Background: The pro-proliferative Krüppel-like factor 5 (KLF5) is posttranslationally regulated.
Results: SMAD ubiquitination regulatory factor 2 (SMURF2) interacts with, ubiquitinates and degrades KLF5.
Conclusion: SMURF2 negatively regulates KLF5.
Significance: The findings increase the understanding of the mechanisms by which KLF5 is regulated posttranslationally.
Keywords: Posttranslational Modification, Proliferation, Proteasome, Ubiquitin Ligase, Ubiquitination, Degradation, KLF5, SMURF2
Abstract
The zinc finger transcription factor Krüppel-like factor 5 (KLF5) is regulated posttranslationally. We identified SMAD ubiquitination regulatory factor 2 (SMURF2), an E3 ubiquitin ligase, as an interacting protein of KLF5 by yeast two-hybrid screen, coimmunoprecipitation, and indirect immunofluorescence studies. The SMURF2-interacting domains in KLF5 were mapped to its carboxyl terminus, including the PY motif of KLF5 and its zinc finger DNA-binding domain. KLF5 protein levels were reduced significantly upon overexpression of SMURF2 but not catalytically inactive SMURF2-C716A mutant or SMURF1. SMURF2 alone reduced the protein stability of KLF5 as shown by cycloheximide chase assay, indicating that SMURF2 specifically destabilizes KLF5. In contrast, KLF5(1–165), a KLF5 amino-terminal construct that lacks the PY motif and DNA binding domain, was not degraded by SMURF2. The degradation of KLF5 by SMURF2 was blocked by the proteasome inhibitor MG132, and SMURF2 efficiently ubiquitinated both overexpressed and endogenous KLF5. In contrast, knocking down SMURF2 by siRNAs significantly enhanced KLF5 protein levels, reduced ubiquitination of KLF5, and increased the expression of cyclin D1 and PDGF-A, two established KLF5 target genes. In consistence, SMURF2, but not the E3 ligase mutant SMURF2-C716A, significantly inhibited the transcriptional activity of KLF5, as demonstrated by dual luciferase assay using the PDGF-A promoter, and suppressed the ability of KLF5 to stimulate cell proliferation as measured by BrdU incorporation. Hence, SMURF2 is a novel E3 ubiquitin ligase for KLF5 and negatively regulates KLF5 by targeting it for proteasomal degradation.
Introduction
Protein ubiquitination is a key form of posttranslational modification central to eukaryotic regulation (1, 2). As a main mechanism of controlling the stability and turnover of transcription factors, proteasomal degradation triggered by ubiquitination is pivotal to transcriptional control (1, 2). The specific effects from ubiquitination-triggered degradation are mainly achieved by E3 ubiquitin ligases, of which there are hundreds, and often determine the substrate availability and specificity of the proteasomal destruction (1). A given protein can be targeted by multiple E3 ubiquitin ligases, whereas the same E3 ubiquitin ligase can target multiple substrates, demonstrating a highly complex and dynamic regulation. Hence, identifying new targets for these ubiquitin ligases and, conversely, new ubiquitin ligases for a given target, will improve our understanding of the dynamic regulation of cellular functions by ubiquitination.
SMURF2 is an E3 ubiquitin ligase recently grouped into the Nedd4 family of HECT ubiquitin ligases (2). It contains WW domains, which directly bind to a PPXY motif (also known as PY motif) in its targets (2). This interaction is further stabilized by the PY tail, a six-amino acid stretch immediately carboxyl-terminal to the PPXY motif, although additional interactions exist (3–5). As a HECT E3 ubiquitin ligase, SMURF2 catalyzes ubiquitination at specific lysine residues in its targets, which triggers subsequent degradation by proteasomes (2). SMURF2 has a relatively broad spectrum of targets and is involved in diverse signal pathways and cellular processes (2, 6–12). Thus, the identification of new targets for SMURF2 may provide further insights into the mechanisms by which the SMURF family of ubiquitin ligases regulates cellular functions.
KLF5 is a zinc finger-containing transcription factor that promotes cell proliferation and plays important roles in development, differentiation, tumorigenesis, and embryonic stem cell renewal (13–16). The expression and protein activity of KLF5 are tightly regulated at both transcriptional and posttranscriptional levels (5, 7, 17–20). A primary mechanism by which KLF5 is posttranslationally regulated is through ubiquitination and subsequent degradation of KLF5, as mediated by a number of E3 ubiquitin ligases (5, 7, 17, 19), including WWP1 and FBW7 (5, 7, 19). The interaction between KLF5 and WWP1 involves the PPXY motif of KLF5 (PPPSY) and the WW domains in WWP1 (5). However, whether additional ubiquitin ligases for KLF5 exist and how KLF5 is regulated by various ubiquitin ligases are not clearly defined. Here we present evidence for a novel interaction between KLF5 and SMURF2 and demonstrate that SMURF2 negatively regulates KLF5 by targeting KLF5 for ubiquitination and degradation. This report therefore presents KLF5 as a target for SMURF2 and SMURF2 as an ubiquitin ligase that regulates KLF5.
EXPERIMENTAL PROCEDURES
Plasmids
pMT3-HA-KLF5, PMT3-KLF5, pCMV-Myc-SMURF2, pCMV-FLAG-SMURF2, pCMV-FLAG-SMURF2-C716A, pCMV-FLAG-SMURF1, pCMV-FLAG-SMURF1-C699A, the PDGF-A luciferase reporter plasmid, and full-length or truncation yeast two-hybrid constructs have all been described (8, 14, 21–23). KLF5 lysine-to-arginine mutants were constructed with the QuikChange site-directed mutagenesis kit (Stratagene) to replace each lysine site with arginine in the PMT3-HA-KLF5 construct. pMT3-KLF5(1–165), which encodes the amino-terminal 165 residues of KLF5, was constructed by digestion of pMT3-HA-KLF5 with SalI, followed by self-ligation.
Yeast Two-hybrid Screen and Assay
A yeast two-hybrid screen was performed as described previously (21). A yeast two-hybrid assay was performed at extremely high stringency with the Matchmaker Gold Yeast two-hybrid system (Clontech). Briefly, the indicated KLF5, SMURF2, or vector control constructs were transformed in the Saccharomyces cerevisiae Y2HGold strain, and specific interaction was verified under selection with leucine, tryptophan, adenine, histidine, and aureobasidin A in the absence or presence of X-α-Gal according to the manufacturer's instructions.
Small Interfering RNA
siRNA against SMURF2, in the form of either a mixture of three siRNAs targeting different regions of SMURF2 (Origene, company-guaranteed Trilencer-27 siRNA duplex kit, catalog no. SR312096), two individual siRNAs (Origene, catalog nos. SR312096A/452087 and SC312096B/452091), or the negative control siRNA included in the kit (Origene, catalog no. SR30004) was transfected into 25% confluent COS-1 cells with Lipofectamine RNAiMAX (Invitrogen, catalog no. 13778-150) according to the manufacturer's instructions. Three days later, cells were subjected to Western blotting, immunoprecipitation, or quantitative RT-PCR analysis.
