Background: We sought to identify novel peptide sequences that interact at the BH3 binding groove of Mcl-1.
Results: We identified a peptide that binds to Mcl-1 in a reverse orientation compared with canonical BH3 peptides.
Conclusion: The peptide binds to Mcl-1 selectively over other Bcl-2 proteins.
Significance: The novel sequence is useful in identifying Mcl-1 binding partners and for developing possible therapeutics.
Keywords: Apoptosis, Bacteriophage, Bcl-2, Biophysics, NMR, Peptide Interactions, Mcl-1, Sabutoclax
Abstract
Recent characterization of Mcl-1 as the primary anti-apoptotic Bcl-2 family member expressed in solid tumors, coupled with its ability to enable therapeutic resistance, has provided the impetus for further study into how Mcl-1 is involved in apoptosis signaling. Here, we employ Sabutoclax, a potent and effective Mcl-1 antagonist, as a competing agent to screen a randomized 12-residue phage display library for peptides that bind strongly to the Bcl-2 homology 3 (BH3) binding groove of Mcl-1. Although the screen identified a number of α-helical peptides with canonical BH3 domain sequences, it also isolated a pair of unique peptide sequences. These sequences exhibit a reverse organization of conserved hydrophobic and acidic residues when compared with canonical BH3 sequences, and we therefore refer to them as reverse BH3 (rBH3) peptides. Furthermore, studies of the rBH3 peptides using NMR spectroscopy, fluorescence polarization displacement assays, and alanine scanning data all suggest that they bind to the BH3 binding groove of Mcl-1 selectively over Bcl-xL. A search for proteins containing the rBH3 motif has identified a number of interesting Mcl-1 protein partners, some of which have previously been associated with apoptosis regulation involving Mcl-1. These findings provide insights into the development of more specific Mcl-1 antagonists and open the way to the identification of a previously unknown family of apoptosis-regulating and Mcl-1 interacting proteins.
Introduction
During oncogenesis and subsequent chemotherapeutic treatment, the ability of a cancer cell to manipulate and disable the natural processes of programmed cell death (apoptosis) largely determines its ability to survive. One event that enables cancer cells to evade natural cellular regulation and chemotherapeutic induced cell-death is the up-regulation of anti-apoptotic proteins (1). In mammals, the Bcl-2 (B-cell lymphoma 2) family of pro- and anti-apoptotic proteins lie at the center of apoptosis regulation. These proteins are related by a series of Bcl-2 homology (BH)2 domains, and their function as either a pro- or anti-apoptotic regulator is determined by the composition of their BH domains (2, 3). The functional interplay of the Bcl-2 proteins is centered on the ability of the six anti-apoptotic family members (Bcl-2, Bcl-W, Bcl-xL, Mcl-1, Bfl-1/A1, and Bcl-B) to bind to the BH3 helices of the proapoptotic effectors Bak and Bax and thereby suppress the oligomerization of Bak and Bax in the mitochondrial outer membrane (4). This suppression is overcome selectively through activation of a third group of Bcl-2 proteins, the BH3-only apoptotic activators, whose BH3 helices bind to the six anti-apoptotic Bcl-2 family proteins through their BH3 binding grooves and release Bak or Bax, thereby initiating mitochondrial outer membrane permeabilization, cytosolic release of cytochrome c, and ultimately cell death (3, 5).
The BH3 domain consists of an amphipathic helix with a strictly conserved C-terminal aspartic acid residue that is often preceded by both an isoleucine residue and a glycine residue (see Fig. 1, top panel). In addition, a series of hydrophobic residues, which form the interaction surface with the BH3 binding groove, serve to selectively target the BH3 helices toward one or a subset of the six anti-apoptotic Bcl-2 proteins (6, 7). For instance, the Noxa A BH3 sequence is able to bind selectively to Mcl-1 to the exclusion of the other five proteins, whereas the BH3 sequence from Bim shows strong binding to all six anti-apoptotic Bcl-2 proteins. The sequence characteristics that dictate selectivity for individual or sets of the anti-apoptotic proteins have recently been examined and a series of BH3-derived peptides specific for either Mcl-1 or the closely related proteins, Bcl-2, Bcl-xL, and Bcl-W, have been identified (8, 9).
