Background: Three mutations in the NAGPA gene encoding uncovering enzyme (UCE) have been found in some individuals with persistent stuttering.
Results: All three mutations lead to lower UCE activity.
Conclusion: NAGPA mutations present in individuals with persistent stuttering have negative effects on the enzyme.
Significance: These findings extend the genetic data implicating NAGPA mutations in the persistent stuttering phenotype.
Keywords: ER-associated Degradation, Glycobiology, Lysosomes, Proteasome, Protein Misfolding, NAGPA Gene, Stuttering, Uncovering Enzyme
Abstract
GlcNAc-1-phosphodiester-N-acetylglucosaminidase (“uncovering enzyme” (UCE); EC 3.1.4.45) is a Golgi enzyme that mediates the second step in the synthesis of the mannose 6-phosphate lysosomal targeting signal on acid hydrolases. Recently, three mutations (two missense and one deletion/frameshift) in the NAGPA gene that encodes UCE have been identified in individuals with persistent stuttering. We now demonstrate that each mutation leads to lower cellular UCE activity. The p.R328C mutation impairs folding in the endoplasmic reticulum, resulting in degradation of a significant portion by the proteasomal system. The p.H84Q mutation also impairs folding and, in addition, decreases the specific activity of the enzyme that folds sufficiently to traffic to the Golgi. The p.F513SfsX113 frameshift mutation adds 113 amino acids to the C terminus of the cytoplasmic tail of the protein, including a VWLL sequence that causes rapid degradation via the proteasomal system. These biochemical findings extend the genetic data implicating mutations in the NAGPA gene in the persistent stuttering phenotype.
Introduction
Stuttering is a speech disorder characterized by repetitions, prolongations, and interruptions in the flow of speech. It is estimated to affect ∼1% of the population (1). Twin studies, adoption studies, and family studies have indicated that genetic factors contribute to the etiology of the disorder (2). Recently, Kang et al. (3) reported 10 different mutations (nine missense) in the GNPTAB, GNPTG, and NAGPA genes in 25 of 393 alleles of unrelated Pakistani and North American stutterers, whereas only 1 of 372 alleles of control individuals had a mutation in any of these genes. These genes encode the two enzymes that generate the Man-6-P recognition marker on lysosomal acid hydrolases. GNPTAB and GNPTG encode the subunits of UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-1-phosphotransferase), which transfers GlcNAc-1-P from UDP-GlcNAc to selected mannose residues on N-linked high mannose glycans of newly synthesized lysosomal acid hydrolases (4). NAGPA encodes GlcNAc-1-phosphodiester N-acetylglucosaminidase (“uncovering enzyme” (UCE)),2 which removes the GlcNAc to produce a Man-6-P monoester that functions as a high affinity ligand for Man-6-P receptors in the trans-Golgi network (5). This is followed by packaging of the ligand-receptor complex into clathrin-coated carriers for delivery to the endosomal/lysosomal system. Inactivating mutations in GNPTAB and GNPTG are known to cause severe forms of autosomal recessive lysosomal storage diseases termed I-cell disease (mucolipidosis II (ML II)) and pseudo-Hurler polydystrophy (ML III) (4, 6, 7). Mutations in the NAGPA gene have not previously been associated with any clinical disorder.
Kang et al. (3) pointed out three findings that support a causative role for these mutations in stuttering: (a) only one mutation in the GNPTAB, GNPTG, and NAGPA genes was detected in 372 alleles of control individuals; (b) all of the mutations occur at positions with a high degree of amino acid conservation across species, although none were found at these locations in individuals with ML II or ML III; and (c) the mutations involve genes that function sequentially in a well defined metabolic pathway. However, the effect of the mutations on the function of GlcNAc-1-phosphotransferase or UCE was not examined, leaving open the possibility that the mutations were not detrimental.
UCE is a type I homotetrameric transmembrane glycoprotein that is mainly localized to the trans-Golgi network (TGN) (8). It is synthesized as a 515-amino acid preproprotein with a 24-amino acid signal sequence and a luminal domain of 423 residues, including a 24-residue prosequence, a 27-residue transmembrane region, and a 41-residue cytoplasmic tail (5). The signal sequence is excised in the ER, and upon folding, the catalytically inactive protein is transported to the TGN, where it is activated by furin cleavage of the prosequence (9). In addition, two splice forms have been found. One of these isoforms lacks 102 base pairs corresponding to exon 8 of the 10 exons present in the genomic DNA and is predominantly expressed in the brain (5).
In this report, we have analyzed the consequences of the three mutations in the NAGPA gene (two missense in the luminal domain and one 16-base pair deletion at the C terminus of the cytoplasmic tail) identified in individuals with persistent stuttering on the synthesis, half-life, and activity of UCE. We found that all three mutations diminish total cellular UCE activity, but each impacts the enzyme in a different manner.
