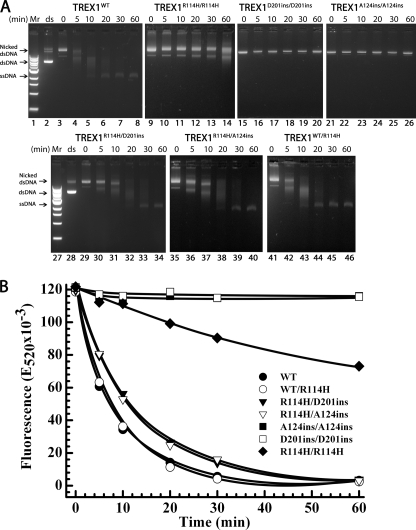
FIGURE 5.
The dsDNA degradation activities of TREX1 R114H-containing mutants. The supercoiled dsDNA plasmid 1 (A, lanes 2 and 28) was nicked and purified as described under “Experimental Procedures.” Exonuclease time course reactions (200 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml) and 15 nm of the indicated TREX1 enzyme. Samples (20 μl) were removed after incubation for the indicated times and subjected to electrophoresis on agarose gels (A) or quenched in 20 μl of 15× aqueous SYBR Green I dye (B) (Invitrogen). The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), and circular ssDNA (ssDNA) are indicated. Lanes 1 and 27 contain the 1-kb ladder (Invitrogen). The SYBR Green-quenched reaction products were quantified by excitation at 480 nm and emission was measured at 520 nm. The plot of fluorescence (E520 × 10−3) versus time was generated using SigmaPlot 8.02 (SPSS Science, Inc.).
FIGURE 6.
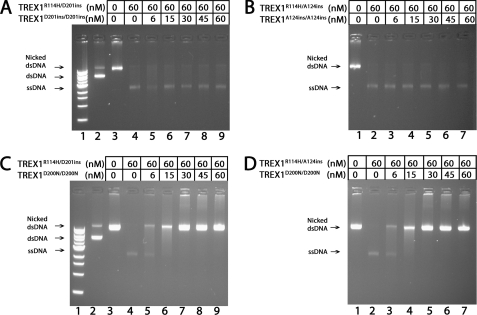
The TREX1D201ins and TREX1A124ins mutants are recessive to TREX1R114H compound heterodimer dsDNA degradation. The supercoiled dsDNA plasmid 1 (A and C, lane 2) was nicked and purified as described under “Experimental Procedures.” Exonuclease reactions (20 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml) and no enzyme (A and C, lane 3; and B and D, lane 1), or 60 nm TREX1R114H/D201ins (A and C, lanes 4–9) or TREX1R114H/A124ins (B and D, lanes 2–7), and the indicated concentrations of TREX1 D201ins/D201ins (A, lanes 5–9), TREX1A124ins/A124ins (B, lanes 3–7), or TREX1D200N/D200N (C, lanes 5–9; and D, lanes 3–7). The reactions were incubated 60 min at 25 °C, and the products were separated by electrophoresis on agarose gels stained with ethidium bromide. The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), and circular ssDNA (ssDNA) are indicated. Lane 1 (A and B) contains the 1-kb ladder (Invitrogen).
For visualization of dsDNA reaction products, assays were resuspended in 20 μl of TAE-agarose gel running solution and electrophoresed on 0.8% agarose gels containing ethidium bromide. DNA was visualized using a FluorChem 8900 imaging system (Alpha Innotech). Our dsDNA assay (23) was modified to precisely quantify the TREX1 dsDNA degradation using fluorescence emission. Samples (20 μl) were removed at the indicated times and quenched in wells of a 384-well plate containing 20 μl of 15× SYBR Green I dye (Invitrogen). Fluorescent emission at 520 nm was determined using a POLARstar Omega microplate reader (BMG LABTECH). The amount of dsDNA remaining was determined by comparing fluorescence values to those obtained from a standard curve of fluorescence emission using varied plasmid 1 concentrations (0.1–15 μg/ml) stained with SYBR Green. The amount of dsDNA degraded was used to calculate dNMPs excised and activities for TREX1WT and variants (femtomole of dNMP/s/fmol of enzyme).