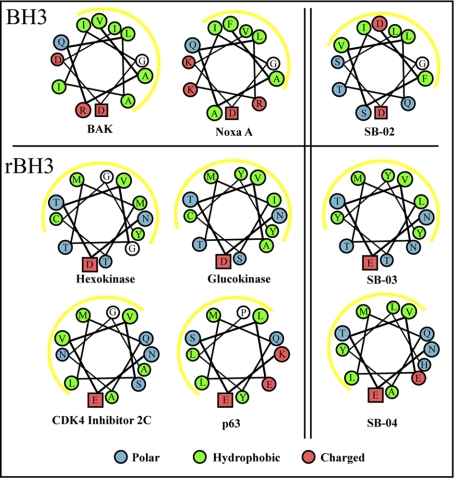
FIGURE 1.
Helical wheel plots of BH3 and rBH3 peptide sequences. Helical wheel plots of the BH3 peptide sequences from BH3-only proteins, the BH3-like peptide SB-02, the rBH3 peptides SB-03 and SB-04, and a selection of rBH3 peptides from native proteins, highlight the similarities between the chemical nature of BH3 and rBH3 helices. Of significance, the location of the conserved acidic residue, shown in a box, and the cluster of hydrophobic side chains, depicted by the yellow line, for the rBH3 peptides is oriented in a inverted position to that seen in the rBH3 sequences.
Because of the ability of the Bcl-2 family to aid in cancer cell survival, development of anti-cancer therapeutics targeting the BH3 groove of anti-apoptotic Bcl-2 proteins has emerged as a promising but difficult goal (10, 11). The first compounds, targeted to the structure of Bcl-xL and thereby able to bind to Bcl-2, Bcl-xL, and Bcl-W, have shown promise in the clinic for the treatment of chronic lymphocytic leukemia, but recent studies have shown that Mcl-1 overexpression allows cancers to resist their treatment (12). In addition, Mcl-1 is emerging as an important apoptotic and cell growth regulator as it has been implicated in taxol resistance and as a key component of glucose metabolism (13, 14). The importance of specifically targeting Mcl-1 or developing pan-active Bcl-2 inhibitors has been further highlighted by our recent study of the mRNA expression of all six anti-apoptotic Bcl-2 subfamily proteins in 68 human cancer-derived cell lines, which identified Mcl-1 as the most highly expressed anti-apoptotic Bcl-2 family member (15). To this end, recent success at the development of stable and cell-permeable stapled BH3 peptides has provided the impetus to identify short sequences capable of selectively binding to Mcl-1 (16, 17).
To design Mcl-1-selective agents, a number of studies have been carried out to identify novel Mcl-1-selective peptide sequences as well as to determine what amino acid compositions of the BH3 helix can specifically target Mcl-1 over Bcl-xL (8, 9). Thus far, all studies have been restricted to traditional BH3 peptides to probe for the association of other peptides or small molecules. Recently, we developed the pan-active Bcl-2 inhibitor, Sabutoclax (“BI97C1”), which is a potent small molecule antagonist of the Mcl-1-BH3 interaction (18). We used Sabutoclax to screen a randomized 12-residue N-terminal phage display library that revealed a pair of non-BH3 peptide sequences that specifically bind to Mcl-1. These sequences exhibit characteristics of amphipathic α-helices (see Fig. 1) and require the presence of a conserved glutamic acid residue and a conserved methionine residue to bind to Mcl-1. Due to the reverse orientation of the conserved acidic and hydrophobic residues in these sequences, when compared with typical BH3 peptides, we refer to these two new peptides as having a reverse BH3 (rBH3) motif. BLAST analysis of these sequences against the human genome identified similar sequences in a selection of interesting proteins including glucokinase, hexokinase, as well as a number of tumor suppressors. A short peptide derived from the glucokinase rBH3 motif exhibits binding to Mcl-1 comparable to that seen for a 12-residue Noxa A peptide (Table 1). We believe that this sequence represents a unique peptide motif present in native proteins that serves as a new class of Mcl-1-specific binding proteins, much as the BH3-only proteins are able to interact with various anti-apoptotic Bcl-2 family members. This finding will allow for development of improved Mcl-1 specific small molecule and stapled peptide-based therapeutics (16, 17). Furthermore, its identification may provide the basis for increased understanding of possible cross-talk taking place between a number of divergent cellular signaling and homeostatic processes and the regulation of apoptosis through Mcl-1.
TABLE 1.
