Background: The role of Merlin in breast cancer is unknown.
Results: Merlin protein is degraded in advanced breast cancer due to osteopontin-initiated signaling.
Conclusion: Merlin is regulated at the post-translational level in breast tumors.
Significance: We have defined a functional role for Merlin in limiting breast tumor growth and elucidated the utility of Merlin as an important biomarker in breast cancer.
Keywords: Akt PKB, Breast Cancer, Protein Degradation, Tumor Promoter, Tumor Suppressor Gene, Merlin, NF2, Osteopontin, Post-translational Regulation, Ubiquitination
Abstract
Unlike malignancies of the nervous system, there have been no mutations identified in Merlin in breast cancer. As such, the role of the tumor suppressor, Merlin, has not been investigated in breast cancer. We assessed Merlin expression in breast cancer tissues by immunohistochemistry and by real-time PCR. The expression of Merlin protein (assessed immunohistochemically) was significantly decreased in breast cancer tissues (although the transcript levels were comparable) simultaneous with increased expression of the tumor-promoting protein, osteopontin (OPN). We further demonstrate that the loss of Merlin in breast cancer is brought about, in part, due to OPN-initiated Akt-mediated phosphorylation of Merlin leading to its proteasomal degradation. Restoring expression of Merlin resulted in reduced malignant attributes of breast cancer, characterized by reduced invasion, migration, motility, and impeded tumor (xenograft) growth in immunocompromised mice. The possibility of developing a model using the relationship between OPN and Merlin was tested with a logistic regression model applied to immunohistochemistry data. This identified consistent loss of immunohistochemical expression of Merlin in breast tumor tissues. Thus, we demonstrate for the first time a role for Merlin in impeding breast malignancy, identify a novel mechanism for the loss of Merlin protein in breast cancer, and have developed a discriminatory model using Merlin and OPN expression in breast tumor tissues.
Introduction
Merlin (Moesin-Ezrin-Radixin-like protein), encoded by the NF2 gene, is a tumor suppressor that is frequently inactivated in tumors of the nervous system (1–7). Merlin complexes with ERM (Ezrin-Radixin-Moesin) proteins that link the cytoskeleton to glycoproteins in the cell membrane (7). Merlin is critically involved in regulating cell growth and proliferation. In vitro, Merlin mediates contact inhibition and inhibits invasiveness (8, 9). Underlying the tumor suppressor function of Merlin is likely a combination of the signaling pathways that attribute its ability to suppress Ras and Rac (9–11), negatively regulate FAK, down-regulate expression of cyclin D1 (12), inhibit the p21-activated kinase, Pak1 (13), and interfere with the interaction between CD44 and hyaluronan (10, 14). The stability of Merlin protein is regulated, in part, by Akt-mediated phosphorylation at threonine 230 and serine 315 (15). Phosphorylation at these amino acids leads to Merlin degradation by ubiquitination. The reduced levels of Merlin in tumors of the nervous system are predominantly brought about by mutations or loss of heterozygosity (4, 16–18). However, the role of Merlin in breast cancer has been largely ignored due to early, sporadic studies that did not detect mutations in tumor tissues (19, 20).
OPN3 is a secreted phosphoglycoprotein (21) that acts as an effector of tumor progression and metastasis at several levels (22, 23). Elevated OPN is a marker for advanced breast cancer and multiple other cancer histotypes (24–30). OPN-initiated signaling activates NF-κB, PI 3-kinase, and Akt pathways (31–33) and manifests as enhanced cell proliferation and survival, migration, and adhesion (30).
We report here that although the transcript levels of Merlin are unaltered in breast cancer tissues, there is loss of Merlin expression at the protein level in breast tumors, concomitant with an increase in OPN expression. Our studies revealed that OPN-initiated signaling induced Akt-mediated phosphorylation and degradation of Merlin in breast cancer cells. Further, restoration of Merlin in breast cancer cells functionally impeded their malignant behavior. Logistic regression consistently identified decreased Merlin staining intensity in tumor tissues. It also showed that given the Merlin intensity, OPN ameliorates discrimination between normal and tumor tissue. Thus, our studies provide evidence that the availability of Merlin in breast tumors is regulated at the post-translational level. This is exciting from the perspective that Merlin was not found to be mutated or compromised at the transcript level in breast cancers. We have also defined a functional role for Merlin in limiting breast tumor growth and elucidated the utility of Merlin as an important biomarker in breast cancer.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF10AT, MCF7, MDA-MB-231, and MDA-MB-435 cells were cultured as described previously (34). SUM159 cells were grown in DMEM/F-12 supplemented with FBS, insulin, and hydrocortisone in a humidified 5% CO2 environment. The lineage infidelity of MDA-MB-435 cells has been discussed in several studies (35–37). We used this cell line as a model due to the fact that it naturally expresses copious OPN. Stable Merlin-expressing transfectants of MDA-MB-231 and SUM159 cells were generated by transfecting a Merlin-expressing construct. Empty vector was transfected as control; stable transfectants were selected on G418 (Invitrogen).