EXPERIMENTAL PROCEDURES
Materials
UDP-6-[3H]GlcNAc (20–35 Ci/mmol) and d-[2-3H]mannose (20–30 Ci/mmol) were obtained from PerkinElmer Life Sciences. Tran35S-labelTM was purchased from MP Biomedicals (Irvine, CA). UDP-GlcNAc, cycloheximide (CHX), α-methylmannoside, and QAE-Sephadex were from Sigma. MG132 was obtained from Calbiochem.
Monoclonal anti-HA Ab was obtained from Covance and monoclonal anti-FLAG Ab was obtained from Stratagene. Polyclonal anti-HA Ab and polyclonal anti-FLAG Ab were obtained from Sigma.
Cell Lines
CRL-1509, CRL-1474, and HeLa cells were obtained from ATCC and human dermal fibroblasts was purchased from Invitrogen (catalog no. C-013-5C). Skin fibroblasts from one affected female (NAGPA F513Sfsx113; no. 229) and one affected male (NAGPA R328C; no. 288) were obtained under a National Institutes of Health institutional review board-approved protocol 01-HG-0106. The cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 μg/ml penicillin, and 100 units/ml streptomycin.
Assay of Fibroblasts for UCE Activity
Synthesis of the [3H]GlcNAc-α-Me-P-Man substrate was performed as described previously (10) except that purified recombinant human GlcNAc-1-phosphotransferase provided by W. Canfield (Genzyme) was used instead of Acanthamoeba castellanii GlcNAc-1-phosphotransferase. The cultured fibroblasts were solubilized with a buffer containing 0.1 m Tris, pH 8.0, 150 mm NaCl, and 1% Triton X-100. Aliquots (100 μg) were assayed for UCE activity as described (10).
Measurement of Phosphorylated Oligosaccharide Synthesis and Efficiency of Phosphomonoester Formation
Confluent fibroblast cultures in 60-mm dishes were incubated for 4 h in low glucose (1 mm) medium with 150 μCi/ml of [2-3H]mannose and chased for 2 h in complete medium. Oligosaccharide samples were prepared, and the degree of phosphorylation was measured as described previously (11) except that aliquots were treated with 10 units of calf alkaline phosphatase (Promega Protégé, Madison, WI) overnight prior to analysis by QAE-Sephdex.
UCE Constructs
The p.R328C and the p.H84Q UCE mutant constructs were generated using the QuikChange site-directed mutagenesis protocol (Stratagene). The full-length human WT UCE construct and the WT UCE construct lacking bases 1176–1275 (“brain form”) served as the templates. The constructs were subcloned into the pcDNA 4/TO expression vector (Invitrogen) using EcoRI and AflII sites. The HA tag (YPYDVPDYA) or FLAG tag (DYKDDDDK) was introduced at the C terminus of the UCE constructs.
To generate the p.F513SfsX113 UCE construct, which has a 16-base pair deletion (residues 1535–1550), an oligonucleotide was synthesized from nucleotide 539 to 1885 with the 1535–1550 deletion and an HA tag in front of the TGA stop codon and an AflII site (IDT, Coralville, IA). The fragment between the KpnI (nucleotide 539) and AflII sites of the WT UCE construct was then replaced with the synthetic oligonucleotide. Subsequent deletion and amino acid substitution mutants of the p.F513SfsX113 UCE construct were generated using the QuikChange site-directed mutagenesis protocol.
Transfection and Expression
TREx-HeLa cells (Invitrogen) were grown at 37 °C with 5% CO2 in DMEM supplemented with 10% tetracycline-free FBS (Clontech) in 6-well plates until they reached 80–90% confluence. The plates were then washed with serum-free OPTI-MEM and transfected with UCE cDNA constructs using Lipofectamine 2000 (Invitrogen). Six hours later, the plates were supplemented with additional OPTI-MEM and tetracycline-free FBS to bring the volume to 2.5 ml and 10% FBS. To test expression, tetracycline was added at a concentration of 300 ng/ml 24 h post-transfection. After an additional 24 h, the cells were solubilized with a buffer composed of 0.1 m Tris, pH 8.0, 150 mm NaCl, 1% Triton X-100, and protease inhibitor mixture (Complete, Roche Applied Science) and analyzed for UCE expression by Western blot. To obtain stably transfected cell lines, a selection medium containing zeocin (100 μg/ml) and blasticidin (5 μg/ml) was applied 48 h post-transfection.
