Background: Triplet repeat expansions in myotonic dystrophy reduce muscle expression of myotonic dystrophy protein kinase (DMPK).
Results: DMPK localizes to the nuclear envelope, and DMPK depletion disrupts the nuclear envelope.
Conclusion: DMPK is critical to maintain nuclear envelope integrity.
Significance: Reduced DMPK expression in myotonic dystrophy could contribute to nuclear instability, a common mechanism of muscle wasting in muscular dystrophies.
Keywords: Lamin, Muscular Dystrophy, Nuclear Membrane, Nucleus, Skeletal Muscle, Myogenesis, Myotonic Dystrophy, Myotonic Dystrophy Protein Kinase
Abstract
Myotonic dystrophy 1 (DM1) is a multisystemic disease caused by a triplet nucleotide repeat expansion in the 3′ untranslated region of the gene coding for myotonic dystrophy protein kinase (DMPK). DMPK is a nuclear envelope (NE) protein that promotes myogenic gene expression in skeletal myoblasts. Muscular dystrophy research has revealed the NE to be a key determinant of nuclear structure, gene regulation, and muscle function. To investigate the role of DMPK in NE stability, we analyzed DMPK expression in epithelial and myoblast cells. We found that DMPK localizes to the NE and coimmunoprecipitates with Lamin-A/C. Overexpression of DMPK in HeLa cells or C2C12 myoblasts disrupts Lamin-A/C and Lamin-B1 localization and causes nuclear fragmentation. Depletion of DMPK also disrupts NE lamina, showing that DMPK is required for NE stability. Our data demonstrate for the first time that DMPK is a critical component of the NE. These novel findings suggest that reduced DMPK may contribute to NE instability, a common mechanism of skeletal muscle wasting in muscular dystrophies.
Introduction
Myotonic dystrophy 1 (DM1)2 is an autosomal dominant genetic disease leading to a variety of neuromuscular disorders such as myotonia, muscle weakness/wasting, and cardiac conduction defects (1). The genetic basis for DM1 is expansion of an unstable CTG triplet repeat sequence within the 3′ untranslated region of the myotonic dystrophy protein kinase gene (DMPK) (2–4). DMPK is a serine/threonine protein kinase containing coiled-coil, C-terminal membrane association, and autoregulatory domains. DMPK mRNA transcripts containing CUG expansions accumulate as nuclear foci (5), reducing the expression of DMPK in affected skeletal muscle and altering trans mRNA processing (6–8). However, trans RNA processing defects alone do not explain the pathophysiology of DM1 because animal models on the basis of defective mRNA processing do not recapitulate symptoms of the DM1 (9–13). Until the function of DMPK is understood, a role for reduced DMPK in DM1 pathophysiology must be considered.
Previous muscular dystrophy research has revealed the nuclear envelope (NE) to be a key determinant of nuclear structure, gene regulation, and muscle function. The nuclear lamina is a nuclear meshwork composed of A- and B-type lamins. Lamins interact with inner NE proteins and chromatin. A subset of lamins is found throughout the nucleoplasm in distinct nuclear structures termed speckles. A-type lamins, Lamin-A, and Lamin-C are encoded from a single gene, Lmna (14). B-type lamins, Lamin-B1, and Lamin-B2 are encoded by two separate genes (15). The critical NE proteins Emerin, Nesprin-1, and Nesprin-2 form a network linking the inner NE to the cytoskeleton (16). NE defects have been linked to muscle wasting and weakening. Mutations in inner NE proteins are the genetic cause of Emery-Dreifuss muscular dystrophy (EDMD) and limb girdle muscular dystrophy 1b (LGMD1B, reviewed in Ref. 17). Mutations in Emerin are attributed to the X-linked forms of EDMD, whereas mutations in Lamin-A/C and Nesprin are linked to autosomal dominant forms of EDMD (16). Lamin-A/C mutations are also linked to LGMD1B (18). Thus, nuclear envelope defects may represent a common mechanism of muscle wasting in muscular dystrophies (17).
We have reported previously that DMPK is required for C2C12 myoblast differentiation into myotubes (19). Depletion of DMPK specifically inhibits myogenin expression. Other laboratories have demonstrated that muscle differentiation is associated with structural changes in intranuclear Lamin-A/C (20–22). During C2C12 myoblast differentiation into myotubes, intranuclear Lamin-A/C undergoes reorganization and intranuclear Lamin-A/C speckles change conformation. Disruption of Lamin-A/C organization inhibits differentiation, specifically reducing expression of myogenin (21). Expression of a Lamin-A/C mutation linked to EMDM in C2C12 cells halted myotube formation (20). Thus, reorganization of intranuclear lamins is a key aspect of muscle differentiation. We hypothesized that DMPK and Lamin-A/C may cooperatively regulate myogenic differentiation.
Although the physiologic function of DMPK is unknown, decreased DMPK expression is apparent in DM1 (23–28). DMPK RNA transcripts containing CUG expansions accumulate as nuclear foci (5), reducing the expression of DMPK in affected skeletal muscle. DMPK mRNA transcripts with expanded CUG repeats sequester mRNA transcripts containing CAG repeats and essential RNA binding proteins such as CUG triplet repeat RNA binding protein 1 (CUG-BP1) and the human muscleblind protein homolog MBNL and mediate the alternative splicing of numerous genes (6–8). However, RNA processing defects cannot explain the complete pathophysiology of DM1. Animal models on the basis of altered expression of CUG-BP1, MBNL, or expanded triplet repeats do not exhibit many hallmark symptoms of the disease (9–13).
A reduction in DMPK mRNA abundance has been observed in skeletal muscle of DM1 patients (23, 26, 28). RNA studies have demonstrated that in heterozygous DM1 individuals with one normal and one expanded allele, both DMPK alleles were transcribed into pre-mRNA. However, when mature poly(A) mRNA was examined, DMPK mRNA abundance was only 10–20% of normal (27). The most severe form of myotonic dystrophy, congenital DM1, is associated with numerous developmental defects, including delayed muscle development. Cultured myoblasts from congenital DM1 fetuses have a reduced capacity to fuse and differentiate and produce less than 50% of the normal levels of DMPK protein (24, 25), suggesting that DMPK might have a specific role in embryonic myocyte development. The comparison of DM1 and DM2 (an expansion of repeats in the zinc finger protein 9 gene (29)) further suggests a key role for DMPK activity. DM1 and DM2 are caused by expanded nucleotide repeats in two different genes (30). Both diseases exhibit RNA processing defects that account for a majority of DM symptoms (6, 12, 13, 31). However, only DM1 manifests as a congenital disease (32), suggesting a specific role for DMPK in embryonic myogenesis.
To investigate the role of DMPK in NE stability, we analyzed DMPK in HeLa cells. HeLa cells were chosen as a standard and easily manipulated model system. We found that DMPK localizes to the NE and coimmunoprecipitates with Lamin-A/C. Overexpression of DMPK in HeLa cells disrupts Lamin-A/C and Lamin-B1 localization and causes nuclear fragmentation. Depletion of DMPK also disrupts NE lamina. We followed these studies with examination of the NE in mouse myoblasts after DMPK over- or underexpression. We found that tight regulation of DMPK levels is necessary to maintain NE stability in C2C12 myoblasts. Our data demonstrate that DMPK is a critical component of the NE and that reduced DMPK may contribute to skeletal muscle wasting in DM1.