Western Blotting Analysis
Immunoblotting was done with anti-Merlin (Santa Cruz Biotechnology, Santa Cruz, CA), phospho-Ser473-Akt (Cell Signaling, Danvers, MA), total Akt (Cell Signaling), anti-mouse HA (Santa Cruz Biotechnology), anti-phospho-Ser315 Merlin (provided by Dr. Keqiang Ye), and anti-GAPDH (Cell Signaling). Anti-rabbit or anti-mouse HRP-conjugated secondary antibody was used for detection, and blots were developed with SuperSignal substrate (Pierce) and exposed using a Fuji LAS3000 imager.
Transfection and Drug Treatment
Cells were transfected with empty vector, Merlin (WT; wild-type), or T230A/S315A Merlin mutant and treated with clasto-lactacystin β-lactone (Sigma) for 2 h. Recombinant OPN (100 ng/ml) (R&D Systems, Minneapolis, MN) was added, and cells were lysed after 6 h. Where indicated, cells were first treated with Akt inhibitor IV (Calbiochem) in serum-free medium for 30 min followed by 100 ng/ml human recombinant OPN for 16 h.
Immunoprecipitation
Cells were transfected with pcDNA3.1 HA-ubiquitin alone or in combination with pIRES2-Myc-Merlin and incubated for 24 h. Cells were treated with 10 μm lactacystin, 100 ng/ml OPN, and 10 μm Akt inhibitor IV for 12 h and lysed in Nonidet P-40 buffer. The lysate was immunoprecipitated with anti-Merlin antibody, and the immunoprecipitate was assessed by immunoblotting.
Real-time Quantitative PCR of Tissue Array
TissueScan plates (OriGene, Rockville, MD) were assessed for the expression of OPN and Merlin transcripts using the manufacturer's protocol. The reaction was carried out in a Bio-Rad iCycler iQ5 using the following program: activation step of 50 °C for 2 min, then 42 cycles of 95 °C for 5 min, 95 °C for 15 s, and 60 °C for 1 min. Data were expressed as -fold change (2−ΔΔCt). Statistical analysis was conducted using JMP version 7.0.1 (SAS, Inc., Cary, NC). A 5% level of significance was used to determine the significance of results. The data were summarized using mean, S.D., and S.E. The Pearson's correlation coefficient was used to determine the correlation between numerical variables such as age. The Wilcoxon test was used to compare CT levels of Merlin and OPN by group (normal or tumor), grade, and stage. p value of < 0.05 was considered significant between groups.
Soft Agar Colonization Assay
Cells were seeded in soft agar in triplicate in a 6-well plate, allowed to grow for 2–3 weeks, and stained with crystal violet solution. Colonies with >50 cells were microscopically counted.
Foci Formation Assay
Cells were transfected with empty vector or pcDNA3.1-Merlin or pcDNA3.1-T230A/S315A-Merlin, detached, and reseeded in medium containing selection antibiotics. Foci formed were counted after 10–14 days.
Animal Studies
Cells (1 million) suspended in Hanks' balanced salt solution (Invitrogen) were injected into the exposed third mammary fat pad of female athymic nude mice. Orthogonal tumor measurements were recorded twice weekly. Mean tumor diameter was calculated as the square root of the product of orthogonal measurements. These studies were conducted under Institutional Animal Care and Use Committee-approved protocols.
Immunohistochemistry
Breast tumor tissue microarrays from the NCI Cooperative Breast Cancer Tissue Resource through the National Institutes of Health (supplemental Data 1) were immunohistochemically stained for OPN (AKm2A1; Santa Cruz Biotechnology) and Merlin (A-19; Santa Cruz Biotechnology) using the streptavidin biotin complex method. Staining intensity was quantitated with computer-assisted image analysis in a Dako ACIS III image analysis system (Glostrup, Denmark).
Statistical Analyses
Associations between intensities of Merlin and OPN expressions and patient clinicopathologic data were assessed using the Wilcoxon rank test for categorical data and the Pearson's correlation coefficient for numerical data. The percentages of normal and tumor tissues expressing Merlin or OPN were compared using a Chi-square test. The significance of percentages of samples expressing Merlin or OPN as compared with the chance occurrence was determined using the exact binomial test. The univariate and multiple logistic regression models were fit to a binary variable normal versus tumor with Merlin and OPN as possible predictors. The possibility of developing a model using the relationship between OPN and Merlin was tested with a logistic regression model on a selected cohort of the data, scoring only the positive staining events from normal tissues for Merlin and the positive staining events from tumor tissues for OPN. The selection criteria were based on the fact that Merlin is a tumor suppressor, with a strong expression in normal tissue, whereas OPN, a tumor-promoting protein, is known to be overexpressed in tumor tissue. The Chi-square test was used to assess the usefulness of the model for prediction of likelihood of tumor. The effect likelihood ratio test was used to assess the usefulness of predictor variables in the model. The area under the receiver operating characteristic (ROC) curve was used to determine the predictive ability of models and in model selection. All statistical analyses were performed using software JMP version 7 (SAS Inc.). All results with p value < 0.05 were considered statistically significant.