UCE Pulse-Chase Analysis
Confluent cultures of TREx-HeLa cell lines expressing non-brain or brain forms of UCE in 60-mm dishes were incubated for 1 h with 1 ml of Cys/Met-free α-MEM containing 70 μCi of Tran35S-label and either harvested or chased for 3–24 h with 1.2 ml of complete medium supplemented with 20 mm HEPES, 10% dialyzed FBS, and 20 mm methionine. At the indicated time points, the chase medium was removed and replaced with a buffer composed of 0.1 m Tris, pH 8.0, 150 mm NaCl, 1% Triton X-100, protease inhibitor mixture, and 1 mm DTT, followed by sonication and centrifugation to yield cell lysates. The UCE-HA was immunoprecipitated with Protein A-agarose (IPA300, RepliGen) conjugated with polyclonal anti-HA antibody. The immunoprecipitates were eluted with 50 μl of 3× SDS-PAGE sample buffer containing β-mercaptoethanol, subjected to 8% SDS-PAGE, and visualized by autoradiography. The bands were excised, rehydrated with 100 μl of water, and incubated with 200 μl of 60% perchloric acid and 400 μl of 30% H2O2 overnight at 55 °C. Scintillation fluid was added, and the radioactivity was determined in a scintillation counter (Beckman LS6500).
Endo Hf and PNGase F Digestion
Twenty micrograms of transfected HeLa cell lysates was treated with 2,000 units of Endo Hf or 1,000 units of PNGase F overnight at 37 °C following the recommended procedures of the vendor (New England Biolabs, Ipswich, MA). The samples were then subjected to SDS-PAGE and Western blot analysis.
RESULTS
Fibroblasts of Individuals with Persistent Stuttering Have Decreased UCE Activity
Skin fibroblasts from two individuals with persistent stuttering and mutations in the NAGPA gene, one with a p.R328C (homozygous) mutation and the other with a p.F513SfsX113 (heterozygous) deletion resulting in a frameshift, along with WT fibroblasts were assayed for UCE activity. As shown in Table 1, the fibroblasts expressing the p.R328C mutant protein exhibited 54% of WT activity, whereas the fibroblasts with the p.F513Sfsx113 deletion mutant had 40% WT activity.
TABLE 1.
UCE activity of fibroblast lines
Two control fibroblast lines were utilized: CRL-1509 (n = 6) and CRL-1474 (n = 1). The values were combined and averaged. n, number of experiments.
| Cell line | n | UCE activity | Percentage of WT |
|---|---|---|---|
| nmol/h/mg | % | ||
| WT | 7 | 1.20 ± 0.39 | |
| R328C | 6 | 0.65 ± 0.33 | 54.2 (p < 0.01) |
| F513SfsX113 | 6 | 0.48 ± 0.09 | 40.0 (p < 0.001) |
We next determined the ability of the various fibroblast lines to generate the Man-6-P monoester recognition marker on lysosomal hydrolases. Cells were incubated with [2-3H]mannose for 4 h to label N-linked glycans, followed by a 2-h chase. The cells were then harvested, and following extraction of the lipids, the protein pellets were solubilized with 0.5% SDS and incubated with Endo Hf to release the high mannose N-linked oligosaccharides (neutral and phosphorylated with one or two Man-6-P diesters or monoesters), which were separated from Endo Hf-resistant complex-type N-linked glycans using a Centricon 10 filtration unit. One half of the Endo Hf-released material was applied directly to a 2-ml QAE-Sephadex column and eluted as described previously (11). Under these conditions, neutral high mannose oligosaccharides pass through the column; species with 1-phosphodiester bind and are eluted with 20 mm NaCl, those with 2-phosphodiesters or 1-phosphomonoester are eluted with 70 mm NaCl, and species with 2-phosphomonoesters are eluted with 140 mm NaCl. The other half of the released material was incubated overnight with alkaline phosphatase to cleave the phosphate from the phosphomonoesters. This material was then analyzed on a QAE-Sephadex column using the same elution procedures. Quantitation of the radioactivity in the various fractions allowed determination of the percentage of Man-6-P phosphodiesters that were converted to phosphomonoester. By analyzing the amount of the Endo Hf-resistant material, the percentage of total N-linked glycans that contain Man-6-P residues, either monoesters or diesters, could be calculated.
The results of these experiments are summarized in Table 2. All of the fibroblast lines synthesized similar amounts of phosphorylated high mannose glycans (1.88–1.38% of total N-linked glycans). However, the fibroblasts obtained from the two individuals with persistent stuttering showed only 37% conversion of the Man-6-P diesters to monoesters compared with 51% conversion by the WT cells (p = 0.01), reflecting diminished UCE activity.
TABLE 2.