Peptide binding constants to Bcl-2 family proteins
| Peptide name | Sequence | FPA (IC50)a |
NMRb |
ITC (Kd) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mcl-1 | Bcl-W | Bcl-xL | Bfl-1 | Bcl-2 | Mcl-1 | Bcl-xL | Mcl-1 | ||
| μm | μm | μm | μm | μm | μm | ||||
| Noxa A22 | ELPPEFAAQLRKIGDKVYCTWS-NH2 | 0.3 ± 0.1 | >75 | >75 | >75 | >75 | + | + | 0.2 ± 0.1 |
| Noxa A12 | EFAAQLRKIGDK-NH2 | 49.7 ± 10 | >75 | >75 | >75 | >75 | − | − | NDc |
| Bak | GQVGRQLAIIGDDINR-NH2 | 3.4 ± 0.4 | 1 ± 0.2 | 0.2 ± 0.1 | 0.3 ± 0.1 | 6.5 ± 2 | + | + | 0.7 ± 0.2 |
| SB-02 | DFSVLQTIGDSL-NH2 | 0.22 ± 0.04 | >75 | >75 | >75 | 28 ± 5 | + | Weak | 1.8 ± 0.6 |
| SB-03 | NETVNTMLTYYY-NH2 | 1.6 ± 0.2 | >75 | >75 | >75 | >75 | + | − | 0.5 ± 0.2 |
| SB-04 | NETVELMQAYLH-NH2 | 0.69 ± 0.02 | >75 | >75 | >75 | >75 | + | − | 1.2 ± 0.6 |
| SB-04M7A | NETVELAQAYLH-NH2 | >75 | ND | >75 | ND | ND | − | − | ND |
a FPA analysis was performed using N-terminal His-tagged mMcl-1, Bcl-xL, and Bcl-2 and N-terminal GST-tagged Bfl-1 and Bcl-W.
b For NMR analysis, + denotes δΔ of >0.25 ppm; weak denotes δΔ of 0.1–0.25 ppm; − denotes δΔ < 0.1 ppm.
c ND, not determined.
EXPERIMENTAL PROCEDURES
Protein Sample Preparation
Unlabeled and 15N-labeled recombinant GST-Bcl-W (18), His-mMcl-1 (19), GST-Bfl-1 (18), His-Bcl-2 (7), and His-Bcl-xL (20) were expressed and purified as described previously. After purification, the samples were exchanged into PBS (0.02 m sodium phosphate, 0.14 m NaCl, pH 6.7) or TBS (0.02 m Tris-HCl, 0.14 m NaCl, pH 7.4) and concentrated to 0.02 m. The 7- and 12-mer phage display libraries (New England Biolabs) were enriched for mMcl-1 binding sequences during three sequential pan and amplification steps following the manufacturer's instructions using either 150 μl of a 26-amino acid Bim peptide or the pan-active Bcl-2 inhibitor Sabutoclax in TBS buffer as competitive agents. To avoid hydrophobic interference, we expressed and purified N-terminally His-tagged recombinant mMcl-1 and chelated the protein to nickel coated 96-well plates (Sigma). After the final panning step, 20 plaques from each phage fraction were sequenced (Genewiz), and the resulting sequences were analyzed for similarity using Unipro UGENE 1.7.2. BLAST software was used to search for homologous human peptide sequences to SB-03 and SB-04 in the NCBI gene database. Identified sequences were aligned with a BLOSUM62 matrix, using default optimized parameters for small sequences.
Peptide Analysis
Identified peptides were synthesized (Abgent) and NMR, fluorescent polarization displacement assay (FPA), and ITC studies of the interactions were carried out in PBS at pH 7.4 for FPA and ITC or pH 6.7 for NMR. NMR spectra were collected at 298 K on a Bruker Avance 600 MHz spectrometer equipped with a 5-mm cryogenic inverse gradient triple resonance probe. The spectra were processed using TOPSPIN 1.3 (Bruker) and analyzed with CARA (21). FPA analysis was performed as described previously (18). ITC measurements were carried out at 25 °C with a VP-ITC instrument (Microcal). ITC samples were equilibrated into uniform PBS buffer at pH 7.4 and degassed. For each experiment, 500 μl of a 150 μm solution of each peptide was titrated into a 2-ml solution of 10 μm Mcl-1. Each ITC experiment was repeated twice to evaluate reproducibility, and the data were fit to a standard one-site binding model using Origin as supplied by Microcal.
