Background: Arabidopsis plants with a knock-out in UDP-glucose dehydrogenase provide less nucleotide sugars for cell wall biosynthesis.
Results: Mutants with reduced UDP-glucuronic acid show developmental defects and changes in the pectic network.
Conclusion: Pectins are important for plant cell walls. Alternative pathways to UDP-glucuronic acid are unable to compensate the mutation and limited by the inositol supply.
Significance: Pectic polymers are more important for cell wall integrity and development than previously thought.
Keywords: Carbohydrate Biosynthesis, Carbohydrate Metabolism, Cell Wall, Development, Nucleoside Nucleotide Biosynthesis, Plant, Rhamnogalacturonan II, UDP-glucose Dehydrogenase
Abstract
UDP-glucose dehydrogenase (UGD) plays a key role in the nucleotide sugar biosynthetic pathway, as its product UDP-glucuronic acid is the common precursor for arabinose, xylose, galacturonic acid, and apiose residues found in the cell wall. In this study we characterize an Arabidopsis thaliana double mutant ugd2,3 that lacks two of the four UGD isoforms. This mutant was obtained from a cross of ugd2 and ugd3 single mutants, which do not show phenotypical differences compared with the WT. In contrast, ugd2,3 has a strong dwarfed phenotype and often develops seedlings with severe root defects suggesting that the UGD2 and UGD3 isoforms act in concert. Differences in its cell wall composition in comparison to the WT were determined using biochemical methods indicating a significant reduction in arabinose, xylose, apiose, and galacturonic acid residues. Xyloglucan is less substituted with xylose, and pectins have a reduced amount of arabinan side chains. In particular, the amount of the apiose containing side chains A and B of rhamnogalacturonan II is strongly reduced, resulting in a swollen cell wall. The alternative pathway to UDP-glucuronic acid with the key enzyme myo-inositol oxygenase is not up-regulated in ugd2,3. The pathway also does not complement the ugd2,3 mutation, likely because the supply of myo-inositol is limited. Taken together, the presented data underline the importance of UDP GlcA for plant primary cell wall formation.
Introduction
Nucleotide sugars are fundamental components for the biosynthesis of the plant cell wall. Using the energy liberated by the split of nucleotides, glycosyltransferases mainly located in the lumen of the Golgi coordinately work to generate cell wall polysaccharides (1). The glycosyltransferases needed by the plant are numerous because of the high complexity and variability of cell wall glycans made up of at least 14 different monosaccharides connected by various glycosidic linkages (2, 3). After their assembling, hemicellulosic and pectic components are transported into the wall where they are modified and assembled together in interaction with the cellulose fibrils. These are produced by plasma membrane-associated cellulose synthase complexes using cytosolic UDP-glucose as donor substrates.
Major sugar moieties are synthesized as UDP- or GDP-sugars by interconverting enzymes located in the cytosol and Golgi apparatus. Arabidopsis thaliana possesses about 10 different types of epimerases, decarboxylases, and dehydrogenases involved in the biosynthesis of the nucleotide sugars. They are all encoded by small gene families with typically two to six isoforms. UDP-glucuronic acid (UDP-GlcA)2 is derived from an NAD+-dependent 2-fold oxidation of UDP-Glc catalyzed by the enzyme UDP-glucose dehydrogenase (UGD). The two oxidation steps correspond to a net four-electron oxidation without released intermediates. Therefore, the reaction is irreversible under physiological conditions. UDP-GlcA acts as an important precursor in the synthesis of many different polysaccharides contributing to up to 50% of A. thaliana cell wall biomass. Xylose, arabinose, apiose, and GalA residues in plant polymers are synthesized from it in plants (4).
The first UGD from eucaryotes was cloned from soybean (5). Although it was initially believed to be a single-copy gene, data from genomic sequencing clearly indicate the presence of small gene families in plants (6–9). Within the plant kingdom, the protein sequences of UGDs are highly conserved, showing typically more than 90% sequence conservation between the isoforms. A. thaliana has four functional UGDs that differ in their biochemical properties. In particular the affinity to the substrate UDP-Glc is highly variable, ranging from 15 to 335 μm (9, 10). Higher values up to 900 μm have been reported for two maize UGDs (11). The enzyme UGD directly competes with sucrose-6-phosphate synthase for the substrate UDP-glucose and thus affects channeling of carbohydrates between the targets sucrose and cell wall precursors including cellulose synthesis. A further competing reaction is performed by the UDP-glucose-4-epimerase, which converts UDP-Glc into UDP-Gal.
The UGD product UDP-GlcA can also be synthesized via ring cleavage of myo-inositol by the enzyme myo-inositol oxygenase (MIOX) (pathway in Fig. 1). The pathway is long known (for review, see Ref. 12) and functional in A. thaliana (13, 14). However, biochemical labeling experiments with A. thaliana cell cultures suggest a dominance of the UGD pathway for UDP-GlcA formation (15). MIOX genes are expressed in a tissue-specific manner and harbor high differences in their expression levels.
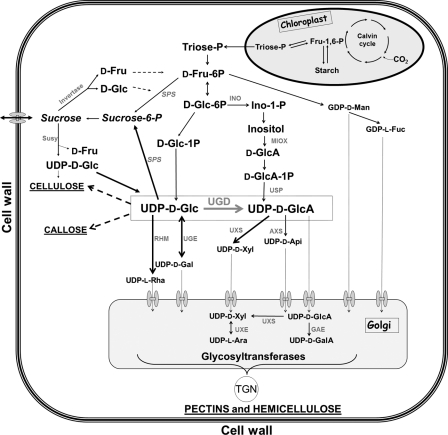
FIGURE 1.
Biochemical pathway to illustrates the central role of UDP-GlcA for cell wall biosynthesis. The reaction of the enzyme UDP-glucose dehydrogenase (UGD is boxed) is shown. The product UDP-GlcA is the direct precursor for xylose, arabinose, apiose (Api), and galacturonic acid residues of the cell wall. The uptake of nucleotide sugars into the Golgi by (mostly unknown) transporters is indicated by dotted arrows and transmembrane uptake systems. AXS, UDP-apiose/UDP-xylose synthase; GAE, UDP-glucuronic acid epimerase; INO, inositol-1-phosphate synthase; RHM, UDP-rhamnose synthase; SPS, sucrose-phosphate synthase; SUSY, sucrose synthase; UGE, UDP-glucose epimerase; UXE, UDP-xylose epimerase; UXS, UDP-xylose synthase; USP, UDP-sugar pyrophosphorylase).
Here we investigate a knock-out mutant for two UGD isoforms to determine the role of individual UGD enzymes and to test the flexibility of the UGD network in plants. The UGD2 and UGD3 isoforms were chosen because they contribute mostly to the total UGD activity, and they seem to act synergistically to provide plants with nucleotide sugars as cell wall precursors. Furthermore, the interaction between the UGD and MIOX pathway for the synthesis of cell wall precursors is addressed. The A. thaliana ugd2,3 double mutant has an altered metabolite composition and architecture of the cell wall causing developmental defects. The reduction in different sugars is mainly present in pectic polymers. The role of UGD2 and UGD3 is the efficient conversion of carbohydrates into nucleotide sugar for cell wall polymers biosynthesis. This may compete with sucrose synthesis and thereby limit the amount of soluble sugars.