RESULTS AND DISCUSSION
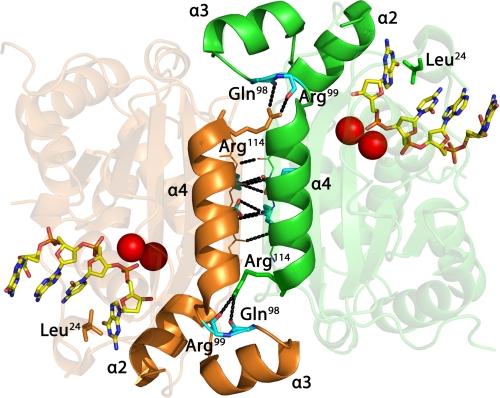
A series of TREX1 R114H-containing mutants was prepared to test the hypothesis that the Arg-114 residue of one protomer positioned at the dimer interface contributes to the catalytic function of the opposing protomer within a TREX1 dimer. Gel filtration analysis of TREX1 purified as an endogenous protein from mammalian cells (1, 28) and as a recombinant enzyme expressed in bacterial cells (3) indicate a native molecular mass consistent with a dimer structure. There is no evidence of a TREX1 protein as a monomer in these enzyme preparations. The TREX1 structure confirms the dimeric nature of the exonuclease (20). TREX1 protomers form a dimer through extensive interactions between the β3 strands of the central β-sheet and the α4 helices of each protomer (Fig. 1). The Arg-114 residues are about 15 Å away from their respective active sites positioned at opposite edges of the dimer interface. This considerable distance from the catalytic center raises questions about how mutation of Arg-114 to histidine causes reduced ss- and dsDNA degradation activities in the TREX1R114H/R114H homodimer (20, 23). The TREX1 dimer 2-fold axis of symmetry positions the active sites on opposite edges of the same face. The 3′ nucleotide base of the DNA substrate is stabilized in a hydrophobic pocket between Leu-24 and the hydrophobic face of helix α2. The helix-loop-helix motifs formed by helices α2 and α3 are positioned on opposite edges at the dimer interface (Fig. 1). The Arg-114 side chain extends from the α4 helix of one TREX1 protomer and hydrogen bonds with the backbone carbonyl oxygens of residues Gln-98 and Arg-99 positioned on a loop after the α3 helix in the opposing protomer. Mutation of Arg-114 to histidine could disrupt critical contacts between Arg-114 and the opposing protomer altering position of the α2 helix to perturb positioning of the 3′ terminal nucleotide for cleavage.
FIGURE 1.
The TREX1 dimer with bound ssDNA. The interface between TREX1 protomers (orange and green) at the α4 helices and β3 strands is shown. Arg-114 residues (orange and green) extend from the ends of the α4 helices and form hydrogen bonds with the backbone carbonyl oxygens of Gln-98 and Arg-99 (cyan) positioned on a loop between the α3 and α4 helices of the opposing protomer. The nitrogenous base of the 3′ ssDNA nucleotide interacts with the hydrophobic face of the α2 helix and Leu-24 to maintain proper positioning for catalysis. Hydrogen bonding (black dotted lines) at the dimer interface is indicated. The metal ions (red spheres) and 4-mer ssDNA (as sticks) are shown bound in the active sites. The figure was prepared using PyMol (Schrodinger, LLC).
Stability of the TREX1 Dimer
TREX1 expressed as a recombinant enzyme in bacterial cells generates kinetically stable dimers that do not dissociate and re-equilibrate at measurable rates. An experimental strategy was developed to measure the stability of the TREX1 dimer (Fig. 2). Equimolar mixtures of His-MBP-TREX1WT and TREX1WT homodimers were incubated for up to 4 h at 4 and 25 °C. The MBP-tagged enzyme was recovered using amylose resin and visualized by SDS-PAGE in an attempt to provide evidence for dissociation and reassociation of TREX1 protomers originating from the different homodimer populations. The His-MBP-TREX1WT was recovered from the mixtures with no detectable TREX1WT protomer that would have been present in His-MBP-TREX1WT/TREX1WT heterodimers generated during a dissociation and re-equilibration process (Fig. 2A, lanes 2–5). In a further attempt to detect TREX1 dimer re-equilibration, the concentration of TREX1WT was increased in the mixture to a 8-fold molar excess over the His-MBP-TREX1WT protein. The mixture was incubated for 16 h at 4 °C, and the His-MBP-TREX1WT protein was again recovered with no evidence for the generation of His-MBP-TREX1WT/TREX1WT heterodimers (Fig. 2A, lanes 6 and 7). To demonstrate that stable TREX1 dimers are recovered using the amylose resin, the His-NusA-TREX1WT protein present in the His-NusA-TREX1WT/His-MBP-TREX1WT heterodimer was recovered from a mixture of His-NusA-TREX1WT homodimers and His-NusA-TREX1WT/His-MBP-TREX1WT heterodimers (Fig. 2A, lanes 8 and 9). This result contrasts with the recovery of only the His-MBP-TREX1WT protein using the amylose resin from a mixture of His-NusA-TREX1WT and His-MBP-TREX1WT homodimers (Fig. 2A, lanes 10 and 11). These results demonstrate that the TREX1WT protomers form stable dimers that do not dissociate and re-equilibrate at detectable levels under the conditions routinely used in our exonuclease activity assays.