Statistical analyses of in vitro data were determined as follows. Statistical differences between groups were assessed using the Mann-Whitney test, Student's t test, or analysis of variance, using GraphPad Prism 4 software. Statistical significance was determined if the analysis reached 95% confidence. The precise p values are listed in the corresponding figure legends.
RESULTS
Merlin and OPN Are Inversely Expressed in Breast Cancer Tissues
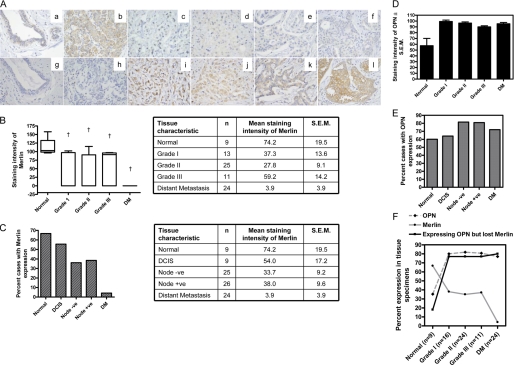
We probed a breast tumor tissue microarray comprising 75 invasive breast cancer cases and nine normal breast tissues for Merlin by immunohistochemistry. The expression of Merlin did not change significantly with respect to ethnicity, age, estrogen receptor or progesterone receptor status, or tumor size. However, there was an overall decrease in Merlin staining in invasive breast cancer tissues grades I–III (infiltrating ductal carcinoma) as compared with normal tissues (Fig. 1A, images c–f (tumor) relative to images a and b (normal)). The expression of Merlin was significantly lower regardless of the nodal involvement. Notably, of the 75 carcinoma tissues, 56 tissues (75%) had lost expression of Merlin (p = 0.0000097) (Fig. 1, B and C).
FIGURE 1.
Merlin and OPN are inversely expressed in breast cancer tissues. A, Merlin is expressed in normal breast tissue (images a and b) but is lost in invasive breast cancer (images c–f). Conversely, OPN is expressed at very low levels in normal breast (images g and h) but is up-regulated in invasive breast cancer (images i–l). Immunohistochemical staining was performed for Merlin and OPN on serial sections from 75 cases of invasive breast cancer and nine normal breast tissues. We recorded loss of Merlin in 75% (56 cases) of invasive breast cancer cases. Of these 56 tissues, 43 (77%) showed concomitant increased OPN expression. Shown are representative photomicrographs of the results. Images a and g; b and h; c and i; d and j; e and k; and f and l represent serial tissue sections. B, the staining intensity of Merlin is reduced in breast cancer tissues (Grades I, II, and III) and in tumors showing distant metastasis (DM). The box and whiskers plot shows the range of staining intensities for the tissues. † indicates statistically significant difference relative to normal breast tissues. Relative to normal breast tissue, Merlin expression is significantly lower in Grade I (p = 0.0026), Grade II (p = 0.0005), and Grade III (p = 0.0017) tumors and in tumors with distant metastasis (p 0.0001). Error bars indicate S.E. C, a greater proportion of normal breast tissues expresses Merlin in contrast to breast cancer tissues (node-negative and node-positive) as well as those showing distant metastasis (p = 0.0005). Relative to normal breast tissue, the levels of Merlin are significantly lower in node-negative (p = 0.0171) and node-positive (p = 0.0457) tumors and in tumors with distant metastasis (p < 0.0001). The levels of Merlin in ductal carcinoma in situ (DCIS) tissues are not significantly different from normal tissue (p = 0.2026). D, the staining intensity of OPN is significantly increased (†, p < 0.0001) in breast cancer tissues (Grades I, II, and III) and in tumors showing distant metastasis relative to normal breast tissue. Error bars indicate S.D. E, a greater proportion of breast cancer tissues (node-negative and node-positive as well as those showing distant metastasis) express Merlin in contrast to normal breast tissues. Error bars indicate S.D. F, 77% of patients (across all grades and with distant metastasis) show loss of Merlin expression with concomitant increase in OPN expression. Merlin is expressed in normal breast tissue; its expression is decreased in breast cancer and is negligible in cases with distant metastases. Conversely, OPN expression increases in breast cancer patients. Error bars indicate S.D.