EXPERIMENTAL PROCEDURES
Plant Material and Molecular Analysis
A. thaliana was either germinated on soil or on solid Murashige Skoog-phytagel medium. If required, seeds were surface-sterilized and plated on Petri dishes (0.5× MS-salts (Duchefa; The Netherlands), 0.5% (w/v) sucrose, 0.8% (w/v) Phytagel, adjusted to pH 5.7 with KOH) in a vertical position. In both cases, seeds were cold-treated for 2 days to synchronize germination, and plants were grown in a 10-h light/14-h dark cycle at 22 °C.
The double mutant ugd2,3 was generated by crossing two homozygous single T-DNA insertion mutants in UGD2 (At3g29630) and UGD3 (At5g15490) (ugd2, SAIL_1144_F10; ugd3, SALK_006234) and confirmed by PCR. Col-0 (NASC N60000) was used as the WT control.
Semiquantitative PCR was initially used to test for UGD transcripts in ugd knock-out lines. The PCR program was 94 °C for 3 min then 29× (94 °C for 15 s, 55 °C for 30 s, and 72 °C for 1 min). Ubiquitin samples were removed after 24 cycles. Quantitative RT-PCR was later used to ensure that the transcript in the insertion lines was absent. Seven day-old seedlings were harvested and processed according to Klinghammer and Tenhaken (9), but new optimized primers (supplemental Table S1) were used. Quantitative RT-PCR was performed on a MX3000 cycler (Stratagene) using the SYBR Green method and the PCR program (94 °C for 3 min, 40× (94 °C for 15s, 55 °C for 30 s, and 72 °C for 1 min)) followed by a product dissociation melting curve. The locus At4g05320 for ubiquitin was used as a housekeeping gene. MIOX transcripts were quantified as described in Endres and Tenhaken (16). The UGD enzyme assay was performed with desalted leaf extracts according to Seitz et al. (13).
RNA Hybridization Microarray Analysis
RNA from WT and ugd2,3 seedlings (6 days old) were isolated with the Nucleo Spin RNA Plant kit (Macherey-Nagel). The experiment was repeated once with an independently grown batch of seedlings. cDNA labeling, hybridization, and bioinformational analysis of an Agilent 4 × 44K Arabidopsis oligonucleotide array was performed by a commercial service (ImaGenes; Berlin, Germany).
Complementation
A 3.2-kb genomic fragment of the native UGD2 gene cassette was amplified by PCR with primers Ugd2-genome-F/Ugd2-genome-R using the Phusion proof reading polymerase (Finnzymes) and cloned into the vector pGreen 0179 (17). The construct was introduced into Agrobacterium tumefaciens GV3101 and then transformed into ugd2,3 by floral dip.
Transmission Electron Microscopy (TEM) and Immunohistological Analysis
Seven-day-old cotyledons were cryofixed in a Leica EMPACT high pressure freezer (Leica Mikrosysteme GmbH, Vienna, Austria) and freeze-substituted in 1% osmium and 0.05% uranyl acetate in acetone for 73 h in a Leica EM AFS freeze substitution apparatus (Leica Microsysteme GmbH, Vienna, Austria) following the method of Lütz-Meindl and Aichinger (18). Samples were rinsed in acetone, then transferred into ethanol and embedded in LR-White Resin. Ultrathin sections oriented in the longitudinal plane were viewed in a LEO 912 AB TEM (ZEISS, Oberkochen, Germany) operated at 80 kV (19).
Immunolabeling for Confocal Laser Scanning Microscopy and TEM
Fixation, infiltration, and embedding of seedlings in Steedman's wax (PEG 400 distearate and 1-hexadecanol, 9:1 (w/w)) was made as previously described in Vitha et al. (20). 8-μm-thick transversal sections were cut with a rotation microtome (Microm HM355S, Histocom, Neudorf, Austria), then mounted on polylysine-coated microscope slides (Menzel GmbH, Braunschweig, Germany). Sections were dewaxed in 97% ethanol and rehydrated in an ethanol series (97 to 50% ethanol in PBS). For immunostaining, sections were first blocked 30 min in PBST-BSA (PBS + 3% BSA (w/v) + 0.05% Tween 20 (v/v)). Primary antibodies were diluted in PBST and incubated for 2 h followed by 5 washes with PBST. Secondary antibody (Cy3-labeled goat anti-rat; Dianova) was diluted 1:500 in PBST and incubated for a further 2 h. Sections were washed with PBS three times to remove unbound antibodies. Sections were mounted in Mowiol4.88 (Roth, Karlsruhe, Germany) then examined in Zeiss LSM 510 confocal laser microscope (Jena). Sections were excited with a helium laser (543 nm line) and viewed using a 560–615-nm band pass filter. Images were processed and analyzed with Zeiss LSM 510 software.
Immunogold labeling was performed according to Oertel et al. (21). Sections mounted on gold grids were preincubated in 0.05 m Tris-buffered saline (pH 7.5) containing 1% BSA and 0.1% Tween (TBST-BSA) for 1 h at room temperature and were then transferred onto 15-μl droplets of the primary antibody (diluted 1:10 in TBST-BSA) for 14–16 h at 4 °C in a moist chamber. After 4 washes on 20-μl droplets of TBST-BSA, 15 min each, sections were stained with 10-nm gold-conjugated anti-rat IgG antibodies (Sigma G7035) diluted 1:40 in TBST-BSA. Last, grids were transferred onto droplets of TBST-BSA and rinsed thoroughly with a soft spray of double-distilled water.
Low Temperature Scanning Electron Microscopy
Seven-day-old plants were mounted on an aluminum stub using a cryo-adhesive (Bio-Rad) and were rapidly frozen by plunging into liquid propane to preserve the material integrity. Samples were then partially freeze-dried in high vacuum (<2 × 10−4 pascal) at −80 °C for 10 min and coated with platinum in a preparation chamber before being transferred onto a temperature-controlled cryostage in the scanning electron microscope. Low temperature scanning electron microscopy was then performed as previously described by Frey et al. (22).
Analysis of Monosaccharide Composition, Arabinan, and Xyloglucan Fingerprints
Cell wall material was prepared from Arabidopsis leaves or seedlings (∼150 mg fresh weight) by homogenization in liquid nitrogen followed by 2 extractions in 70% ethanol, 1 extraction in methanol/chloroform, and 1 extraction with acetone. The dried powder was gelatinized in sodium acetate buffer (50 mm (pH 4)) for 20 min at 80 °C and freed of starch by amylase and pullulanase incubation, washed to remove soluble sugars, and finally, hydrolyzed in 2 m TFA (90 min; 119 °C) and vacuum-dried. Monosaccharides were separated on a CarboPac-PA20 column (Dionex ICS-3000) (15 mm NaOH, 0.45 ml min−1 for 0–12 min; 50 mm NaOH + 200 mm sodium acetate for 12–22 min).
Xyloglucan fingerprints were generated with a xyloglucan-specific endo-glucanase (Novozyme) according to Lerouxel et al. (23). Fragments were separated on a Dionex ICS3000 system using a CarboPac PA200 column using a gradient from 100 mm NaOH to 100 mm NaOH plus 100 mm sodium acetate within 30 min and a flow rate of 0.45 ml min−1. A glucan polymer standard was obtained by partial α-amylase digestion of soluble starch and separated with the same gradient.
Arabinan side chains were fingerprinted with a recombinant endo-arabinase from Aspergillus aculeatus, expressed in Aspergillus oryzae. Both enzymes were kindly provided by Dr. Kirk Matthew Schnorr, Novozymes, Denmark. Crude cell wall material was incubated with 0.5 μg of purified arabinase in 50 mm sodium acetate buffer (pH 5.5) for 12 h. Products were separated on a CarboPac PA20 column.