Efficiency of Man-6-P monoester formation by fibroblast lines
Three control fibroblast lines were assayed: CRL-1509 (n = 5), CRL-1474 (n = 2), and human dermal fibroblasts (n = 1). The values were combined and averaged. HM, high mannose; n, number of experiments.
| Cell line | n | Percentage of phosphorylated HM glycans | Percentage of conversion of diesters to monoesters |
|---|---|---|---|
| % | % | ||
| WT | 8 | 1.63 ± 0.34 | 51.0 ± 9.5 |
| R328C | 4 | 1.38 ± 0.13 (p > 0.1) | 37.1 ± 6.5 (p = 0.01) |
| F513SfsX113 | 4 | 1.88 ± 0.29 (p > 0.1) | 37.4 ± 3.5 (p = 0.01) |
Effect of Mutations in NAGPA on UCE Biosynthesis
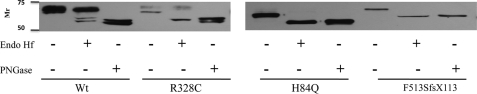
To study the effect of the three NAGPA mutations on UCE biosynthesis, we prepared HA-tagged human WT and mutant UCE (non-brain type) constructs in the expression vector pcDNA 4/TO, transfected HeLa cells, and selected stable expressors. Extracts of the transfected cells were then subjected to SDS-PAGE and Western blotting, with or without prior treatment with Endo Hf or PNGase F to determine the localization of the UCE mutants. The Endo Hf releases high mannose units, whereas the PNGase F cleaves both high mannose and complex-type N-linked glycans. At steady state, the WT UCE migrated as a broad band that was mostly resistant to Endo Hf but sensitive to PNGase F, consistent with oligosaccharide processing in the Golgi and localization of the UCE in the TGN (Fig. 1). By contrast, the p.R328C mutant exhibited two bands, with the faster migrating major band being sensitive to Endo Hf and both being acted upon by PNGase F. This indicated that the majority of the p.R328C molecules were still in the ER. The p.H84Q and p.F513Sfsx113 mutants exhibited single bands that were completely sensitive to both Endo Hf and PNGase F, indicating that the bulk of these molecules were retained in the ER. The accumulation of the mutant proteins in the ER indicated that they were probably impaired in folding.
FIGURE 1.
Sensitivity of WT and mutant UCEs to deglycosylating enzymes. Twenty micrograms of transfected HeLa cell lysates was treated with denaturing buffer containing 0.5% SDS. The samples were then incubated with or without 2,000 units of Endo Hf or 1,000 units of PNGase F overnight at 37 °C, followed by SDS-PAGE and Western blot analysis.
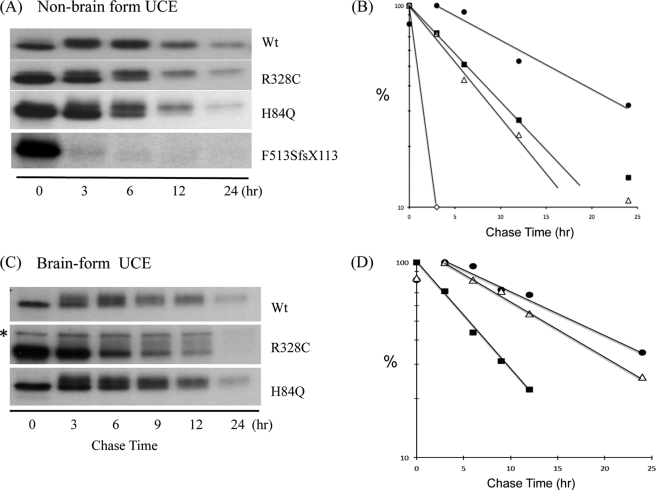
We next performed [35S]Met/Cys pulse/chase experiments with the transfected HeLa cells to study the half-life of the various forms of UCE. The cells were labeled for 1 h, and following chase periods of 3–24 h, the tagged UCE was immunoprecipitated and analyzed by SDS-PAGE and autoradiography. As seen in Fig. 2A, after a 3-h chase, almost all of the WT UCE exhibited a slower migration compared with the 0 h chase sample, consistent with the conversion of its high mannose units to complex-type species. This was shown to be the case because only the rapidly migrating band was sensitive to Endo Hf digestion (Fig. 1). This indicates that the newly synthesized UCE had folded properly, exited the ER, and trafficked to the TGN by this time point. By contrast, the p.R328C and p.H84Q mutant proteins were retained in the ER for an extended period and significantly degraded with less than 50% of the molecules trafficking to the Golgi. By excising and counting the radioactive bands, we determined that the t½ of the WT UCE is 14 h versus 7 and 6 h for the p.R328C and p.H84Q mutants, respectively (Fig. 2B). The most striking phenotype was observed with the p.F513SfsX113 mutant, which was rapidly degraded with a t½ of <30 min (Figs. 2, A and B, and 4A). In these experiments no Golgi form could be detected.
FIGURE 2.
