Abstract
Nuclear factor I-X3 (NFI-X3) is a newly identified splice variant of NFI-X that regulates expression of several astrocyte-specific markers, such as glial fibrillary acidic protein. Here, we identified a set of genes regulated by NFI-X3 that includes a gene encoding a secreted glycoprotein YKL-40. Although YKL-40 expression is up-regulated in glioblastoma multiforme, its regulation and functions in nontransformed cells of the central nervous system are widely unexplored. We find that expression of YKL-40 is activated during brain development and also differentiation of neural progenitors into astrocytes in vitro. Furthermore, YKL-40 is a migration factor for primary astrocytes, and its expression is controlled by both NFI-X3 and STAT3, which are known regulators of gliogenesis. Knockdown of NFI-X3 and STAT3 significantly reduced YKL-40 expression in astrocytes, whereas overexpression of NFI-X3 dramatically enhanced YKL-40 expression in glioma cells. Activation of STAT3 by oncostatin M induced YKL-40 expression in astrocytes, whereas expression of a dominant-negative STAT3 had a suppressive effect. Mechanistically, NFI-X3 and STAT3 form a complex that binds to weak regulatory elements in the YKL-40 promoter and activates transcription. We propose that NFI-X3 and STAT3 control the migration of differentiating astrocytes as well as migration and invasion of glioma cells via regulating YKL-40 expression.
Keywords: Brain, Gene Expression, Inflammation, STAT Transcription Factor, Transcription, Astrocytes, Migration
Introduction
Astrocytes, the most abundant CNS cells, are critical for many functions of normal and pathological brains (1, 2). Generation and differentiation of astrocytes from neural progenitors is controlled by activation of the JAK-STAT3, BMP-SMAD, and Notch-HES pathways in vivo (3, 4) and promoted by cytokines of the IL-6 family that activate STAT3 in vitro (5, 6). In addition to these pathways, evolutionarily conserved NFI3 transcription factors, consisting of NFI-A, -B, -C, and -X, regulate astrocyte differentiation (7, 8). NFIs are expressed in overlapping patterns during embryogenesis, with high expression levels of NFI-A, -B, and -X found in the developing neocortex (9, 10). Consequently, Nfia, Nfib, and Nfix knock-out mice show severe brain anatomical defects, including agenesis of corpus callosum (9, 10), whereas NPs from Nfix knock-out mice show defects in proliferation and migration (9, 10). NFI-A and -B control gliogenesis in chick embryonic spinal cord (7), whereas NFI-X and -C regulate the expression of late astrocyte markers during the differentiation of human NPs in vitro (7, 8). Expression of NFI-A is induced by Notch signaling in NP and leads to the demethylation of astrocyte-specific genes (11), as well as down-regulation of Notch signaling via repression of Notch effector Hes1 (12). Each of the NFI transcripts undergoes alternative splicing, generating as many as nine different NFI splice variants (13), likely possessing distinctive functions. We have recently characterized a novel human NFI-X3 splice variant, which is a potent activator of gene expression in astrocytes (14). NFI-X3 contains a unique transcriptional activation domain, completely conserved in primates, which efficiently stimulates expression of astrocyte-specific markers, such as GFAP and secreted protein acidic and rich in cysteine-like protein 1 (SPARCL1). However, the other targets of NFI-X3 have yet to be indentified.
YKL-40, also known as chitinase 3-like 1, human cartilage glycoprotein 39, and breast regression protein 39, is a secreted glycoprotein expressed in both vertebrates and invertebrates (15, 16) that belongs to the 18-glycosyl-hydrolase family of proteins but lacks glycolytic activity. YKL-40 is produced by many cell types, including macrophages, neutrophils, chondrocytes, synovial, smooth muscle, and endothelial cells, as well as several solid tumors, such as GBM, osteosarcoma, and ovarian carcinoma (17, 18). YKL-40 acts as an angiogenic factor for breast and colon cancer cells (19) and also induces VEGF expression in GBM cell lines, promoting angiogenesis and radioresistance of GBM tumors (20). Conversely, neutralizing YKL-40 antibodies block growth, angiogenesis, and progression of xenografted tumors (21). YKL-40 levels are also elevated in patients with a wide array of pathologies characterized by tissue destruction, ongoing inflammation, and development of fibrosis, such as rheumatoid arthritis, atherosclerosis, osteoarthritis, giant cell arthritis, and sarcoidosis (17), implicating its role in inflammatory processes and tissue remodeling. Increased levels of YKL-40 have also been reported in CNS pathologies, including Alzheimer disease, HIV-associated dementia, and multiple sclerosis (22–24). Although YKL-40 is produced by activated microglia (23), it has recently been shown that it is also secreted by reactive astrocytes (25).
In contrast to multiple reports implicating YKL-40 with various pathologies, its biological functions and its regulation in normal physiological conditions are only partially explored. YKL-40 stimulates proliferation of chondrocytes and synovial cells (26), and it is an adhesion and migratory factor for vascular smooth muscle and endothelial cells (17, 27, 28). Accordingly to the predicted role of YKL-40 in inflammation, YKL-40 knock-out mice have impaired antigen-induced Th2-dependent immune responses (29). The proinflammatory cytokines, such as IL-1, TNF-α, and IL-6, modulate YKL-40 expression in chondrocytes, macrophages, and also GBM cells (30, 31).
Here we report that expression of YKL-40 in astrocytes is controlled by a complex of NFI-X3 and STAT3, which are both important gliogenic factors. YKL-40 expression is also activated during postnatal brain development and differentiation of NP toward astrocytes in vitro. Functionally, YKL-40 is a migration factor for astrocytes that also regulates migration and invasion of glioma cells.
EXPERIMENTAL PROCEDURES
Cell Culture
Human BG01V embryonic stem cells (ATCC, Manassas, VA) were cultured on mitomycin C-inactivated mouse embryonic fibroblast layer. Cells were cultured in DMEM/F-12 medium supplemented with 20% knock-out serum replacement (Invitrogen), 1 mm l-glutamine, 0.1 mm nonessential amino acids, 50 units/ml penicillin, 50 μg/ml streptomycin, 4 ng/ml basic fibroblast growth factor (PeproTech, Rocky Hill, NJ), and 0.1 mm β-mercaptoethanol. The cells were propagated in 4 day cycles and enzymatically passaged with collagenase and trypsin. Human cortical astrocyte cultures were obtained from Dr. Sarah Wright (Elan Pharmaceuticals, South San Francisco, CA) and cultured as previously described (32). Human glioblastoma U373-MG cells were obtained from American Type Culture Collection (Manassas, VA), whereas human glioma LN229 cells were obtained from Dr. Dmitri Kapitonov (Virginia Commonwealth University, Richmond, VA). The cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, antibiotics, sodium pyruvate, and nonessential amino acids.
Generation of Neural Progenitors and Astrocyte Differentiation
BG01V neural progenitors were generated exactly as described previously (8). The cells were plated on laminin and poly-l-ornithin-coated dishes and propagated in NB27 medium consisting of neurobasal medium, B27 supplement, 2 mm l-glutamine, antibiotics, 20 ng/ml basic fibroblast growth factor, and 10 ng/ml leukemia inhibitory factor. The cells were maintained by passaging using trypsin. To generate astrocytes, neural progenitors were cultured on laminin- and poly-l-ornithin-coated dishes in DMEM, supplemented with 10% FBS, 2 mm l-glutamine, antibiotics, and nonessential amino acids for 21–24 days.
Cytokines and Cell Stimulation
The cells were stimulated with 25 ng/ml OSM, 25 ng/ml IL-6, and 25 ng/ml sIL-6R (all from R & D, Systems, Inc., Minneapolis, MN) or 200 ng/ml purified YKL-40 (Quidel Corp., San Diego, CA).