In contrast, the expression of OPN was increased in breast cancer cases as compared with normal breast tissues (Fig. 1A, images i–l (tumor) relative to images g and h (normal)) (p = 0.0097) (Fig. 1D). Relative to normal tissues, a greater proportion of the tumor tissues showed OPN expression (Fig. 1E). Overall, of all the three grades combined, 43 tissues out of the 56 tissues showed no staining for Merlin simultaneous with increased staining for OPN. Thus, 77% (43 out of 56 Merlin-negative tissues) of the tissues that had lost Merlin expression showed increased OPN expression (p = 0.000031). Specifically, the primary tumors from 23 out of the 24 cases with distant metastasis showed no staining for Merlin (p = 0.000001). Of these, 20 cases (80%) showed increased OPN staining (p = 0.00077) (Fig. 1F). Thus, our studies showed that Merlin protein expression is lost in invasive breast cancer and that the loss of Merlin is accompanied by an increased expression of OPN.
The Transcript Levels of Merlin Are Unaltered in Breast Cancer Tissues, whereas Those of OPN Are Increased
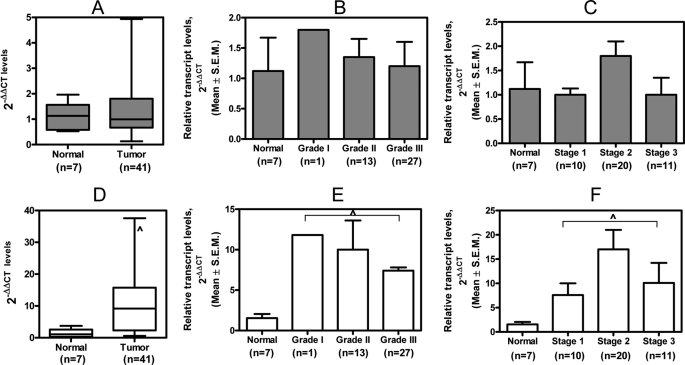
We queried the expression of Merlin in breast tumor tissues at two levels: the amount of the transcript and the extent of protein expression. We assessed the transcript levels in tissues from 41 breast cancer patients and seven normal control tissues. The transcript levels of Merlin did not show any appreciable changes (p > 0.05) between normal and breast tumor-derived tissues; there was also no change in the Merlin transcript levels across the different grades of tumors or the disease stage (Fig. 2, A–C). In contrast, the transcript levels of OPN were significantly (p < 0.01) greater in the tumor tissues relative to normal tissues. The OPN transcript levels also increased significantly in tissues derived from grades II and III tumors and with progression of the disease stage (Fig. 2, D–F).
FIGURE 2.
Breast tumor tissues have increased OPN transcript levels but comparable Merlin transcript levels relative to normal breast tissues. A, the overall levels of Merlin are not significantly reduced (p = 0.82) in tumor tissues relative to normal breast tissues. B and C, there is no appreciable change in the levels of Merlin across the grade (p = 0.6) of the breast tumor tissues and the disease stage (p = 0.15). D, the transcript levels of OPN are significantly increased (^, p = 0.0028) in breast tumor tissues relative to normal breast tissues. E and F, the transcript levels of OPN are notably increased with the advance in the grade of the breast tumor (^, p = 0.04) and disease stage (^, p = 0.01). Error bars in all panels indicate S.D.
Merlin Suppresses Malignant Behavior of Breast Cancer Cells
The role of Merlin as a tumor suppressor is characterized in tumors of the nervous system. To determine the role of Merlin in impacting malignant behavior of breast cancer cells, we restored the expression of Merlin in two human breast cancer cell lines, SUM159 (Fig. 3A) and MDA-MB-231 (Fig. 3B). We assessed the malignant attributes of the resultant Merlin-expressing transfectants. Expression of Merlin caused a significant reduction in the ability of breast cancer cells to form foci (Fig. 3, C and D), invade through Matrigel (Fig. 3, E and F), laterally migrate (Fig. 3G), and grow under anchorage-independent conditions (Fig. 3H). When injected into the mammary fat pad of female athymic nude mice, the Merlin-expressing SUM159 cells showed notable (p < 0.05) latency in the appearance of palpable tumors (Fig. 3I). Although the tumors formed by vector control cells were evident beginning at 10 days after injection, those formed by the mixed pool and clone 6 were palpable 19 and 54 days after injection, respectively. The Merlin-transfectant A1 and A2 clones of MDA-MB-231 also demonstrated a significantly (p < 0.05) reduced growth rate (Fig. 3F). The mixed pool of Merlin transfectants of both SUM159 and MDA-MB-231 cells showed a modest but significant reduction on tumor growth rate. This may likely be due to a mixed population of Merlin-expressing and non-expressing cells. Cumulatively, restoration of Merlin expression in both breast cancer cell lines resulted in reduced malignant behavior.
FIGURE 3.