EXPERIMENTAL PROCEDURES
Cell Culturing and Transfection
HeLa and C2C12 cells were purchased from the ATCC and cultured in DMEM (ATCC) supplemented with 10% FBS and penicillin/streptomycin (100 units/ml) in 5% CO2 at 37 °C. For microscopic visualization, cells were grown on glass coverslips coated with 0.1% gelatin. HeLa cells were transfected with FuGENE6 (Roche) at a 3:1 ratio (FuGENE6:DNA), and C2C12 cells were transfected with FuGENE HD (Roche) at a 4:2 ratio. Transfection constructs DMPKΔMA-GFP, FL-DMPK-GFP, and FL-DMPK-mCherry were created by cloning DMPKΔMA or FL-DMPK (33) into pEGFP-C2 or pCherry (BD Biosciences) at EcoR1 restriction sites. pEGFP-C2 was transfected in parallel as a control of GFP-tagged DMPK constructs. We introduced a point mutation in the ATP binding site of DMPK (K100R) to create an enzymatically inactive form of the kinase, DMPKΔMA-100R-GFP (7) using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with forward primer sequence 5′-AT GCC ATG AGG ATC ATG AAC-3′. Short hairpin RNA plasmids (shControl, shDMPK) were purchased from SuperArray (Frederick, MD).
Immunofluorescent Imaging
Before staining, cells were fixed in 4% paraformaldehyde for 5 min and permeabilized in methanol for 5 min. Cells were blocked for 1 h in 10% horse serum, 1% BSA, and 0.02% sodium azide in PBS before incubation with antibodies in 3.0% BSA in PBS: rabbit anti-β-tubulin (1:100, NeoMarkers, Fremont, CA), rabbit anti-Lamin-A/C (1:100, Cell Signaling Technology, Inc., Danvers, MA), rabbit anti-Lamin-B1 (1:100, Abcam, Cambridge, MA), mouse anti-Emerin (1:40, Novocastra, Buffalo Grove, IL), and mouse anti-DMPK (1:100). Secondary anti-rabbit Texas Red or Alexa Fluor 488 antibodies (Invitrogen) were diluted 1:100. Cells were incubated with DAPI (0.5 μg/ml bisBenzimide; Sigma) for 5 min before mounting in Fluoromount (Electron Microscopy Sciences, Hatfield, PA). All fluorescent images were obtained on an Olympus FV1000 confocal microscope with an Olympus ×60 PlanApo ×60/1.40 oil immersion objective using Olympus FV10-ASW (version 2.1) acquisition software. To quantify events documented by confocal imaging, total cells and any cells containing at least one disruption in nuclear envelope continuity were counted from 8 to 10 high-power (×60) microscopic fields for each experiment. Each experiment was replicated three to six times (n), as noted. Statistics are represented as mean ± S.E. Statistical significance was determined by paired Student's t test and reported as a p value.
Real-time PCR
Real-time PCR was conducted on an ABI 7500 (Applied Biosystems, Inc., Carlsbad CA). RNA was extracted using the RNeasy kit (Qiagen) and reverse-transcribed to cDNA (Taqman reverse transcription kit, Applied Biosystems, Inc.). Real-time PCR was performed using probe primer sets Mm00446261_m (mouse DMPK) and Hs01094324_g1 (human DMPK) from Applied Biosystems, Inc. Mouse or human GAPDH was used as an internal control.
Coimmunoprecipitation and Western Blotting
HeLa cells were transfected and harvested in radioimmune precipitation assay buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 1% Triton-X, 0.5% deoxycholate, 0.1% SDS, and complete protease inhibitor mixture (Roche)). Protein concentration was assayed using the DC Lowry assay kit (Bio-Rad). For coimmunoprecipitation, 500 μg of protein was aliquoted, and 5 μg of antibodies against GFP (Santa Cruz Biotechnology, Inc.), DMPK, Lamin-B1 (Abcam), or Lamin A/C (Novocastra) was added. Samples were rocked overnight at 4 °C. One ml of protein G beads (Amersham Biosciences) was centrifuged at 8000 × g for 5 min. The supernatant was discarded, and beads were washed four times with 500 μl of PBS. Beads were resuspended in 500 μl of PBS, and 20 μl of washed beads was added to the lysate/antibody mixture and allowed to rock at 4 °C for 2 h to overnight. The mixture was spun at 8000 × g for 5 min, and the supernatant was removed. The remaining beads were washed twice with 500 μl of radioimmune precipitation assay buffer, twice with 500 μl of 1 m NaCl/ radioimmune precipitation assay buffer, and once with 500 μl of PBS. The beads were then resuspended in LDS sample buffer (Invitrogen) and boiled for 5 min. The supernatant was analyzed by Western blotting using a standard protocol (19). Blots were probed with 1:4000 anti-DMPK, 1:1000 anti-Lamin-B1(Abcam), or 1:1000 anti-Lamin-A/C (Cell Signaling Technology, Inc.). TrueBlot anti-mouse or anti-rabbit secondary antibody was used at a 1:1000 dilution (eBioscience, Inc., San Diego, CA). To confirm DMPK knockdown, transfected cells were sorted on a BD Biosciences FACSVantage SE flow cytometry system. Sorted GFP-positive cells were lysed in radioimmune precipitation assay buffer and analyzed by Western blot analysis using the Western Breeze kit (Invitrogen). Blots were probed with mouse monoclonal antibodies against DMPK (1:4000) and GAPDH (1:8000, Ambion, Austin, TX). Blots were visualized using the Western Breeze detection system (Invitrogen). Chemiluminescent images were captured and quantified using a VersaDoc 3000 (Bio-Rad).
RESULTS
DMPK Localizes to the Nuclear Envelope
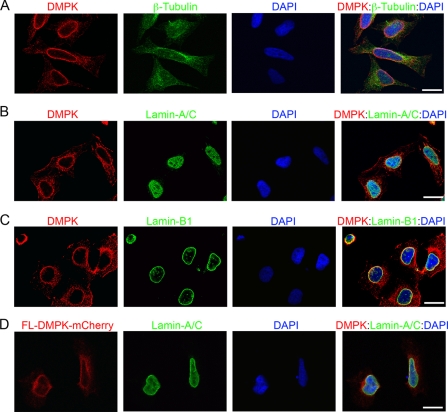
To elucidate the cellular role of DMPK, we analyzed the subcellular localization of DMPK in HeLa cells. We have produced a highly specific and sensitive monoclonal antibody against the coiled-coil region of DMPK. The precise specificity of this monoclonal antibody was validated previously (19, 34). Immunofluorescent imaging revealed specific localization of DMPK at the NE in interphase HeLa cells (Fig. 1A). To confirm NE localization, we colabeled HeLa cells with DMPK and a polyclonal Lamin-A/C antibody. The Lamin-A/C antibody labels NE and intranuclear Lamin-A/C proteins. Colabeling revealed DMPK localization to the Lamin-A/C proteins at the NE (Fig. 1B). We colabeled HeLa cells with DMPK and the NE protein Lamin-B1 (Fig. 1C). We observed specific colocalization of DMPK with Lamin-B1 protein at the NE.
FIGURE 1.