Quantitative RT-PCR
siRNAs against SMURF2 (Origene, catalog no. SR312096) or the negative control siRNA (Origene, catalog no. SR30004) was transfected into 25% confluent COS-1 cells with Lipofectamine RNAiMAX. Three days later, total RNA was isolated with TRIzol (Ambion/Invitrogen), and quantitative real-time RT-PCR was performed in four triplicates with primer sets specific for SMURF2, SMURF1, KLF5, cyclin D1, PDGF-A (Qiagen, QT00079961, QT00031689, QT00074676, QT00495285, and QT01664488), and the control gene GAPDH (forward, ACCCAGAAGACTGTGGATGG and reverse, TTCTAGACGGCAGGTCAGGT). Products were amplified and detected with the Power SYBR Green RNA-to-CT 1-Step kit (Applied Biosystems) on an Eppendorf REALPLEX epgradient S real-time PCR Mastercycler according to the manufacturer's instructions. Relative changes in expression were calculated based on the comparative CT (−ΔΔCT) method (32) after normalization with the GAPDH control.
Ubiquitination Assay
For ubiquitination of overexpressed KLF5, HEK293T cells were transfected with the indicated plasmids, treated with 20 μm MG132 (Sigma) for 2.5 h, and disrupted in lysis buffer (10 mm Tris-HCl (pH7.4), 1% SDS, 150 mm NaCl, 50 mm sodium fluoride, complete protease inhibitor mixture (Roche)). The lysates were denatured by boiling for 10 min, diluted in five volumes of dilution buffer (10 mm Tris-HCl (pH7.4), 150 mm NaCl, 1% Triton X-100, 2 mm EDTA, complete protease inhibitor mixture), and immunoprecipitated with the mixture of a rabbit KLF5 antibody (18) and commercial rabbit KLF5 antibody from Santa Cruz (SC22797), followed by incubation with protein A beads (Upstate). The immune complexes were washed with 10 mm Tris-HCl (pH7.4), 135 mm NaCl, 1 mm EDTA, and 1% Nonidet P-40 for three times, followed by washing once with 10 mm Tris-HCl (pH6.8) and 1 mm EDTA, and Western blotting with mouse HA (Covance), Myc (Sigma), and β-actin (Sigma), and rabbit KLF5 antibodies.
For ubiquitination of endogenous KLF5, COS-1 cells were transfected with HA-ubiquitin and either vector alone or Myc-SMURF2, and disrupted in lysis buffer (10 mm Tris-HCl (pH7.4), 1% SDS, 150 mm NaCl, 50 mm sodium fluoride, and complete protease inhibitor mixture (Roche)). The lysates were denatured by boiling for 10 min, diluted in five volumes of dilution buffer (10 mm Tris-HCl (pH7.4), 150 mm NaCl, 1% Triton X-100, 2 mm EDTA, complete protease inhibitor mixture), and immunoprecipitated with either control rabbit IgG (Bethyl Laboratories) or the mixture of rabbit KLF5 antibody (18) and commercial KLF5 antibody (Santa Cruz Biotechnology, Inc.), followed by incubation with protein A beads. The immune complexes were washed with 10 mm Tris-HCl (pH7.4), 135 mm NaCl, 1 mm EDTA, and 1% Nonidet P-40 for four times, followed by washing once with 10 mm Tris-HCl (pH6.8) and 1 mm EDTA, and Western blotting with mouse HA, Myc, and β-actin, and rabbit KLF5 antibodies.
For ubiquitination of endogenous KLF5 after SMURF2 knockdown, siRNAs against SMURF2 (Origene, catalog no. SR312096) or the control siRNA (Origene, catalog no. SR30004) were transfected into 25% confluent COS-1 cells with Lipofectamine RNAiMAX. Two days later, cells were transfected with HA-ubiquitin. The next day, cells were treated with 20 μm MG132 (Sigma) for 1 h and disrupted in the lysis buffer. The lysates were denatured by boiling, diluted in the dilution buffer, and immunoprecipitated with either control rabbit IgG or the mixture of rabbit KLF5 antibody (18) and commercial KLF5 antibody (Santa Cruz Biotechnology), followed by incubation with protein A beads. The immune complexes were washed for four times, and Western blotted with mouse HA and β-actin, and rabbit SMURF2 (Upstate, catalog no. 07-249) and KLF5 antibodies.
Fluorescence Microscopy
COS-1 cells were transfected with pMT3-HA-KLF5 and pCMV-Myc-SMURF2 at 10:1 in plasmid ratio, treated with 20 μm MG132 (Sigma) for 6 h to stabilize KLF5, fixed with 4% formaldehyde, and permeabilized/blocked with 0.2% Triton X-100 and 2% BSA in PBS. Cells were then incubated with chicken HA (Chemicon) and rabbit Myc (Upstate) antibodies, followed by incubation with FITC-conjugated donkey α-chicken and Cy5-conjugated donkey α-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). For immunofluorescence of endogenous KLF5 and SMURF2, DLD-1 cells were treated with 20 μm MG132 (Sigma) for 6 h to stabilize proteins, fixed with 4% formaldehyde, and permeabilized/blocked with 0.2% Triton X-100 and 2% BSA in PBS. Cells were then incubated with rabbit SMURF2 (Upstate) and mouse KLF5 (Santa Cruz Biotechnology, Inc., catalog no. SC69906) antibodies, followed by incubation with FITC-conjugated donkey α-mouse and Rhodamine Red-X-conjugated donkey α-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). Cells were also stained with Hoechst dye to reveal nuclei. Immunofluorescence was monitored by both conventional and confocal microscopy as described previously (18, 21).
Immunoprecipitation
COS-1 cells were cotransfected with HA-KLF5 and Myc-SMURF2 at 4:1 in plasmid ratio, treated with 40 μm MG132 (Sigma) for 4 h, and lysed with 50 mm Tris-HCl (pH7.4), 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, and complete protease inhibitor mixture (Roche). Lysates were immunoprecipitated with a mouse HA antibody (Covance) and EZ protein G beads (Sigma), preblocked with BSA and salmon sperm DNA, and washed with lysis buffer five times. Western blotting was then preformed with rabbit Myc (Chemicon) and HA (Sigma) and mouse β-actin antibodies.
For coimmunoprecipitation of endogenous SMURFs with KLF5, COS-1 cells were treated with 40 μm MG132 (Sigma) for 3 h and lysed with 20 mm Tris-HCl (pH7.4), 135 mm NaCl, 1 mm EDTA, 1% Triton X-100, 50 mm sodium fluoride, and complete protease inhibitor mixture (Roche). Lysates were immunoprecipitated with a mixture of rabbit KLF5 antibody (18) and commercial KLF5 antibody (Santa Cruz Biotechnology, Inc.) and protein A beads (Upstate) and washed with lysis buffer three times, followed by washing once with 10 mm Tris-HCl (pH 6.8) and 1 mm EDTA. Western blotting was then preformed with rabbit SMURF2 (Upstate) and mouse SMURF1 (Novus Biologicals, catalog no. H00057154-M01, clone 1D7), KLF5 (Santa Cruz Biotechnology, Inc., catalog no. SC69906), and β-actin antibodies.