Starch and Soluble Sugars
50 mg of leaf material of adult plant were frozen, homogenized, and resuspended in 650 μl of 80% ethanol and incubated at 80 °C for 20 min before being centrifuged. The supernatants of two consecutive alcohol extractions were pooled and constitute the soluble sugars. Samples were analyzed on a PA 20 column using the method described above for monosaccharide analysis.
Quantification of RGII Chains A and B
Cell wall material was prepared from Arabidopsis plant material (7-day-old seedlings or 4-week-old leaves) by homogenization in liquid nitrogen followed by two extractions in 70% ethanol, 1 extraction in methanol/chloroform, and 1 extraction with acetone. RGII chains were released from dry cell wall material by mild acid hydrolysis with 0.1 m TFA (24), borohydrate-reduced and -enriched by solid phase extraction (25). Identification and quantification of the structures was made by LC-ESI-MS-MS (ESI, electrospray ionization) using an SPE-PGC cartridge (Hypercarb 10-mg column, Thermo scientific). Both plant cell wall preparations showed a similar reduction in the side chains A and B in ugd2,3 compared with wild type. The data shown are from the 4-week-old plant material. The experiment was performed twice independently in duplicate.
RESULTS
Isolation of a ugd2,3 Double Mutant
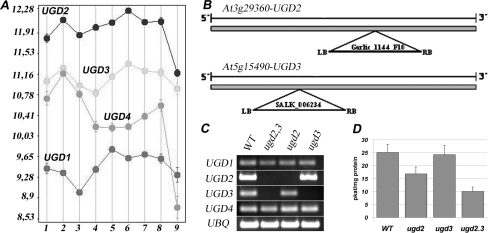
The UGD gene family has four members, all of which were previously shown to be true UDP-glucose dehydrogenases (9). UGD2 and UGD3 showed the highest expression among the isoforms during plant development (Fig. 2A), as indicated by the microarray data visualized with Genevestigator (26) and in agreement with RT-PCR and β glucuronidase-reporter gene analyses performed by Klinghammer and Tenhaken (9). To investigate the role of these isoforms, we isolated T-DNA insertion mutants in the UGD2 (At3g29360) and UGD3 gene (At5g15490). As all of the UGD genes have a single intron-less open reading frame, the insertion of a T-DNA within the coding region might result in a loss of function of the respective gene (Fig. 2B).
FIGURE 2.
Gene expression and T-DNA insertional mutants of UGD2 and UGD3. A, shown is gene expression level (log2 scale) of all four UGD genes in different tissues from Arabidopsis based on public available microarray data using Genevestigator. Germinated seed (1), seedling (2), young rosette (3), developed rosette (4), bolting (5), young flower (6), developed flower (7), flowers and siliques (8), mature siliques (9) are shown. B, schematic representation of the positions of the T-DNA insertions in the UGD2 and UGD3 genes is shown. Both T-DNAs, flanked by left border (LB) and right border (RB) sides, are located in the single exon, thus, disrupting the open reading frame for UGD. C, an RT-PCR analysis of transcript levels in WT and the ugd mutant lines is shown. Semiquantitative PCR was performed on first-strand cDNA made from 10-day-old seedlings. D, shown is UGD enzyme activity in WT and ugd mutants.
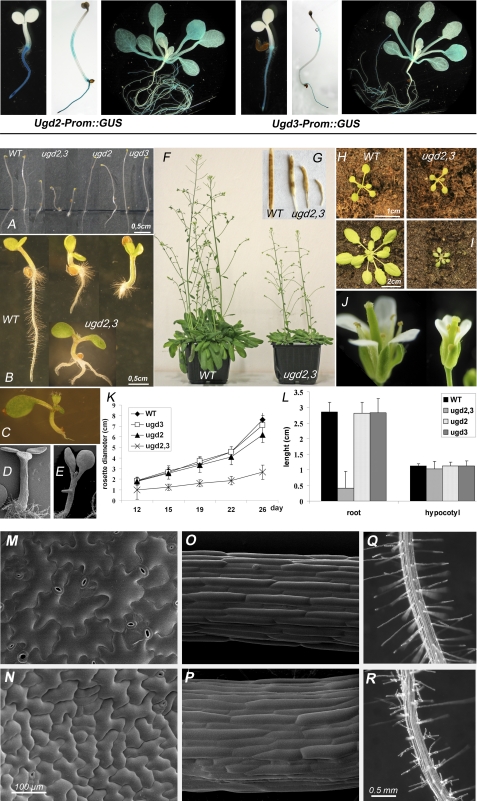
ugd2 and ugd3 single mutant plants showed no visible phenotypes, indicating genetic redundancy (Fig. 3, A, K, and L). The very similar promoter activity pattern observed with UGD2::GUS and UGD3::GUS reporter gene lines (Fig. 3, upper panel) suggested already a redundant function of both UGD isoforms. We, therefore, tried to obtain a double knock-out mutant for both UGD2 and UGD3. A cross of ugd2 and ugd3 was performed, and individual siblings were genotyped by PCR to identify homozygous ugd2,3 plants (Fig. 2C). In accordance to the Mendelian law, we frequently observed homozygous ugd2 and heterozygous ugd3 and vice versa. Within ∼200 individual plants we found no homozygous double mutant. To increase the probability for such a double knock-out, we tested siblings after selfing ugd2ugd2/UGD3ugd3 plants. A genotyping of random seedlings resulted in ∼1 double knock-out among 25 seedlings. Quantitative RT-PCR confirmed the absence of transcripts for UGD2 and UGD3 in the ugd2,3 mutant lines. The lack of these two functional UGD genes compromises the UGD enzyme activity in plants (Fig. 2D). The total UGD activity found in leaves from mature plants of ugd2,3 dropped to 40% of the WT level. The reduction of UGD enzyme activity in ugd2,3 is higher than the cumulative effect of the two single knockouts.
FIGURE 3.
Upper panel, examples of UGD2-prom::GUS and UGD3-prom::GUS stainings illustrate the coexpression of both isoforms in Arabidopsis. Lower panel, phenotype of ugd2,3 is shown. A, shown are 5-day-old etiolated seedlings. B, shown are developmental defects in 4-day-old light-grown WT and ugd2,3 seedlings. C–E, shown are examples of developmental defects in seedlings with one (C) or three cotyledons (D). E, development of a “secondary root” out of the hypocotyl is shown. F, shown are adult plants grown for 8 weeks in the greenhouse. ugd2,3 remains much smaller. G, siliques of WT and ugd2,3 are shown. H and I, 2-week-old (H) and 3-week-old (I) WT and ugd2,3 rosettes are shown. J, flowers of WT and ugd2,3 are shown. K, rosette diameters were determined over a 4-week period for WT, ugd2, ugd3, and ugd2,3. L, lengths of roots and hypocotyls of 5-day-old seedlings (S.E.) are shown. M–Q, shown are low temperature scanning electron micrographs of WT (M and O) and ugd2,3 (N and P). Epidermal cells from cotyledons (M and N) and hypocotyls (O and P) of 7-day-old seedlings show abnormal pattern in ugd2,3 mutants. Q, shown are root hairs of 7-day-old wild type plants. R, disturbed root hair development in ugd2,3 is shown.