DMPK localizes to the nuclear envelope. A–C, immunofluorescent labeling of DMPK (red) confirmed localization to the NE in HeLa cells. Scale bar = 10 μm in all panels. Staining of β-tubulin (A, green) showed cell size and shape. Colocalization with Lamin-A/C (B, green) and Lamin-B1 (C, green) confirmed DMPK at the NE. D, FL-DMPK-mCherry in HeLa cells (16-h transfection, red) colocalizes with Lamin-A/C (green). DAPI (blue) stained the nucleus in all overlay panels.
To confirm the colocalization of DMPK and lamin proteins at the NE, we directly visualized subcellular localization of fluorescent-tagged, full-length DMPK (FL-DMPK-mCherry) in HeLa cells. After a short transfection (16 h), we observed tagged DMPK colocalization with Lamin-A/C at the NE (Fig. 1D). Similar localization was observed after a short transfection with FL-DMPK-GFP (data not shown). These analyses revealed specific localization of DMPK at the NE in interphase HeLa cells. NE localization of DMPK was reported previously in C2C12 mouse myoblasts and neonatal rat cardiac myocytes (19, 35).
DMPK Overexpression Causes Nuclear Fragmentation
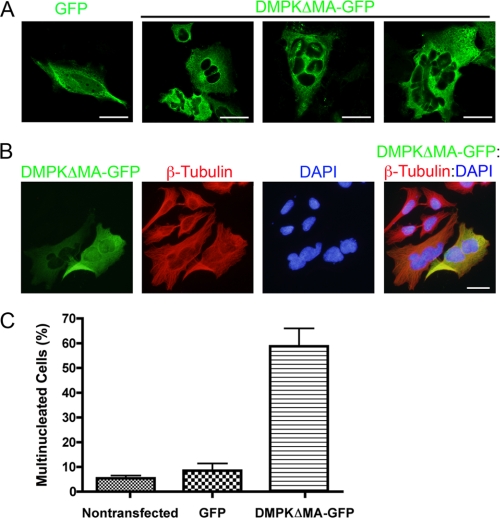
We investigated the role of DMPK in human epithelial cells by overexpressing DMPK in HeLa cells. HeLa cells were transfected with a GFP-tagged DMPK lacking C-terminal membrane association and autoinhibitory domains (DMPKΔMA-GFP). DMPKΔMA corresponds to endogenous DMPK2 and possesses increased enzymatic activity as compared with full-length DMPK (33). We examined HeLa cells expressing either DMPKΔMA-GFP or GFP alone by confocal microscopy. We found that overexpression of DMPKΔMA-GFP in HeLa cells led to nuclear fragmentation (Fig. 2). Nuclear fragmentation resulted in the appearance of multiple micronuclei in the cell. GFP-expressing cells were primarily mononucleated, whereas the majority of the DMPKΔMA-GFP-expressing cells contained multiple nuclei (Fig. 2A).
FIGURE 2.
DMPK over-expression (48 h) causes nuclear fragmentation. A, overexpression of GFP (green, first panel) and DMPKΔMA-GFP (green, second through fourth panels) was compared by confocal microscopic analysis. Scale bars = 10 μm. B, overexpression of DMPKΔMA-GFP (green). β-tubulin (red) is shown together with DAPI DNA staining (blue) by standard fluorescent microscopic analysis. Scale bar = 10 μm. C, multinucleated cells were counted 48 h after transfection. Multinucleation was observed in 5.5% (non-transfected), 7% (GFP), and 58% (DMPKΔMA-GFP) of cells (n = 3 replications).
DNA staining with DAPI confirmed nuclear fragmentation in transfected cells (Fig. 2B). Cells expressing either high or low levels of GFP-tagged DMPK fluorescence were multinucleated, demonstrating the significant effects of minimal DMPK overexpression. Cell counting revealed that a majority of cells transfected with DMPKΔMA-GFP were multinucleated cells as compared with untransfected control cells that remained mononucleated (Fig. 2C). We used transiently transfected HeLa cells to demonstrate that approximately 58% of the cells transfected with the DMPKΔMA-GFP construct contained a fragmented nucleus by 48 h post-transfection. In contrast, multinucleation was observed in only 5.5 and 7% of the cells treated with transfection reagent alone or transfected with GFP, respectively (n = 3 replications). In summary, overexpression of DMPK led to nuclear fragmentation and the formation of cells containing multiple micronuclei.
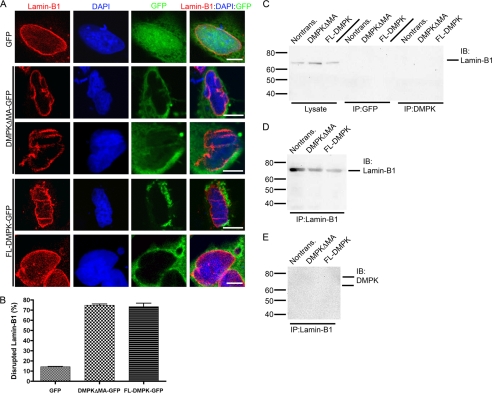
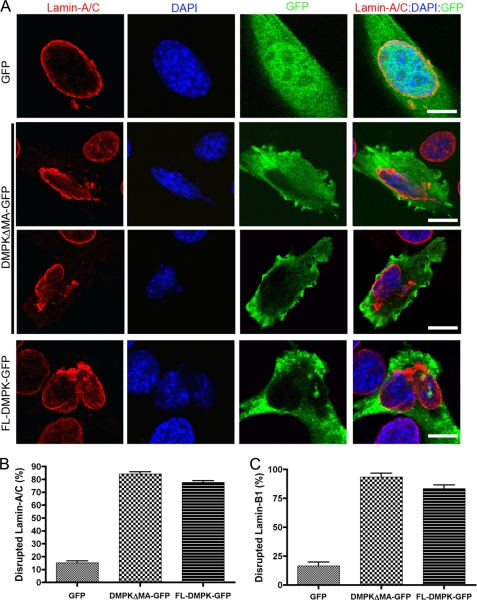
DMPK Overexpression Disrupts Lamin-A/C
Disruptions in NE proteins, including Lamin-A/C, have been linked previously to fragmentation of the nucleus (16, 17). We hypothesized that the nuclear defects caused by DMPK overexpression are linked to Lamin-A/C disruption. We therefore analyzed Lamin-A/C by immunofluorescent imaging in HeLa cells after a 24-h transfection with GFP, DMPKΔMA-GFP, or full-length (FL) DMPK-GFP (Fig. 3A). In cells transfected with GFP, Lamin-A/C localized to the NE. However, transfection with DMPKΔMA-GFP or FL-DMPK-GFP disrupts the normal localization of Lamin-A/C. Transfection of either DMPK construct causes a loss of Lamin-A/C in distinct segments around the nucleus. In addition, DMPK overexpression causes a distinctive honeycombing localization of Lamin-A/C into the nucleus itself. Transfection with GFP disrupts the NE localization of Lamin-A/C in only 13.6% ± 2.2% (n = 5) of cells (Fig. 3B). Transfection with DMPKΔMA-GFP or FL-DMPK-GFP disrupted Lamin-A/C in 78.3% ± 1.6% (n = 5, p < 0.01) and 68.0% ± 2.8% (n = 5, p < 0.01) of cells, respectively.