Cycloheximide Chase Assay
A cycloheximide chase assay was performed as described (5, 7, 17, 19). Briefly, COS-1 cells were transfected with the indicated plasmids or vector alone, treated with 100 μg/ml cycloheximide for the indicated time, lysed, boiled in Laemmli buffer containing complete protease inhibitor mixture, and subjected to SDS-PAGE and Western blotting with rabbit KLF5 and mouse Myc, FLAG (Sigma), and β-actin antibodies.
Luciferase Reporter Assay
The dual luciferase reporter assay was performed as described (21, 22). Briefly, RKO cells were transfected with equal amounts of vector alone, pMT3-KLF5, pCMV-FLAG-SMURF2, pCMV-FLAG-SMURF2-C716A, pMT3-KLF5 and pCMV-FLAG-SMURF2, pMT3-KLF5 and pCMV-FLAG-SMURF2-C716A, or pMT3-KLF5(1–165) (KN), together with PDGF-A luciferase reporter. A Renilla luciferase control vector was cotransfected to normalize the transfection efficiency. The assay was performed with a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions.
BrdU Incorporation Assay
The BrdU incorporation assay was performed as described (21, 22). Briefly, COS-1 cells were transfected overnight with pMT3-HA-KLF5 and pCMV-Myc-SMURF2 at 10:1 in plasmid ratio to ensure cotransfection and detection, under which HA-KLF5 was not completely degraded by Myc-SMURF2 in the majority of cells. Cells were fixed and permeabilized with methanol, treated with HCl, neutralized, and blocked with 2% BSA in PBS. Cells were then incubated with mouse BrdU (BD Pharmingen), chicken HA (Chemicon), and rabbit Myc (Upstate) antibodies, and with Cy5-conjugated α-mouse, donkey FITC-conjugated α-chicken, and RRX-conjugated α-rabbit antibodies (Jackson ImmunoResearch Laboratories, Inc.). The percentages of transfected cells stained positive for BrdU were then calculated.
RESULTS
KLF5 Interacts with SMURF2
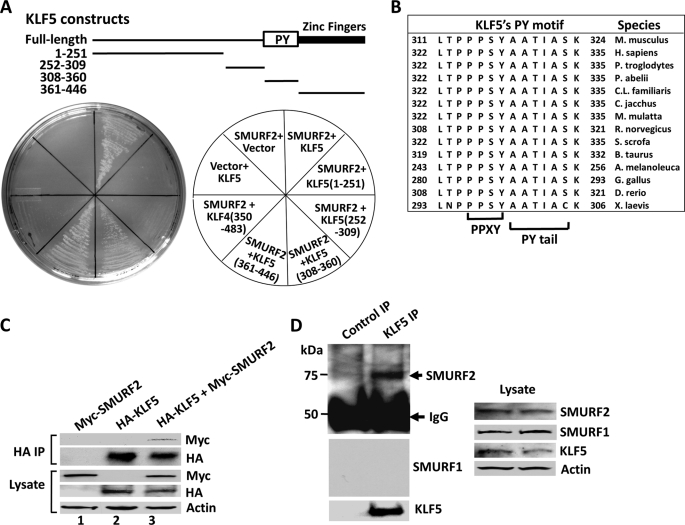
A previous yeast two-hybrid screen with KLF5 as bait revealed a number of proteins that interact with KLF5 (21). A repeat screen identified an additional protein, SMURF2, a WW domain-containing E3 ubiquitin ligase that interacts with KLF5. Individual yeast two-hybrid assays confirmed this physical interaction (Fig. 1A). KLF5 binds to SMURF2 in the two-hybrid assay under highly stringent conditions with at least five selection markers, leucine, tryptophan, adenine, histidine, and Aureobasidin A (Fig. 1A). The interaction was also demonstrated by coimmunoprecipitation (Fig. 1C). When HA-tagged KLF5 and Myc-tagged SMURF2 were cotransfected in COS-1 cells stabilized with MG132, a proteasome-specific inhibitor, immunoprecipitation with a HA antibody followed by Western blotting against Myc indicated that Myc-SMURF2 coimmunoprecipitated with HA-KLF5 (Fig. 1C). This interaction was not detected without MG132 treatment, presumably because of constant degradation of KLF5 in the immune complexes (data not shown). Endogenous KLF5 and SMURF2 also interacted with each other, as demonstrated by coimmunoprecipitation of endogenous SMURF2 with KLF5 immunoprecipitated from COS-1 cells treated with MG132 (Fig. 1D). This interaction is specific for SMURF2, as the interaction of endogenous SMURF1 with KLF5 was not detected (Fig. 1D). Thus, KLF5 specifically interacts with SMURF2.
FIGURE 1.
KLF5 interacts with SMURF2. A, SMURF2 binds KLF5 in a yeast two-hybrid assay through the carboxyl terminus of KLF5. The indicated SMURF2 or full-length or truncated KLF5 constructs (or the corresponding vector alone) were transformed in yeast and selected with leucine, tryptophan, adenine, histidine, and aureobasidin A. The top panel is a schematic showing the various KLF5 constructs and relative location of the PY motif and DNA-binding domain of KLF5 (Zinc Fingers). B, a high degree of conservation of the PY motifs of KLF5 from different species. The six-residue PY tail immediately following the PPXY core is also shown. C, coimmunoprecipitation of SMURF2 with KLF5. COS-1 cells were cotransfected with HA-KLF5 and Myc-SMURF2, treated with MG132, and immunoprecipitated (IP) with a mouse HA antibody. The coprecipitated Myc-SMURF2 was revealed by Western blotting with a rabbit Myc antibody. The proteins in lysates and immunoprecipitates were revealed by blotting with rabbit Myc and HA and mouse β-actin antibodies. D, KLF5 coimmunoprecipitates with endogenous SMURF2 but not SMURF1. COS-1 cells were treated with MG132 and immunoprecipitated with rabbit KLF5 or control antibodies. The coprecipitated SMURF2 was revealed by Western blotting with a rabbit SMURF2 antibody. The proteins in lysates and immunoprecipitates were also revealed by blotting with rabbit SMURF2 and mouse SMURF1, KLF5, and β-actin antibodies.
We also mapped the domains that mediate the association of KLF5 with SMURF2 using the two-hybrid system and deletion constructs of KLF5. KLF5 has a known PPXY motif (codons 314–317 in mouse and 325–328 in human KLF5), which is absolutely conserved across all the available species (Fig. 1B) (5). Its PY tail, the six residues immediately following the PPXY motif that help stabilize SMURF2 binding (3), is also highly conserved (Fig. 1B). Consistent with the presence of this SMURF2-interacting PY motif in KLF5, SMURF2 binds efficiently to a portion (amino acids 308 to 360) of the mouse KLF5 that spans the PPXY motif and PY tail (Fig. 1A). In addition, SMURF2 interacts with the flanking zinc finger DNA-binding domain at the very carboxyl terminus of the mouse KLF5 (amino acids 361 to 446) (Fig. 1A). This interaction is specific, as it prefers the zinc finger DNA-binding domain of KLF5 to that of KLF4 (amino acids 350–483 in mouse KLF4) (Fig. 1A), although both KLF5 and KLF4 contain similar C2H2-type zinc fingers (Fig. 7A) (15). Therefore, the minimal domains in KLF5 that mediate its association with SMURF2 are localized to the carboxyl terminus of KLF5, including its PY motif and DNA-binding domain.