Wound Healing Assay
The cells were plated in 6-well plates (3 × 105/well for U373 cells and 1.5 × 105/well for astrocytes) to obtain a single confluent monolayer. Twenty-four hours later, media from each well were removed and stored in labeled Eppendorf tubes. Subsequently, fresh media were added to the cells, and scratches were made using 20-μl sterile pipette tips. Cell debris from scratch were removed by washing cells once with fresh media followed by adding back the original media from each well. Pictures of the wounded area (eight fields/well) were taken at time points mentioned in the respective figure legends. The number of cells migrated in the wound area are counted in each field, and the data are presented as the cells migrated per field. The experiments were done twice. In the case of siRNA experiments, 2 × 105 cells/well were plated followed by siRNA treatment for 30 h, and subsequently scratches were made.
Transwell Migration Assay
Transwell migration was measured in a modified Boyden chamber using polycarbonate filters (25 × 80 mm, 12-μm pore size) coated with fibronectin. The cells were added to the upper chamber at 5 × 104/well. After 8 or 12 h, as indicated in figure legends, the cells on the upper membrane surface were removed using cotton swabs, and the cells that migrated through the pores and spread on the lower surface of the filter were fixed and stained with Diff-Quik (Fisher). The migrated cells were counted using an inverted microscope and a 10× objective. Each data point is the average number of cells in five random fields and is the mean ± S.D. of five individual wells.
Invasion Assay
The assays were performed exactly as described in the Transwell migration assay section except that the filters were coated with Matrigel.
Down-regulation of Target Genes
Expression of NFI, NFI-X, STAT3, STAT1, and YKL-40 mRNAs was down-regulated using SmartPool siRNAs from Dharmacon (Lafayette, CO). The NFI-X3-specific siRNAs (si-NFI-X3) was designed to specifically target mRNA encoded by exon 9 (5′-UCCUAUGCCUGAUUCCAAAUU-3′) (software provided on Dharmacon's website) and transfected into astrocytes using Dharmafect 1.
Quantitative PCR
Total RNA was prepared by TRIzol (Invitrogen) and 1 μg of RNA was reverse-transcribed using the high capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). YKL-40, NFI-X, STAT3, STAT1, and GAPDH mRNA levels were examined using premixed primer-probe sets and TaqMan Universal PCR Master Mix (Applied Biosystems). The cDNAs were diluted 10-fold (for the target genes) or 100-fold (for GAPDH) and amplified using the ABI 7900HT cycler. The expression of NFI-X3 splice isoform was analyzed using the Power SYBRGreen PCR kit (Applied Biosystems) as previously described (14). Gene expression levels were normalized to GAPDH and presented as a fold induction with the mean values ± standard deviation. Statistical analysis was performed by one-way analysis of variance. Differences were considered statistically significant when the p values were <0.05.
Microarray Analysis
Total RNA was isolated by TRIzol and extracted with the MagMAXTM-96 for microarray total RNA isolation kit (Ambion, Austin, TX) in the automated magnetic particle processor MagMAX Express (Applied Biosystems). RNA purity was judged by spectrophotometry at 260, 270, and 280 nm, and RNA integrity was judged by running 1-μl samples in RNA 6000 Nano LabChips® on the 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). Microarray-based gene expression analyses were performed using the Affymetrix® protocol as previously described (33). Briefly, starting from 500 ng of total RNA, cDNA synthesis and cRNA labeling were performed using the GeneChip® 3′ IVT express kit (Affymetrix, Santa Clara, CA). 10 μg of fragmented cRNA were hybridized on the GeneChip® HG-U133A 2.0 array for 16 h at 60 rpm in a 45 °C hybridization oven. The arrays were washed and stained with streptavidin phycoerythrin (Molecular Probes, Eugene, OR) in the Affymetrix fluidics work station. Every chip was scanned at a high resolution, with pixelations ranging from 2.5 μm down to 0.51 μm, by the Affymetrix GeneChip® Scanner 3000. After scanning, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip® operating software (v1.4; Affymetrix). The overall quality of each array was assessed by monitoring the 3′/5′ ratios for a housekeeping gene (GAPDH) and the percentage of “Present” genes (%P), where arrays exhibiting GAPDH 3′/5′ < 3.0 and %P > 40% were considered good quality arrays.
Microarray Statistical Analysis
Background correction, normalization, and estimation of probe set expression summaries were performed using the log-scale robust multiarray analysis method (34). Identification of altered gene expression among the U373-vec and U373-NFI-X3 cells was assessed by using the Significance score (S-score) method (35). The S-score method uses an error-based model to determine the variances for probe pair signals and follows a normal standard distribution. The procedure produces scores centered around 0 (no change) with a standard deviation of 1. Thus, scores >2 or <−2 from a single comparison have, on average, a 95% chance of being significant hybridization changes, at a univariate level, corresponding to a p value of less than 0.05. Because this approach does not take into account the number of multiple comparisons inherent to any microarray experiment, we used the Benjamini-Hochberg correction method (36) to correct for multiple testing and obtained adjusted alpha-levels for each probe set.
Western Blotting
The cells were lysed in 10 mm Tris, pH 7.4, 150 mm sodium chloride, 1 mm EDTA, 0.5% Nonidet P-40, 1% Triton X-100, 1 mm sodium orthovanadate, 0.2 mm PMSF, and protease inhibitor mixture (Roche Applied Science). The samples were resolved using SDS-PAGE and electroblotted onto nitrocellulose membranes (Schleicher & Schuell). The anti-YKL-40, anti-NFI and anti-β-tubulin antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and anti-Stat3 antibody from Cell Signaling Technology (Danvers, MA). Antigen-antibody complexes were visualized by enhanced chemiluminescence using Immobilon Western blotting kit (Millipore, Temecula, CA).
Coimmunoprecipitation
Protein lysates were prepared as described in Western blotting section. Briefly, lysates containing 200–300 μg of protein were precleared with 25 μl of the protein G-Sepharose beads (GE Healthcare) for 1 h. The lysates were then incubated with 2 μg of anti-NFI antibody overnight at 4 °C. Subsequently, 30 μl of protein G-Sepharose beads were added and incubated for 1 h at 4 °C. The beads were extensively washed with the lysis buffer, and immunoprecipitated proteins were eluted in sample buffer at 95 °C for 5 min.
Synthetic Oligonucleotides
The following oligonucleotides were synthesized to introduce mutations into the regulatory elements of the YKL-40 promoter: distal STAT, 5′-ACATGCTAGCAAGCCCCCATCATCACCC-3′ and 5′-GCTTGCTAGCATGTGACTCAGCCGCATTTCC-3′; proximal STAT, 5′-CAGTTGGTACCAAAGGGCTGGTTTGCCGCG-3′ and 5′-CCTTTGGTACCAACTGAGCTATGTGTCAATGAAG-3′, distal NFI, 5′-TCCACGAGCTCAAGGGGCCTGGAG TGAATGC-3′ and 5′-CCCTGAGCTCGTGGAAACTGGGCTCAATTTAG-3; proximal NFI, 5′-CTTCAGCATGCCATAGCTCAGTTCCCATAA-3′ and 5′-CTATGGCATGCTGAAGGAATCACGAGGC-3′; and AP-1, 5′-CGGCAAGCTTACATCTCCAGAAGCCCCCC-3′ and 5′-ATGTAAGCTTGCCGCATTTCCCAGCCTTC-3′. Following oligonucleotides were used in EMSA: STAT probe, 5′-GGGCTCAGTTCCCATAAAAGGG-3′ and 5′-GGCCCTTTTATGGGAACTGAGC-3′; NFI-STAT probe, 5′-GGCTTCATTGACACATAGCTCAGTTCCCATAAAAGGG-3′ and 5′-GGCCCTTTTATGGGAACTGAGCTATGTGTCAATGAAG-3′; NFI probe, 5′-GGCTTCATTGACACATAGCTCAGTT-3′ and 5′-GGAACTGAGCTATGTGTCAATGAA-3′; and AP-1 probe, 5′-GATCTCGGCTGAGTCACATCA-3′ and 5′-GATCTGATGTGACTCAGCCGA-3′.