FIGURE 3.
DMPK over-expression (24 h) disrupts Lamin-A/C. A, 24 h transfection with DMPKΔMA-GFP or FL-DMPK-GFP disrupts localization of Lamin-A/C (red). Control transfection with GFP reveals normal localization at the NE. Staining with DAPI (blue) shows the nuclear shape. Transfected GFP-tagged constructs are shown in green. Scale bars = 5 μm. B, cell counting confirmed Lamin-A/C disruption in a significant number of nuclei after DMPK overexpression (n = 5). C–E, immunoprecipitations (IP) and immunoblotting (IB) of HeLa cell lysates revealed an interaction between DMPK and Lamin-A/C. Demarcations along the top of the blots indicate transfected plasmid. C, non-IP lysate detected Lamin-A/C at the expected molecular weight. No changes in Lamin-A/C expression were observed after DMPK transfection. Lamin-A/C immunoprecipitated with DMPK. IP with GFP served as a negative control. D, anti-Lamin-A/C antibody immunoprecipitates Lamin-A/C itself. E, conversely, DMPK immunoprecipitates with Lamin-A/C.
It was unknown whether the effects of DMPK overexpression were caused by increased kinase activity or increased expression of the protein itself. A point mutation in the ATP binding domain of DMPK (K100R) abolishes all enzymatic activity (7). We transfected HeLa cells with GFP-tagged kinase-inactive DMPK (DMPKΔMA-100R-GFP), active DMPK (DMPKΔMA-GFP), or GFP. Identical to DMPKΔMA-GFP, DMPKΔMA-100R-GFP disrupted Lamin-A/C at the NE (supplemental Fig. 1). Both constructs induced a loss of Lamin-A/C in segments around the nucleus and a honeycombing of Lamin-A/C into the nucleus. These results showed that kinase activity is not required for Lamin-A/C disruption. Overexpression of the protein itself is sufficient to disrupt the NE.
Our immunolocalization demonstrated colocalization of both DMPK and Lamin-A/C at the NE. To determine whether these two proteins interact, we tested whether DMPK and Lamin-A/C coimmunoprecipitated together. Probing with an antibody against Lamin-A/C revealed an expected double band in non-transfected HeLa cell lysate (Fig. 3C). The amount of Lamin-A/C did not change after a 24 h transfection with DMPKΔMA or FL-DMPK (non-tagged). Lamin-A/C coimmunoprecipitated with our monoclonal anti-DMPK antibody in transfected and non-transfected HeLa cells. However, Lamin-A/C did not coimmunoprecipitate with a negative control anti-GFP antibody. Using a monoclonal antibody against Lamin-A/C, we immunoprecipitated Lamin-A/C from non-transfected or DMPK-overexpressing HeLa cells (Fig. 3D). Finally, we were able to confirm that DMPK coimmunoprecipitates with Lamin-A/C (Fig. 3E). These results confirmed a coassociation of DMPK and Lamin-A/C in the same protein complex.
DMPK Overexpression Disrupts Lamin-B1
Our data suggested that DMPK overexpression caused instability in the nuclear envelope. To confirm these findings, we analyzed the localization of another nuclear envelope protein, Lamin-B1. In cells transfected with GFP, Lamin-B1 localized specifically to the NE (Fig. 4A). Transfection with DMPKΔMA-GFP or FL-DMPK-GFP induced a radical disruption in Lamin-B1 localization. Similar to Lamin-A/C, Lamin-B1 localization became intermittent and honeycombed throughout the nucleus. Only 14.3% ± 0.3% (n = 3) of HeLa nuclei had disruptions in Lamin-B1 after GFP transfection (Fig. 4B). However, transfection with DMPKΔMA-GFP or FL-DMPK-GFP caused Lamin-B1 disruption in 74.8% ± 1.3% (n = 3, p < 0.01) and 73.4% ± 3.4% (n = 3, p < 0.01) of nuclei, respectively.
FIGURE 4.
DMPK overexpression (24 h) disrupts Lamin-B1. A, 24 h transfection with DMPKΔMA-GFP or FL-DMPK-GFP disrupts localization of Lamin-B1 (red). Control transfection with GFP reveals normal localization at the NE. Staining with DAPI (blue) shows the nuclear shape. Transfected GFP-tagged constructs are shown in green. Scale bars = 5 μm. B, cell counting confirmed Lamin-B1 disruption in a significant number of nuclei after DMPK overexpression (n = 3). C–E, immunoprecipitations (IP) and immunoblotting (IB) of HeLa cell lysates suggests no direct interaction between DMPK and Lamin-B1. Demarcations along the top of the blots indicate transfected plasmid. C, non-IP lysate detected Lamin-B1 at the expected molecular weight. Lamin-B1 expression and stability do not change after DMPK overexpression. Lamin-B1 does not immunoprecipitate with DMPK. IP with GFP served as a negative control. D, anti-Lamin-B1 antibody immunoprecipitates Lamin-B1 itself. E, conversely, DMPK does not immunoprecipitate with Lamin-B1.
DMPK enzymatic activity is not required for the disruption of Lamin-B1 at the NE. Expression of kinase inactive DMPK (DMPKΔMA-100R-GFP) mimics the effects of active DMPK (DMPKΔMA-GFP) in HeLa cells (supplemental Fig. 2). Therefore, the NE instability caused by DMPK overexpression is not due to increased kinase activity.
To determine whether DMPK directly interacts with Lamin-B1, we performed coimmunoprecipitation analyses of HeLa cell lysates. Western blotting of non-transfected or DMPK-transfected HeLa cells revealed no change in Lamin-B1 expression levels (Fig. 4C). Unlike Lamin-A/C, Lamin-B1 did not coimmunoprecipitate with DMPK. Furthermore, DMPK did not coimmunoprecipitate with Lamin-B1 (Fig. 4E). However, we confirmed immunoprecipitation of Lamin-B1 itself with anti-Lamin-B1 antibody (Fig. 4D). Therefore, DMPK overexpression disrupts Lamin-B1 localization, yet DMPK does not directly interact with Lamin-B1 in HeLa cells.
To confirm the disruption of the Lamin network after DMPK overexpression, we analyzed the localization of Emerin, a lamin-associated component of the NE (36, 37). Transfection with DMPKΔMA-GFP or FL-DMPK-GFP caused a disruption in Emerin at the NE (supplemental Fig. 3). Similar to Lamin-A/C and Lamin-B1, Emerin became intermittent at the NE and honeycombed in the nucleus. Therefore, DMPK overexpression disrupts the NE Lamin network, affecting both lamins and Emerin.