Merlin can functionally suppress the malignant behavior of breast cancer cells. A and B, stable Merlin-expressing transfectants were derived from SUM159 and MDA-MB-231 cells. Vec, empty vector. C and D, restoration of Merlin significantly reduces the foci formation ability (^, p = 0.005 for SUM159 and ^, p = 0.003 for MDA-MB-231), E and F, invasive properties of SUM159 (^, p < 0.0001) and MDA-MB-231 cells (^, p < 0.0001). G and H, restoration of Merlin in SUM159 cells significantly (p = 0.014) reduces their ability to laterally migrate (in a wound healing assay) (G) and grow under anchorage-independent conditions (H) (^, p = 0.02). I, expression of Merlin in SUM159 cells results in increased latency and reduced tumor growth of the xenografts. The tumor size is represented as mean tumor diameter (^, p < 0.0001 relative to vector controls; four mice were assessed per group). J, MDA-MB-231 cells restored for Merlin expression show significantly slower growth of the cells as xenografts. (^, p < 0.016 relative to vector controls; four mice were assessed per group). Error bars in panels C–J indicate S.D.
OPN Targets Merlin for Akt-mediated Proteasomal Degradation
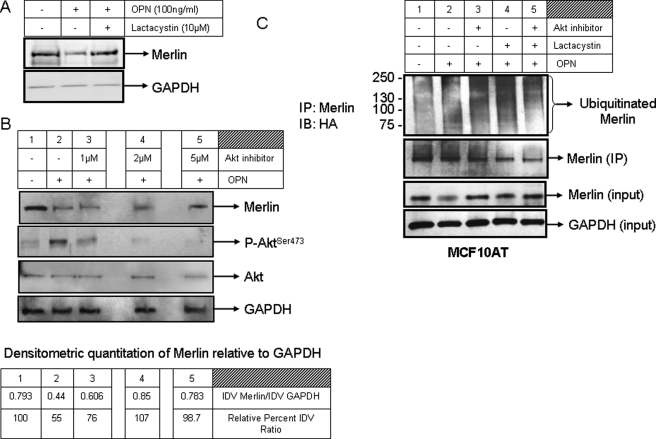
We hypothesized that Akt signaling initiated downstream of OPN could regulate Merlin. Thus, to directly test the effects of OPN on the post-translational regulation of Merlin, specifically the stability of Merlin protein, we transfected SUM159 breast cancer cells with Merlin cDNA and treated the cells with recombinant OPN. OPN causes a decrease in the protein levels of Merlin (Fig. 4A). Treatment with the proteasome inhibitor, lactacystin, rescued the levels of Merlin in OPN-treated cells, suggesting that OPN-initiated signaling targeted Merlin for proteasome-mediated degradation. OPN interacts with a variety of cell surface receptors including CD44 and multiple integrins to activate signaling via the Akt pathway (31, 38, 39). To assess the role of Akt in OPN-initiated degradation of endogenous Merlin, we treated MCF7 cells (which express Merlin but do not express detectable levels of OPN) with recombinant OPN. Treatment with OPN caused phosphorylation of Akt concomitant with a decrease in the levels of endogenous Merlin, suggesting that degradation of Merlin can be initiated by signaling downstream of OPN via Akt (Fig. 4B, lane 3). MCF7 cells were also treated with Akt inhibitor IV in addition to OPN (lanes 3–5). Although the levels of Akt phosphorylation predictably decreased after treatment, the levels of endogenous Merlin were restored by the inhibition of Akt phosphorylation even in the presence of OPN (lanes 4 and 5), suggesting that inhibition of Akt activation downstream of OPN blocks the effects on degradation of Merlin. As seen in the accompanying table, co-treatment with the Akt inhibitor blocks the effects of OPN, allowing for a total recovery of endogenous Merlin (lanes 4 and 5).
FIGURE 4.
OPN targets Merlin for Akt-mediated proteasomal degradation. A, OPN (100 ng/ml) causes a decrease in the levels of Merlin in SUM159 cells. Treatment with lactacystin (10 μm) rescues Merlin in the presence of OPN. B, in MCF7 cells that express endogenous Merlin, OPN treatment activates Akt (increased levels of phospho-Ser473-Akt (P-AktSer473, lane 2). The levels of total Akt remain unaltered. Akt inhibitor IV reduces levels of phospho-Akt and restores endogenous Merlin levels (lanes 3–5). The levels of phospho-Akt were assessed 8 h after treatment with recombinant human OPN and Akt inhibitor, whereas Merlin levels were scored 16 h after the treatment. IP, immunoprecipitation; IB, immunoblotting. C, OPN increases the levels of ubiquitinated endogenous Merlin (lane 2). The smear represents ubiquitinated Merlin. Lactacystin arrests the ubiquitinated Merlin, resulting in increased intensity of the signal (lane 4). Inhibition of Akt reduces the intensity of ubiquitinated Merlin (lane 5). MCF10AT cells were transfected with HA-ubiquitin and treated with OPN, lactacystin, and Akt inhibitor IV. Cell lysate (2 mg) harvested in Nonidet P-40 buffer was immunoprecipitated overnight for endogenous Merlin. The immunoprecipitate was immunoblotted with anti-HA antibody and anti-Merlin antibody. Merlin and GAPDH levels from the cell lysates are represented as inputs for the experiment. The band intensity of Merlin in all the lanes was quantitated relative to the respective GAPDH bands and is depicted in the accompanying table and represented as the ratio of integrated density values (IDV) and also as a relative percentage of integrated density value ratio (relative to lane 1, untreated cells). Densitometry was done using the AlphaEase program.