Nuclear Extract Preparation and EMSA
Nuclear extract from astrocytes were prepared as described (37). Double-stranded DNA fragments were labeled by filling in the 5′-protruding ends with Klenow enzyme using [α-32P]dCTP (3000 Ci/mmol). EMSA was carried out according to the published procedures (37). Briefly, 5 μg of nuclear extracts and ∼10 fmol (10,000 cpm) of probe were used. The competition experiment was performed in the presence of a 100-fold excess of the cold oligonucleotides. Anti-NFI, anti-c-Fos, anti-FosB, anti-Fra-1, anti-Fra-2, anti-c-Jun, anti-JunB, anti-JunD (Santa Cruz Biotechnology, Santa Cruz, CA), anti-STAT3, anti-STAT1 (Cell Signaling Technology, Danvers, MA), and anti-FLAG (M2) (Sigma-Aldrich) antibodies were used in the supershift studies.
ChIP Assay
The cells were cross-linked with 1% formaldehyde for 10 min at 37 °C and then washed with ice-cold PBS containing 125 mm glycine and 1 mm PMSF. Chromatin was sonicated and immunoprecipitated using specific antibodies exactly as described in the chromatin immunoprecipitation protocol from Upstate Inc. (Charlottesville, VA). The following antibodies were used: anti-acetyl-H3 (06–599, Millipore, Billerica, MA), anti-FLAG (M2) (Sigma), and anti-STAT3 (9132, Cell Signaling Technology, Danvers, MA). The following primers were used in the qPCR: 5′-CTCTGCGGTTTCTCAACCC-3′ and 5′-CCAGGCCCTGTACTTCCTTT-3′ for the YKL-40 promoter.
Plasmids
The pYKL(−1300)Luc reporter was provided by Dr. Michael Rehli (University of Regensberg, Regensberg, Germany) (38). Reporter plasmids containing mutations introduced into the putative binding sites were generated by QuikChange II site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) using the oligonucleotides described above. The generation of plasmids expressing NFI-X1 and NFI-X3 were described (14).
Transfections
The cells were transiently transfected in 12-well clusters using FuGENE6 transfection reagent (Roche Applied Science). Eight hours post-transfection, the cells were stimulated by OSM for 24 h, as indicated in the figures, followed by cell extract preparation, and the protein concentration was determined by the BCA method (Sigma). Luc assays were performed using a dual luciferase reporter assay kit (Promega Corporation, Madison, WI). Luc activities were normalized to Renilla activity and are the means ± S.E. (five to seven determinations). Stable cells overexpressing NFI-X1 and NFI-X3 were described previously (14).
RESULTS
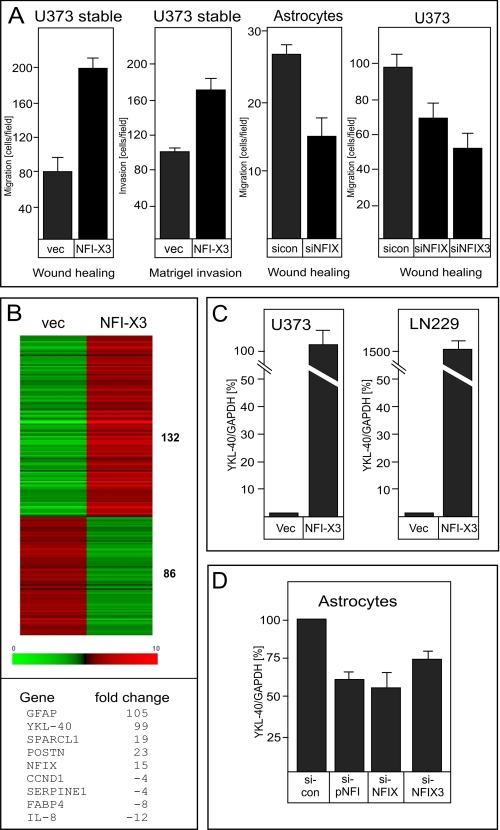
NFI-X3 Affects Expression of a Set of Genes and Enhances Migration of Glioma Cells and Astrocytes
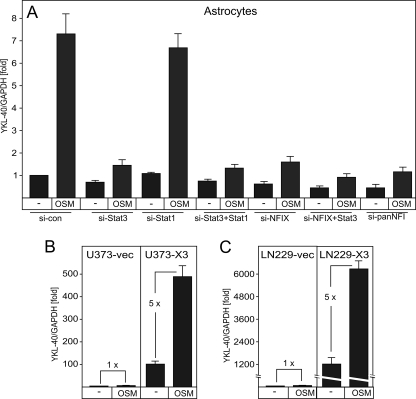
We have recently characterized a novel NFI-X3 splice variant, which regulates the expression of GFAP and SPARCL1 in both primary human astrocytes and human NP differentiating toward astrocytes in vitro (8, 14). NFI-X3 also up-regulates these astrocyte-specific markers when overexpressed in U373 glioma cells (14). In the course of our experiments, we noticed that overexpression of NFI-X3 in glioma cells significantly induced their migration (Fig. 1A, left panel) and invasion (Fig. 1A, second panel). To test whether NFI-X3 controls migration under normal physiological conditions, we knocked down expression of NFI-X in primary human astrocytes and found that migration of these cells was substantially diminished (Fig. 1A, third panel). In addition, knockdown of either NFI-X or NFI-X3 in U373 cells inhibited migration of these cells (Fig. 1A, right panel and supplemental Fig. S1A). Because NFI-X3 is a potent transcriptional activator, we hypothesized that it up-regulates expression of gene(s) that promote cell migration and invasion. To identify novel targets of NFI-X3 that promote migration, we analyzed global gene expression in glioma cell pools stably expressing NFI-X3 using microarray analysis. This analysis identified 132 genes whose expression was strongly up-regulated and 86 genes significantly repressed by NFI-X3 (Fig. 1B). The genes encoding GFAP and SPARCL1, which we have previously reported as NFI-X3-regulated (14), were among the most up-regulated by NFI-X3 (Fig. 1B, lower panel). Interestingly, gene encoding YKL-40, a previously identified marker of invasive glioma (39, 40), was the second most up-regulated gene by NFI-X3. To exclude the possibility of cell line-specific effects, we also analyzed the expression of YKL-40 in LN229 glioma cell pools overexpressing NFI-X3. Similarly to U373 cells, NFI-X3 strongly induced YKL-40 expression in LN229 cells (Fig. 1C). In contrast, overexpression of NFI-X1, another splice variant of NFI-X lacking the long proline-rich transcription activation domain, in either U373 or LN229 cells did not significantly induce YKL-40 expression (data not shown). We have also verified that NFI-X3 regulates YKL-40 in primary human astrocytes. Significantly, a knockdown of either all of the NFIs or NFI-X reduced the expression of YKL-40 by ∼45% in primary astrocytes, whereas a specific knockdown of NFI-X3 reduced YKL-40 expression by 30% (Fig. 1D and supplemental Fig. S1B).
FIGURE 1.