Molecular and Protein Interaction Modeling
The NMR structure of mMcl-1 in complex with the Noxa A peptide (Protein Data Bank code 2ROD) was used as a reference structure for chemical shift mapping with VMD (version 1.8.7) (22). Chemical shift perturbation data were used to define the binding site for docking of a three-dimensional ideal α-helical model of SB-04 to mMcl-1 using Haddock (version 1.8) (23). The most energetically favorable solution was energy minimized using 1000 steps of steepest descent minimization followed by 1000 steps of conjugate gradient energy minimization in an explicit box water. An isothermal-isobaric ensemble was used to equilibrate the explicit solvent with 100 ps of position restrained molecular dynamics of the complex and finally, a 20-ns long molecular dynamics simulation was carried out. Energy minimization and molecular dynamics were conducted in Gromacs (version 4.0) (24); V-rescale temperature coupling and a Berendsen barostat were applied to control the temperature and pressure at 300 K and 1 atm. Particle mesh Ewald method laws were used for long range electrostatic interactions, and a cut-off of 0.9 nm was applied to short range non-bonded interactions. Molecular analysis and images of the complex structure were prepared in SYBYL (version 8.0, Tripos). The STRING analysis of Mcl-1-rBH3 protein interaction was performed using the software website and searching the software using the following protein list: Mcl-1, HK1, HK2, GLK, TP73, TP73L (p63), PTBP1, and CDKN2C. After four steps of network expansion, the interactions leading to Mcl-1 coordination to the rBH3 proteins were highlighted and expected interactions between BH3 proteins that did not bind to the other proteins were removed for clarity.
Cell Viability Assays
Cell lines were cultured, and cell viability was monitored using the ATP-Lite 1Step assay (Perkin Elmer) as described previously (15).
RESULTS
Identification of Mcl-1-specific rBH3 Peptides
We screened randomized 7- and 12-mer phage display libraries (New England Biolabs) associated to the N terminus of the minor coat protein (pIII) of M13 phage to identify peptide sequences capable of binding to the BH3 groove of recombinant mouse Mcl-1 (mMcl-1). These libraries were enriched for mMcl-1 binding sequences during three sequential pan and amplification steps using either a 26-amino acid Bim peptide (Bim peptide) or the pan-active Bcl-2 inhibitor Sabutoclax as competitive agents. To avoid hydrophobic interference, we expressed and purified N-terminally His-tagged recombinant mMcl-1 and chelated the protein to nickel coated 96-well plates (Sigma). The 12-mer library screened with the Bim peptide yielded a population of sequences that was 40% enriched with a novel Mcl-1-specific BH3-like peptide SB-02 (DFSVLQTIGDSL). However, use of the pan-active anti-apoptotic Bcl-2 inhibitor Sabutoclax as a displacer resulted in a sequence population that was 20% enriched with SB-02, 20% enriched with the non-BH3 peptide sequence SB-03 (NETVNTMLTYYY), and 40% enriched with the non-BH3 peptide sequence SB-04 (NETVELMQAYLH).
Alignment of the non-BH3 sequences SB-03 and SB-04 (Table 1) with other BH3 sequences showed no direct similarity, but reverse alignment of the SB-03 and SB-04 sequences successfully positions the Glu-2 residue and a series of hydrophobic residues with the conserved aspartic acid and hydrophobic residues, respectively, observed in BH3 sequences (Fig. 1). Because of this reverse alignment of SB-03 and SB-04 with other BH3 sequences we refer to them as having a rBH3 motif. Helical wheel plotting of the rBH3 peptides, the BH3-like peptide SB-02 and other known BH3 peptides shows that they position a selection of hydrophobic residues opposite of the conserved acidic residue (Fig. 1). In addition, positioning the conserved acidic residue at the bottom of the wheel shows that the rBH3 peptide sequences have an inverted orientation to that seen in BH3 peptides.
Analysis of SB-02 and SB-04 Binding to mMcl-1
To confirm that the new sequences, SB-02 and SB-04, interact with the BH3 binding groove of mMcl-1, we mapped the chemical shift perturbation of mMcl-1 two-dimensional [1H,15N] HSQC spectra after addition of either the BH3 like peptide SB-02 or the rBH3 peptides SB-03 and SB-04. The interaction of the peptides to mMcl-1 was observed to take place in slow exchange on the NMR timescale and therefore accurate perturbation analysis required the reassignment of the backbone 1HN and 15N resonances for mMcl-1 in the presence of both peptides based on a prior backbone assignment published by Day et al. (19). Considerable chemical shift perturbation was observed and the changes were saturated at a 1:1 ratio with 20 μm mMcl-1 and 20 μm peptide (Fig. 2a). Similar test for binding of either peptide to the related anti-apoptotic Bcl-2 proteins Bcl-xL showed very weak or no interaction with SB-02 or SB-04, respectively (Table 1, data not shown).