The TREX1 R114H-containing mutants also form stable dimers. A series of TREX1 R114H mutant enzymes was prepared to determine whether mutation of Arg-114 positioned at the dimer interface affects TREX1 dimer stability. The His-MBP-TREX1R114H homodimer was mixed with equimolar amounts of the TREX1R114H homodimer (Fig. 2B, lanes 2–5), TREX1R114H/D201ins heterodimer (Fig. 2B, lanes 6–9), and TREX1R114H/A124ins heterodimer (Fig. 2B, lanes 10–13) and incubated for up to 4 h at 4 and 25 °C. The MBP-tagged enzyme was recovered using amylose resin and visualized by SDS-PAGE. The His-MBP-TREX1R114H was recovered from the mixtures with no detectable untagged TREX1 protomer. Increased concentrations of untagged TREX1R114H homodimer in the mixtures up to a 8-fold molar excess over the His-MBP-TREX1R114H protein indicated no evidence for TREX1 dissociation and re-equilibration in the protein mixtures (data not shown). Thus, the TREX1 R114H mutation does not measurably effect the formation of stable TREX1 dimers.
The TREX1 3′ ssDNA Exonuclease Activities of Arg-114-containing Mutants
The TREX1R114H/D201ins compound heterozygous genotype has been identified in AGS (8). Because TREX1 is a stable dimer, cells of an individual with this genotype could contain TREX1R114H/R114H and TREX1D201ins/D201ins homodimers and TREX1R114H/D201ins heterodimers. The TREX1R114H/R114H and TREX1D201ins/D201ins homodimers exhibit about 23- and 19,000-fold reduced ssDNA exonuclease activities, respectively (Fig. 3), consistent with our previous studies (20, 23). The greatly reduced ssDNA exonuclease activity of the TREX1D201ins/D201ins homodimer is likely due to the aspartate insertion mutation acutely disrupting the α8-helix and preventing proper positioning of the catalytic residues for nucleotide hydrolysis (20). The TREX1R114H/D201ins heterodimer was prepared, and the ssDNA exonuclease activity was 2.2-fold reduced compared with TREX1WT (Fig. 3). This modest reduction in activity indicates that the TREX1R114H/D201ins heterodimer is about 20-fold more active than predicted if each protomer in the heterodimer acted independently. The TREX1R114H/D201ins compound heterodimers would be expected to exhibit a 46-fold reduced 3′ exonuclease activity compared with TREX1WT. Instead, this 20-fold higher activity measured in the TREX1R114H/D201ins heterodimer is attributed to Arg-114 of the TREX1D201ins protomer contributing to ssDNA degradation in the TREX1R114H protomer. Similarly, Arg-114 from the TREX1WT protomer in the TREX1WT/R114H heterodimer contributes to activity in the TREX1R114H protomer (Fig. 3), and the measured activities of the TREX1R114H/D201ins and TREX1WT/R114H heterodimers are attributed to the TREX1R114H protomer active site. These data provide direct evidence for contributions of Arg-114 to ssDNA exonuclease activity across the dimer interface in the TREX1 dimer.
A compound heterozygous AGS individual with a TREX1R114H/A124ins genotype has also been identified.3 The TREX1A124ins/A124ins homodimer and TREX1R114H/A124ins heterodimer exhibit about 39,000- and 2.4-fold reduced ssDNA exonuclease activities, respectively (Fig. 3). The additional Ala-124 insertion in the TREX1A124ins/A124ins homodimer likely alters positioning of the Arg-128 DNA-binding residue and the Asp-130 metal-binding residue resulting in a dramatic reduction in ssDNA degradation activity. Similar to the TREX1R114H/D201ins heterodimer, the TREX1R114H/A124ins heterodimer ssDNA degradation activity is about 20-fold more active than predicted based on the activities of the TREX1A124ins/A124ins and TREX1R114H/R114H homodimers, further supporting the concept that Arg-114 from the TREX1A124ins protomer contributes to ssDNA exonuclease activity in the opposing TREX1R114H protomer.
The TREX1WT/R114H, TREX1R114H/D201ins, and TREX1R114H/A124ins heterodimers have nearly identical ssDNA exonuclease activities that are reduced about 2-fold compared with TREX1WT (Fig. 3). Replacing the TREX1WT protomer with a TREX1D201ins or TREX1A124ins protomer does not further reduce the DNA degradation activity compared with the TREX1WT/R114H heterodimer. This is despite the nearly complete lack of catalytic potential in the Asp-201ins and Ala-124ins mutants apparent from the 104-fold reduced activities of the TREX1D201ins/D201ins and TREX1A124ins/A124ins homodimers. These data indicate that the TREX1R114H protomer active site is responsible for the ssDNA exonuclease activities measured in these TREX1 R114H-containing heterodimers. It is likely that the R114H mutation results in destabilization of the flexible loop between the α3 and α4 helices in the opposing protomer preventing the α2 helix from appropriately interacting with the nitrogenous base of the incoming 3′ nucleotide. Thus, the effect of the R114H mutation is transferred to the active site of the opposing protomer in the TREX1 dimer.