NFI-X3 is a critical regulator of migration and invasion of glioma cells and astrocytes. A, migration of U373 cell pools overexpressing vector (vec) or NFI-X3 expression plasmid (NFI-X3). Migrated cells were counted after 24 h as described under “Experimental Procedures” (left). The indicated U373 cell pools were allowed to invade through the Matrigel-coated membranes for 8 h (second panel). Primary human astrocytes were transfected with control siRNA (si-con) or siRNA targeting NFI-X (si-NFI-X) and wounded 48 h post-transfection. Migrated astrocytes were counted after 15 h (third panel). U373 cells were transfected with control siRNA (si-con), siRNAs specifically targeting NFI-X (si-NFIX), or NFI-X3 (si-NFI-X3) for 48 h. Migration of cells was analyzed after 30 h as described under “Experimental Procedures” (right). B, microarray-based gene expression analyses (U373 cells expressing either vector or NFI-X3) was performed as described under “Experimental Procedures.” Heat map of 132 up-regulated and 86 repressed NFI-X-responsive genes. A list of the most affected (up-regulated or repressed) genes and the fold change is shown (lower panel). C, RNA was isolated from stable pools of U373 (left panel) or LN229 (right panel) expressing vector or NFI-X3 expression plasmid. YKL-40 expression was analyzed by qPCR. D, primary human astrocytes were transfected with control siRNA (si-con), siRNAs specifically targeting all of the NFI transcripts (si-panNFI), NFI-X (si-NFIX), or NFI-X3 (si-NFI-X3). 48 h post-transfection, RNA was isolated, and expression of YKL-40 was examined by qPCR.
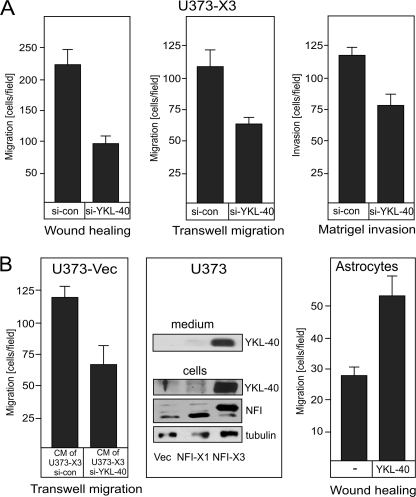
YKL-40 Affects Migration and Invasion of Glioma Cells
Because overexpression of NFI-X3 up-regulates glioma cell migration and invasion (Fig. 1A), NFI-X3 regulates YKL-40 expression (Fig. 1, B–D), and YKL-40 is associated with invasive GBM tumors (39), we explored whether NFI-X3 controls migration and invasion of glioma cells via the YKL-40-dependent mechanism. Knockdown of YKL-40 expression in U373-X3 cells (supplemental Fig. S2) drastically reduced migration of these cells in both wound healing (Fig. 2A, left panel) and Transwell migration assays (Fig. 2A, center panel) and also significantly diminished their invasion into Matrigel (Fig. 2A, right panel). Because YKL-40 is a secreted protein, we further explored its paracrine role in migration of glioma cells. For this purpose, we collected the conditioned media of U373-X3 cells transfected with either control or YKL-40 siRNAs, added these media to U373-vec cells, and analyzed their migration. Significantly, U373-vec cells migrated twice slower in the presence of medium of si-YKL-40-treated than si-con-treated U373-X3 cells (Fig. 2B, left panel). Medium of U373-X3 cells contained high levels of YKL-40 in comparison with cells expressing either the vector or NFI-X1 (Fig. 2B, center panel). Moreover, exogenous purified YKL-40 significantly promoted migration of astrocytes (Fig. 2B, right panel). These results suggest that NFI-X3-induced YKL-40 regulates migration of astrocytes and migration and invasion of glioma cells.
FIGURE 2.
YKL-40 affects migration and invasion of glioma cells and astrocytes. A, U373-X3 cells were transfected with si-con or siRNA targeting YKL-40 (si-YKL-40), and their migration (left and center panels, 30 and 12 h, respectively) and Matrigel invasion (right panel, 12 h) were analyzed as described under “Experimental Procedures.” B, conditioned medium (CM) of si-con- or si-YKL-40-treated U373-X3 cells was added to U373-vec cells, and migration was analyzed after 12 h (left). YKL-40 protein was detected by Western blotting in conditioned medium and whole cell lysates of U373 cells overexpressing either vector, NFI-X1, or NFI-X3 expression plasmids. Tubulin was used as a loading control (center). Primary human astrocytes were treated with 200 ng/ml purified YKL-40 for 24 h, and their migration was analyzed as above (right).
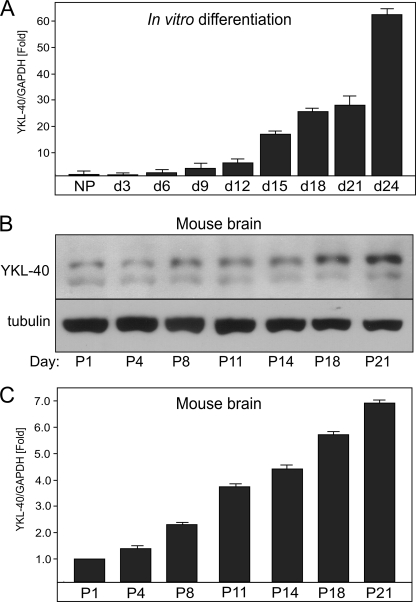
YKL-40 Is Expressed during Both Brain Development and Astrocyte Differentiation in Vitro
YKL-40 is expressed during development of tissues such as cartilage (17). Because YKL-40 is expressed in GBMs, we hypothesized that it may also be expressed by astrocytes that are generated during the late embryonic and early postnatal brain development (41). First, we examined the expression of YKL-40 during the in vitro differentiation of human NPs into astrocytes (8). Interestingly, the expression of YKL-40 was dramatically increased (∼60-fold) during astrocyte differentiation (Fig. 3A). Second, we could easily detect YKL-40 in primary cultures of human embryonic astrocytes (Figs. 1D, 4, and 5). YKL-40 expression also gradually increases during the postnatal mouse brain development at both RNA and protein levels (Fig. 3, B and C) with YKL-40 levels increased ∼7-fold at postnatal day 21 and relatively unchanged until 6 months (data not shown). These data suggest that astrocytes, at least in part, account for the YKL-40 expression in the brain during its development.
FIGURE 3.
YKL-40 is expressed during mouse brain development in vivo and human astrocyte differentiation in vitro. A, human NPs were differentiated into astrocytes. RNA was isolated every 3 days (d), and the expression of YKL-40 was analyzed by qPCR. Target gene expression was normalized to GAPDH mRNA and is represented as a fold induction in reference to NPs. B and C, YKL-40 expression in total brain homogenates was analyzed by Western blotting (B) and qPCR (C) at the indicated times.
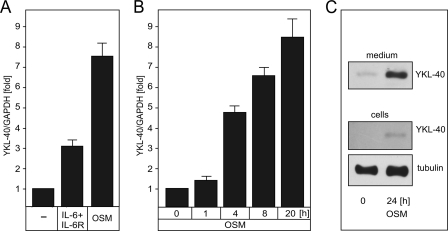
FIGURE 4.
OSM and IL-6/sIL-6R stimulate YKL-40 expression in astrocytes. A, astrocytes were stimulated with IL-6/sIL-6r or OSM for 24 h, and YKL-40 expression was analyzed by qPCR. B, astrocytes were stimulated with OSM for the indicated times, and YKL-40 expression was analyzed by qPCR. C, astrocytes were stimulated with OSM for 24 h, and YKL-40 was detected in medium and cells by Western blotting. Tubulin was used as a loading control.
FIGURE 5.
STAT3 and NFI-X are essential for the activation of YKL-40 expression by OSM. A, astrocytes were transfected with the indicated siRNAs. Forty hours post-transfection, the cells were stimulated with OSM for 24 h, and YKL-40 expression was analyzed by qPCR. B and C, U373 (B) and LN229 (C) cell pools overexpressing vector or NFI-X3 were stimulated with OSM for 24 h. YKL-40 expression was analyzed by qPCR. YKL-40 expression in unstimulated U373 (B) and LN229 (C) cells transfected with vector was set to 1.