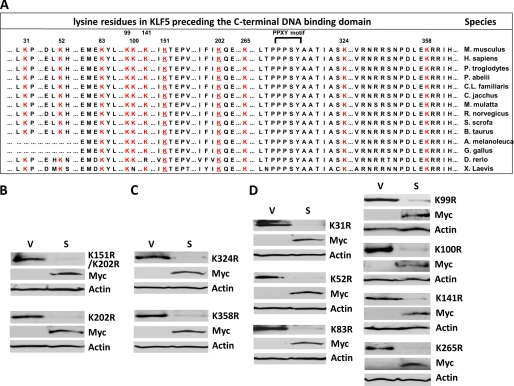
FIGURE 7.
Mutation at each lysine residue preceding the DNA binding domain of KLF5 does not prevent SMURF2 from degrading KLF5. A, alignment of all the lysine residues preceding the DNA binding domain of KLF5 from various species. Numbers indicate positions of these lysine residues, which are shown in red. Conserved residues adjacent to these lysines are also shown. The two SUMOylation site lysine residues are underlined (18). The PPXY (PPPSY) motif is bracketed (5). B, the two SUMOylation site lysines are not targets of SMURF2. HEK293T cells were transfected with the indicated KLF5 SUMOylation mutants and either Myc-SMURF2 (S) or vector alone (V), and whole cell lysates were subjected to Western blotting with mouse HA, Myc, and β-actin antibodies. K, lysine; r = arginine, Myc, Myc-SMURF2. C, individual mutations at Lys-324 and Lys-358, the two lysine lysines adjacent to the PY motif of KLF5, do not block the degradation KLF5 by SMURF2. HEK293T cells were transfected with K324R or K358R and either Myc-SMURF2 or vector alone, and whole cell lysates were subjected to Western blotting. D, mutation at each of the other lysine residues at the N terminus of KLF5 does not block the degradation of KLF5 by SMURF2. HEK293T cells were transfected with each indicated mutant and either Myc-SMURF2 or vector alone and whole cell lysates subjected to Western blotting.
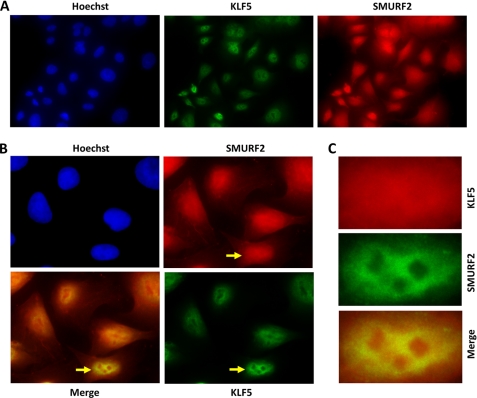
Consistent with the interaction between KLF5 and SMURF2, the two proteins colocalize in both the nucleus and cytoplasm. After COS-1 cells were transfected with HA-KLF5 and Myc-SMURF2 and treated with MG132 to stabilize HA-KLF5, indirect immunofluorescence demonstrated that both HA-KLF5 and Myc-SMURF2 were primarily colocalized to the nucleus (Fig. 2, A and B). Endogenous KLF5 and SMURF2 also colocalized primarily in the nucleus (Fig. 3, A and B), and the colocalization was especially visible under higher magnification and better resolution, often excluding nucleolar-shaped subnuclear structures (Figs. 2A and 3C). We reported previously that a fraction of KLF5 was also localized to cytoplasm (18). For the small fraction of cells that had considerable cytoplasmic localization of KLF5, the cytoplasmic KLF5 also colocalized with SMURF2 (Fig. 2A, center and right columns). Combining the results of all the experiments, KLF5 apparently interacts with SMURF2 in cells.
FIGURE 2.
KLF5 colocalizes with SMURF2. COS-1 cells were transfected with HA-KLF5 and Myc-SMURF2, treated with MG132 to stabilize HA-KLF5, and stained with chicken anti-HA and rabbit anti-Myc, followed by FITC-conjugated donkey α-chicken and Cy5-conjugated donkey α-rabbit secondary antibodies. A, representative staining of three different cells, including two with both nuclear and cytoplasmic KLF5. B, representative staining of multiple cells in the same field.
FIGURE 3.
Endogenous KLF5 colocalizes with SMURF2. DLD-1 cells were treated with MG132 to stabilize cellular proteins and stained with rabbit anti-SMURF2 and mouse anti-KLF5, followed by RRX-conjugated donkey α-rabbit and FITC-conjugated donkey α-mouse secondary antibodies. A, the staining pattern of multiple cells in a single field. B, a magnified view of several cells imaged in A. The arrows in B point to a single cell with nuclear KLF5, which is further magnified in C.
SMURF2 Degrades KLF5
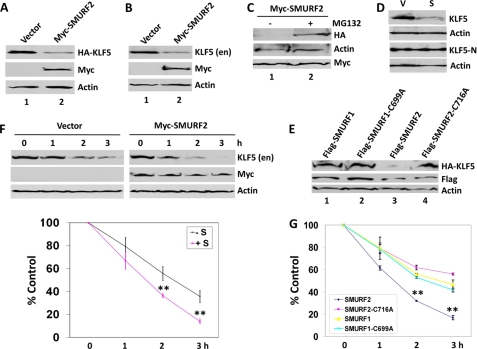
A consequence of the physical interaction between SMURF2 and KLF5 is the ability of SMURF2 to degrade both exogenous and endogenous KLF5. In cells cotransfected with HA-KLF5 and Myc-SMURF2, the level of HA-KLF5 was considerably lower compared with cells transfected with HA-KLF5 and vector (Fig. 4A), suggesting that SMURF2 triggers KLF5 degradation. Similarly, when cells were transfected with Myc-SUMRF2 alone, the abundance of endogenous KLF5 was lower than that in vector-transfected cells (Fig. 4B), indicating that SMURF2 degrades endogenous KLF5 as well. Lending further support that SMURF2 facilitates KLF5 degradation, treatment with MG132 prevented the ability of Myc-SMURF2 to degrade HA-KLF5 (Fig. 4C, compare lanes 1 and 2). Furthermore, a KLF5 deletion construct (KLF5-N) that contains only the amino terminus of KLF5 and lacks the PY motif and DNA-binding domain was unresponsive to Myc-SMURF2-mediated degradation (Fig. 4D), indicating that the interaction between KLF5 and SMURF2 is important for SMURF2 to degrade KLF5.