Arg-114 Is Required for Full Activity in the TREX1 Dimer
Arg-114 in TREX1 (Arg-107 in TREX2) is conserved in the mammalian dimeric 3′ exonucleases (19, 20). TREX1 mutants R114A and R114K were generated to further test the requirement for arginine at this position. The R114A mutation eliminates the positively charged arginine side chain and replaces it with the short neutral side chain of alanine. This amino acid substitution was used to determine whether the loss of catalytic activity in the TREX1R114H could be attributed to the loss of the arginine or to the presence of histidine at this position. The R114K mutation was prepared to determine whether the long, basic side chain of lysine could substitute for arginine at this position. The ssDNA degradation activities of the TREX1R114A and TREX1R114K homodimers are reduced by 22- and 11-fold, respectively, compared with TREX1WT (Fig. 4). The similarly reduced activities of TREX1R114H and TREX1R114A homodimers compared with TREX1WT indicate that it is the loss of arginine at this position that contributes mostly to the reduced catalytic function. This conclusion is further supported by the 11-fold reduced ssDNA degradation activity detected in the TREX1R114K homodimer compared with TREX1WT. This rather dramatic loss of catalytic function is detected despite the relatively conserved substitution of lysine for arginine at position 114. Together, the loss of ssDNA exonuclease activities in these Arg-114 mutations demonstrates the requirement for arginine at position 114 in TREX1.
The R114A and R114K Mutations Contribute to Reduced ssDNA Exonuclease Activity in the Opposing TREX1 Protomer
The TREX1WT/R114A, TREX1R114A/A124ins, TREX1WT/R114K, and TREX1R114K/A124ins heterodimers were prepared and the ssDNA exonuclease activities were measured (Fig. 4). The TREX1WT/R114A heterodimer exhibits ssDNA exonuclease activity 1/3.8 of that measured for the TREX1WT enzyme. The activity of the TREX1R114A/A124ins compound heterodimer is 1/5.8 of TREX1WT. Similar ssDNA exonuclease activities in TREX1WT/R114A and TREX1R114A/A124ins heterodimers support the concept that mutations in TREX1 at Arg-114 predominantly affect activity in the opposing protomer of the TREX1 dimer and contribute less to activity in the TREX1 protomer containing the Arg-114 mutation. Similarly, the TREX1WT/R114K heterodimer exhibits ssDNA exonuclease activity 1/2.5 of that measured for the TREX1WT enzyme. The activity of the TREX1R114K/A124ins compound heterodimer is 1/3.2 of TREX1WT. The TREX1WT/R114K and TREX1R114K/A124ins heterodimers exhibit ssDNA degradation activities greater than that of the TREX1R114K homodimer (Fig. 4). Thus, the TREX1A124ins protomer contributes to exonuclease activity in the TREX1R114K/A124ins heterodimer just as the TREX1WT protomer contributes to activity in the TREX1R114K/WT heterodimer despite the nearly complete loss of catalytic activity in the TREX1A124ins protomer. These data further indicate that TREX1 Arg-114 of one protomer contributes to the ssDNA exonuclease activity in the opposing protomer.
The TREX1 3′ dsDNA Exonuclease Activities of R114H-containing Mutants
Arg-114 in the TREX1D201ins and TREX1A124ins protomers contributes to dsDNA exonuclease activities of the TREX1R114H protomer in the TREX1R114H/D201ins and TREX1R114H/A124ins compound heterodimers. The dsDNA degradation activities of the R114H-containing mutants were directly compared in time course reactions using a nicked DNA plasmid (Fig. 5). The products were visualized on agarose gels (Fig. 5A) and quantified using SYBR Green fluorescence (Fig. 5B). Incubation of the nicked DNA with TREX1WT results in >97% complete degradation of the nicked polynucleotide strand and the accumulation of the un-nicked ssDNA (Fig. 5A, lanes 3–8). The TREX1R114H/R114H homodimer degrades the nicked DNA less efficiently compared with TREX1WT with ∼40% of the nicked DNA degraded and no detectible accumulation of the ssDNA (Fig. 5A, lanes 9–14). The TREX1D201ins/D201ins (Fig. 5A, lanes 15–20) and TREX1A124ins/A124ins (Fig. 5A, lanes 21–26) exhibit no detectable dsDNA degradation activity. However, the TREX1R114H/D201ins (Fig. 5A, lanes 29–34) and TREX1R114H/A124ins (Fig. 5A, lanes 35–40) compound heterodimers exhibit dramatically higher dsDNA degradation activities compared with the TREX1R114H/R114H homodimer with obvious accumulation of the unnicked ssDNA. The activities of the TREX1R114H/D201ins and TREX1R114H/A124ins compound heterodimers are similar to the TREX1WT/R114H (Fig. 5A, lanes 41–46). Thus, despite the lack of dsDNA degradation activities in the TREX1D201ins and TREX1A124ins protomers, these mutant protomers contribute to dsDNA degradation activity in the TREX1 R114H-containing dimer nearly as well as the TREX1WT protomer.