STAT3 and NFI-X3 Are Critical for OSM-stimulated YKL-40 Expression
Because signaling of the IL-6 family of cytokines through the JAK-STAT pathway is critical for astrocyte differentiation (4), we explored whether YKL-40 is regulated by this pathway. To this end, we stimulated astrocytes with OSM and IL-6 together with soluble IL-6 receptor (sIL-6R) (because astrocytes express a limited amount of IL-6R (32)). Expression of YKL-40 mRNA was activated 8–10-fold by OSM and 3-fold by IL-6/sIL-6R (Fig. 4A), and this time-dependent stimulation on the mRNA level (Fig. 4B) was followed by the subsequent secretion of YKL-40 into the medium (Fig. 4C). To identify which of the STATs controls YKL-40 expression, we down-regulated the expression of STAT1 and STAT3, which are known to be activated by OSM. Knockdown of STAT3 alone or together with STAT1 (supplemental Fig. S3) reduced basal expression of YKL-40 by 30–40% and almost abolished OSM stimulation (Fig. 5A). In contrast, knockdown of STAT1 had no effect on YKL-40 expression (Fig. 5A). These data suggest that STAT3, but not STAT1, controls OSM-induced YKL-40 expression in astrocytes.
Because NFI-X3 regulates basal expression of YKL-40 in astrocytes and glioma cells (Fig. 1), we asked whether NFI-X3 is also critical for the OSM-induced expression. We knocked down the expression of NFI-X, as well as all NFIs, in astrocytes (supplemental Fig. S3) and analyzed both basal and OSM-induced YKL-40 expression. Surprisingly, knockdown of NFIs and NFI-X diminished not only the basal but also OSM-induced YKL-40 expression (Fig. 5A), implying that NFI-X3 is needed for OSM responsiveness in astrocytes. In another approach, we utilized glioma cells overexpressing NFI-X3 and examined YKL-40 expression upon OSM treatment. The expression of YKL-40 was induced by ∼5-fold in NFI-X3-overexpressing cells but not in cells transfected with empty vector (Fig. 5, B and C). Collectively, these data demonstrate that both STAT3 and NFI-X3 are needed for OSM-induced YKL-40 expression.
Identification of Regulatory Elements within the YKL-40 Promoter Critical for Its Activation by OSM
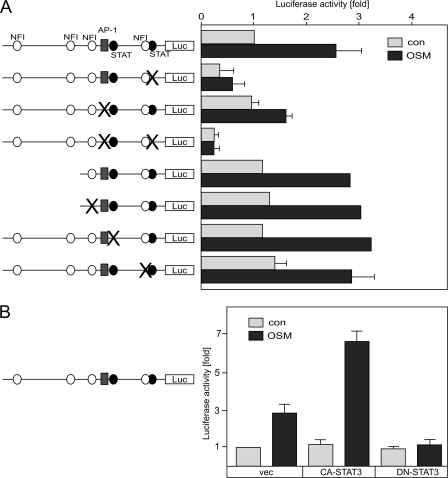
To further characterize the mechanism of YKL-40 regulation by STAT3 and NFI-X, the 1.3-kb upstream region of the human YKL-40 gene was screened for the putative regulatory elements using the MatInspector and Transfac programs. One AP-1, two STAT and four NFI binding elements were identified within the 1.3-kb-long 5′-flanking region of YKL-40 (Fig. 6A). To asses the importance of these elements, we introduced mutations into the proximal and distal STAT-binding sites within the −1.3-kb YKL-40 reporter and analyzed these reporters in astrocytes. OSM activated both the wild-type and the mutated distal STAT reporters by 3-fold (Fig. 6A). In contrast, mutation of the proximal STAT element at −78 to −69 dramatically reduced both basal and OSM-induced reporter activity (Fig. 6A), suggesting that the proximal STAT element is critical for the full basal activity and OSM responsiveness. To validate that STAT3 is important for OSM-stimulated YKL-40 expression, we cotransfected astrocytes with the wild-type YKL-40 reporter together with plasmids expressing constitutively active or dominant-negative STAT3. Accordingly, CA-STAT3 enhanced response to OSM, while DN-STAT3 abolished this response (Fig. 6B).
FIGURE 6.
STAT and AP-1 elements are critical for OSM-induced YKL-40 reporter activity in astrocytes. A, point mutations were introduced into the putative STAT, AP-1, and NFI elements of the 1.3-kb-long YKL-40 promoter. A deletion of the fragment containing two distal NFI elements was generated as described under “Experimental Procedures.” Astrocytes were transfected with the indicated reporters and stimulated with OSM for 24 h, and luciferase and Renilla activities were determined. The data are shown as fold induction to the wild-type reporter (luciferase/Renilla). B, astrocytes were transfected with pYKL(−1300)Luc and plasmids expressing either vector (vec), constitutively active (CA-STAT3), or dominant-negative (DN-STAT3). The cells were stimulated with OSM for 24 h and processed as described above. con, control.
Because OSM activates AP-1 family members (42) and an AP-1 element is localized within the YKL-40 promoter, we explored whether this element contributes to OSM responsiveness. Mutation of this element alone markedly reduced OSM-induced YKL-40 reporter activity, whereas mutations of both STAT and AP-1 elements completely abolished this activity (Fig. 6A), suggesting that both STAT and AP-1 elements are necessary for full OSM responsiveness. To evaluate the effects of the four putative NFI elements, we made a deletion removing the two distal elements and utilized this reporter to mutate the remaining two NFI elements. Surprisingly, these mutations had no effect on either basal or OSM-induced reporter activity (Fig. 6A), suggesting that these elements are not needed for activation. In sum, we conclude that STAT and AP-1 elements mediate YKL-40 activation in response to OSM, whereas the NFI elements are dispensable.
OSM-induced Binding of STAT3, NFI-X3, and AP-1 to the YKL-40 Promoter
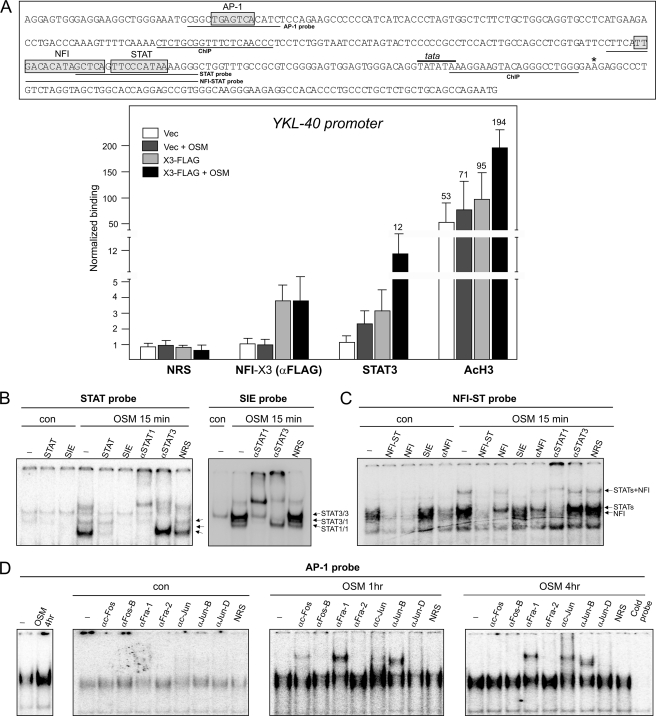
We have analyzed the binding of NFI-X3 and STAT3 to the YKL-40 promoter in OSM-stimulated cells by ChIP (Fig. 7A). We could detect binding of NFI-X3 in nonstimulated U373-X3 cells, and this binding was not changed after OSM stimulation. Significantly, STAT3 binding was promoted by overexpression of NFI-X3 and further enhanced by OSM stimulation. In agreement with the low expression of YKL-40 in U373-vec cells (Fig. 1C), binding of STAT3 was very low and only marginally induced in these cells (Fig. 7A). Subsequently, we analyzed the in vitro binding of OSM-activated proteins to the YKL-40 regulatory elements. OSM induced both STAT3 and STAT1 binding to the proximal STAT element of YKL-40 in both U373 cells (Fig. 7, B and C) and primary astrocytes (supplemental Fig. S4, A and B); however, this binding was relatively weak in comparison with their binding to the SIS-inducible element of the c-fos gene (32, 43). Although mutation of the NFI elements had no effect on the YKL-40 reporter activity (Fig. 6A), we could detect a binding of NFIs to the probe containing both the STAT and NFI elements in U373 cell and astrocyte extracts (Fig. 7C and supplemental Fig. S4B). This NFI binding was out-competed by the excess of the cold probe and was also inhibited by the anti-NFI antibodies (Fig. 7C). We also observed a very strong protein binding to the AP-1 element in untreated astrocytes, which was further enhanced by OSM. The OSM-induced AP-1 complexes consisted of c-Fos, Fra-1, c-Jun, and JunB (Fig. 7D).