The Presence of Either UGD2 or UGD3 Isoform Is Required for Normal Growth and Development
The ugd2,3 double mutant is characterized by a dwarfed phenotype and a retarded development. At the adult stage, all organs of ugd2,3 were shorter than the WT organs (Fig. 3, F–J). The rosette leaves were ∼75% smaller, and the inflorescence stems were ∼25% shorter compared with WT (Fig. 3F). A reduction in the number of side shoots was also observed that were in addition longer and thinner than those of the WT.
Flowers of ugd2,3 were also significantly smaller than the WT flowers (Fig. 3J). All sexual organs were present; nevertheless, they were shorter. Flowers has difficulties fully opening their petals. Many siliques remained tiny and contained no seeds (Fig. 3G). The number of siliques that contained fertile seeds was strongly reduced. When seedlings were grown on MS plates, ugd2,3 appeared with different phenotypes. Some seedlings were dwarfed but had normal anatomy; roots from 4-day-old seedlings were shorter (Fig. 3L), but cotyledons had the same color and shape as in WT. Secondary roots appeared about 2 days later as in the WT. The majority of seedlings possess significantly reduced roots (Fig. 3B). The most pronounced phenotype for abnormal development (10–20% of seedlings) is the absence of a primary root (Fig. 3, B and C). In this case, at the age of 4 to 5 days after germination, seedlings formed secondary roots directly from the base of the root (Fig. 3B) or from the hypocotyl (Fig. 3E). Additional examples are shown in supplemental Fig. S1. Most ugd2,3 seedlings exhibited two cotyledons, but a few bore one or three cotyledons (Fig. 3, C and D). The majority of ugd2,3 seedlings were glassy and light green-colored. Dark-grown ugd2,3 seedlings also showed a strong reduced elongation of the hypocotyl and root (Fig. 3A).
We further analyzed developmental defects of epidermal cells of ugd2,3 by low temperature scanning electron microscopy. The shape of epidermal cells from ugd2,3 cotyledons and hypocotyls differ from those of the WT. The pavement cells of the mutant are much smaller and do not show the normal jigsaw puzzle form (Fig. 3, M and N). Epidermal cells of mature leaves remain smaller during maturation. The height of trichomes is not affected, but as a consequence of the reduced epidermal cell size they have a higher density than in WT plants. Stomata in ugd2,3 are slightly smaller (17.8 ± 2.25 μm in ugd2,3 versus 19.1 ± 1.79 μm in WT) and appear at a higher density (278 ± 52 mm−2 in ugd2,3 versus 212 ± 45 mm−2 in WT). The stomata distribution pattern in ugd2,3 is somewhat irregular, and stomata seem to be clustered, whereas the WT shows a regular distribution pattern (Fig. 3, M and N).
ugd2,3 hypocotyls have a larger diameter compared with the WT. In the middle of the hypocotyl the epidermal cells are organized in linear cell files, and each cell normally has a similar cell length (Fig. 3O). ugd2,3 has a different characteristic. The cells in the middle of the hypocotyl show heterogeneity in cell length and no perfect linear cell files, indicating that the pattern development of the hypocotyl was affected (Fig. 3P). Root hairs in ugd2,3 show branched, deformed hairs with bulbous tips. Their distribution along the rhizodermis of roots is more irregular when compared with the pattern of WT root hairs (Fig. 3, Q and R).
Biochemical Analysis of ugd2,3 Cell Wall Reveals Complex Alterations in the Sugar Composition
The loss of two of the four UGD genes will likely reduce the rate of UDP-GlcA synthesis and affects the availability of those UDP sugars that are directly derived from UDP-GlcA (UDP-GalA, UDP-Ara, UDP-Xyl, UDP-apiose (Api), compare the scheme in Fig. 1). We, therefore, analyzed the sugar monomer composition of cell walls from mature leaves of WT and mutant plants by high performance anion-exchange chromatography and electrochemical pulsed amperometric detection.
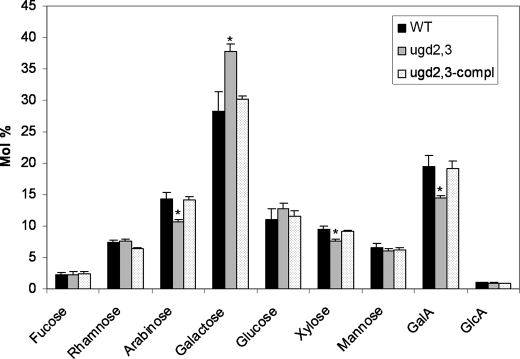
The single mutant ugd2 or ugd3 unexpectedly did not show any major quantitative differences in the sugar composition compared with the WT (not shown). In contrast, ugd2,3 exhibits significant differences (p < 0.05; Student's t test) in the amount of four sugars (Fig. 4). GalA was ∼27% reduced when compared with the WT. The neutral sugars arabinose and xylose were also reduced (20–25% each), which is consistent with their biosynthetic origin from the precursor UDP-GlcA. Galactose (+30%) is increased presumably as a result of the increased availability of UDP-Glc/UDP-Gal and also possibly as a compensatory mechanism.
FIGURE 4.
Cell wall composition of 5-week-old WT, ugd2,3 mutants, and complemented ugd2,3 plant. The sugar composition of cell wall was determined after TFA hydrolysis and separation on anion-exchange column (CarboPac PA20, Dionex ICS300 System). Values are (mol%) ± S.E. from five independent biological samples. The amount of Ara, Xyl, and GalA is statistically significantly reduced (p < 0.05) and marked by an asterisk.
The Phenotype of ugd2,3 Can Be Restored by Genetic and Biochemical Complementation
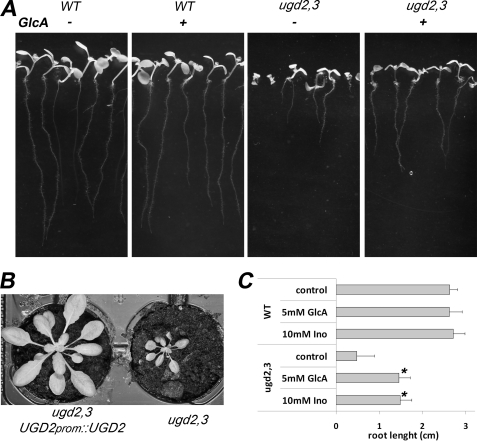
To prove that the observed developmental and growth defects are caused by the T-DNA insertions in ugd2,3, we performed biochemical and genetic complementation experiments. We hypothesized that exogenous GlcA should restore normal growth. Biochemical complementation of mutant phenotypes could indeed be partially achieved by growing ugd2,3 double mutant seedlings on MS plates supplemented with the sugar GlcA (5 mm). All seedlings developed roots of equal length that are significantly longer than those of seedlings grown on MS plates without GlcA addition (Fig. 5A). The seedlings looked much healthier, revealing that feeding a precursor of UDP-GlcA facilitates partial biochemical complementation of ugd2,3 knock-out. Because the product of the UGD reaction is UDP-GlcA, we did not expect that it would be possible to completely recover the WT phenotype by GlcA feeding. To synthesize UDP-GlcA from the fed GlcA, two enzymatic reactions are necessary, a glucuronokinase (27, 28) and a pyrophosphorylase (29). Feeding of arabinose gives a lesser complementation as compared with GlcA, whereas the addition of xylose does not complement. ugd2,3 seedlings sewn on plates with higher concentrations of GlcA (25 mm) or a combination of added GlcA, GalA, arabinose, and xylose do not grow significantly better than seedlings few with GlcA alone. As controls, we fed sugars like glucose or galactose, requiring UGD activity to contribute to the biosynthesis of UDP-GlcA. These sugars show no beneficial effect on root growth in ugd2,3 (data not shown).