[35S]Met/Cys pulse-chase analysis of WT and mutant UCEs. Stably transfected TREx-HeLa cell lines were metabolically labeled with [35S]Met/Cys for 1 h, followed by chase periods of 3–24 h. Lysates were incubated with polyclonal anti-HA Ab to immunoprecipitate the UCEs, followed by SDS-PAGE and visualization by autoradiography as described under “Experimental Procedures.” The t½ values for the non-brain form of UCE (A and B) were as follows: WT, 14 h; R328C, 7 h; H84Q, 6 h; F513SfsX113, <30 min. The values for the brain forms (C and D) were as follows: WT, 18 h; R328C, 6 h; H84Q, 14 h. ●—●, WT; ■—■, R328C; △—△, H84Q; ♢—♢, F513SfsX113. *, nonspecific band.
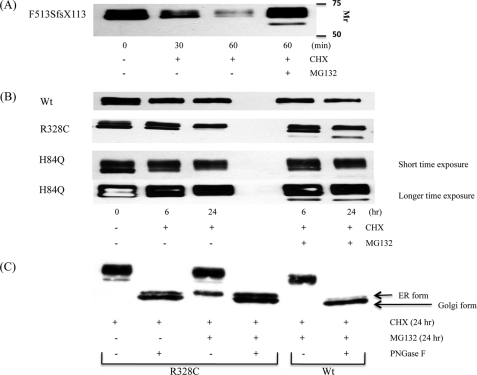
FIGURE 4.
Effect of proteasome inhibition on UCE degradation. TREx-HeLa cells stably expressing F513SfsX113 (A) or WT, R328C, or H84Q UCEs (B) were treated with 120 μg/ml CHX for either 30 or 60 min (A) or 6 or 24 h (B) in the presence or absence of 10 μm MG132. Cell lysates were prepared and subjected to immunoblotting with monoclonal anti-HA Ab. Either 50 μg (A) or 5 μg (B) of cell lysates was used for the immunoblots. In C, aliquots of cell lysates from the 24-h CHX samples were incubated with or without 1,000 units of PNGase F overnight at 37 °C, followed by SDS-PAGE and Western blot analysis.
Because the stuttering phenotype is presumed to arise from neuron dysfunction, we next performed similar experiments with constructs encoding human UCE that lacks the 102 base pairs derived from exon 8 (“brain form”). We found that the WT species matured somewhat more slowly than the non-brain type, requiring 6–9 h to completely exit the ER (Fig. 2C). As noted with the full-length R328C construct, the brain form p.R328C mutant was retained in the ER for an extended period and degraded more rapidly than the WT species (a t½ of 6 h versus 18 h) (Fig. 2D). Interestingly, the brain form of the H84Q mutant exited the ER only modestly slower than its WT counterpart, and its t½ was longer than its non-brain form (14 h versus 6 h).
A VWLL Sequence in the Cytoplasmic Tail of F513SfsX133 Causes Rapid Degradation of the Protein
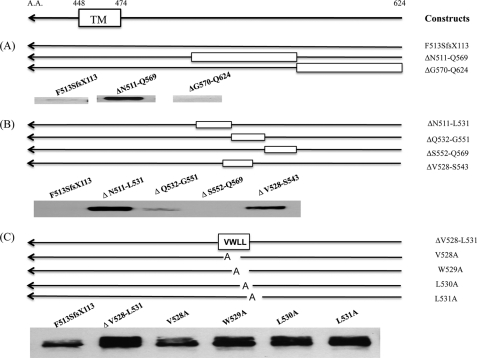
The mutation in p.F513SfsX113 is a 16-base deletion near the C terminus of the protein that results in the deletion of the last three amino acids and the stop codon and generates a frameshift with a run-on of 113 additional amino acids. To determine if the cause of the rapid degradation of the mutant protein resided in a specific region of the additional 113 residues, we constructed deletion mutants and determined their level of expression in transfected HeLa cells. The cells were treated with CHX for 3 h prior to harvest to stop protein synthesis and allow the fate of the various UCEs to be followed. Fig. 3A shows that deletion of residues 511–569 rescued the protein from rapid degradation, whereas deletion of residues 570–624 had no effect. This localized the signal for rapid degradation to residues 511–569. In the second round of analysis, we found that the mutants with deletion of residues 511–531 and 528–543 did not undergo rapid degradation, whereas deletion mutants 532–551 and 552–569 were highly degraded (Fig. 3B). This indicated that the residues leading to rapid degradation were probably localized between positions 528 and 531. This was shown to be the case as deletion of residues 528–531 resulted in a high level of protein expression and mutation of the residues individually to alanine partially rescued the proteins from degradation (Fig. 3C). Together, these findings demonstrate that residues 528VWLL in the cytoplasmic tail of the mutant UCE serve as a signal for the rapid degradation of the protein.
FIGURE 3.