FIGURE 4.
SMURF2 degrades and destabilizes KLF5. A, reduced level of HA-KLF5 upon Myc-SMURF2 cotransfection. HEK293T cells were cotransfected with HA-KLF5 and either vector alone or Myc-SMURF2. Whole cell lysates were immunoblotted with mouse HA, Myc, or β-actin antibodies. B, reduced level of endogenous KLF5 after Myc-SMURF2 transfection. COS-1 cells were transfected with either vector alone or Myc-SMURF2. Lysates were probed with rabbit KLF5, or mouse Myc or β-actin antibodies. Myc, Myc-SMURF2; en, endogenous. C, the proteasomal inhibitor MG132 inhibits the ability of SMURF2 to degrade KLF5. HEK293T cells were cotransfected with HA-KLF5 and Myc-SMURF2 and treated with 20 μm MG132 for 3 h (+) or mock-treated (-). Whole cell lysates were probed with mouse HA, Myc, or β-actin antibodies. D, Myc-SMURF2 reduced the level of wild-type KLF5 but not its truncation fragment, KLF5-N. HEK293T cells were cotransfected with pMT3-KLF5 or pMT3-KLF5(1–165) (KLF5-N) and either vector alone (V) or Myc-SMURF2 (S). Whole cell lysates were immunoblotted with rabbit KLF5 (18) or β-actin antibodies. E, reduced level of HA-KLF5 upon cotransfection of FLAG-SMURF2 but not FLAG-SMURF2-C716A, FLAG-SMURF1, or FLAG-SMURF1-C699A. HEK293T cells were cotransfected with HA-KLF5 and the indicated plasmid. Whole cell lysates were immunoblotted with mouse HA, FLAG, or β-actin antibodies. F, COS-1 cells were transfected with vector alone or Myc-SMURF2 and treated with 100 μg/ml cycloheximide for 1, 2, and 3 h. Whole cell lysates were subjected to Western blotting with rabbit KLF5 or mouse Myc or β-actin antibodies. Degradation of KLF5 over the 3 h period was quantified with a chemiluminescence imager as the percentage of the remaining KLF5 proteins (KLF5 chemiluminescent intensity over that of β-actin) at each time point relative to that for 0 h, which was set as 100% (n = 4). **, p < 0.01 by two-tailed Student's t test. G, COS-1 cells were transfected with FLAG-SMURF2, FLAG-SMURF2-C716A, FLAG-SMURF1, or FLAG-SMURF1-C699A, and treated with 100 μg/ml cycloheximide for 1, 2, and 3 h. Whole cell lysates were subjected to Western blotting with rabbit KLF5, or mouse FLAG or β-actin antibodies. Degradation of KLF5 over the 3-h period was quantified with the chemiluminescence imager and illustrated in the chart (n = 4). **, p < 0.01 by two-tailed Student's t test compared with cells transfected with SMURF2-C716A, SMURF1, or SMURF1-C699A.
The degradation of KLF5 by SMURF2 requires the E3 ubiquitin ligase activity of SMURF2 and is highly specific because the catalytically inactive SMURF2-C716A mutant failed to reduce the steady-state protein level (Fig. 4E, compare lanes 3 and 4) of KLF5, and overexpression of SMURF1 or SMURF1-C699A did not significantly affect the steady-state protein level (Fig. 4E, lanes 1 and 2) of KLF5. To reinforce the conclusion that SMURF2 specifically degrades KLF5, the effect of SMURF2, SMURF1, and their catalytically inactive mutants on the stability of endogenous KLF5 was investigated by cycloheximide chase assay, a standard method for measuring the stability of proteins, including KLF5 (5, 7, 19). The half-life of KLF5 in COS-1 cells was significantly reduced upon transfection of SMURF2 as compared with vector-transfected cells (Fig. 4F). In addition, the half-life of KLF5 in cells transfected with wild-type SMURF2 was significantly lowered than those transfected with SMURF2-C716A, wild-type SMURF1, or SMURF1-C669A (Fig. 4G). Taken together, these results clearly indicate that SMURF2 specifically destabilizes KLF5 in a manner that is dependent on the E2 ubiquitin ligase activity of SMURF2.
KLF5 Is Ubiquitinated by SMURF2
Consistent with our observation that SMURF2 degrades KLF5 and acts as an E3 ligase to ubiquitinate target proteins, SMURF2 promotes the ubiquitination of KLF5. We overexpressed KLF5 together with Myc-SMURF2 and HA-tagged ubiquitin in HEK293T cells. Immunoprecipitation under denaturing condition with rabbit KLF5 antibodies followed by Western blotting with mouse HA demonstrated ubiquitination of immunoprecipitated KLF5 (Fig. 5A). Relatively little ubiquitination was detected in control immunoprecipitation, where KLF5 was not included in the transfection (Fig. 5A, lane 1) because of the relatively low amount of endogenous KLF5 in these cells. However, transfection of KLF5 and HA-ubiquitin resulted in a detectable ladder of ubiquitinated KLF5 (Fig. 5A, lane 2), and this ladder of ubiquitinated KLF5 was further enhanced when Myc-SMURF2 was included in the transfection (Fig. 5A, lane 3), despite the amount of input KLF5 in the presence of Myc-SMURF2 cotransfection being lower than that in the absence of Myc-SMURF2 (Fig. 2A, compare KLF5 in lanes 2 and 3). These results indicate that SMURF2 catalyzes the ubiquitination of KLF5.
FIGURE 5.
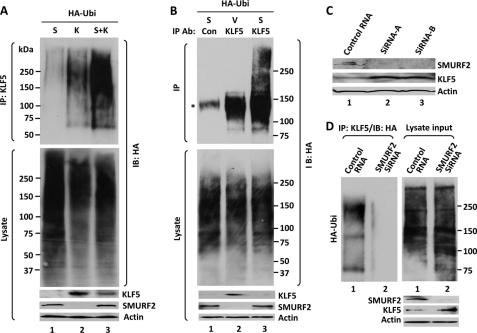
SMURF2 ubiquitinates KLF5. A, ubiquitination of overexpressed KLF5 with HA-tagged ubiquitin. HEK293T cells were cotransfected with HA-ubiquitin (HA-Ubi) and either Myc-SMURF2 (S), KLF5 (K), or both (S+K) and treated with MG132. The corresponding lysates were denatured and immunoprecipitated (IP) with rabbit KLF5 antibodies, followed by Western blotting (IB) of the immune complexes with a mouse HA antibody. The input proteins in cell lysates were also probed by the indicated antibodies. B, ubiquitination of endogenous KLF5 by SMURF2. COS-1 cells were cotransfected with HA-ubiquitin and either vector (V) or Myc-SMURF2, lysed, denatured, and immunoprecipitated with either control rabbit IgG (Con) or rabbit KLF5 (KLF5) antibodies. Western blotting was performed with mouse HA and β-actin and rabbit KLF5 antibodies. The asterisk indicates a nonspecific band in the immunoprecipitates from COS-1 cell lysates in the absence of MG132 treatment, presumably from slight sticking to beads. C, siRNA interference of endogenous SMURF2. COS-1 cells were transfected with either control siRNA or two individual SMURF2 siRNAs (Origene, catalog nos. SR312096A and SC312096B). Lysates from the transfected cells were subjected to Western blotting with rabbit antibodies against SMURF2 and KLF5 and a mouse β-actin antibody. D, ubiquitination of endogenous KLF5 was significantly reduced after SMURF2 depletion. COS-1 cells were transfected with either control siRNA or the Trilencer siRNAs against SMURF2 (Origene, catalog no. SR312096). Two days later, cells were transfected with HA-ubiquitin. The next day, cells were lysed, denatured, and immunoprecipitated with rabbit KLF5 (KLF5) antibodies, followed by immunoblotting with HA antibodies. Western blotting was also performed on the lysate input with mouse HA and β-actin and rabbit SMURF2 and KLF5 antibodies.