FIGURE 5.
Biochemical and genetic complementation of ugd2,3. A, seedlings of WT and ugd2,3 were grown on MS-phytagel plates supplemented with or without 5 mm GlcA. The developmental defects in ugd2,3 roots can be partially rescued. All seedlings develop a normal root which, however, remains shorter than in the WT. B, genetic complementation was done by introducing UGD2prom::UGD2 into the ugd2,3 double mutant. The complemented mutant (left) grows like WT plants and is far bigger than ugd2,3. C, root length of seedlings on MS medium supplemented with 5 mm GlcA or 10 mm inositol (Ino). WT roots show no change in length, whereas roots from ugd2,3 elongate in supplemented media.
We initially hypothesized that a partial reduction in UGD activity should be compensated by the alternative MIOX pathway (see Fig. 1), also providing UDP-GlcA as a final product. The observed mutant phenotype in ugd2,3 already indicates that the expectation was wrong and that the MIOX pathway at most partially compensates for a reduction in UGD activity. To test whether the availability of inositol or the conversion into UDP-GlcA is limiting, we fed myo-inositol to ugd2,3 seedlings. Application of myo-inositol partially complements the ugd2,3 mutant phenotype similarly to the addition of GlcA (Fig. 5C).
We also performed a genetic complementation of ugd2,3 by expressing the UGD2 gene under control of its own promoter in the mutant. As the ugd2 or ugd3 single mutant does not significantly exhibit a phenotype different to WT plants, we decided to complement ugd2,3 double mutant with only one of the two missing UGD isoforms. We obtained three transformants showing a WT-like phenotype (Fig. 5B). Biochemical analyses revealed that the reintroduction of the UGD2 gene into the ugd2,3 mutant also rescues the cell wall composition (Fig. 4). The successful complementation to a normal WT-like phenotype by introduction of an intact copy of the UGD2 gene confirms that the ugd2,3 phenotype is a direct effect of the T-DNA insertions in the respective UGD gene and that the observed phenotypes reflect the influence of these mutations.
Microarray Analysis Reveals No Drastic Change in Expression of Developmental Regulatory Genes
To exclude that the developmental defects are caused by sugar-mediated changes in the expression of developmental key regulatory genes, we did a microarray hybridization experiment with RNA samples from WT and ugd2,3 seedlings using the Agilent 44K Arabidopsis array. Only 31 genes were significantly up- or down-regulated (>2 log2 scale units) (supplemental Table S2) with no evidence for altered gene expression of developmental regulatory genes.
Xyloglucan, RGI, and RGII Structures Are Affected by Mutation of UGD2 and UGD3
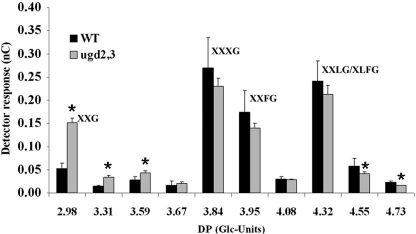
To address the question of which polymers are affected in ugd2,3, we analyzed hemicellulosic and pectic components. Xyloglucans from ugd2,3 were investigated by an enzymatic fingerprint method (23), in which a xyloglucan-specific endo-glucanase cuts xyloglucan polymer after unsubstituted glucose residues of the backbone. The fingerprint patterns of xyloglucans from WT and ugd2,3 are similar when separated on CarboPacPA200 column. However, we noted an overrepresentation of shorter (earlier eluting) fragments (e.g. XXG) in ugd2,3 and an underrepresentation of larger (later eluting) fragments (e.g. XXXG, XXLG) as shown in Fig. 6. To identify some peaks, tamarind xyloglucan fragments were used as a reference, and data were further compared with previous studies performed by Cavalier et al. (30). The Fry nomenclature (31) was used to label the major peaks.
FIGURE 6.
Xyloglucan fragments, released from seedling cell walls by incubation with a xyloglucan-specific endo-glucanase, were separated on a CarboPac PA200 anion-exchange column. The peak areas of xyloglucan-specific fragments was compared between WT and mutant plants in five independent samples. A glucan size standard was separated under identical conditions, and the degree of Glc-polymerization (DP) is indicated in the numbers on the x-axes. The major xyloglucan peaks were assigned by comparison with tamarind xyloglucan standard fragments. Shorter fragments, which are less substituted with xylose, are overrepresented in ugd2,3, whereas longer fragments are slightly underrepresented in ugd2,3. Statistically significant data (p < 0.05) are marked by an asterisk.
The data suggest a modification of xyloglucans toward less xylose substitution of the glucan backbone chain, whereas the overall amount remains unchanged. Xylose is also used for protein glycosylation. We analyzed the N-glycosylation pattern by immunoblotting of protein extracts using an anti-xylose antibody (Agrisera AS07 267) but found no differences in the pattern of the Western blot (data not shown), suggesting that changes in glycoproteins are not likely to be responsible for the phenotype of ugd2,3.
The lower amount of arabinose in ugd2,3 was also further investigated with an enzymatic fingerprint method using an endo-arabinase from A. aculeatus, which cuts arabinan side chains typically found in the pectic RGI polymer. Products of the enzymatic digest were separated on a CarboPac PA20 column. The amount of released arabinan oligosaccharides was strongly reduced (∼60%) in cell walls from ugd2,3 compared with WT cell walls (data not shown).
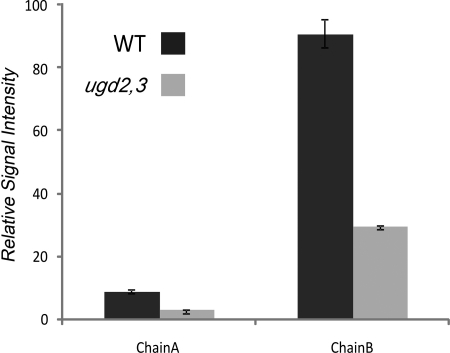
When we performed transmission electron microscopy, we noticed that the thickness of the cell wall in ugd2,3 is increased compared with the WT. As the amount of cell wall per gram of fresh weight is the same in WT and ugd2,3, the increased thickness points to a swollen cell wall as previously found in RGII mutants or boron-depleted plants. We analyzed the amount of RGII by mass spectroscopy coupled to liquid chromatography. Both side chains A and B are attached to the polygalacturonan backbone via an apiose residue, allowing their release by mild acidic treatment. The structures of side chains A and B found in WT are also detectable in ugd2,3. However, the amount of both those side chains is strongly reduced in the mutant by about 65% (Fig. 7).
FIGURE 7.
Quantification of RGII side chains A and B by mass spectroscopy. Counts obtained for WT in the measurement performed by mass spectroscopy coupled to liquid chromatography were set to 100%. Error bars represent S.E. values. Each sample was independently prepared twice (2× WT and 2× ugd2,3). Each measurement was averaged over two separate infusions (2 × 2 runs). Chain A and chain B in ugd2,3 were both found to be reduced to about 35% of WT values (WT chain A, 9.12 ± 0.65; ugd2,3 chain A, 3.08 ± 0.12; WT chain B, 90.88 ± 4.48; ugd2,3 chain B, 29.55 ± 0.60). Chain A: (apiose), Rha, (GalA)2, Fuc-2O-Me-Xyl, GlcA, Gal). Chain B: (Api, Rha, aceric acid, Gal, 2O-Me-Fuc, Ara (Rha)).