A VWLL motif in the cytoplasmic tail of F513SfsX113 UCE accounts for the rapid degradation. HeLa cells were transfected with mutant UCE constructs containing an HA tag at the C terminus. After 24 h, the transfected cells were treated with 120 μg/ml CHX for 3 h, and then cell lysates were subjected to SDS-PAGE and immunoblotting with monoclonal anti-HA Ab. Either 15 μg (A and B) or 25 μg (C; 2 times longer exposure than A or B) of cell lysate was used for the immunoblots.TM, transmembrane domin.
Mutant Forms of UCE Are Degraded by the Proteasomal System
The effect of the proteasomal inhibitor MG 132 on the degradation of the non-brain mutant forms of UCE is shown in Fig. 4. In this experiment, the cells were treated with CHX with or without 10 μm MG132 for various times, and the fate of the UCE was followed by Western blotting. Fig. 4A shows that in the absence of MG132, the F513SfsX113 mutant protein was rapidly degraded, as noted before. The MG132 greatly decreased this degradation. Note that a faster migrating band appeared in the MG132-treated cells. This most likely represents mutant UCE that has been released back into the cytosol and deglycosylated, as occurs with other glycoproteins subjected to degradation via the proteasomal system (12, 13).
Similar findings were obtained with the p.R328C and p.H84Q mutants (Fig. 4B). In both instances, a faster migrating form of the mutant protein appeared in the MG132-treated samples but not in the cells treated with CHX alone. The cells expressing WT UCE did not generate a rapidly migrating species even after 24-h treatment with MG132.
To establish that the faster migrating bands present in MG132-treated cells expressing the mutant proteins truly represent deglycosylated ER forms of these proteins, extracts of the cells expressing WT and R328C forms of UCE were treated with PNGase F and analyzed by SDS-PAGE (Fig. 4C). The R328C mutant ran as two bands, the slower of which corresponds to the band that appears with MG132 treatment. The WT UCE exhibited primarily the faster migrating band, representing the Golgi form of the protein that had been cleaved by furin. This supports the conclusion that the slower migrating bands seen with MG132 treatment of the mutant UCEs represent the deglycosylated ER form. These data indicate that the UCE mutant proteins are degraded primarily via the proteasomal system.
Effect of the p.R328C and p.H84Q Mutations on UCE Activity
UCE is synthesized as an inactive proenzyme that is activated by furin in the TGN (8). Therefore, to assess the activity of the p.R328C and p.H84Q mutants, we treated HeLa cells expressing the various forms of UCE with CHX to allow the molecules in the ER to either be degraded via the proteasomal pathway or move to the TGN, where they would be activated. After 6 h of CHX exposure, aliquots of cell extracts were either assayed for UCE activity or subjected to SDS-PAGE followed by quantitative Western blotting to determine the level of expression of the Golgi form of UCE. Setting the WT values of enzyme activity/protein content determined by Western blot to 1.0, both the non-brain and brain forms of the p.R328C mutant were found to have relative specific activities similar to that of the WT enzyme, whereas the H84Q mutant had about half the relative specific activity of WT (Table 3). The level of mature p.F513SfsX113 mutant was too low to evaluate.
TABLE 3.
Relative activities of various UCEs
Cell lines stably expressing non-brain and brain forms of WT and mutant UCEs were treated with 120 μg/ml CHX for 6 h. Aliquots (1.5–5.0 μg) of cell lysates were used for UCE assays as described under “Experimental Procedures.” Separate aliquots were subjected to SDS-PAGE and Western blotting with anti-HA Ab. The density of the bands representing the mature form of UCE was determined using a Kodak Image Station 440. The WT values for units of enzyme activity were divided by the density of the band of mature UCE and set to 1.0. The background level of UCE activity in non-transfected cells was less than 1% of the value obtained with WT UCE-transfected cells. The data are the average of three different sets of experimental results. The level of mature F513SfsX113 mutant was too low to evaluate.
| Cell line | Relative activity |
|
|---|---|---|
| Non-brain form | Brain form | |
| WT | 1 | 1 |
| R328C | 0.91 ± 0.11 | 1.15 ± 0.22 |
| H84Q | 0.45 ± 0.09 | 0.58 ± 0.12 |
The R328C Mutant Forms Oligomers with WT UCE
Because most of the stuttering individuals with the R328C and H84Q mutations are heterozygotes, we asked whether the mutant polypeptides could form oligomers with WT subunits and perhaps function in a dominant negative manner. To test this, HeLa cells were co-transfected with WT UCE-FLAG and R328C-UCE-HA constructs. After 24 h, cell lysates were prepared, and aliquots were immunoprecipitated with either anti-FLAG Ab or anti-HA Ab followed by SDS-PAGE and Western blotting using both antibodies. As controls, HeLa cells were transfected with WT UCE-FLAG or R328C-UCE-HA constructs alone and then worked up in the same manner as the double transfections.