The dsDNA degradation activities of the TREX1 R114H-containing enzymes were precisely quantified using the nicked plasmid DNA (Table 1). The dsDNA exonuclease activity of the TREX1R114H/R114H homodimer is reduced 24-fold compared with TREX1WT and the TREX1D201ins/D201ins and TREX1A124ins/A124ins homodimers exhibit no detectable activity. The TREX1R114H/D201ins, TREX1R114H/A124ins, and TREX1WT/R114H heterodimers exhibit reduced activities of 2.1-, 2.6-, and 1.6-fold, respectively, compared with TREX1WT (Table 1). Thus, the TREX1R114H/D201ins and TREX1R114H/A124ins compound heterodimers are ∼20-fold more active than would be expected based on the dsDNA activities of the TREX1D201ins/D201ins and TREX1A124ins/A124ins homodimers. These dramatically higher heterodimer activities parallel the higher than expected activities for ssDNA degradation by these same enzymes.
TABLE 1.
The dsDNA exonuclease activity of TREX1WT and variants
| TREX1 | dsDNA activitya | Relative activityb |
|---|---|---|
| fmol/s/fmol E | ||
| WT | 8.2 ± 0.31 | 1 |
| R114H/R114H | 0.34 ± 0.028 | 1/24 |
| D201ins/D201ins | NDc | ND |
| A124ins/A124ins | ND | ND |
| R114H/D201ins | 3.9 ± 0.20 | 1/2.1 |
| R114H/A124ins | 3.5 ± 0.33 | 1/2.6 |
| R114H/WT | 5.2 ± 0.49 | 1/1.6 |
| R114A/R114A | 0.45 ± 0.038 | 1/18 |
| R114K/R114K | 0.93 ± 0.051 | 1/8.8 |
| R114A/A124ins | 1.1 ± 0.01 | 1/7.6 |
| R114K/A124ins | 3.2 ± 0.38 | 1/2.6 |
| R114A/WT | 2.0 ± 0.21 | 1/4.2 |
| R114K/WT | 3.2 ± 0.12 | 1/2.6 |
a Exonuclease reactions (200 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml) and TREX1WT at 0.5, 1.0, and 1.5 nm, TREX1R114H/R114H and TREX1R114A/R114A at 40, 60, and 80 nm, TREX1R114K/R114K at 10, 15, and 20 nm, TREX1WT/R114H, TREX1R114H/D201ins, TREX1R114H/A124ins, TREX1WT/R114K, and TREX1R114K/A124ins at 1, 1.5, and 2 nm, and TREX1WT/R114A and TREX1R114A/A124ins at 2, 3, and 4 nm. Samples were removed at 10, 15, 20, 25, and 30 min and dsDNA degradation was quantified as described under “Experimental Procedures.” Plots of activity versus enzyme concentrations confirmed linearity of the assay and were used to generate the enzyme activity values. The average activities and standard errors were determined by regression analysis using SigmaPlot 8.02 (SPSS Science, Inc.).
b The relative activity was calculated as: relative activity = 100 × ((femtomole of dNMP released/s/fmol of mutant enzyme)/(fmol of dNMP released/s/fmol of WT enzyme)).
c ND, not determined.
The TREX1 3′ dsDNA Exonuclease Activities of R114A- and R114K-containing Mutants
The dsDNA exonuclease activities of the TREX1 R114A- and R114K-containing mutants were measured (Table 1). The TREX1R114A/R114A activity is 18-fold lower than TREX1WT and TREX1R114K/R114K is reduced 8.8-fold. The TREX1R114A/A124ins and TREX1R114K/A124ins compound heterodimers are 7.6- and 2.6-fold reduced relative to TREX1WT. The TREX1R114A/WT and TREX1R114K/WT heterodimers exhibit a 4.2- and 2.6-fold reduced activity compared with TREX1WT (Table 1). Thus, the activities of the TREX1R114A/A124ins and TREX1R114K/A124ins heterodimers are about 5- and 7-fold more active than would be expected from the dsDNA degradation activities of their respective homodimers. The dsDNA degradation activities measured in the TREX1 Arg-114 containing mutants support the concept that Arg-114 contributes to the catalytic activity in the active site of the opposing protomer during dsDNA degradation.