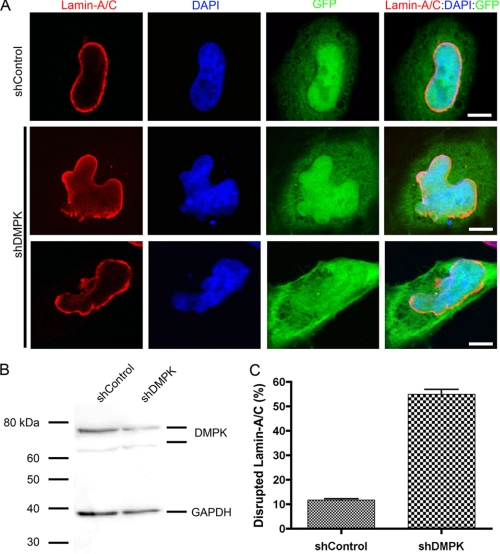
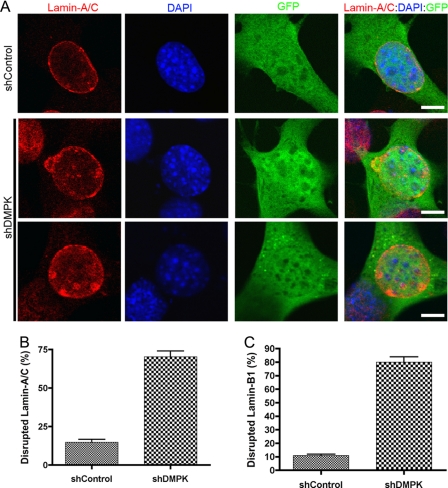
DMPK Depletion Disrupts Lamin-A/C
We have shown that overexpression of DMPK is sufficient to disrupt the NE. To determine whether DMPK is necessary for NE stability, we knocked down DMPK in HeLa cells using a shRNA against DMPK (shDMPK). Within 48 h of shDMPK transfection, DMPK protein levels were decreased to 52.9% ± 8.0% (n = 6) of control (shControl-transfected) HeLa cell protein levels (Fig. 5B). The knockdown of DMPK after shDMPK transfection (48 h) was confirmed by real-time PCR analysis. After shDMPK transfection, DMPK mRNA transcripts were decreased to 60.8% ± 5.6% (n = 6) compared with shControl transfected cells. An IRES-GFP construct in the shRNA plasmids allowed identification of transfected cells. Transfection with a control shRNA (shControl) did not affect Lamin-A/C localization at the NE (Fig. 5A). Only 11.7% ± 0.5% (n = 4) of cells transfected with shControl showed Lamin-A/C disruption (Fig. 5C). However, a depletion of DMPK disrupted normal Lamin-A/C localization (Fig. 5A). When DMPK levels were reduced, Lamin-A/C became intermittent around the nucleus. These disruptions were observed in 55.0% ± 2.0% (n = 4, p < 0.01) of transfected cells (Fig. 5C). The disruptions in Lamin-A/C localization to the NE led to blebbing of the nucleus. Staining DNA with DAPI revealed the blebbing of nuclear material out of the NE. However, DMPK knockdown did not alter Lamin-A/C expression or stability (supplemental Fig. 4). Thus, DMPK is necessary to maintain Lamin-A/C localization and preserve nuclear structure.
FIGURE 5.
DMPK depletion disrupts Lamin-A/C. A, knockdown of DMPK with shDMPK disrupts Lamin-A/C at the NE. This disruption leads to abnormal nuclear shape and blebbing. Lamin-A/C is lost and aggregates at the NE after DMPK knockdown. Scale bars = 5 μm. B, knockdown of DMPK in FACS-sorted HeLa cells by shDMPK was confirmed by Western blotting. The GAPDH protein band demonstrates equal loading (n = 4). C, cell counting confirmed Lamin-A/C disruption in a significant number of nuclei after DMPK depletion.
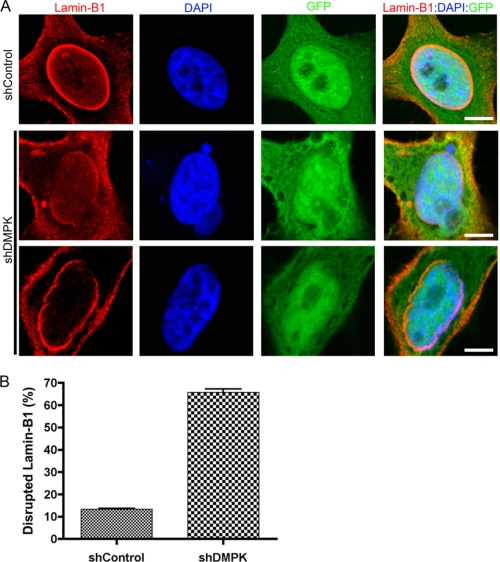
DMPK Depletion Disrupts Lamin-B1
Lamin-A/C localization revealed the necessity of DMPK for maintaining nuclear envelope integrity. To determine whether DMPK is likewise necessary for Lamin-B1 localization, we analyzed Lamin-B1 immunostaining after DMPK depletion. Transfection with shControl did not alter the normal localization of Lamin-B1 at the NE (Fig. 6A). However, DMPK knockdown disrupted this NE pattern. When DMPK levels were reduced, Lamin-B1 was lacking from portions of the NE. Reduction in Lamin-B1 led to obvious nuclear blebbing, revealing a loss of NE integrity. Cell counts revealed Lamin-B1 disruptions in 65.7% ± 1.6% (n = 3) of HeLa cells after shDMPK transfection, whereas shControl transfection only disrupted Lamin-B1 in 13.4% ± 0.3% (n = 3, p < 0.01) of cells. The depletion of DMPK did not affect the cellular levels of Lamin-B1 protein, showing that Lamin-B1 expression and stability were unchanged (supplemental Fig. 4). Therefore, DMPK is necessary for the perinuclear localization of both Lamin-A/C and Lamin-B1.
FIGURE 6.
DMPK depletion disrupts Lamin-B1. A, knockdown of DMPK with shDMPK disrupts Lamin-B1 at the NE. This disruption leads to abnormal nuclear shape and blebbing. Scale bars = 5 μm. B, cell counting confirmed Lamin-B1 disruption in a significant number of nuclei after DMPK depletion (n = 3).
Furthermore, DMPK is also required to maintain the localization of Emerin, a Lamin-associated protein, around the nucleus. Depletion of DMPK in HeLa cells caused the disruption of Emerin, inducing protein aggregation and loss at the NE (supplemental Fig. 5). Therefore, DMPK is required to maintain NE integrity.
DMPK Overexpression Disrupts Lamins in Cultured Myoblasts
Our experiments in HeLa epithelial cells suggested that tight regulation of DMPK levels is required to maintain nuclear envelope integrity. Immunohistochemical staining detected DMPK expression in developing embryonic muscle cells, implying that DMPK may have a role in regulation of myocyte differentiation (19). To investigate the role of DMPK in myocyte development, we manipulated DMPK levels inC2C12 mouse myoblasts. We examined cultured mouse myoblast (C2C12) cells expressing DMPKΔMA-GFP, FL-DMPK-GFP, or GFP alone by confocal microscopy. Similar to observations in HeLa cells, overexpression of DMPK was sufficient to disrupt the nuclear envelope in C2C12 cells. Transfection with either DMPKΔMA-GFP or FL-DMPK-GFP induced loss of Lamin-A/C around portions of the nucleus and aggregation of Lamin-A/C in foci around the nucleus (Fig. 7A). We observed disrupted Lamin-A/C around the nucleus in only 15.3% ± 1.4% (n = 3) of GFP-transfected cells. Transfection with DMPK-MA-GFP disrupted Lamin-A/C pattern in 84.2% ± 1.7% (n = 3, p < 0.01) of cells. Likewise, transfection with FL-DMPK-GFP induced Lamin-A/C disruptions in 77.6% ± 1.4% (n = 3, p < 0.01) of cells.