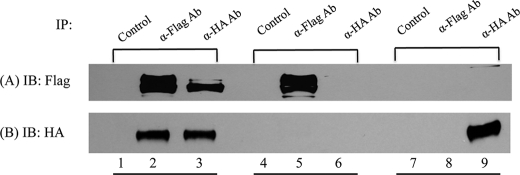
As shown in Fig. 5, in the double transfected cells, anti-HA Ab brought down the WT UCE-FLAG, and anti-FLAG Ab brought down the R328C-UCE-HA. In the single transfected cells, the different forms of UCE were only immunoprecipitated by antibodies directed to their own tag, showing the specificity of the antibodies. These findings establish that the R328C mutant polypeptides can form oligomers with the WT subunits.
FIGURE 5.
In vivo interaction of WT UCE and R328C-UCE. HeLa cells in 100-mm dishes were transiently transfected with 20 μg of WT UCE-FLAG and 10 μg of R328C-UCE-HA (lanes 1–3) or transfected with 20 μg of WT UCE-FLAG only (lanes 4–6) or 10 μg of R328C-UCE-HA only (lanes 7–9). After 24 h, cell lysates were prepared and subjected to immunoprecipitation (IP) with anti-FLAG Ab or anti-HA Ab. The precipitates were subjected to SDS-PAGE and Western blotting with both antibodies.
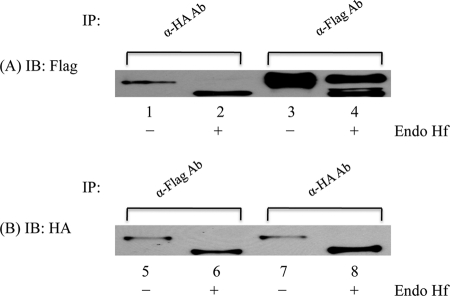
We next analyzed the effect of the R328C mutant on the trafficking of WT UCE to the Golgi. Fig. 6 shows that WT UCE co-expressed with the R328C mutant and immunoprecipitated with antibody to the mutant UCE was completely Endo Hf-sensitive, whereas WT UCE expressed by itself exhibited equal amounts of Endo Hf-sensitive and -resistant species (compare lanes 1 and 2 with lanes 3 and 4). This indicates that interaction of the WT UCE with the R328C mutant impairs its movement to the Golgi, consistent with a dominant negative effect. Essentially all of the R328C mutant was present in the ER, as shown by its complete sensitivity to Endo Hf regardless of whether or not it was co-expressed with WT UCE (lanes 5–8). Thus, oligomerization of the R328C mutant with the WT UCE does not enhance its trafficking to the Golgi under the conditions of this experiment.
FIGURE 6.
Effect of R328C-UCE on maturation of WT UCE. HeLa cells in 100-mm dishes were transiently transfected with 7 μg of WT UCE-FLAG and 7 μg of R328C-UCE-HA (A, lanes 1, 2, 5, and 6) or transfected with 7 μg of WT UCE-FLAG only (A, lanes 3 and 4) or 7 μg of R328C-UCE-HA only (B, lanes 7 and 8). After 24 h, cell lysates were prepared, treated with or without Endo Hf, and immunoprecipitated (IP) with anti-FLAG Ab or anti-HA Ab. The precipitates were subjected to SDS-PAGE and Western blotting (IB) using both antibodies.
DISCUSSION
The findings presented in this study established that the three mutations in the NAGPA gene identified in individuals with persistent stuttering have adverse effects that diminish the total activity of the enzyme in cells. Interestingly, each mutation affects the enzyme in a different manner. The primary effect of the p.R328C mutation is to impair folding of the nascent protein in the ER, resulting in more than half of the newly synthesized polypeptides being degraded by the proteasomal system. This occurred with both the non-brain and brain forms of the mRNA, which differ by the presence or absence of the 102 base pairs encoded by exon 8 in the genomic DNA. The portion of the mutant UCE molecules that successfully trafficked to the TGN appeared to have a normal catalytic function. This can explain how fibroblasts from an individual with persistent stuttering who is homozygous for the p.R328C mutation can have 54% of WT UCE activity. Although the pulse/chase experiments indicate that more than 50% of the newly synthesized mutant UCE was degraded in the ER, these experiments utilized transfected HeLa cells expressing high levels of UCE, and therefore there might have been a greater degree of misfolding and proteasomal degradation than occurs in fibroblasts expressing physiologic levels of the protein. The analysis of HeLa cells co-transfected with the R328C mutant and WT UCE established that the mutant protein oligomerizes with the WT protein and impairs the migration of the latter to the Golgi. Thus R328C has the potential to have a dominant negative effect in the heterozygote state.