Phosphorylation of Merlin via Akt targets it for degradation by the proteasome (15, 40, 41). Thus, to determine whether OPN can induce ubiquitination of endogenous Merlin leading to its proteasomal degradation, MCF10AT cells were transfected with an HA-ubiquitin-expressing construct. In the presence of OPN, endogenous Merlin undergoes some ubiquitination that is evident as a smear (Fig. 4C, lane 2). This smear persisted in the presence of lactacystin (Fig. 4C, lane 3), suggesting that Merlin was likely ubiquitinated in the cells in the presence of OPN. The beads by themselves do not non-specifically bind the HA-tagged ubiquitinated proteins in the cells (supplemental Data 2). To specifically assess the role of activated Akt induced by OPN, we co-treated with an Akt inhibitor. Ubiquitination of endogenous Merlin was abolished in the presence of Akt inhibitor, suggesting that OPN-induced Akt phosphorylation caused degradation of endogenous Merlin via the ubiquitin-proteasome pathway (Fig. 4C, lane 5). Similar results were observed in SUM159 cells constitutively expressing Merlin. Merlin ubiquitination was enhanced when co-treated with OPN and was abolished in the presence of Akt inhibitor, reaffirming the role of Akt downstream of OPN in modulating the stability of Merlin (supplemental Data 3, panel A). The converse was seen when we treated MDA-MB-435 cells with the proteasome inhibitor, lactacystin (10–25 μm), and the PI 3-kinase inhibitor, wortmannin (100 nm). The MDA-MB-435 cells do not express detectable levels of Merlin, but express abundant OPN. Combined treatment with lactacystin and wortmannin restored Merlin expression in the cells, suggesting that the PI 3-kinase/Akt pathway, in conjunction with the activities of the proteasome, regulates the protein levels of Merlin in the cells (supplemental Data 3, panel B). Silencing the expression of OPN reduced the overall levels of ubiquitinated Merlin; in combination with Akt inhibitor and lactacystin, abrogating OPN expression caused a notable decrease in the ubiquitinated Merlin (supplemental Data 3, panel C).
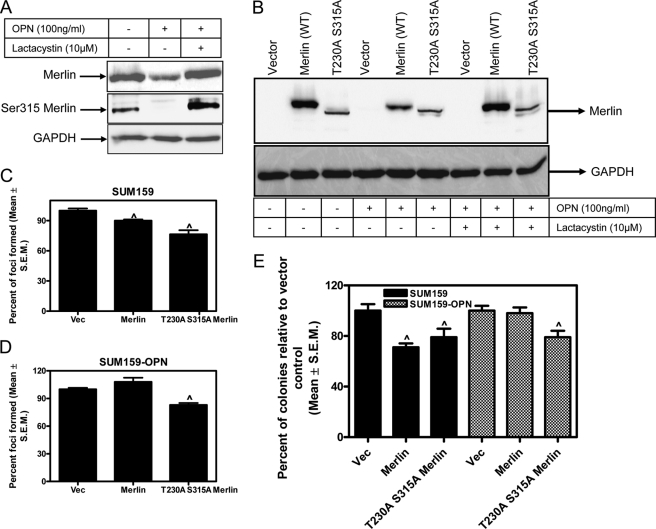
OPN-initiated Signaling Causes Phosphorylation of Merlin at Serine 315
The loss of Merlin in the presence of OPN is caused by the phosphorylation of Merlin at the Ser-315 position (Fig. 5A). Specifically, phosphorylation of Merlin at this residue has been reported to target it for proteasome-mediated degradation (15, 40). This form of Merlin was detectable upon inhibition of proteasomal degradation with lactacystin in the presence of OPN. We further determined that although OPN is able to induce degradation of Merlin, the Merlin mutant T230A/S315A (which cannot be phosphorylated by Akt) is resistant to the effects of OPN (Fig. 5B). Thus, cumulatively, our results suggest that OPN activates Akt-mediated signaling that causes phosphorylation of Merlin at Ser-315. This event targets Merlin for ubiquitin-mediated degradation in breast cancer cells.
FIGURE 5.