The addition of boron often complements RGII mutant phenotypes (32). We, therefore, tested whether exogenous boron (2 mm) can rescue ugd2,3 phenotypes to WT. We did not observe any complementation of ugd2,3 defects by boron, suggesting that the affinity of the cross-linking apiose of chain A is not affected in ugd2,3 phenotypes. This speaks for a reduction of the numbers of sites of cross-linking rather than for a decrease in the affinity of these sites.
Immunolabeling of Pectic Epitopes Differs between WT and ugd2,3 Cell Walls
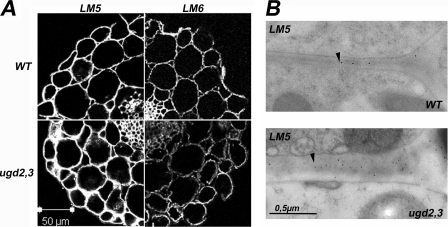
Because of their simple anatomy, we decided to perform labeling on hypocotyls to confirm the high performance anion-exchange chromatography and electrochemical-pulsed amperometric detection sugar composition analysis with an independent method. To investigate the distribution of pectins, we used the monoclonal antibodies JIM5 (partially deesterified homogalacturonan) and JIM7 (partially methylated homogalacturonan) (33). RGI side chains are recognized by LM5 ((14)-β-d-galactan) and LM6 ((15)-α-d-arabinan) (34).
Similar results in the labeling intensity and pattern were observed with the antibodies JIM5, JIM7, LM7, JIM13 (AGP-glycan), LM8 (xylogalacturonan), LM10, LM11 (glucuronoarabinoxylan), CCRC-M1 (fucosylated xyloglucan), CCRC-M23, M48, M77, M85, and CCRC-M109. However, the LM5 antibody shows a much stronger signal in cell walls of ugd2,3 compared with the WT, indicating an increase in RGI-galactan side chains (Fig. 8A). The signal for the LM6 antibody is diminished in ugd2,3, in particular in the parenchyma cell walls, suggesting less abundant arabinan side chains (Fig. 8A). We further used immunotransmission electron microscopy as an additional method to confirm the increased galactan chain signal in cell walls of ugd2,3. Cell walls of both parenchymatic and mesophyll cells show a higher density of gold particles for the galactan epitope in ugd2,3 compared with WT (Fig. 8B). The distribution is random over the entire cell wall. In summary, immunolabeling of transverse sections taken from the elongation part of hypocotyls revealed differences between the mutant and the WT cell walls only with the antibodies recognizing RGI side chains.
FIGURE 8.
A, immunofluorescence labeling of transverse sections of WT and ugd2,3 hypocotyls with LM5 and LM6 monoclonal antibodies is shown. Transverse sections were taken from 7-day-old seedlings. ugd2,3 reveals a higher labeling with galactan specific LM5 antibody but shows a significant reduction in the (15)-α-d-arabinan epitope, recognized by LM6. B, shown are immunotransmission electron microscopy of parenchymatic cells from cotyledons with LM5 antibodies. The cell wall of ugd2,3 is swollen and contains more LM5 epitopes compared with WT.
ugd2,3 Mutant Plants Exhibit an Accumulation of Soluble Sugar
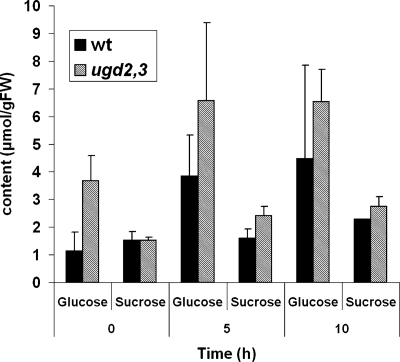
The cell wall is a major sink for carbohydrates in plants. We, therefore, anticipated a role of UGD in limiting free sugar levels in plants. To test this hypothesis, we analyzed soluble sugar contents in wild type and ugd2,3 lines that are generally higher in ugd2,3 (Fig. 9). In particular, the glucose concentration is elevated at the end of the night and remains above the WT level during the day. The same trend was observed for sucrose although the increase is somewhat lower as for glucose. However, the slower growth of ugd2,3 may also contribute to these variations.
FIGURE 9.
Elevated levels of soluble sugars in ugd2,3. Glucose and sucrose content in 6-week-old WT and ugd2,3 plants harvested at three different times during the day. Time 0 h corresponds to the beginning of illumination in the morning. Each value is the mean of five replicate samples.
DISCUSSION
Here we analyze a novel cell wall mutant of Arabidopsis that shows a distinct phenotype resulting from the reduced availability of the glycosyl donor UDP-GlcA. A. thaliana has four UGD genes involved in the biosynthesis of this important cell wall precursor (6, 9). The overlap in gene expression of UGD2 and UGD3, revealed by Genevestigator tool (26), RT-PCR analyses, and promotor-GUS reporter lines (9), suggests genetic redundancy. At least in seedlings, UGD4 is expressed in the same tissues as UGD2 and UGD3, although at a lower level (9). Indeed, when one UGD isoform (ugd2 or ugd3) is missing, the plant grows normally, and adult plants have the same biochemical cell wall composition as the WT, confirming the functional overlap of UGD2 and UGD3. The double mutant has a low residual UGD activity as predicted from the single mutants, suggesting a cooperation of UGD2 and UGD3 in providing the majority of UDP-GlcA. How do these isoforms interact? It was observed that the UGD enzyme is active as a multimeric enzyme complex in some organisms (35, 36). It is tempting to speculate that these complexes are preferentially formed in the vicinity of the Golgi apparatus, where the enzyme product is needed for a more efficient substrate channeling. The question of whether these complexes represent homo- or heteromers has not been addressed yet as the highly similar protein sequence of the isoforms made it impossible to create isoform-specific antibodies. The use of tagged UGD proteins might help to address this question in the future.
Cell Walls of ugd2,3 Have an Altered Composition
Sugar residues derived from UDP-GlcA are abundant components of cell wall polymers including both pectin and hemicellulose. It is, therefore, not surprising that ugd2,3 mutant plants exhibits a strong reduction in the GalA, xylose, arabinose, and apiose content of cell walls, as indicated by biochemical analysis. Enzymatic fingerprints and immunostainings performed on different tissues reveal that RGI side chains and xyloglucan are definitively different in the ugd2,3 mutant. Many of the previously described mur mutants as well as other cell wall mutants have a defect in the biosynthesis pathway of one nucleotide sugar or in a particular glycosyltransferase. For example, in mur4, the arabinose content is reduced by 50%, which is far higher than the reduction in ugd2,3. Nevertheless, mur4 shows no visible phenotype but a reduction in arabinan side chains in RGI and a reduction of the glycosylation in arabinogalactan proteins (37). The arabinan deficient 1 mutant also shows a strong reduction in the arabinose content of about 50% and contains a modified RGI, which has only 30% of the arabinose content of WT plants (38). Despite these striking compositional changes, the mutant is very similar to WT plants, suggesting that a reduction in arabinose is not critical for intact cell walls.