The p.H84Q mutation also impairs folding of the nascent polypeptide, resulting in significant degradation via the proteasomal pathway. However, the folding of the non-brain form of this mutation was more impaired than that of the brain form. It should be pointed out that although the brain form of the UCE mRNA is the predominant species expressed in various regions of the brain, some non-brain form is also present. Because the neurons involved in stuttering have yet to be identified, it is unknown which form of the UCE message is synthesized in these critical cells. Regardless, both forms of the H84Q mutant are impaired in their catalytic function with estimated relative activities of 50% of WT.
The p.F513SfsX113 mutant exhibited the most striking phenotype, being rapidly degraded with a t½ of less than 30 min. This compares with a t½ of 7 h for the R328C mutant and 14 h for the WT enzyme. The basis for the rapid degradation was found to reside in a sequence of four hydrophobic residues, VWLL, present in the 113-amino acid extension of the cytoplasmic tail, that results from the frameshift mutation. The rapid degradation of this mutant UCE is mediated by the proteasomal pathway, as evidenced by the finding that the proteasomal inhibitor MG132 greatly slowed the rate of degradation. It is of interest that Park et al. (13) have reported that a trileucine cluster in the cytoplasmic tail of the human cytomegalovirus US10 membrane glycoprotein is responsible for the rapid degradation of this protein via the proteasomal system. However, in this case, degradation depends on US10 forming a complex with the HLA-G protein.
These data strengthen the genetic arguments that the mutations found in the NAGPA gene in individuals with persistent stuttering play a causative role in this phenotype. The key observation is that each of the three mutations leads to diminished UCE enzyme activity in cells, albeit via different mechanisms. Prior to these studies it was unknown whether the mutations would actually impair UCE activity. However, it is still unclear how a partial deficiency of UCE activity contributes to the stuttering phenotype. The answer to this puzzle will require a greater understanding of the pathophysiology of stuttering, including identification of the critical neurons that are involved in this process.
Acknowledgments
We thank Balraj Doray, Jennifer Govero, Eline van Meel, Rachel Idol, and Yi Qian for suggestions.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NIDCD, Grant Z01-00046-11 (to D. D.) and by NIH Grant CA08759 (to S. K.).
- UCE
- uncovering enzyme
- Man-6-P
- mannose 6-phosphate
- ML
- mucolipidosis
- TGN
- trans-Golgi network
- CHX
- cycloheximide
- Ab
- antibody
- Endo Hf
- endoglycosidase Hf
- PNGase F
- peptide N-glycosidase F
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Bloodstein O., Ratner N. A. (2007) Handbook on Stuttering, 6th Ed., Delmar Cengage Learning, Clifton Park, NY [Google Scholar]
- 2. Kang C., Drayna D. (2011) Annu. Rev. Genomics Hum. Genet. 12, 5.1–5.20 [DOI] [PubMed] [Google Scholar]
- 3. Kang C., Riazuddin S., Mundorff J., Krasnewich D., Friedman P., Mullikin J. C., Drayna D. (2010) New Engl. J. Med. 362, 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kollmann K., Pohl S., Marschner K., Encarnação M., Sakwa I., Tiede S., Poorthuis B. J., Lübke T., Müller-Loennies S., Storch S., Braulke T. (2010) Eur. J. Cell Biol. 89, 117–123 [DOI] [PubMed] [Google Scholar]
- 5. Kornfeld R., Bao M., Brewer K., Noll C., Canfield W. (1999) J. Biol. Chem. 274, 32778–32785 [DOI] [PubMed] [Google Scholar]
- 6. Kornfeld S., Sly W. S. (2000) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R. ed) pp. 3469–3483, McGraw-Hill, New York [Google Scholar]
- 7. Cathey S. S., Leroy J. G., Wood T., Eaves K., Simensen R. J., Kudo M., Stevenson R. E., Friez M. J. (2010) J. Med. Genet. 47, 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rohrer J., Kornfeld R. (2001) Mol. Biol. Cell 12, 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Do H., Lee W. S, Ghosh P., Hollowell T., Canfield W., Kornfeld S. (2002) J. Biol. Chem. 277, 29737–29744 [DOI] [PubMed] [Google Scholar]
- 10. Mullis K. G., Ketcham C. M. (1992) Anal. Biochem. 205, 200–207 [DOI] [PubMed] [Google Scholar]
- 11. Dustin M. L., Baranski T. J., Sampath D., Kornfeld S. (1995) J. Biol. Chem. 270, 170–179 [DOI] [PubMed] [Google Scholar]
- 12. Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 13. Park B., Spooner E., Houser B. L., Strominger J. L., Ploegh H. L. (2010) J. Exp. Med. 207, 2033–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]