The TREX1D201ins and TREX1A124ins Mutants Are Recessive to TREX1R114H Compound Heterodimer dsDNA Degradation
The TREX1 D201ins and A124ins mutations have been identified as homozygous and as compound heterozygous mutations with R114H in recessive AGS (8, 9). An AGS-affected individual with the TREX1R114H/D201ins or TREX1R114H/A124ins genotype could generate TREX1 dimers as mixtures of TREX1R114H/R114H, TREX1R114H/(D201ins or A124ins), and TREX1D201ins/D201ins or TREX1A124ins/A124ins. The TREX1R114H/D201ins and TREX1R114H/A124ins dsDNA degradation activities were measured in the presence of increasing amounts of the TREX1D201ins/D201ins and TREX1A124ins/A124ins homodimers to measure possible inhibitory effects on the compound heterodimers. Incubation of the nicked plasmid DNA with 60 nm TREX1R114H/D201ins (Fig. 6A, lane 4) and TREX1R114H/A124ins (Fig. 6B, lane 2) results in complete degradation of the nicked DNA as evidenced by the accumulation of the unnicked ssDNA. Similarly, upon addition of increased amounts of the TREX1D201ins/D201ins (Fig. 6A, lanes 5–9) or TREX1A124ins/A124ins (Fig. 6B, lanes 3–7), TREX1R114H/D201ins and TREX1R114H/A124ins completely degrade the nicked DNA indicating the lack of inhibition by these enzymes. These results contrast with the complete inhibition of dsDNA degradation by TREX1R114H/D201ins (Fig. 6C, lane 4) and TREX1R114H/A124ins (Fig. 6D, lane 2) upon addition of increased amounts of the TREX1D200N/D200N (Fig. 6, C, lanes 5–9, and D, lanes 3–7). These results are consistent with the recessive genetics of the TREX1 D201ins and A124ins alleles.
The Stable TREX1 Dimer
The strength of the TREX1 dimer has implications for the enzymatic behavior during DNA degradation to prevent nucleic acid-mediated immune activation. The TREX1 dimeric structure is conserved in all species expressing this enzyme indicating that the dimeric architecture is critical for its biologic and enzymatic functions. TREX1 protomers interact in the dimer along the outermost β3-strands forming an extended central antiparallel β-sheet that stretches through the length of the dimer. The remarkable strength of the TREX1 dimer indicated in structural studies (20) is validated here in solution studies demonstrating that TREX1 does not dissociate and re-equilibrate at measurable rates after the dimer is generated when expressed in bacterial cells. It is likely that TREX1 dimers exhibit similar stability properties when expressed in mammalian cells. This concept is supported by the dominant disease genetics exhibited by some TREX1 alleles that parallel precisely the biochemical properties of the TREX1 dimers during dsDNA degradation of plasmid and chromatin DNA in vitro (23, 26). Our finding that Arg-114 in one protomer contributes to the catalytic function in the active site of the opposing protomer provides the first direct evidence for a required TREX1 dimeric structure for full catalytic competency. This “across the dimer interface” action by Arg-114 might help to explain the immune dysfunction in some R114H heterozygous cases (13, 14). In addition, many TREX1 mutations in lupus and retinal vasculopathy with cerebral leukodystrophy (12) are heterozygous and locate outside the catalytic domain in the C-terminal region. The stable TREX1 dimeric structure could lead to dominant noncatalytic dysfunction effects related to cellular localization or protein interactions.
Acknowledgment
We thank Y. Crow for providing unpublished data on TREX1 AGS-causing alleles.
This work was supported, in whole or in part, by National Institutes of Health Grant GM069962 (to F. W. P.), Alliance for Lupus Research Grant 179222 (to F. W. P.), and American Heart Association Grant 10GRNT3650033 (to T. H.).
This article was selected as a Paper of the Week.
Y. Crow, unpublished data.
- ssDNA
- single-stranded DNA
- AGS
- Aicardi-Goutières syndrome
- SLE
- systemic lupus erythematosus
- dsDNA
- double-stranded DNA
- MBP
- maltose-binding protein.