Another class of mutants comprises plants with lower xylose content in the cell walls due to a defect in xylosyltransferases (xxt1, xxt2). A double mutant has ∼50% of the xylose content of WT cell walls, no detectable xyloglucan, and looks quite normal except for root hairs (30). Although a loss in xyloglucans is tolerated in the xxt1,xxt2 mutant, ugd2,3 has a xyloglucan similar to wild type plants but less substituted by xylose. The difference in xyloglucan structure may well be responsible for the defects in root hair development in ugd2,3 (Fig. 3R). The data also suggest that under conditions where UDP-xylose is limiting, the plant has a preference to maintain xyloglucan. The third sugar underrepresented in ugd2,3 is GalA, which shows a reduction of about 27%. A similar lowering of GalA was found in the quasimodo1 mutant (39). In qua1 the amount of homogalacturonan is strongly reduced because the mutant has a defect in a putative galacturonosyltransferase. The major phenotype of qua1 is a reduced cell adhesion of the seedlings that we never observed in ugd2,3, indicating that the phenotypes and the reduction of GalA in ugd2,3 is not solely generated by a decrease in homogalacturonan biosynthesis.
The strong reduction in RGII likely explains the increase in cell wall thickness (swollen cell wall) as observed by TEM analysis. Previously, similar phenotypes were found in other RGII mutants or boron-depleted plants, suggesting that RGII cross-linking is a very critical event when assembling a tightly packed cell wall. Often a defect in RGII could be complemented by high boron concentrations that were supplied externally. In ugd2,3 however, the addition of boron has no beneficial effect to rescue the mutant phenotype. This suggests that the remaining RGII in ugd2,3 is already extensively cross-linked by boron and that the reduction of the total amount of RGII is responsible for the swollen cell wall. ugd2,3, therefore, represents a novel class of RGII mutants that has a WT-like structure in the A and B chain but has a strongly reduced amount of RGII to about ⅓ of the WT amount.
In summary, the (strong) reduction of specific sugars of the cell wall in other mutants leads to no or different phenotypes compared with ugd2,3, which has a moderate reduction in at least four cell wall sugars. Thus ugd2,3 is a novel unique cell wall mutant. The differences are mainly in the pectic polymers and support the recent findings by Zykwinska et al. (40), which propose direct interactions between pectins and cellulose and a more pronounced role of pectic polymers for cell walls. It is important to keep in mind that all subclasses of pectins are reduced along with a change in xyloglucan. Each of these changes by itself does likely not cause developmental problems, but the interplay of several minor changes has drastic consequences for plant development.
Contribution of the Alternative MIOX Pathway to the UDP-GlcA Pool
As outlined in Fig. 1, an alternative pathway is also capable of synthesizing the nucleotide sugar UDP-GlcA through oxygenation of myo-inositol to GlcA, which is then converted to UDP-GlcA via a salvage pathway.
A radiotracer study by Sharples and Fry (15) using A. thaliana cell cultures suggests a minor contribution of the MIOX pathway to primary cell wall precursors. Nevertheless, the MIOX pathway is functional in this plant (13), and a knockdown in MIOX genes leads to a reduced incorporation of myo-inositol derived precursors into cell wall polymers (14, 41). Real-time PCR with ugd2,3 shows only a moderate up-regulation (up to ∼3-fold) of 3 of the 4 MIOX genes with the strongest contribution by the MIOX2 gene (supplemental Fig. S2). MIOX gene induction has been previously observed in other experiments. In a study by Osuna et al. (42) plants were deprived of carbon, which leads to an induction of several MIOX genes. The drop in free sugar levels may be a signal for MIOX gene induction. Similarly nematode-infected A. thaliana roots show a strong gene induction of MIOX genes (43), suggesting a metabolite-dependent gene induction. Taking these studies into account, the increased concentration of glucose and sucrose in ugd2,3 might explain why the MIOX gene induction response of ugd2,3 is lower compared e.g. to the effect of nematode infection.
The complementation experiment (Fig. 5C) with inositol resulted in a partial complementation of ugd2,3 growth defects similar to the feeding of GlcA. This strongly indicates two conclusions; one of the two enzymes downstream of MIOX, the glucuronokinase or the pyrophosphorylase, limits the flux of UDP-GlcA in cells. Otherwise, we would have found a full complementation of the ugd2,3 alterations in the presence of GlcA or myo-inositol. Second, the MIOX activity in ugd2,3 is not limiting in the alternative MIOX pathway to UDP-GlcA, as indicated by the same partial complementation obtained with GlcA and myo-inositol. As we see the complementation of ugd2,3 growth defects upon myo-inositol feeding, we can further conclude that the biosynthesis of myo-inositol, available for the MIOX pathway, is under natural conditions the limiting factor for complementation through the MIOX pathway.
ugd2,3 Shows Multiple Developmental Defects
The ugd2,3 double mutant is clearly affected in growth, development, and biochemical aspects. When germinating seeds on soil we observed that many ugd2,3 seedlings died during the early phase of development due to an unsuccessful formation of primary roots. These seedlings were in the process of forming a secondary root, which starts to grow too late. A likely reason for the accumulation of root defects is the high expression of UGD2 and UGD3 in developing roots (9, 13). Some of the seedling phenotypes (e.g. one or three cotyledons instead of two) have been observed in embryo mutants, which often have a defect in phytohormone signaling. In contrast to traditional embryo mutants, which show the phenotype in all seedlings, ugd2,3 only occasionally exhibits these phenotypes. This argues against a primary phytohormone defect. Another class of mutants shows abnormal seedling development due to a defect in the secretory pathway. The emb30/gnom mutant affected in the Sec7-like protein is such an example (44). The non-coordinated deposition of cell wall material leads to local changes in the pectic material of the cell wall and to abnormal root development. An alternative explanation might be the modified cell wall itself. Ectopic and inducible expression of the cell wall-loosening enzyme expansin leads to morphological changes like leaf lobes or even new organ induction close to the meristem (45). It is well conceivable that a certain threshold in cell wall changes or loosening must be reached during a particular phase of the development and that, therefore, ugd2,3 shows defects only occasionally because the threshold has not been reached. The cell wall itself might also provide signals for development as shown for the axis development of the brown algae Fucus (46).
We observe the developmental defects in siblings of normal growing parental ugd2,3 plants as well as in siblings of ugd2,3 seedlings with developmental defects. We never observed any of these defects in the complemented line, suggesting that the developmental problems are caused by cell wall modifications rather than by second side mutations. The surprisingly low number of up- or down-regulated genes in ugd2,3 revealed no major change in the expression of developmental key genes, suggesting that cell wall modification is the major cause for the observed developmental defects in the mutant.
In summary we have demonstrated that UGD enzymes are of great importance for the integrity of the cell wall. ugd2,3 shows defects in seedling development, slow growth, dwarfism, and very low seed-set. The nucleotide sugars derived from UDP-GlcA have to be present in concentrations high enough to allow the correct biosynthesis of cell wall polymers. Depletion of one or more UDP-sugars combined with a possible increase in others could change the specificity of glycosyltransferases. This may also allow other glycosyltransferases to jump in and attach sugars, for which the precursors are available in sufficient amounts as previously found in mur1 (32). The consequence is a modified polymer that cannot be corrected later in development.
Supplementary Material
Acknowledgments
We thank Gregory Mouille and Christoph Ringli for interesting and helpful discussions, Ancuela Andosch for help with the immuno-TEM method and high pressure freeze fixation, Richard Strasser for the gift of an anti-xylose antibody, and Kirk Matthew Schnorr, Novozymes, for the generous gift of cell wall degrading enzymes.
The project is funded by a grant from the German Science Foundation (Deutsche Forschungsgemeinschaft; Bonn, Germany; to R. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- UDP-GlcA
- UDP-glucuronic acid
- GalA
- d-galacturonic acid
- GlcA
- d-glucuronic acid
- UGD
- UDP-glucose dehydrogenase
- MIOX
- myo-inositol oxygenase
- TEM
- transmission electron microscopy.