FIGURE 7.
Binding of the OSM-induced proteins to the YKL-40 promoter. A, upper panel, YKL-40 proximal promoter. AP1, NFI, STAT3, and TATA box are indicated. The asterisk indicates the transcription start site, and the position of ChIP primers is indicated (ChIP). Lower panel, ChIP was performed using chromatin prepared from U373-vec and U373-NFI-X3-FLAG cells treated with OSM for 15 min (as indicated). The YKL-40 promoter was analyzed for the presence of acetylated-histone H3, NFI-X3 (anti-FLAG) or STAT3 using the antibodies described under “Experimental Procedures.” NRS indicates normal rabbit serum used for immunoprecipitation. The results are shown as normalized binding (anti-FLAG immunoprecipitated samples of U373-vec cells were set as 1). The experiments were performed four times. B–D, nuclear extracts were prepared from control and OSM-treated U373-NFI-X3-FLAG cells as indicated. The binding was then analyzed by EMSA using the 32P-labeled oligonucleotide probes derived from the 5′-flanking region of the YKL-40. Indicated specific antibodies or normal rabbit serum (NRS) were added to the binding reaction. Competition was carried out in the presence of 100-fold excess of the indicated cold probes. B, binding to the YKL-40 proximal STAT element. SIE probe was used as a positive control to visualize STAT3 and STAT1 complexes. C, binding to the probe containing both the NFI and STAT elements. D, binding to the AP-1 probe. Vec, vector; con, control.
NFI-X and STAT3 Form a Complex
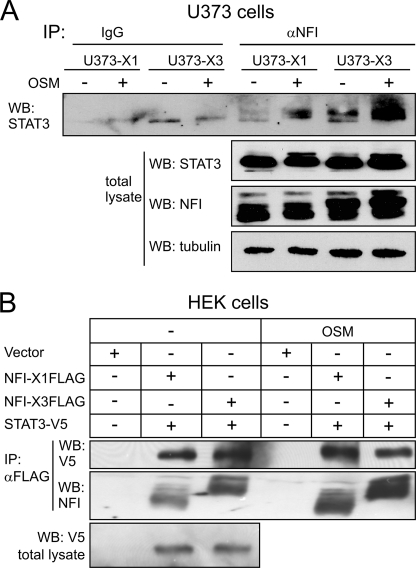
Although both NFI-X3 and STAT3 regulate YKL-40 expression in astrocytes and glioma cells, the YKL-40 reporter lacking four NFI elements was fully activated by OSM (Fig. 6A). Nevertheless, both STAT3 and NFIs were weakly bound in vitro to their proximal elements, which are juxtaposed within the YKL-40 promoter (Fig. 7C and supplemental Fig. S4B) and also were recruited to the YKL-40 promoter in U373-X3 cells (Fig. 7A). We hypothesized that STAT3 may directly interact with NFI-X3, and the complex may more efficiently bind to the relatively weak binding elements within the YKL-40 promoter. Indeed, STAT3 was easily coimmunoprecipitated from U373-NFI-X3 and U373-NFI-X1 cells (Fig. 8A) and U373-NFI-X3FLAG and U373-NFI-X1FLAG cells (supplemental Fig. S5). Remarkably, NFI-X3 interacted more efficiently with STAT3 than NFI-X1, which parallels the strong NFI-X3-dependent activation of YKL-40 expression. The STAT3 interaction with NFI-X was not restricted to glioma cells because we could coimmunoprecipitate STAT3 with NFI-X3 and NFI-X1 from transiently transfected 293 HEK cells (Fig. 8B).
FIGURE 8.
NFI-X3 and STAT3 form a complex. A, NFI-containing complexes were immunoprecipitated from U373-X1 and NFI-X3 cells using anti-NFI antibodies (pan-specific). Immunoprecipitates were resolved by SDS-PAGE, and STAT3 was detected by Western blotting (WB). The levels of STAT3, NFI, and tubulin were determined in total lysates by Western blotting (bottom panel). B, HEK cells were transfected with the indicated plasmids. 24 h post-transfection, the cells were stimulated with OSM for 15 min, and cell lysates were prepared. FLAG-tagged proteins were immunoprecipitated (IP) with using FLAG beads, and STAT3-V5 was detected by Western blotting using anti-V5 antibodies.
DISCUSSION
Although YKL-40 has been detected in various CNS pathologies associated with ongoing inflammation, including Alzheimer's disease (22) and multiple sclerosis (24), as well as in GBM (39, 40), the identity of cells expressing YKL-40 remains debatable. It has been shown that YKL-40 is expressed by activated microglial cells, which are the resident macrophages of the brain, in simian immunodeficiency viral Encephalitis-associated inflammation (23). However, a recent study showed that it is also secreted by reactive astrocytes (25). Because YKL-40 is expressed in GBM cells, which are of glial origin, it could be expected that YKL-40 is expressed by normal astrocytes either during development or in response to their activation. We report that primary human astrocytes express and secret YKL-40 (Figs. 4 and 5). Expression of YKL-40 is activated during the differentiation of NP toward astrocytes in vitro (Fig. 3A) and is also induced during postnatal brain development in vivo (Fig. 3, B and C). Interestingly, expression of YKL-40 during the period of postnatal days 1–21 coincides with the differentiation and maturation of astrocytes in mouse brain (embryonic day 16 to postnatal day 30) (41), suggesting that astrocytes likely contribute to the increased pool of YKL-40 in the developing brain. Our results are further supported by the transcriptome analysis of the mouse neonatal astrocytes, neurons, and oligodendrocytes, which revealed that YKL-40 mRNA is specifically present in astrocytes (41).
NFIs have recently been implicated as critical regulators of gliogenesis with NFI-A and NFI-B controlling early phases of gliogenesis (7), whereas NFI-X and NFI-C are needed at later stages of astrocyte differentiation (8). We report that in human astrocytes, the expression of YKL-40 is primarily regulated by NFI-X and specifically NFI-X3 (Fig. 1D). Moreover, the NFI-X3 splice variant controls YKL-40 expression because its knockdown in astrocytes and overexpression in glioma cells affect YKL-40 expression (Fig. 1, C and D). Accordingly, the expression of NFI-X3 precedes the expression of YKL-40 during differentiation of NP to astrocytes (Fig. 3A) (14). It remains to be established whether activation of YKL-40 expression by NFI-X3 is astrocyte-specific. Because a knockdown of NFI-X does not significantly affect the expression of YKL-40 in THP-1 monocytes (data not shown), cell type-specific regulation is very likely.