OPN-initiated signaling causes phosphorylation of Merlin at serine 315; the degradation-resistant Merlin can reduce malignant attributes of breast cancer cells. A, Merlin is phosphorylated at Ser-315 in response to OPN. Lysate from SUM159 cells transfected with Merlin and treated with OPN was probed for total Merlin and phosphorylated Merlin (Ser315 Merlin). GAPDH was used as a loading control. B, phosphorylation of serine 315 and threonine 230 makes Merlin refractory to OPN. SUM159 cells were transfected with Merlin (WT) or T230A/S315A Merlin mutant and treated with OPN and lactacystin. Cell lysates were probed for total Merlin. GAPDH was used as a loading control. Mutant Merlin (T230A/S315A) is not degraded in response to OPN, whereas wild-type Merlin is degraded by OPN. C, wild-type Merlin and T230A/S315A Merlin mutant can significantly (^, p < 0.05) reduce foci formation ability of SUM159 cells. Plasmids corresponding to empty vector (Vec), wild-type Merlin, and Merlin mutant were transfected into SUM159 cells. Cells were detached and reseeded in medium containing selection antibiotics. Foci were counted after 10–14 days. D, only the degradation-resistant T230A/S315A Merlin mutant can reduce foci formation in the presence of elevated OPN signaling in SUM159 cells (^, p = 0.003, relative to vector control). E, wild-type Merlin and T230A/S315A Merlin mutant can significantly (^, p < 0.05) reduce colony formation in soft agar by SUM159 cells. Only the degradation-resistant T230A/S315A Merlin mutant can reduce the ability to grow under anchorage-independent condition in soft agar in the presence of elevated OPN signaling in SUM159 cells (^, p < 0.05, relative to vector control). Error bars in panels C–E indicate S.D.
Degradation-resistant Merlin Functionally Restricts Malignant Behavior
We assessed the ability of the Merlin mutant T230A/S315A for its ability to impact the properties of breast cancer cells in the perspective of OPN signaling. Both the wild-type Merlin and the T230A/S315A Merlin mutant significantly (p < 0.05) reduced the numbers of foci formed by the SUM159 cells (Fig. 5C). To test the effectiveness of T230A/S315A Merlin mutant under conditions of elevated OPN expression, we tested the ability of Merlin to impact the foci formation capability of SUM159-OPN (stably expressing OPN) cells. Although wild-type Merlin cannot impact the foci formation capability of the SUM159-OPN cells, the T230A/S315A Merlin mutant brings about a significant (p < 0.05) reduction in the numbers of foci formed (Fig. 5D). Similar results were obtained in the assessment of anchorage-independent growth in a soft agar colonization assay (Fig. 5E), suggesting that the degradation-resistant T230A/S315A Merlin mutant retains its ability to effectively blunt malignant attributes in the presence of OPN.
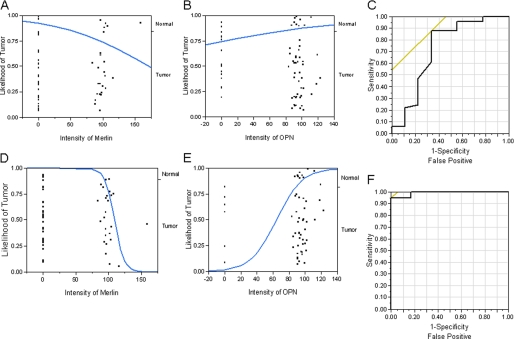
OPN Enhances Tissue Identification and Discriminatory Power of Merlin
To assess the discriminatory power of Merlin and OPN, we applied a logistic regression model to a binary variable of normal and tumor tissue to our data. The Chi-square test for appropriateness of model (p = 0.0448; ROC curve area = 0.7220) indicates that Merlin has a discriminatory power for distinguishing between normal and tumor tissues (Fig. 6A). The logistic regression also showed that OPN by itself is not a good discriminator between normal and tumor tissues (p = 0.2878; ROC area = 0.6040) (Fig. 6B). Further, multiple logistic regression showed that OPN does not increase the discriminatory power of Merlin (p = 0.162; ROC area = 0.723) (Fig. 6C). Toward the possibility of developing a model that uses the unique inverse relationship between OPN and Merlin, we applied a logistic regression model to a selected cohort of the data, scoring only the positive staining events from normal tissues for Merlin and the positive staining events from tumor tissue for OPN. As seen in Fig. 6D, it is apparent that the logistic model for Merlin alone, using this data set, is very good at discriminating between normal and breast tumor tissues (p < 0.0001; R2 = 0.43; ROC area = 0.93). Furthermore, given the Merlin intensity, OPN expression ameliorates tissue identification with increased discriminative power of the model (n = 46; p < 0.0001; R2 = 0.81; ROC area = 0.9917) (Fig. 6E). We then applied a model developed from this training set to our selected data, and we found that out of the 46 samples queried, only two samples were misclassified (Fig. 6F), resulting in 96% probability of correct classification.
FIGURE 6.