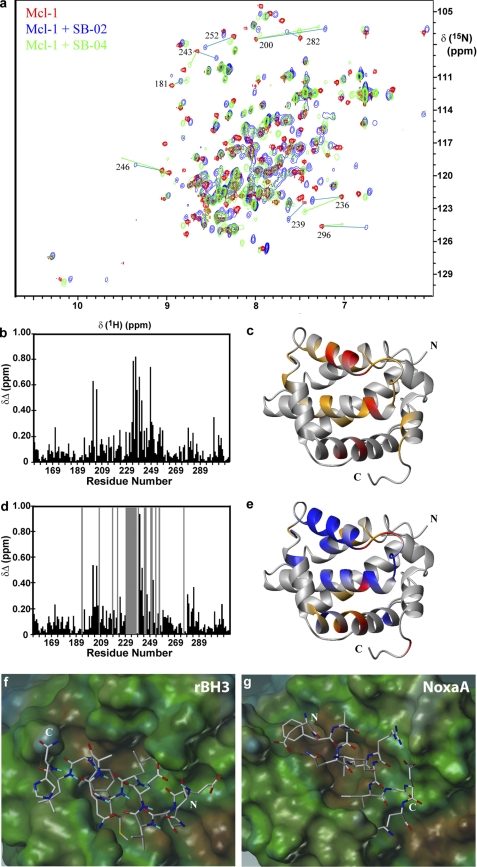
FIGURE 2.
15N and 1H NMR analysis and model of rBH3 binding to mMcl-1. a, superposition of the two-dimensional [15N,1H] HSQC spectra of free mMcl-1 (red), 1:1 mMcl-1 + SB-02 (cyan), and 1:1 mMcl-1+SB-04 (green). A selection of cross-peaks that underwent significant change are labeled. New peak locations are identified using cyan or green lines for SB-02 and SB-04, respectively. b and d, combined chemical shift differences of 1HN and 15N, δΔ = {[(δΔNH)2 + ((δΔN)2/25)]/2}1/2 (ppm), observed upon addition of SB-02 or SB-04, respectively. The location of cross-peaks that disappeared upon addition of SB-04 are identified by gray bars on the chart. c and e, schematic of the mMcl-1 polypeptide backbone, with the BH3 binding groove oriented in front, highlighting in orange and red the locations of residues that exhibit a combined chemical shift difference >0.2 and 0.4 ppm, respectively. Residues that disappear upon addition of SB-04 are colored in blue. f, model of the peptide SB-04 bound to the previously determined mMcl-1 NMR structure (19). g, picture of the corresponding 12 Noxa A residues bound to mMcl-1 as determined by NMR. The surface of mMcl-1 is colored according to the lipophilicity, and the N (N) and C (C) termini of the peptides are labeled.
Sequential mapping of the chemical shift perturbation on the structure of mMcl-1 illustrates that nearly identical regions of the protein are affected after addition of the BH3 and rBH3 peptides, SB-02 and SB-04, respectively (Fig. 2, b and d). Interestingly, although the same cross-peaks were affected upon addition of either peptide, the magnitude and direction of those changes was significantly different. Furthermore, the mMcl-1 amides undergoing the largest chemical shift change or loss of peak intensity upon binding to SB-04 cluster near the BH3 binding groove of mMcl-1 (Fig. 2, c and e). This data supports that SB-02 and SB-04 both bind to the BH3 binding groove of mMcl-1 but that SB-04 has an inherently different binding mode than that of the BH3-like peptide SB-02.
To confirm the selectivity of the SB-02 and SB-04 peptides for Mcl-1 over Bcl-xL, we used a FPA to determine the respective IC50 and dissociation constants for a Bak peptide (GQVGRQLAIIGDDINR), SB-02, SB-03, and SB-04. FPA analysis of the Bak peptide binding to His-Mcl-1 or His-Bcl-xL showed strong binding in the high nanomolar range (Table 1). Each of the three peptides isolated from the phage display screen showed weaker but specific binding to His-mMcl-1 with IC50 values of 0.22, 1.6, and 0.69 μm compared with values >75 μm for Bcl-xL, Bcl-W, and Bfl-1 and >30 μm for Bcl-2 (Table 1). Interestingly, the rBH3 peptide SB-03 showed significantly stronger binding by ITC than FPA, with a Kd similar to the full-length, 22-residue NoxaA (6) peptide (Table 1).