The role of YKL-40 in the developing brain remains speculative because YKL-40 knock-out mice have not been reported to present any obvious brain defects (29). Nevertheless, YKL-40 increased astrocyte migration as well as glioma cell migration and invasion in vitro (Fig. 2) and transformed astrocytes overexpressing YKL-40 are also more invasive (40). Notably, NFI-X knock-out mice exhibit defects in proliferation and migration of the subventricular zone progenitor cells (10). Although neuronal precursors are strongly affected in these mice, the absence of NFI-X also delays differentiation of mature glia (44). Moreover, both NFI-A and NFI-B govern tangential neuronal migration by regulating expression of ephrin-B1 and N-cadherin, which are cell adhesion molecules (45). NFIs are expressed in overlapping patterns during embryonic brain development (9), and their expression coincides with the migration of neural, neuronal, and astrocyte progenitors. Therefore, NFIs emerge as important regulators of migration likely via activating different sets of target genes in unique populations of progenitor cells.
STAT3 and NFI-X are important regulators of gliogenesis (3, 4, 7, 8), and they both control the expression of YKL-40 in primary human astrocytes and glioma cells (Figs. 1, 2, and 5). Both STAT3 and NFI-X also govern the expression of well known astrocyte markers, such as GFAP (5, 8, 46). STAT3 activates the transcription of the gfap gene by binding to the critical STAT element, which is specifically demethylated during gliogenesis (47). This demethylation is induced by Notch signaling and requires NFI-A and NFI-B (11). However, NFI-X3 activates GFAP expression by binding to the GFAP enhancer and inducing changes in chromatin architecture at the gfap gene promoter resulting in +1 nucleosome eviction and subsequent recruitment of RNA polymerase II (14). Similarly to the GFAP activation, STAT3 binds to the proximal STAT element of the YKL-40 promoter (Fig. 7, A and B) and is critical for OSM-induced activation (Fig. 6A). Surprisingly, activation of YKL-40 expression by OSM also requires NFI-X3 (Fig. 5), but the four putative NFI elements of the −1.3 kb YKL-40 promoter were dispensable for the reporter activation (Fig. 6A). In contrast, our ChIP data suggest that NFI-X3 binds to the proximal NFI element in glioma cells, and it may help recruit STAT3 (Fig. 7A). This is further supported by the finding that both NFIs and STAT3 weakly bind to the proximal juxtaposed NFI and STAT elements in vitro (Fig. 7C) and STAT3 forms a complex with NFI-X3 (Fig. 8). Binding of NFI-X3 to the endogenous YKL-40 promoter suggests that NFI-X3 likely affects local chromatin architecture and allows for efficient recruitment of the cytokine-activated STAT3 to the YKL-40 promoter. Furthermore, NFI-X3 may stabilize a weak binding of STAT3 because of its ability to form a complex with STAT3. Interestingly, NFI-C is known to interact with both histone H3 (48) and remodeling complexes containing Brg1 (49), also indicating that NFIs may act by affecting chromatin remodeling. In agreement, NFI-X3 reduces nucleasome occupancy at the GFAP promoter, which increases RNA polymerase II promoter occupancy and GFAP transcription (14). The interaction of STAT3 with NFI-X3 is stronger than with NFI-X1, which lacks a long, proline-rich TA domain (Fig. 8A). Because proline-rich domains are often involved in protein/protein interactions (50), it is very likely that the TA domain of NFI-X3 contributes to the interaction with STAT3. It remains to be explored whether the NFI-X3-STAT3 complexes control a specific subset of the astrocyte-specific genes. Similarly, a specific subset of genes is regulated by the STAT3:NF-κB complexes (51), and these genes differ from those regulated by STAT3 alone (51). Because STAT3 is ubiquitously expressed, it is possible that its interactions with NFI-X3, NF-κB, and other transcription factors allow for the tissue-specific activation of selected subsets of genes.
Our data also suggest that AP-1 is a critical regulator of YKL-40 expression in astrocytes (Figs. 6A and 7D). It has previously been reported that AP-1 and NFI elements are often juxtaposed in regulatory regions of astrocyte-specific genes (52). AP-1 is also indispensable for the full expression of some astrocyte-specific genes, including GFAP (53). Members of the AP-1 family are known to form complexes with NFI-X (54) and STAT3 (55, 56). Thus it is possible that enhanceosome formed at the YKL-40 promoter involves binding of a complex consisting of NFI-X, STAT3, and AP-1; nevertheless, this needs to be analyzed in the future. In conclusion, we propose that NFI-X3 and STAT3 control the migration of differentiating astrocytes, as well as migration and invasion of glioma cells, in part, via regulating YKL-40 expression.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS044118 and R21NS063283. This work was also supported by Massey Cancer Pilot Project no 2009 FPP-02 from Virginia Commonwealth University (to T. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- NFI
- nuclear factor 1
- GFAP
- glial fibrillary acidic protein
- Luc
- luciferase
- NP
- neural progenitors
- OSM
- oncostatin M
- SPARCL1
- secreted protein acidic and rich in cysteine-like protein 1
- GBM
- glioblastoma multiforme
- qPCR
- quantitative PCR.
REFERENCES
- 1. Mucke L., Eddleston M. (1993) FASEB J. 7, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 2. Eddleston M., Mucke L. (1993) Neuroscience 54, 15–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deverman B. E., Patterson P. H. (2009) Neuron 64, 61–78 [DOI] [PubMed] [Google Scholar]
- 4. Miller F. D., Gauthier A. S. (2007) Neuron 54, 357–369 [DOI] [PubMed] [Google Scholar]
- 5. Yanagisawa M., Nakashima K., Taga T. (1999) Neurosci. Lett. 269, 169–172 [DOI] [PubMed] [Google Scholar]
- 6. Barnabé-Heider F., Wasylnka J. A., Fernandes K. J., Porsche C., Sendtner M., Kaplan D. R., Miller F. D. (2005) Neuron 48, 253–265 [DOI] [PubMed] [Google Scholar]
- 7. Deneen B., Ho R., Lukaszewicz A., Hochstim C. J., Gronostajski R. M., Anderson D. J. (2006) Neuron 52, 953–968 [DOI] [PubMed] [Google Scholar]
- 8. Wilczynska K. M., Singh S. K., Adams B., Bryan L., Rao R. R., Valerie K., Wright S., Griswold-Prenner I., Kordula T. (2009) Stem Cells 27, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plachez C., Lindwall C., Sunn N., Piper M., Moldrich R. X., Campbell C. E., Osinski J. M., Gronostajski R. M., Richards L. J. (2008) J. Comp. Neurol. 508, 385–401 [DOI] [PubMed] [Google Scholar]
- 10. Campbell C. E., Piper M., Plachez C., Yeh Y. T., Baizer J. S., Osinski J. M., Litwack E. D., Richards L. J., Gronostajski R. M. (2008) BMC Dev. Biol. 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T., Nakashima K. (2009) Dev. Cell 16, 245–255 [DOI] [PubMed] [Google Scholar]