REFERENCES
- 1. Höss M., Robins P., Naven T. J., Pappin D. J., Sgouros J., Lindahl T. (1999) EMBO J. 18, 3868–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mazur D. J., Perrino F. W. (1999) J. Biol. Chem. 274, 19655–19660 [DOI] [PubMed] [Google Scholar]
- 3. Mazur D. J., Perrino F. W. (2001) J. Biol. Chem. 276, 17022–17029 [DOI] [PubMed] [Google Scholar]
- 4. Mazur D. J., Perrino F. W. (2001) J. Biol. Chem. 276, 14718–14727 [DOI] [PubMed] [Google Scholar]
- 5. Morita M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., Daly G., Lindahl T., Barnes D. E. (2004) Mol. Cell. Biol. 24, 6719–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008) Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Y. G., Lindahl T., Barnes D. E. (2007) Cell 131, 873–886 [DOI] [PubMed] [Google Scholar]
- 8. Crow Y. J., Hayward B. E., Parmar R., Robins P., Leitch A., Ali M., Black D. N., van Bokhoven H., Brunner H. G., Hamel B. C., Corry P. C., Cowan F. M., Frints S. G., Klepper J., Livingston J. H., Lynch S. A., Massey R. F., Meritet J. F., Michaud J. L., Ponsot G., Voit T., Lebon P., Bonthron D. T., Jackson A. P., Barnes D. E., Lindahl T. (2006) Nat. Genet. 38, 917–920 [DOI] [PubMed] [Google Scholar]
- 9. Rice G., Patrick T., Parmar R., Taylor C. F., Aeby A., Aicardi J., Artuch R., Montalto S. A., Bacino C. A., Barroso B., Baxter P., Benko W. S., Bergmann C., Bertini E., Biancheri R., Blair E. M., Blau N., Bonthron D. T., Briggs T., Brueton L. A., Brunner H. G., Burke C. J., Carr I. M., Carvalho D. R., Chandler K. E., Christen H. J., Corry P. C., Cowan F. M., Cox H., D'Arrigo S., Dean J., De Laet C., De Praeter C., Dery C., Ferrie C. D., Flintoff K., Frints S. G., Garcia-Cazorla A., Gener B., Goizet C., Goutieres F., Green A. J., Guet A., Hamel B. C., Hayward B. E., Heiberg A., Hennekam R. C., Husson M., Jackson A. P., Jayatunga R., Jiang Y. H., Kant S. G., Kao A., King M. D., Kingston H. M., Klepper J., van der Knaap M. S., Kornberg A. J., Kotzot D., Kratzer W., Lacombe D., Lagae L., Landrieu P. G., Lanzi G., Leitch A., Lim M. J., Livingston J. H., Lourenco C. M., Lyall E. G., Lynch S. A., Lyons M. J., Marom D., McClure J. P., McWilliam R., Melancon S. B., Mewasingh L. D., Moutard M. L., Nischal K. K., Ostergaard J. R., Prendiville J., Rasmussen M., Rogers R. C., Roland D., Rosser E. M., Rostasy K., Roubertie A., Sanchis A., Schiffmann R., Scholl-Burgi S., Seal S., Shalev S. A., Corcoles C. S., Sinha G. P., Soler D., Spiegel R., Stephenson J. B., Tacke U., Tan T. Y., Till M., Tolmie J. L., Tomlin P., Vagnarelli F., Valente E. M., Van Coster R. N., Van der Aa N., Vanderver A., Vles J. S., Voit T., Wassmer E., Weschke B., Whiteford M. L., Willemsen M. A., Zankl A., Zuberi S. M., Orcesi S., Fazzi E., Lebon P., Crow Y. J. (2007) Am. J. Hum. Genet. 81, 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee-Kirsch M. A., Chowdhury D., Harvey S., Gong M., Senenko L., Engel K., Pfeiffer C., Hollis T., Gahr M., Perrino F. W., Lieberman J., Hubner N. (2007) J. Mol. Med. 85, 531–537 [DOI] [PubMed] [Google Scholar]
- 11. Rice G., Newman W. G., Dean J., Patrick T., Parmar R., Flintoff K., Robins P., Harvey S., Hollis T., O'Hara A., Herrick A. L., Bowden A. P., Perrino F. W., Lindahl T., Barnes D. E., Crow Y. J. (2007) Am. J. Hum. Genet. 80, 811–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richards A., van den Maagdenberg A. M., Jen J. C., Kavanagh D., Bertram P., Spitzer D., Liszewski M. K., Barilla-Labarca M. L., Terwindt G. M., Kasai Y., McLellan M., Grand M. G., Vanmolkot K. R., de Vries B., Wan J., Kane M. J., Mamsa H., Schäfer R., Stam A. H., Haan J., de Jong P. T., Storimans C. W., van Schooneveld M. J., Oosterhuis J. A., Gschwendter A., Dichgans M., Kotschet K. E., Hodgkinson S., Hardy T. A., Delatycki M. B., Hajj-Ali R. A., Kothari P. H., Nelson S. F., Frants R. R., Baloh R. W., Ferrari M. D., Atkinson J. P. (2007) Nat. Genet. 39, 1068–1070 [DOI] [PubMed] [Google Scholar]