Positional Scanning Analysis of SB-04
To determine which residues of the rBH3 peptide affect its stability or the ability of the peptide to interact with Mcl-1, we sequentially scanned the rBH3 peptide SB-04 with alanine. The resulting peptides were screened for binding to mMcl-1 using two-dimensional [15N,1H] HSQC chemical shift perturbation and FPA analysis using GST-mMcl-1 (supplemental Table 1). The results of this data showed that replacement of the Thr-3, Glu-5, Gln-8, or His-12 residues of SB-04 had no effect on binding while replacement of the residues Asn-1, Glu-2, Val-4, Met-7, Tyr-10, and Leu-11 had large effects on the Ki values. These results were mirrored in the NMR analysis as each of the later group of replacements either required a higher ratio of peptide to protein to observe chemical shift changes (weak binding) or caused no change in the two-dimensional [15N,1H] HSQC spectra of mMcl-1 at 10:1 peptide to protein ratios (no binding).
Identification of Native Proteins Containing rBH3 Sequences
We next sought to identify native proteins that may contain rBH3 sequences. For this analysis, we utilized the alanine scanning data to focus on peptides that contained an acidic residue at position 2, and hydrophobic residues at positions 4, 7, 10, and 11, numbered according to the identified 12-residue rBH3 peptides. BLAST analysis of SB-03 and SB-04 sequences against the human genome identified similar sequences in a selection of interesting proteins, including glucokinase, hexokinase I, II, and III, as well as a number of tumor suppressor proteins (Fig. 3a and supplemental Table 2). Structural information of a selection of these proteins shows that the rBH3 motif is present in the form of a surface accessible helix near a functional site of the protein (i.e. at the active site (25, 26) or near a known protein-protein interaction site (27, 28)). Similar to the BH3 motif, we identified that a conserved acidic residue (Glu-2) and a conserved methionine (Met-7) residue constitute a signature for rBH3 containing proteins (Fig. 3a). Although a 12-residue glucokinase rBH3 peptide exhibited weak (50.1 μm) interaction with mMcl-1 using FPA, NMR analysis (5:1 peptide to protein ratio) showed that the peptide could induce similar chemical shift changes as seen with the SB-03 and SB-04 rBH3 peptides.
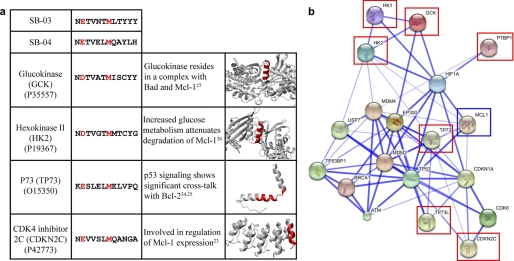
FIGURE 3.
Sequence and STRING analysis of rBH3-containing proteins. a, a table of proteins containing a BLAST identified rBH3-like sequence. For each protein, the rBH3 sequence is shown with the conserved acidic and methionine residues highlighted in red, a short description of prior research identifying each protein interaction with Mcl-1 or the Bcl-2 protein family is highlighted, and a schematic of the three-dimensional structure of each protein, with the rBH3 sequence region colored in red, is presented. b, a STRING-generated interaction network of Mcl-1 (blue box) with a selection of rBH3-containing proteins (red boxes). Each spherical node represents a gene/protein, and the line thickness represents the combined interaction score based on experimental data, established protein network information, text mining, and homology.
Cell Culture Analysis of Peptide Activity
To study the ability of the SB-04 peptide to induce cell death in culture we tested a selection of prostate (PC-3 and DU-145) and lung cancer (Hop-62 and Hop-92) derived cell lines. Although the non-binding variant of the SB-04 peptide, SB-04M7A (Table 1), exhibited no cell death in all cell lines studied up to 10 μm, SB-04, similar to ABT-737 (11), had ED50 values in the low micromolar range for each of the cell lines (Fig. 4).
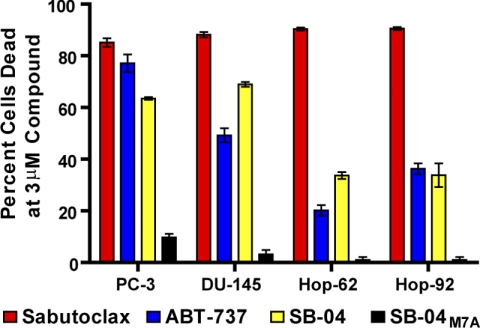
FIGURE 4.
Cell killing by SB-04 in culture. The bar graph shows the percent of cells killed in the four cell lines tested at a dose of 3 μm following a 72-h incubation period. In each cell line, SB-04 shows a level of killing on par with that observed with the small molecule ABT-737, whereas the Mcl-1 inactive SB-04 variant (SB-04M7A) shows no significant cell killing under the same conditions.