The ubiquitination of KLF5 by SMURF2 was also detected for endogenous KLF5 without any MG132 treatment. COS-1 cells in which a reasonable amount of endogenous KLF5 is detected (Fig. 4) were transfected with HA-ubiquitin and either Myc-SMURF2 or vector alone, disrupted under denaturing conditions, followed by immunoprecipitation of endogenous KLF5 (Fig. 5B). In cells transfected with HA-ubiquitin, some ubiquitination of endogenous KLF5 was detectable (Fig. 5B, lane 2). In Myc-SMURF2-cotransfected cells, the ubiquitination was increased further, despite the level of endogenous KLF5 in the presence of Myc-SMURF2 transfection being significantly lower (Fig. 5B, lane 3). The increase was especially evident for the high molecular weight, polyubiquitinated forms of KLF5 and occurred even in the absence of MG132, which inhibits proteasomes and stabilizes highly polyubiquitinated proteins (Fig. 5B, lane 3). Thus, endogenous KLF5 is ubiquitinated by SMURF2. To lend further support that SMURF2 ubiquitinates KLF5, ubiquitination of endogenous KLF5 was reduced significantly (Fig. 5D) when the level of endogenous SMURF2 was diminished by siRNA interference against SMURF2 (C). Taken together, these results indicate that SMURF2 degrades KLF5 through the proteasome-ubiquitin pathway.
SMURF2 Does Not Target Any Single Lysine Site in KLF5
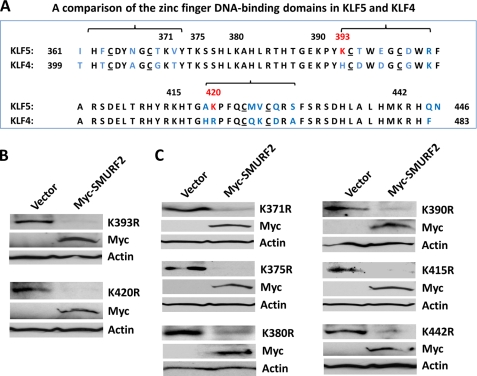
We next determined whether there is a predominant site targeted by SMURF2 in KLF5. As SMURF2 is a HECT ubiquitin ligase known to target lysine residues in its substrates (1, 2), we substituted every lysine residue in mouse KLF5 with arginine. Each KLF5 lysine-to-arginine (K-to-R) single mutant was cotransfected with Myc-SMURF2 to see if any mutant was resistant to the destabilization by SMURF2. We initially tested lysine residues within the zinc finger DNA-binding domain of KLF5 because of its interaction with KLF5, the proximity to the PY motif of KLF5, and the abundance of lysine residues (eight of a total of 19 lysine residues in full-length KLF5) (Fig. 6A). Given the specific interaction of SMURF2 with the DNA-binding domain of KLF5 rather than KLF4, we first tested Lys-393 and Lys-420, the two lysine residues unique to KLF5 and not conversed between KLF5 and KLF4. Both the KLF5-K393R and K420R mutants were efficiently degraded by Myc-SMURF2 (Fig. 6B), indicating that neither Lys-393 nor Lys-420 is the predominant target site. We further tested the other six KLF5 lysine mutants in the DNA binding domain. All of them were efficiently degraded by SMURF2 to extents comparable with wild-type KLF5 (Fig. 6C), indicating that SMURF2 does not target a single lysine site within the carboxyl-terminal DNA-binding domain of KLF5.
FIGURE 6.
Mutation at single lysine residues within KLF5's DNA binding domain does not prevent SMURF2 from degrading KLF5. A, comparison of the zinc finger DNA-binding domains of mouse KLF5 and KLF4. The numbers indicate the positions of lysine residues. The two lysine residues not conserved between KLF5 and KLF4 are indicated in red, and other residues not conserved between KLF5 and KLF4 are blue. In addition, the three divergent areas around the C2 parts of the three C2H2 zinc fingers are bracketed. The two cysteine residues that form the C2 part of each C2H2 zinc finger are underlined. B, mutation at Lys-393 and Lys-420, the two lysines not conserved between KLF5 and KLF4, do not block the degradation KLF5 by SMURF2. HEK293T cells were transfected with HA-KLF5/K393R or K420R and either Myc-SMURF2 or vector alone, and whole cell lysates were subjected to Western blotting with mouse HA, Myc, and β-actin antibodies. K, lysine; r, arginine; Myc, Myc-SMURF2. C, mutation at each of the other lysines within the DNA binding domain of KLF5 does not block the degradation KLF5 by SMURF2. HEK293T cells were transfected with each indicated mutant and either Myc-SMURF2 or vector alone and whole cell lysates subjected to Western blotting.
We also tested whether SMURF2 primarily targets a lysine site amino-terminal to the DNA-binding domain of KLF5. We first tested the two SUMOylation sites in KLF5, Lys-151 and Lys-202 (18), given that SUMOylation and ubiquitination may interplay and share identical target lysine residues (24). However, both the partially SUMOylated K202R and SUMOylation-deficient K151R/K202R double mutants were efficiently degraded by SMURF2 (Fig. 7B), indicating that the SUMOylation sites in KLF5 are not targeted by SMURF2. We next tested Lys-324 and Lys-358, given their proximity to the PY motif and the high degree of conservation of residues around them, especially Lys-324, which is immediately juxtaposed to the PPXY motif and PY tail of KLF5 (Fig. 7A). However, both K324R and K358R were effectively degraded by SMURF2 (Fig. 7C). Lastly, we tested the remaining seven lysine residues within the amino terminus of KLF5, and none of these K-to-R mutants resisted SMURF2-triggered degradation (Fig. 7D). Altogether, these results indicate that SMURF2 does not target a single lysine residue in KLF5.