- 13. Lee-Kirsch M. A., Gong M., Chowdhury D., Senenko L., Engel K., Lee Y. A., de Silva U., Bailey S. L., Witte T., Vyse T. J., Kere J., Pfeiffer C., Harvey S., Wong A., Koskenmies S., Hummel O., Rohde K., Schmidt R. E., Dominiczak A. F., Gahr M., Hollis T., Perrino F. W., Lieberman J., Hübner N. (2007) Nat. Genet. 39, 1065–1067 [DOI] [PubMed] [Google Scholar]
- 14. Namjou B., Kothari P. H., Kelly J. A., Glenn S. B., Ojwang J. O., Adler A., Alarcón-Riquelme M. E., Gallant C. J., Boackle S. A., Criswell L. A., Kimberly R. P., Brown E., Edberg J., Stevens A. M., Jacob C. O., Tsao B. P., Gilkeson G. S., Kamen D. L., Merrill J. T., Petri M., Goldman R. R., Vila L. M., Anaya J. M., Niewold T. B., Martin J., Pons-Estel B. A., Sabio J. M., Callejas J. L., Vyse T. J., Bae S. C., Perrino F. W., Freedman B. I., Scofield R. H., Moser K. L., Gaffney P. M., James J. A., Langefeld C. D., Kaufman K. M., Harley J. B., Atkinson J. P. (2011) Genes Immun. 12, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kavanagh D., Spitzer D., Kothari P. H., Shaikh A., Liszewski M. K., Richards A., Atkinson J. P. (2008) Cell Cycle 7, 1718–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kolivras A., Aeby A., Crow Y. J., Rice G. I., Sass U., André J. (2008) J. Cutan. Pathol. 35, 774–778 [DOI] [PubMed] [Google Scholar]
- 17. Crow Y. J., Rehwinkel J. (2009) Hum. Mol. Genet. 18, R130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramantani G., Kohlhase J., Hertzberg C., Innes A. M., Engel K., Hunger S., Borozdin W., Mah J. K., Ungerath K., Walkenhorst H., Richardt H. H., Buckard J., Bevot A., Siegel C., von Stülpnagel C., Ikonomidou C., Thomas K., Proud V., Niemann F., Wieczorek D., Häusler M., Niggemann P., Baltaci V., Conrad K., Lebon P., Lee-Kirsch M. A. (2010) Arthritis Rheum. 62, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 19. Perrino F. W., Harvey S., McMillin S., Hollis T. (2005) J. Biol. Chem. 280, 15212–15218 [DOI] [PubMed] [Google Scholar]
- 20. de Silva U., Choudhury S., Bailey S. L., Harvey S., Perrino F. W., Hollis T. (2007) J. Biol. Chem. 282, 10537–10543 [DOI] [PubMed] [Google Scholar]
- 21. Parra D., Manils J., Castellana B., Viña-Vilaseca A., Morán-Salvador E., Vázquez-Villoldo N., Tarancón G., Borràs M., Sancho S., Benito C., Ortega S., Soler C. (2009) Cancer Res. 69, 6676–6684 [DOI] [PubMed] [Google Scholar]
- 22. Chowdhury D., Beresford P. J., Zhu P., Zhang D., Sung J. S., Demple B., Perrino F. W., Lieberman J. (2006) Mol. Cell 23, 133–142 [DOI] [PubMed] [Google Scholar]
- 23. Lehtinen D. A., Harvey S., Mulcahy M. J., Hollis T., Perrino F. W. (2008) J. Biol. Chem. 283, 31649–31656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Günther C., Meurer M., Stein A., Viehweg A., Lee-Kirsch M. A. (2009) Dermatology 219, 162–166 [DOI] [PubMed] [Google Scholar]
- 25. Haaxma C. A., Crow Y. J., van Steensel M. A., Lammens M. M., Rice G. I., Verbeek M. M., Willemsen M. A. (2010) Am. J. Med. Genet. A 152A, 2612–2617 [DOI] [PubMed] [Google Scholar]
- 26. Fye J. M., Orebaugh C. D., Coffin S. R., Hollis T., Perrino F. W. (2011) J. Biol. Chem. 286, 32373–32382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Higuchi R. (1990) PCR Protocols: A Guide to Methods and Applications, Academic Press, San Diego [Google Scholar]
- 28. Perrino F. W., Miller H., Ealey K. A. (1994) J. Biol. Chem. 269, 16357–16363 [PubMed] [Google Scholar]