DISCUSSION
Our phage display screen of mouse Mcl-1 using the small molecule Sabutoclax as a competing agent had two significant results. First, the identification of the peptide SB-02, which is the shortest BH3-like sequence able to bind specifically to the BH3 groove of Mcl-1. This sequence should aid in the development of stapled BH3 peptides that target the Mcl-1 BH3 binding groove.
Secondly, our screen identified the two rBH3 peptides SB-03 and SB-04, which have low micromolar affinities to Mcl-1 and display chemical shift perturbation data consistent with their binding to the BH3 groove of Mcl-1. To better illustrate this interaction, we derived a model (Fig. 2f) of a helical SB-04 peptide bound to mMcl-1 based on our experimental NMR data, the current NMR structure of mMcl-1 in complex with the BH3 domain of Noxa A (6) and molecular dynamics simulations. This model positions the conserved glutamic acid residue (Glu-2) and retains the hydrophobic interactions with the binding groove, both commonly seen in BH3-Mcl-1 interactions (8). In addition, this model positions the conserved rBH3 methionine residue (Met-7) into a hydrophobic cleft at the bottom of the Mcl-1 binding groove that has previously been recognized (8) as an area that is occluded in Bcl-xL and may allow Mcl-1 specificity to be elicited.
With the identification of the rBH3 peptides, we sought to find native proteins that may contain similar sequences that therefore may be identified as potential Mcl-1 binding partners. BLAST analysis of the human genome identified a large group of rBH3 containing proteins with highly divergent sequences (supplemental Table 2). Similar to the BH3 motif, we identified that a conserved acidic residue, most often glutamic acid, and a conserved methionine residue constitute a signature for rBH3 containing proteins (Table 2). Available structural data for several rBH3 containing proteins, e.g. glucokinase, hexokinase 1, hexokinase 2, polypyrimidine tract binding protein 1, MEK5, and CDK4 inhibitor 2C, show that the rBH3 motif is present in helical regions on the surface of each of these proteins.
Interestingly, a number of the identified rBH3 containing proteins have been previously implicated as having significant interactions with Bcl-2 family proteins (Fig. 3a) (29). For instance, members of the p53 family have been shown to bind to both pro- and anti-apoptotic Bcl-2 family proteins and in particular can cause the disruption of the Bak - Mcl-1 complex (30, 31). Additionally, recent evidence has found that Mcl-1 protein levels can be affected significantly by glucose metabolism through an unknown mechanism (14, 32). To quantify the strength of association of Mcl-1 with a number of rBH3-containing proteins, we used the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database of physical and functional interactions (33). The STRING-generated protein-protein interaction network shows very close association of Mcl-1 with a number of the rBH3-containing proteins, often requiring only a single gene/protein node to bridge a direct protein-protein connection (Fig. 3b).
To preliminarily prove that SB-04 is capable to antagonize Mcl-1 in cells, we tested its cell killing ability against a variety of cell lines that are known to depend on Bcl-2 family proteins for survival (15). As controls, we used the Bcl-xL/Bcl-2 antagonist ABT-737 (11), the pan-Bcl-2 family antagonist Sabutoclax (18) and an inactive SB-04 mutant (SB-04M7A) (Table 1). The killing activity of SB-04 parallels well with the reference molecules, whereas the Mcl-1 inactive mutant peptide SB-04M7A is not active under the same experimental conditions (Fig. 4). Given the success at developing stapled and stabilized BH3 helical peptides as therapeutic agents (16, 17), this result provides a promising first step that the SB-04 motif may also yield a therapeutic benefit in developing Mcl-1-specific inhibitors.