FIGURE 7.
DMPK overexpression disrupts myoblast Lamins. A, in C2C12 myoblasts, a 24-h transfection with DMPKΔMA-GFP or FL-DMPK-GFP disrupts localization of Lamin-A/C (red). Control transfection with GFP reveals normal localization at the NE. Staining with DAPI (blue) shows the nuclear shape. Transfected GFP-tagged constructs are shown in green. Scale bars = 5 μm. B, cell counting confirmed Lamin-A/C disruption in a significant number of nuclei after DMPK overexpression (n = 3). C, cell counting confirmed Lamin-B1 disruption in a significant number of nuclei after DMPK overexpression (n = 3).
Similarly, elevated levels of DMPK disrupted Lamin-B1 in C2C12 cells (data not shown and Fig. 7C). Transfection with DMPKΔMA-GFP (24 h) disrupted nuclear envelope Lamin-B1 in 93.3% ± 3.3% (n = 3) of cells, and transfection with FL-DMPK-GFP disrupted Lamin-B1 in 83.3% ± 3.2% (n = 3). However, transfection with GFP alone only disrupted Lamin-B1 in 16.7% ± 3.2% (n = 3, p < 0.01). Similar to observations in HeLa cells, DMPK is sufficient to disrupt the NE in myoblasts.
DMPK Depletion Disrupts Lamins in Cultured Myoblasts
Reduced DMPK mRNA transcripts have been recorded in the skeletal muscle of congenital and adult-onset DM1 (see above). Our immunohistochemical staining detected DMPK expression in developing embryonic muscle cells (19). To observe whether DMPK was necessary for myoblast differentiation, we conducted RNAi-mediated knockdown experiments in C2C12 cells. After a 48-h transfection with a shRNA plasmid against DMPK (shDMPK), we observed a reduction of DMPK protein levels in FACS-sorted GFP-positive cells to 55.3% ± 10.5% (n = 3) of control (shControl-transfected) cell levels. This depletion is similar to our previous findings that transfection of C2C12 cells with a shRNA plasmid against DMPK could reduce protein levels to 63.3% ± 5.5% (n = 4) of control cell levels (19). We confirmed these results by real-time PCR and found a comparable DMPK transcript depletion in FACS-sorted GFP-positive cells to 72.7% ± 3.6% (n = 4) of shControl-transfected cells. Depleting proliferating C2C12 myoblasts of DMPK caused disruptions in Lamin-A/C (Fig. 8A). Similar to observations in HeLa cells, DMPK knockdown led to obvious disruptions in the normal NE localization of Lamin-A/C. Again, these disruptions were accompanied by nuclear blebbing. Lamin-A/C was disrupted at the NE in 70.4% ± 3.7% (n = 3) of shDMPK-transfected cells. However, Lamin-A/C was disrupted in only 14.8% ± 1.8% (n = 3, p < 0.01) of shControl-transfected cells (Fig. 8B).
FIGURE 8.
DMPK depletion disrupts myoblast Lamins. A, in C2C12 myoblasts, a 24-h transfection with DMPKΔMA-GFP or FL-DMPK-GFP disrupts localization of Lamin-B1 (red). Control transfection with GFP reveals normal localization at the NE. Staining with DAPI (blue) shows the nuclear shape. Transfected GFP-tagged constructs are shown in green. Scale bars = 5 μm. B, cell counting confirmed Lamin-A/C disruption in a significant number of nuclei after DMPK depletion (n = 4). C, cell counting confirmed Lamin-B1 disruption in a significant number of nuclei after DMPK depletion (n = 4).
We observed similar disruptions in Lamin-B1 localization after DMPK depletion in C2C12 cells (data not shown and Fig. 8C). Knockdown caused a loss of Lamin-B1 around portions of the nucleus, aggregation of Lamin-B1 at the NE, and nuclear blebbing. Transfection with a control plasmid induced Lamin-B1 disruption in 10.9% ± 1.0% (n = 3) of C2C12 cells. Transfection with a DMPK shRNA plasmid induced Lamin-B1 in 80.1% ± 3.8% (n = 3, p < 0.01) of cells. Therefore, DMPK is necessary for the maintenance of nuclear lamina in proliferating myoblasts.
DISCUSSION
Our studies have identified a novel role for DMPK in maintenance of NE stability. We report that DMPK localizes to the NE and coimmunoprecipitates with Lamin-A/C. Overexpression of DMPK in HeLa epithelial cells or C2C12 myoblasts disrupts Lamin-A/C and Lamin-B1 localization and causes nuclear fragmentation. Depletion of DMPK also disrupts NE lamina. Our data demonstrate that DMPK is critical to maintain NE stability. Because NE integrity is necessary for myocyte stability and function, these novel findings suggest that reduced DMPK could contribute to skeletal muscle wasting in DM1.
Our experiments suggest that tight regulation of DMPK levels is required to maintain NE integrity. A similar effect, nuclear lamina disruption, was observed after DMPK overexpression or depletion. Experiments in animal models have also shown the requirement for tight regulation of DMPK levels in mouse muscle. Interestingly, a similar muscle phenotype was observed in both transgenic DMPK overexpression and DMPK knockout models (for a review, see Ref. 38). Aged DMPK knockout mice develop a mild progressive myopathy, sharing minor pathological similarities with myotonic dystrophy. Transgenic mice expressing DMPK under the control of the M creatine kinase promoter and enhancer exhibit minor skeletal muscle defects similar to those observed in DMPK knockout animals (39). Therefore, our in vitro results are consistent and in agreement with in vivo studies suggesting that slight increases or decreases in DMPK levels can have similar effects in muscle cells.
Our data demonstrates that DMPK localizes to the NE and that any alteration in DMPK levels can cause NE disruption. However, the precise mechanism by which DMPK maintains nuclear stability is still unknown. Here we report that DMPK kinase activity is not required for the effects of DMPK overexpression. Therefore, we suggest that DMPK is a structural component of the NE that complexes with Lamin-A/C. It is likely that any change in the stoichiometric ratio of DMPK to another NE protein induces a breakdown of the lamin network. This weakening of NE architecture leads to nuclear fragmentation and the appearance of multiple micronuclei. Previous work has demonstrated that NE disruption is sufficient to cause nuclear fragmentation and the formation of micro-nuclei in the absence of cell division (16, 18), but the possibility remains that the multinucleation observed after DMPK overexpression is caused by abnormal cell division or cell fusion.
We have described DMPK complex formation with Lamin-A/C and localization at the NE. However, a portion of DMPK also localizes throughout the cytoplasm of epithelial cells and myoblasts (as seen by antibody or tagged DMPK localization). In addition to its role at the NE, it is likely that DMPK also has cellular roles in the cytoplasm. These cytoplasmic functions of DMPK could also contribute to NE stability or serve additional, unrelated roles in cellular biology.