SMURF2 Negatively Regulates the Biological Activities of KLF5
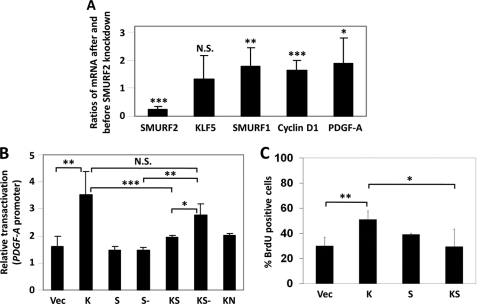
Consistent with the degradation of KLF5, SMURF2 inhibits the transcriptional and pro-proliferative activities of KLF5. Endogenously, SMURF2 depletion inhibited the expression of two major KLF5 target genes, cyclin D1 and PDGF-A. This was accomplished by determining the relative transcript levels of cyclin D1 and PDGF-A following SMURF2 knockdown. As seen in Fig. 8A, siRNA directed against SMURF2 specifically reduced the transcript levels of SMURF2 but not SMURF1 (Fig. 8A). In fact, the SMURF1 level actually increased after SMURF2 knockdown, suggesting that SMURF2 negatively regulates SMURF1 expression, a result consistent with a previous report (25). After the SMURF2 siRNA treatment, the transcript levels of two major KLF5 target genes, cyclin D1 and PDGF-A, were both increased (Fig. 8A), indicating that SMURF2 negatively regulates the activity of KLF5. The knockdown did not affect KLF5 mRNA expression, reinforcing the conclusion that the reduction in KLF5 levels occurs at the posttranslational level. The effect of SMURF2 on the transcriptional activity of KLF5 was also examined by dual luciferase reporter assay using the PDGF-A promoter. The ability of KLF5 to transactivate PDGF-A was suppressed significantly by SMURF2, although this inhibitory effect was abolished when the inactive SMURF2-C716A mutant was used instead (Fig. 8B). KLF5-N (KN), which lacks the PY motif and DNA-binding domain that interact with SMURF2, failed to transactivate PDGF-A (Fig. 8B). Finally, we examined the effect of SMURF2 on the pro-proliferative activity of KLF5 using a BrdU incorporation assay, a typical assay used previously used to study the ability of KLF5 to stimulate cell proliferation (21). Although KLF5 significantly stimulated COS-1 cell proliferation, this activity was suppressed by SMURF2 (Fig. 8C). These results clearly indicate that SMURF2 inhibits the transcriptional and pro-proliferative activities of KLF5.
FIGURE 8.
SMURF2 inhibits the transcriptional and pro-proliferative activities of KLF5. A, depletion of SMURF2 increases the expression of KLF5 target genes. COS-1 cells were transfected with either control siRNA or the Trilencer SMURF2 siRNA mixture from Origene (catalog no. SR312096). Three days later, total RNA was isolated, and quantitative RT-PCR was performed for the SMURF2, SMURF1, KLF5, and KLF5 target genes cyclin D1 and PDGF-A (n = 12). Shown are the average ratios of mRNA levels after and before SMURF2 knockdown. N.S., not significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by two-tailed Student's t test. B, SMURF2 suppresses the transcriptional activity of KLF5. The ability of KLF5 to transactivate PDGF-A luciferase reporter was inhibited by SMURF2 but not the catalytically inactive SMURF2-C716A mutant. RKO cells were transfected with PDGF-A reporter and Renilla control plasmids plus either vector alone (Vec), pMT3-KLF5 (K), pCMV-FLAG-SMURF2 (S), pCMV-FLAG-SMURF2-C716A (S-), pMT3-KLF5 and pCMV-FLAG-SMURF2 (KS), pMT3-KLF5 and pCMV-FLAG-SMURF2-C716A (KS-), or pMT3-KLF5 (1–165) (KN), and dual luciferase reporter assays were performed. Shown are the mean ± S.D. of four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001, by two-tailed Student's t test. C, SMURF2 suppresses the pro-proliferative activity of KLF5. COS-1 cells were transfected with vector alone, pMT3-HA-KLF5, pCMV-Myc-SMURF2, or both pMT3-HA-KLF5 and pCMV-Myc-SMURF2, and a BrdU incorporation assay was performed as described under “Experimental Procedures.” Shown are the mean ± S.D. of four independent experiments. *, p < 0.05; **, p < 0.01 by two-tailed Student's t test.
DISCUSSION
In this report, we demonstrate a novel interaction between KLF5 and SMURF2. Consistent with the presence of the SMURF2-interacting PY motif in KLF5, SMURF2 binds to a region of KLF5 that contains the PPXY motif and PY tail (Fig. 1A). Interestingly, SMURF2 also efficiently interacts with KLF5's DNA-binding domain. This interaction is specific, as it prefers the DNA-binding domain of KLF5 to that in KLF4 (Fig. 1A). Sequence alignment shows that the main discrepancies between KLF5 and KLF4's DNA-binding domains lie in the three areas covering the amino-terminal half, i.e. C2 parts, of the three C2H2 zinc fingers (Fig. 6A). Because these areas coordinate the binding to zinc metal cofactor and mediate direct DNA binding (15), it remains to be determined whether SMURF2 interaction directly affects the ability of KFL5 to recognize its DNA targets and activate transcription. Recently, another SMURF family protein, SMURF1, was reported to bind and degrade KLF2 (26), and the zinc finger DNA-binding domain of KLF2 is sufficient and efficient for SMURF1 binding (26). It is also of interest to note that although it interacts with SMURF2, KLF5 does not bind to SMURF1 (Fig. 1D). Hence, specific SMURFs can recognize specific Krüppel-like factors despite the highly conserved nature of the DNA-binding domains of Krüppel-like factors (27). These results also provide evidence of KLFs as a novel family of transcription factors regulated by SMURFs and demonstrate a new structural basis of substrate recognition for the SMURF family of ubiquitin ligases.
Herein, we present SMURF2 as an ubiquitin ligase that degrades KLF5 but does not degrade KLF5 at a single lysine site (Figs. 6 and 7). SMURF2 therefore likely targets multiple lysine sites within KLF5. This is not only supported by the mutagenesis studies in this work but also consistent with the observation that SMURF2 predominantly polyubiquitinates KLF5 to high molecular weight forms (Fig. 5). Thus, effects from loss in ubiquitination at one site may be largely compensated by ubiquitination at another site. The requirement of multiple lysines in KLF5 for degradation hints at the potential complexity of SMURF2 in regulating KLF5.
The interaction and degradation of KLF5 are quite specific for the SMURF family, as they are highly preferred by SMURF2 rather than the closely related homolog, SMURF1 (Figs. 1 and 4). Hence, the posttranslational regulation of KLF5 by E3 ubiquitin ligases appears to be highly complex and dynamic. For instance, besides SMURF2, KLF5 is degraded by a related ubiquitin ligase, WWP1 (5), that also targets KLF2 (28, 29). WWP1 is a Nedd4 family of HECT ubiquitin ligases that recently included SMURFs (2). WWP1 and SMURF2 share identical types and orientation of major structural domains (2). The destabilization of KLF5 by WWP1 and SMURF2 is highly specific, as most Nedd4 family members, including Nedd4–1, Nedd4–2, AIP4/Itch, WWP2/AIP2, and SMURF1, failed to degrade KLF5 (5). It is currently unknown how KLF5 is selectively regulated by these two E3 ligases. WWP1 and SMURF2 exhibit similar inhibitory activity toward KLF5 (2). For instance, both utilize similar binding domains and catalytic mechanism to degrade KLF5, both are blocked to similar extent by the proteasomal inhibitor MG132, and both exhibit highly comparable activity in suppressing the PDGF-A promoter (5). Theoretically WWP1 and SMURF2 can compete for KLF5. Thus, it would be interesting to determine whether these two ubiquitin ligases act at different stages of growth and development or in response to distinct signaling pathways triggered.