We therefore believe that the rBH3 sequence is a new motif, which is present in native proteins and allows these proteins to make significant physiological interactions with Mcl-1. This finding serves as the basis for identifying a new class of Mcl-1 binding proteins and further develops our understanding of how other systems communicate with the regulation of apoptosis in mammalian cellular systems. Furthermore, the recent success in development of potent, cell-permeable stapled peptides (16, 17) could provide an excellent platform for the study of these interactions and for the development of specific Mcl-1 inhibitors.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA149668 (to M. P.) and NCI T32 Fellowship Grant CA121949 (to W. J. P.). This work was also supported by the Department of Chemical Sciences, University of Padova, Italy (to M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- BH
- Bcl-2 homology
- FPA
- fluorescent polarization displacement assay
- rBH3
- reverse BH3
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Reed J. C. (1999) J. Clin. Oncol. 17, 2941–2953 [DOI] [PubMed] [Google Scholar]
- 2. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 4. Liang H., Fesik S. W. (1997) J. Mol. Biol. 274, 291–302 [DOI] [PubMed] [Google Scholar]
- 5. Green D. R., Kroemer G. (2004) Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 6. Day C. L., Smits C., Fan F. C., Lee E. F., Fairlie W. D., Hinds M. G. (2008) J. Mol. Biol. 380, 958–971 [DOI] [PubMed] [Google Scholar]
- 7. Petros A. M., Medek A., Nettesheim D. G., Kim D. H., Yoon H. S., Swift K., Matayoshi E. D., Oltersdorf T., Fesik S. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dutta S., Gullá S., Chen T. S., Fire E., Grant R. A., Keating A. E. (2010) J. Mol. Biol. 398, 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee E. F., Fedorova A., Zobel K., Boyle M. J., Yang H., Perugini M. A., Colman P. M., Huang D. C., Deshayes K., Fairlie W. D. (2009) J. Biol. Chem. 284, 31315–31326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fesik S. W. (2005) Nat. Rev. Cancer 5, 876–885 [DOI] [PubMed] [Google Scholar]
- 11. Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 12. Vogler M., Butterworth M., Majid A., Walewska R. J., Sun X. M., Dyer M. J., Cohen G. M. (2009) Blood 113, 4403–4413 [DOI] [PubMed] [Google Scholar]
- 13. Harley M. E., Allan L. A., Sanderson H. S., Clarke P. R. (2010) EMBO J. 29, 2407–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao Y., Altman B. J., Coloff J. L., Herman C. E., Jacobs S. R., Wieman H. L., Wofford J. A., Dimascio L. N., Ilkayeva O., Kelekar A., Reya T., Rathmell J. C. (2007) Mol. Cell Biol. 27, 4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Placzek W. J., Wei J., Kitada S., Zhai D., Reed J. C., Pellecchia M. (2010) Cell Death Dis. 1, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart M. L., Fire E., Keating A. E., Walensky L. D. (2010) Nat. Chem. Biol. 6, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walensky L. D., Kung A. L., Escher I., Malia T. J., Barbuto S., Wright R. D., Wagner G., Verdine G. L., Korsmeyer S. J. (2004) Science 305, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei J., Stebbins J. L., Kitada S., Dash R., Placzek W., Rega M. F., Wu B., Cellitti J., Zhai D., Yang L., Dahl R., Fisher P. B., Reed J. C., Pellecchia M. (2010) J. Med. Chem. 53, 4166–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Day C. L., Chen L., Richardson S. J., Harrison P. J., Huang D. C., Hinds M. G. (2005) J. Biol. Chem. 280, 4738–4744 [DOI] [PubMed] [Google Scholar]
- 20. Rega M. F., Leone M., Jung D., Cotton N. J., Stebbins J. L., Pellecchia M. (2007) Bioorg. Chem. 35, 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keller R. (2005) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer-aided Resonance Assignment. Doctoral dissertation, Swiss Federal Institute of Technology, Zurich, Switzerland [Google Scholar]
- 22. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 23. Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 24. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 25. Kamata K., Mitsuya M., Nishimura T., Eiki J., Nagata Y. (2004) Structure 12, 429–438 [DOI] [PubMed] [Google Scholar]
- 26. Aleshin A. E., Kirby C., Liu X., Bourenkov G. P., Bartunik H. D., Fromm H. J., Honzatko R. B. (2000) J. Mol. Biol. 296, 1001–1015 [DOI] [PubMed] [Google Scholar]
- 27. Jeffrey P. D., Tong L., Pavletich N. P. (2000) Genes Dev. 14, 3115–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joerger A. C., Rajagopalan S., Natan E., Veprintsev D. B., Robinson C. V., Fersht A. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eguchi T., Itadani H., Shimomura T., Kawanishi N., Hirai H., Kotani H. (2009) Mol. Cancer Ther. 8, 1460–1472 [DOI] [PubMed] [Google Scholar]
- 30. Leu J. I., Dumont P., Hafey M., Murphy M. E., George D. L. (2004) Nat. Cell Biol. 6, 443–450 [DOI] [PubMed] [Google Scholar]
- 31. Sheikh M. S., Fornace A. J., Jr. (2000) J. Cell Physiol. 182, 171–181 [DOI] [PubMed] [Google Scholar]
- 32. Danial N. N., Gramm C. F., Scorrano L., Zhang C. Y., Krauss S., Ranger A. M., Datta S. R., Greenberg M. E., Licklider L. J., Lowell B. B., Gygi S. P., Korsmeyer S. J. (2003) Nature 424, 952–956 [DOI] [PubMed] [Google Scholar]
- 33. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) Nucleic Acids Res. 37, D412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.