Nuclear envelope defects represent a common mechanism of skeletal muscle wasting in muscular dystrophies. Mutations in inner NE proteins were previously found to be the genetic cause of EDMD and LGMD1B (reviewed in Ref. 17). Here, we report that DMPK is a novel component of the NE. Depleting or overexpressing DMPK levels in HeLa cells leads to disrupted Lamin localization and causes nuclear instability. Thus, manipulation of DMPK disrupts the localization of lamins, which are necessary for nuclear stability and the maintenance of myocyte structure and function. This novel function for DMPK possibly links the pathology muscle wasting in DM1 to laminopathies, including EDMD and LGMD1B. Previous research on DM1 has focused on abnormal RNA processing caused by expanded DMPK triplet repeats. These studies have ignored the well established depletion of DMPK in DM1 pathology. The data presented here supports the ongoing analysis of DMPK function in DM1 pathology and treatment.
Expression of DMPK, Emerin, Lamin-A/C, and Nesprin is widespread. However, mutations in these genes are specifically linked to muscle disease. There are two prevailing models to explain why cardiac and skeletal muscles are disproportionately affected by nuclear envelope defects: mechanical stress and gene expression. The first model suggests that muscle cell nuclei with defective Emerin, Lamin-A/C, or Nesprin are sensitive to mechanical damage caused by contraction. Depletion of or point mutations in nuclear envelope proteins cause structural abnormalities in the nucleus (40). These abnormalities are observed as changes in nuclear shape, nuclear blebbing, or complete nuclear fragmentation. Mechanotransduction analyses of isolated nuclei from Lmna-null mice have directly shown structural weakness. Nuclei lacking Lamin-A/C are prone to deformation after direct mechanical stress (41, 42). Because muscle actin filaments link directly to nuclear envelope Nesprin and Sun proteins (16), mechanical forces caused by muscle contraction would immediately stress the nuclear envelope. Subsequent mechanical stress to the nucleus would be detrimental to a weakened nuclear envelope.
Another model linking nuclear defects with muscle tissue is made on the basis of changes in gene expression. This model proposes that changes in nuclear envelope organization lead to specific changes in muscle gene expression. Emerin and Lamin-A/C directly interact with transcriptional regulators, including GCL, Btf, SREBP, Rb and MOK2 (43–47). In mice lacking Emerin, myogenic gene transcription is delayed during adult muscle regeneration (48). This model is consistent with data showing that myogenic gene expression is linked with specific nuclear rearrangements.
We have previously reported that DMPK promotes myogenin expression in C2C12 skeletal myoblast differentiation (19). A majority of myoblasts transfected with shDMPK did not fuse into myotubes, but instead exhibited a disorganized cytoskeleton and abnormal cellular processes. The morphological changes observed after shDMPK transfection correlate with reduced expression of myogenin in C2C12 cells. The disruption of NE observed after DMPK depletion reveals a possible mechanism for the myogenic role of DMPK in differentiation. DMPK is required to maintain the nuclear lamina structure. As stated above, disruption of NE proteins Lamin-A/C or Emerin has been previously linked with delays in nuclear differentiation. Here we show that DMPK interacts with Lamin-A/C at the NE and is required to maintain Lamin-A/C localization.
A plausible molecular mechanism for DM1 is a general defect of RNA metabolism. This would explain the pleiotropic effects and dominant inheritance pattern of the disease. However, a loss of DMPK expression might be responsible for some DM1 symptoms, with haploinsufficiency of DMPK producing dominant inheritance. It is possible that most DM1 symptoms result from dominant negative effects upon mRNA processing and others from loss of DMPK activity. Current DM1 research has focused on aberrant mRNA processing in DM1 muscle, yet animal models of DM1 on the basis of defective splicing through MBNL or CUG-BP1 cannot fully explain the pathophysiology of DM1. This suggests that a decrease in the abundance of DMPK mRNA in the skeletal muscle of DM1 patients may account for some DM1 symptoms. Our data have revealed that DMPK is critical to maintain the NE. DMPK is necessary for NE integrity and is sufficient to cause nuclear fragmentation after transient overexpression. Therefore, the reduction of DMPK in DM1 could contribute to nuclear instability, a common mechanism of muscle wasting in muscular dystrophies.
Supplementary Material
Acknowledgments
We thank Satoshi Nagata and Tomoko Ise at the Sanford Research/USD Flow Cytometry Core Facility and Kelly Graber at the Sanford Research/USD Imaging Core Facility for technical assistance. We also thank Timothy O'Connell, William Harris, Qiangrong Liang, Gregory Shearer, and Casey Wright for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL064136 (to M. B. P.) and P20-RR-017662 (to E. B. H.). This work was also supported by the South Dakota 2010 Initiative Research Center Program and by Sanford Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- DM1
- Myotonic dystrophy 1
- DMPK
- myotonic dystrophy protein kinase
- NE
- nuclear envelope
- EDMD
- Emery-Dreifuss muscular dystrophy
- LGMD1B
- Limb girdle muscular dystrophy 1b
- CUG-BP1
- CUG triplet repeat RNA binding protein 1
- MBNL
- human muscleblind protein homolog
- FL
- full-length.
REFERENCES
- 1. Harper P. S. (2001) Myotonic Dystrophy, 3rd ed., W. B. Saunders, London [Google Scholar]
- 2. Brook J. D., McCurrach M. E., Harley H. G., Buckler A. J., Church D., Aburatani H., Hunter K., Stanton V. P., Thirion J. P., Hudson T. (1992) Cell 68, 799–808 [DOI] [PubMed] [Google Scholar]
- 3. Fu Y. H., Pizzuti A., Fenwick R. G., Jr., King J., Rajnarayan S., Dunne P. W., Dubel J., Nasser G. A., Ashizawa T., de Jong P. (1992) Science 255, 1256–1258 [DOI] [PubMed] [Google Scholar]
- 4. Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O'Hoy K. (1992) Science 255, 1253–1255 [DOI] [PubMed] [Google Scholar]
- 5. Taneja K. L., McCurrach M., Schalling M., Housman D., Singer R. H. (1995) J. Cell Biol. 128, 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fardaei M., Rogers M. T., Thorpe H. M., Larkin K., Hamshere M. G., Harper P. S., Brook J. D. (2002) Hum. Mol. Genet. 11, 805–814 [DOI] [PubMed] [Google Scholar]