- 12. Piper M., Barry G., Hawkins J., Mason S., Lindwall C., Little E., Sarkar A., Smith A. G., Moldrich R. X., Boyle G. M., Tole S., Gronostajski R. M., Bailey T. L., Richards L. J. (2010) J. Neurosci. 30, 9127–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mason S., Piper M., Gronostajski R. M., Richards L. J. (2009) Mol. Neurobiol. 39, 10–23 [DOI] [PubMed] [Google Scholar]
- 14. Singh S. K., Wilczynska K. M., Grzybowski A., Yester J., Osrah B., Bryan L., Wright S., Griswold-Prenner I., Kordula T. (2011) J. Biol. Chem. 286, 7315–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hakala B. E., White C., Recklies A. D. (1993) J. Biol. Chem. 268, 25803–25810 [PubMed] [Google Scholar]
- 16. Kawamura K., Shibata T., Saget O., Peel D., Bryant P. J. (1999) Development 126, 211–219 [DOI] [PubMed] [Google Scholar]
- 17. Kzhyshkowska J., Gratchev A., Goerdt S. (2007) Biomark Insights 2, 128–146 [PMC free article] [PubMed] [Google Scholar]
- 18. Ling H., Recklies A. D. (2004) Biochem. J. 380, 651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shao R., Hamel K., Petersen L., Cao Q. J., Arenas R. B., Bigelow C., Bentley B., Yan W. (2009) Oncogene 28, 4456–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Francescone R. A., Scully S., Faibish M., Taylor S. L., Oh D., Moral L., Yan W., Bentley B., Shao R. (2011) J. Biol. Chem. 286, 15332–15343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Faibish M., Francescone R., Bentley B., Yan W., Shao R. (2011) Mol. Cancer Ther. 10, 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Colton C. A., Mott R. T., Sharpe H., Xu Q., Van Nostrand W. E., Vitek M. P. (2006) J. Neuroinflammation 3, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonneh-Barkay D., Bissel S. J., Wang G., Fish K. N., Nicholl G. C., Darko S. W., Medina-Flores R., Murphey-Corb M., Rajakumar P. A., Nyaundi J., Mellors J. W., Bowser R., Wiley C. A. (2008) Am. J. Pathol. 173, 130–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Comabella M., Fernández M., Martin R., Rivera-Vallvé S., Borrás E., Chiva C., Julià E., Rovira A., Cantó E., Alvarez-Cermeño J. C., Villar L. M., Tintoré M., Montalban X. (2010) Brain 133, 1082–1093 [DOI] [PubMed] [Google Scholar]
- 25. Bonneh-Barkay D., Wang G., Starkey A., Hamilton R. L., Wiley C. A. (2010) J. Neuroinflammation 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Ceuninck F., Gaufillier S., Bonnaud A., Sabatini M., Lesur C., Pastoureau P. (2001) Biochem. Biophys. Res. Commun. 285, 926–931 [DOI] [PubMed] [Google Scholar]
- 27. Malinda K. M., Ponce L., Kleinman H. K., Shackelton L. M., Millis A. J. (1999) Exp. Cell Res. 250, 168–173 [DOI] [PubMed] [Google Scholar]
- 28. Nishikawa K. C., Millis A. J. (2003) Exp. Cell Res. 287, 79–87 [DOI] [PubMed] [Google Scholar]
- 29. Lee C. G., Hartl D., Lee G. R., Koller B., Matsuura H., Da Silva C. A., Sohn M. H., Cohn L., Homer R. J., Kozhich A. A., Humbles A., Kearley J., Coyle A., Chupp G., Reed J., Flavell R. A., Elias J. A. (2009) J. Exp. Med. 206, 1149–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhat K. P., Pelloski C. E., Zhang Y., Kim S. H., deLaCruz C., Rehli M., Aldape K. D. (2008) FEBS Lett. 582, 3193–3200 [DOI] [PubMed] [Google Scholar]
- 31. Recklies A. D., Ling H., White C., Bernier S. M. (2005) J. Biol. Chem. 280, 41213–41221 [DOI] [PubMed] [Google Scholar]
- 32. Kordula T., Rydel R. E., Brigham E. F., Horn F., Heinrich P. C., Travis J. (1998) J. Biol. Chem. 273, 4112–4118 [DOI] [PubMed] [Google Scholar]
- 33. Dumur C. I., Sana S., Ladd A. C., Ferreira-Gonzalez A., Wilkinson D. S., Powers C. N., Garrett C. T. (2008) Diagn. Mol. Pathol. 17, 200–206 [DOI] [PubMed] [Google Scholar]
- 34. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 35. Zhang L., Wang L., Ravindranathan A., Miles M. F. (2002) J. Mol. Biol. 317, 225–235 [DOI] [PubMed] [Google Scholar]
- 36. Benjamini H. Y. (1995) J R Stat Soc Series B Stat Methodol 57, 289–300 [Google Scholar]
- 37. Paugh B. S., Bryan L., Paugh S. W., Wilczynska K. M., Alvarez S. M., Singh S. K., Kapitonov D., Rokita H., Wright S., Griswold-Prenner I., Milstien S., Spiegel S., Kordula T. (2009) J. Biol. Chem. 284, 3408–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rehli M., Niller H. H., Ammon C., Langmann S., Schwarzfischer L., Andreesen R., Krause S. W. (2003) J. Biol. Chem. 278, 44058–44067 [DOI] [PubMed] [Google Scholar]
- 39. Tanwar M. K., Gilbert M. R., Holland E. C. (2002) Cancer Res. 62, 4364–4368 [PubMed] [Google Scholar]
- 40. Nigro J. M., Misra A., Zhang L., Smirnov I., Colman H., Griffin C., Ozburn N., Chen M., Pan E., Koul D., Yung W. K., Feuerstein B. G., Aldape K. D. (2005) Cancer Res. 65, 1678–1686 [DOI] [PubMed] [Google Scholar]
- 41. Cahoy J. D., Emery B., Kaushal A., Foo L. C., Zamanian J. L., Christopherson K. S., Xing Y., Lubischer J. L., Krieg P. A., Krupenko S. A., Thompson W. J., Barres B. A. (2008) J. Neurosci. 28, 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Underhill-Day N., Heath J. K. (2006) Cancer Res. 66, 10891–10901 [DOI] [PubMed] [Google Scholar]
- 43. Kasza A., Kiss D. L., Gopalan S., Xu W., Rydel R. E., Koj A., Kordula T. (2002) J. Neurochem. 83, 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Piper M., Harris L., Barry G., Heng Y. H., Plachez C., Gronostajski R. M., Richards L. J. (2011) J. Comp. Neurol. 519, 3532–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang W., Mullikin-Kilpatrick D., Crandall J. E., Gronostajski R. M., Litwack E. D., Kilpatrick D. L. (2007) J. Neurosci. 27, 6115–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gopalan S. M., Wilczynska K. M., Konik B. S., Bryan L., Kordula T. (2006) J. Biol. Chem. 281, 13126–13133 [DOI] [PubMed] [Google Scholar]
- 47. Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M., Taga T. (2001) Dev. Cell 1, 749–758 [DOI] [PubMed] [Google Scholar]
- 48. Müller K., Mermod N. (2000) J. Biol. Chem. 275, 1645–1650 [DOI] [PubMed] [Google Scholar]
- 49. Zhao L. H., Ba X. Q., Wang X. G., Zhu X. J., Wang L., Zeng X. L. (2005) Acta Biochim. Biophys. Sin. 37, 440–446 [DOI] [PubMed] [Google Scholar]
- 50. Chen H. I., Sudol M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang J., Chatterjee-Kishore M., Staugaitis S. M., Nguyen H., Schlessinger K., Levy D. E., Stark G. R. (2005) Cancer Res. 65, 939–947 [PubMed] [Google Scholar]
- 52. Amemiya K., Traub R., Durham L., Major E. O. (1992) J. Biol. Chem. 267, 14204–14211 [PubMed] [Google Scholar]
- 53. Gopalan S. M., Wilczynska K. M., Konik B. S., Bryan L., Kordula T. (2006) J. Biol. Chem. 281, 1956–1963 [DOI] [PubMed] [Google Scholar]
- 54. Ravichandran V., Sabath B. F., Jensen P. N., Houff S. A., Major E. O. (2006) J. Virol. 80, 10506–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schuringa J. J., Timmer H., Luttickhuizen D., Vellenga E., Kruijer W. (2001) Cytokine 14, 78–87 [DOI] [PubMed] [Google Scholar]
- 56. Nishikawa T., Hagihara K., Serada S., Isobe T., Matsumura A., Song J., Tanaka T., Kawase I., Naka T., Yoshizaki K. (2008) J. Immunol. 180, 3492–3501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.