REFERENCES
- 1. Scheible W. R., Pauly M. (2004) Curr. Opin. Plant Biol. 7, 285–295 [DOI] [PubMed] [Google Scholar]
- 2. Ridley B. L., O'Neill M. A., Mohnen D. (2001) Phytochemistry 57, 929–967 [DOI] [PubMed] [Google Scholar]
- 3. Willats W. G., McCartney L., Mackie W., Knox J. P. (2001) Plant Mol. Biol. 47, 9–27 [PubMed] [Google Scholar]
- 4. Reiter W. D. (2008) Curr. Opin. Plant Biol. 11, 236–243 [DOI] [PubMed] [Google Scholar]
- 5. Tenhaken R., Thulke O. (1996) Plant Physiol. 112, 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reiter W. D., Vanzin G. F. (2001) Plant Mol. Biol. 47, 95–113 [PubMed] [Google Scholar]
- 7. Seifert G. J. (2004) Curr. Opin. Plant Biol. 7, 277–284 [DOI] [PubMed] [Google Scholar]
- 8. Seifert G. J., Barber C., Wells B., Roberts K. (2004) Plant Cell 16, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klinghammer M., Tenhaken R. (2007) J. Exp. Bot. 58, 3609–3621 [DOI] [PubMed] [Google Scholar]
- 10. Oka T., Jigami Y. (2006) FEBS J. 273, 2645–2657 [DOI] [PubMed] [Google Scholar]
- 11. Kärkönen A., Fry S. C. (2006) Planta 223, 858–870 [DOI] [PubMed] [Google Scholar]
- 12. Loewus F. A. (2006) Subcell Biochem. 39, 21–45 [DOI] [PubMed] [Google Scholar]
- 13. Seitz B., Klos C., Wurm M., Tenhaken R. (2000) Plant J. 21, 537–546 [DOI] [PubMed] [Google Scholar]
- 14. Kanter U., Usadel B., Guerineau F., Li Y., Pauly M., Tenhaken R. (2005) Planta 221, 243–254 [DOI] [PubMed] [Google Scholar]
- 15. Sharples S. C., Fry S. C. (2007) Plant J. 52, 252–262 [DOI] [PubMed] [Google Scholar]
- 16. Endres S., Tenhaken R. (2009) Plant Physiol. 149, 1042–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M. (2000) Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- 18. Lütz-Meindl U., Aichinger N. (2004) Protoplasma 223, 155–162 [DOI] [PubMed] [Google Scholar]
- 19. Eder M., Lütz-Meindl U. (2008) J. Microsc. 231, 201–214 [DOI] [PubMed] [Google Scholar]
- 20. Vitha S., Baluska F., Braun M., Samaj J., Volkmann D., Barlow P. W. (2000) Histochem. J. 32, 457–466 [DOI] [PubMed] [Google Scholar]
- 21. Oertel A., Aichinger N., Hochreiter R., Thalhammer J., Lütz-Meindl U. (2004) J. Phycol. 40, 711–720 [Google Scholar]
- 22. Frey B., Keller C., Zierold K. (2000) Plant Cell Environ. 23, 675–687 [Google Scholar]
- 23. Lerouxel O., Choo T. S., Séveno M., Usadel B., Faye L., Lerouge P., Pauly M. (2002) Plant Physiol. 130, 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Séveno M., Voxeur A., Rihouey C., Wu A. M., Ishii T., Chevalier C., Ralet M. C., Driouich A., Marchant A., Lerouge P. (2009) Planta 230, 947–957 [DOI] [PubMed] [Google Scholar]
- 25. Packer N. H., Lawson M. A., Jardine D. R., Redmond J. W. (1998) Glycoconj. J. 15, 737–747 [DOI] [PubMed] [Google Scholar]
- 26. Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004) Plant Physiol. 136, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leibowitz M. D., Dickinson D. B., Loewus F. A., Loewus M. (1977) Arch. Biochem. Biophys. 179, 559–564 [DOI] [PubMed] [Google Scholar]
- 28. Pieslinger A. M., Hoepflinger M. C., Tenhaken R. (2010) J. Biol. Chem. 285, 2902–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kotake T., Yamaguchi D., Ohzono H., Hojo S., Kaneko S., Ishida H. K., Tsumuraya Y. (2004) J. Biol. Chem. 279, 45728–45736 [DOI] [PubMed] [Google Scholar]
- 30. Cavalier D. M., Lerouxel O., Neumetzler L., Yamauchi K., Reinecke A., Freshour G., Zabotina O. A., Hahn M. G., Burgert I., Pauly M., Raikhel N. V., Keegstra K. (2008) Plant Cell 20, 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fry S. C., York W. S., Albersheim P., Darvill A., Hayashi T., Joseleau J. P., Kato Y., Lorences E. P., Maclachlan G. A., McNeil M., Mort A. J., Grant Reid J. S., Seitz H. U., Selvendran R. R., Voragen A. G. J., White A. R. (1993) Physiol. Plant. 89, 1–3 [Google Scholar]
- 32. O'Neill M. A., Eberhard S., Albersheim P., Darvill A. G. (2001) Science 294, 846–849 [DOI] [PubMed] [Google Scholar]
- 33. Clausen M. H., Willats W. G., Knox J. P. (2003) Carbohydr. Res. 338, 1797–1800 [DOI] [PubMed] [Google Scholar]
- 34. Willats W. G., Marcus S. E., Knox J. P. (1998) Carbohydr. Res. 308, 149–152 [DOI] [PubMed] [Google Scholar]
- 35. Jaenicke R., Rudolph R., Feingold D. S. (1986) Biochemistry 25, 7283–7287 [DOI] [PubMed] [Google Scholar]
- 36. Stewart D. C., Copeland L. (1998) Plant Physiol. 116, 349–355 [Google Scholar]
- 37. Burget E. G., Reiter W. D. (1999) Plant Physiol. 121, 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harholt J., Jensen J. K., Sørensen S. O., Orfila C., Pauly M., Scheller H. V. (2006) Plant Physiol. 140, 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouton S., Leboeuf E., Mouille G., Leydecker M. T., Talbotec J., Granier F., Lahaye M., Höfte H., Truong H. N. (2002) Plant Cell 14, 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zykwinska A. W., Ralet M. C., Garnier C. D., Thibault J. F. (2005) Plant Physiol. 139, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Endres S., Tenhaken R. (2011) Planta 234, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Osuna D., Usadel B., Morcuende R., Gibon Y., Bläsing O. E., Höhne M., Günter M., Kamlage B., Trethewey R., Scheible W. R., Stitt M. (2007) Plant J. 49, 463–491 [DOI] [PubMed] [Google Scholar]
- 43. Siddique S., Endres S., Atkins J. M., Szakasits D., Wieczorek K., Hofmann J., Blaukopf C., Urwin P. E., Tenhaken R., Grundler F. M., Kreil D. P., Bohlmann H. (2009) New Phytol. 184, 457–472 [DOI] [PubMed] [Google Scholar]
- 44. Shevell D. E., Kunkel T., Chua N. H. (2000) Plant Cell 12, 2047–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pien S., Wyrzykowska J., McQueen-Mason S., Smart C., Fleming A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kropf D. L., Coffman H. R., Kloareg B., Glenn P., Allen V. W. (1993) Dev. Biol. 160, 303–314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.