- 7. Sasagawa N., Kino Y., Takeshita Y., Oma Y., Ishiura S. (2003) J. Biochem. 134, 537–542 [DOI] [PubMed] [Google Scholar]
- 8. Timchenko N. A., Cai Z. J., Welm A. L., Reddy S., Ashizawa T., Timchenko L. T. (2001) J. Biol. Chem. 276, 7820–7826 [DOI] [PubMed] [Google Scholar]
- 9. Ho T. H., Bundman D., Armstrong D. L., Cooper T. A. (2005) Hum. Mol. Genet. 14, 1539–1547 [DOI] [PubMed] [Google Scholar]
- 10. Kanadia R. N., Johnstone K. A., Mankodi A., Lungu C., Thornton C. A., Esson D., Timmers A. M., Hauswirth W. W., Swanson M. S. (2003) Science 302, 1978–1980 [DOI] [PubMed] [Google Scholar]
- 11. Timchenko N. A., Patel R., Iakova P., Cai Z. J., Quan L., Timchenko L. T. (2004) J. Biol. Chem. 279, 13129–13139 [DOI] [PubMed] [Google Scholar]
- 12. Orengo J. P., Chambon P., Metzger D., Mosier D. R., Snipes G. J., Cooper T. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yadava R. S., Frenzel-McCardell C. D., Yu Q., Srinivasan V., Tucker A. L., Puymirat J., Thornton C. A., Prall O. W., Harvey R. P., Mahadevan M. S. (2008) Nat. Genet. 40, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin F., Worman H. J. (1993) J. Biol. Chem. 268, 16321–16326 [PubMed] [Google Scholar]
- 15. Lehner C. F., Kurer V., Eppenberger H. M., Nigg E. A. (1986) J. Biol. Chem. 261, 13293–13301 [PubMed] [Google Scholar]
- 16. Zhang Q., Bethmann C., Worth N. F., Davies J. D., Wasner C., Feuer A., Ragnauth C. D., Yi Q., Mellad J. A., Warren D. T., Wheeler M. A., Ellis J. A., Skepper J. N., Vorgerd M., Schlotter-Weigel B., Weissberg P. L., Roberts R. G., Wehnert M., Shanahan C. M. (2007) Hum. Mol. Genet. 16, 2816–2833 [DOI] [PubMed] [Google Scholar]
- 17. Roux K. J., Burke B. (2007) Biochim. Biophys. Acta 1772, 118–127 [DOI] [PubMed] [Google Scholar]
- 18. Muchir A., Worman H. J. (2004) Physiology 19, 309–314 [DOI] [PubMed] [Google Scholar]
- 19. Harmon E. B., Harmon M. L., Larsen T. D., Paulson A. F., Perryman M. B. (2008) Dev. Dyn. 237, 2353–2366 [DOI] [PubMed] [Google Scholar]
- 20. Markiewicz E., Ledran M., Hutchison C. J. (2005) J. Cell Sci. 118, 409–420 [DOI] [PubMed] [Google Scholar]
- 21. Mariappan I., Parnaik V. K. (2005) Mol. Biol. Cell 16, 1948–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muralikrishna B., Dhawan J., Rangaraj N., Parnaik V. K. (2001) J. Cell Sci. 114, 4001–4011 [DOI] [PubMed] [Google Scholar]
- 23. Carango P., Noble J. E., Marks H. G., Funanage V. L. (1993) Genomics 18, 340–348 [DOI] [PubMed] [Google Scholar]
- 24. Furling D., Lam le T., Agbulut O., Butler-Browne G. S., Morris G. E. (2003) Am. J. Pathol. 162, 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furling D., Lemieux D., Taneja K., Puymirat J. (2001) Neuromuscul. Disord. 11, 728–735 [DOI] [PubMed] [Google Scholar]
- 26. Krahe R., Ashizawa T., Abbruzzese C., Roeder E., Carango P., Giacanelli M., Funanage V. L., Siciliano M. J. (1995) Genomics 28, 1–14 [DOI] [PubMed] [Google Scholar]
- 27. Morrone A., Pegoraro E., Angelini C., Zammarchi E., Marconi G., Hoffman E. P. (1997) J. Clin. Invest. 99, 1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Pegoraro E., Menegazzo E., Gennarelli M., Hoop R. C., Angelini C., Hoffman E. P. (1995) Hum. Mol. Genet. 4, 599–606 [DOI] [PubMed] [Google Scholar]
- 29. Liquori C. L., Ricker K., Moseley M. L., Jacobsen J. F., Kress W., Naylor S. L., Day J. W., Ranum L. P. (2001) Science 293, 864–867 [DOI] [PubMed] [Google Scholar]
- 30. Cho D. H., Tapscott S. J. (2007) Biochim. Biophys. Acta 1772, 195–204 [DOI] [PubMed] [Google Scholar]
- 31. Mankodi A., Urbinati C. R., Yuan Q. P., Moxley R. T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M. S., Thornton C. A. (2001) Hum. Mol. Genet. 10, 2165–2170 [DOI] [PubMed] [Google Scholar]
- 32. Day J. W., Ricker K., Jacobsen J. F., Rasmussen L. J., Dick K. A., Kress W., Schneider C., Koch M. C., Beilman G. J., Harrison A. R., Dalton J. C., Ranum L. P. (2003) Neurology 60, 657–664 [DOI] [PubMed] [Google Scholar]
- 33. Bush E. W., Helmke S. M., Birnbaum R. A., Perryman M. B. (2000) Biochemistry 39, 8480–8490 [DOI] [PubMed] [Google Scholar]
- 34. Helmke S. M., Lu S. M., Harmon M., Glasford J. W., Larsen T. D., Kwok S. C., Hodges R. S., Perryman M. B. (2006) J. Mol. Recognit. 19, 215–226 [DOI] [PubMed] [Google Scholar]
- 35. Kaliman P., Catalucci D., Lam J. T., Kondo R., Gutierrez J. C., Reddy S., Palacin M., Zorzano A., Chien K. R., Ruiz-Lozano P. (2005) J. Biol. Chem. 280, 8016–8021 [DOI] [PubMed] [Google Scholar]
- 36. Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D. K., Solimando L., Goldman R. D. (2008) Genes Dev. 22, 832–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holaska J. M. (2008) Circ. Res. 103, 16–23 [DOI] [PubMed] [Google Scholar]
- 38. Wansink D. G., Wieringa B. (2003) Cytogenet. Genome Res. 100, 230–242 [DOI] [PubMed] [Google Scholar]
- 39. Jansen G., Groenen P. J., Bächner D., Jap P. H., Coerwinkel M., Oerlemans F., van den Broek W., Gohlsch B., Pette D., Plomp J. J., Molenaar P. C., Nederhoff M. G., van Echteld C. J., Dekker M., Berns A., Hameister H., Wieringa B. (1996) Nat. Genet. 13, 316–324 [DOI] [PubMed] [Google Scholar]
- 40. Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C. L., Burke B. (1999) J. Cell Biol. 147, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Broers J. L., Peeters E. A., Kuijpers H. J., Endert J., Bouten C. V., Oomens C. W., Baaijens F. P., Ramaekers F. C. (2004) Hum. Mol. Genet. 13, 2567–2580 [DOI] [PubMed] [Google Scholar]
- 42. Lammerding J., Schulze P. C., Takahashi T., Kozlov S., Sullivan T., Kamm R. D., Stewart C. L., Lee R. T. (2004) J. Clin. Invest. 113, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dreuillet C., Tillit J., Kress M., Ernoult-Lange M. (2002) Nucleic Acids Res. 30, 4634–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haraguchi T., Holaska J. M., Yamane M., Koujin T., Hashiguchi N., Mori C., Wilson K. L., Hiraoka Y. (2004) Eur. J. Biochem. 271, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 45. Holaska J. M., Lee K. K., Kowalski A. K., Wilson K. L. (2003) J. Biol. Chem. 278, 6969–6975 [DOI] [PubMed] [Google Scholar]
- 46. Lloyd D. J., Trembath R. C., Shackleton S. (2002) Hum. Mol. Genet. 11, 769–777 [DOI] [PubMed] [Google Scholar]
- 47. Ozaki T., Saijo M., Murakami K., Enomoto H., Taya Y., Sakiyama S. (1994) Oncogene 9, 2649–2653 [PubMed] [Google Scholar]
- 48. Melcon G., Kozlov S., Cutler D. A., Sullivan T., Hernandez L., Zhao P., Mitchell S., Nader G., Bakay M., Rottman J. N., Hoffman E. P., Stewart C. L. (2006) Hum. Mol. Genet. 15, 637–651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.