KLF5 was also targeted by another E3 ligase, FBW7, an F-box ubiquitin ligase (7, 19). FBW7 utilizes very different mechanism for substrate recognition and catalysis. Although SMURF2 and WWP1 bind to KLF5 through WW domains and PY motifs, FBW7 binds to KLF5 through the WD40 domain of FBW7 and the phosphor-binding motifs, called CDC4 phosphodegrons, of KLF5 (7, 19). Thus, KLF5 undergoes multiple layers of regulation by identical or diverse families of ubiquitin ligases. These results reflect the highly complex and dynamic regulation of KLF5 by ubiquitination.
Among the multiple functions attributed to SMURF2 (2, 6–12), it regulates cell polarity (10–12). The intestinal epithelium is a highly polarized system containing terminally differentiated epithelial cells at villi and proliferating and progenitor cells in crypts (14). As the KLF5 protein is highly abundant in the crypt cell population with a diminishing gradient as cells migrate toward the villus (27, 30, 31), SMURF2 may form an opposite gradient along the crypt/villus axis or contribute to establishing the polarity of the intestinal tract.
In summary, we illustrate a novel and specific interaction between KLF5 and SMURF2 and expand the list of ubiquitin ligases that dynamically control the turnover and activity of KLF5 by demonstrating that SMURF2 ubiquitinates, destabilizes, and negatively regulates KLF5. These results endorse KLFs as a new family of targets by the SMURF family of ubiquitin ligases and SMURFs as a new group of ubiquitin ligases that regulate KLF transcription factors.
Acknowledgments
We thank Jerry Lingrel, Jeff Wrana, Chris Yun, and David Kaetzel for KLF5 cDNA, SMURF, HA-ubiquitin, and PDGF-A luciferase reporter plasmids.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grants DK052230, DK064399, and CA084197.
REFERENCES
- 1. Ardley H. C., Robinson P. A. (2005) Essays Biochem. 41, 15–30 [DOI] [PubMed] [Google Scholar]
- 2. Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 3. Chong P. A., Lin H., Wrana J. L., Forman-Kay J. D. (2006) J. Biol. Chem. 281, 17069–17075 [DOI] [PubMed] [Google Scholar]
- 4. Kanelis V., Bruce M. C., Skrynnikov N. R., Rotin D., Forman-Kay J. D. (2006) Structure 14, 543–553 [DOI] [PubMed] [Google Scholar]
- 5. Chen C., Sun X., Guo P., Dong X. Y., Sethi P., Cheng X., Zhou J., Ling J., Simons J. W., Lingrel J. B., Dong J. T. (2005) J. Biol. Chem. 280, 41553–41561 [DOI] [PubMed] [Google Scholar]
- 6. Lin X., Liang M., Feng X. H. (2000) J. Biol. Chem. 275, 36818–36822 [DOI] [PubMed] [Google Scholar]
- 7. Liu N., Li H., Li S., Shen M., Xiao N., Chen Y., Wang Y., Wang W., Wang R., Wang Q., Sun J., Wang P. (2010) J. Biol. Chem. 285, 18858–18867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Narimatsu M., Bose R., Pye M., Zhang L., Miller B., Ching P., Sakuma R., Luga V., Roncari L., Attisano L., Wrana J. L. (2009) Cell 137, 295–307 [DOI] [PubMed] [Google Scholar]
- 11. Schwamborn J. C., Khazaei M. R., Puschel A. W. (2007) J. Biol. Chem. 282, 35259–35268 [DOI] [PubMed] [Google Scholar]
- 12. Schwamborn J. C., Muller M., Becker A. H., Puschel A. W. (2007) EMBO J. 26, 1410–1422 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Sun R., Chen X., Yang V. W. (2001) J. Biol. Chem. 276, 6897–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conkright M. D., Wani M. A., Anderson K. P., Lingrel J. B. (1999) Nucleic Acids Res. 27, 1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swamynathan S. K. (2010) Hum. Genomics 4, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nandan M. O., Yang V. W. (2009) Histol. Histopathol. 24, 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen C., Sun X., Ran Q., Wilkinson K. D., Murphy T. J., Simons J. W., Dong J. T. (2005) Oncogene 24, 3319–3327 [DOI] [PubMed] [Google Scholar]
- 18. Du J. X., Bialkowska A. B., McConnell B. B., Yang V. W. (2008) J. Biol. Chem. 283, 31991–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao D., Zheng H. Q., Zhou Z., Chen C. (2010) Cancer Res. 70, 4728–4738 [DOI] [PubMed] [Google Scholar]
- 20. Dong J. T., Chen C. (2009) Cell Mol. Life Sci. 66, 2691–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du J. X., Yun C. C., Bialkowska A., Yang V. W. (2007) J. Biol. Chem. 282, 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Du J. X., McConnell B. B., Yang V. W. (2010) J. Biol. Chem. 285, 28298–28308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q., Pedigo N., Shenoy S., Khalili K., Kaetzel D. M. (2005) Gene 348, 25–32 [DOI] [PubMed] [Google Scholar]
- 24. Wilson V. G., Heaton P. R. (2008) Expert Rev Proteomics 5, 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fukunaga E., Inoue Y., Komiya S., Horiguchi K., Goto K., Saitoh M., Miyazawa K., Koinuma D., Hanyu A., Imamura T. (2008) J. Biol. Chem. 283, 35660–35667 [DOI] [PubMed] [Google Scholar]
- 26. Xie P., Tang Y., Shen S., Wang Y., Xing G., Yin Y., He F., Zhang L. (2011) Biochem. Biophys. Res. Commun. 407, 254–259 [DOI] [PubMed] [Google Scholar]
- 27. McConnell B. B., Yang V. W. (2010) Physiol. Rev. 90, 1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang X., Srinivasan S. V., Lingrel J. B. (2004) Biochem. Biophys. Res. Commun. 316, 139–148 [DOI] [PubMed] [Google Scholar]
- 29. Conkright M. D., Wani M. A., Lingrel J. B. (2001) J. Biol. Chem. 276, 29299–29306 [DOI] [PubMed] [Google Scholar]
- 30. Ghaleb A. M., Nandan M. O., Chanchevalap S., Dalton W. B., Hisamuddin I. M., Yang V. W. (2005) Cell Res 15, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McConnell B. B., Ghaleb A. M., Nandan M. O., Yang V. W. (2007) Bioessays 29, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmittgen T. D., Livak K. J. (2008) Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]