OPN ameliorates tissue identification and discriminatory power of Merlin. The reciprocal relationship between Merlin and OPN was assessed by logistic regression and ROC curve analyses. A, logistic plot using Merlin as a predictor variable to distinguish between normal and tumor tissues. The effect likelihood test (p = 0.0448; ROC curve area = 0.722) indicates that Merlin has a discriminatory power for distinguishing between normal and tumor tissues. B, logistic plot of OPN as a predictor variable shows that OPN by itself is not reliably able to discriminate between normal and tumor tissues (p = 0.2872; ROC area = 0.6040). C, ROC curve for logistic model with Merlin and OPN as predictor variables to distinguish between normal and tumor tissues indicates that OPN does not augment the discriminatory power of Merlin (whole model test, p = 0.0517; ROC area = 0.7234). The effect likelihood tests show that although Merlin contributes significantly (p = 0.0286) to the prediction of tumor tissue, OPN does not (p = 0.1682). D, logistic plot using data from only the normal tissues that stained for Merlin and the entire dataset of tumor tissue staining for Merlin show that Merlin has a very high discriminatory power for distinguishing between normal and tumor tissues (p < 0.0001; ROC area = 0.93). E, logistic plot using data from only from the tumor tissues that stained for OPN and the entire dataset of normal tissue staining for Merlin show that OPN has discriminatory power for distinguishing between normal and tumor tissues (p < 0.0007; ROC area = 0.7023). F, ROC curve for logistic model utilizing non-zero Merlin values for normal tissues and non-zero OPN values for tumor tissues as predictor variables to distinguish between normal and tumor tissues indicates that OPN augments the discriminatory power of Merlin (whole model test, p < 0.0001; R2 = 0.81; ROC area = 0.9917). The effect likelihood tests show that both Merlin (p < 0.0001) and OPN (p = 0.0001) contribute significantly to the prediction of tumor tissue.
DISCUSSION
Although Merlin has been extensively explored in tumors arising from the nervous system, its role in breast cancer is understudied. Early studies reported that mutations in Merlin were not detected in breast cancer (19). In a separate study, Yaegashi et al. (20) reported infrequent involvement of mutations in the NF2 gene (encoding for Merlin) in an independent cohort of 60 breast cancer patients. Dai et al. (42) reported that the estrogen-response gene and tumor suppressor, Na+/H+ exchange regulatory factor (NHERF) likely acts in conjunction with Merlin to transduce a growth suppressive signal. Thus, although there are sporadic references regarding Merlin in breast cancer, the functional and biological roles of Merlin in breast cancer have largely been ignored due to the absence of detectable mutations and the lack of studies of change at the transcript level.
In this study, we have seen that the level of Merlin transcript does not appreciably change in breast tumor tissues. Thus, it was intriguing to note a significant decrease in the immunohistochemical staining for Merlin, suggestive of the fact that Merlin protein expression is lost in breast cancer. In contrast, the oncoprotein, OPN, showed an increase in expression at the transcript levels as well as at the protein level. OPN binding to cell surface receptors, such as the integrins, causes several signal transduction pathways to turn on, culminating in enhanced proliferation, migration, and survival (22). Our studies demonstrate that OPN induces Akt-mediated phosphorylation of Merlin that targets Merlin for ubiquitin-mediated degradation in breast cancer cells, resulting in decreased overall cellular pools of endogenous Merlin.
Ubiquitin-mediated degradation of tumor suppressors such as p53, promyelocytic leukemia protein (PML), phosphatase and tensin homolog (PTEN), and Von Hippel–Lindau tumor suppressor (VHL) has also been documented to be responsible for the decreased availability of the respective proteins in tumor cells (43, 44). We showed that degradation of endogenous Merlin is one of the ways by which OPN-initiated signaling removes the check of this tumor suppressor. OPN is a secreted protein. Hence, it is available to the tumor cells in their microenvironment. Given this fact, the implications of our findings can have important considerations for understanding and appreciating the effects that OPN can have on tumor cells. OPN levels increase during pathogenesis of breast cancer. OPN is also available to the tumor cells from the surrounding stromal and inflammatory cells that infiltrate the tumor. OPN-initiated signaling via Akt results in phosphorylation of Merlin and its subsequent degradation. Being a secreted protein that utilizes a variety of receptors, OPN can influence signaling in surrounding tumor cells, causing a reduction in Merlin protein levels as a “bystander effect” resulting in a widespread degradation-induced loss of Merlin. As such, although OPN has been reported to induce ubiquitin-mediated degradation of Stat1 (45), ours is the first study to report that OPN causes degradation of a tumor suppressor protein.
Although in breast cancer Merlin may not be a prototypic tumor suppressor gene that conforms to the classic definition of Knudsen's two-hit hypothesis, our study clearly demonstrates that Merlin has a tumor suppressor activity in breast cancer. Restoration of Merlin in breast tumor cells (less than 2-fold up-regulated relative to normal tissues; supplemental Data 4) functionally blunted their malignant properties. As such, the inverse relationship between Merlin and OPN that we observed in clinical specimens is far from just coincidental. Logistic regression identified Merlin intensity as a good predictor for immunohistochemical identification of tumor tissue. It also showed that enhanced staining intensity of OPN ameliorates tissue identification when combined with the staining intensity of Merlin in breast tumor tissues.
The significance of Merlin expression and its function in breast cancer have been ignored thus far. As such, this is the first study reporting a functional role for Merlin in breast cancer, and it is also the first report of OPN causing the degradation of a tumor suppressor protein. Thus, our studies elucidate the utility of Merlin and OPN as important biomarkers in breast cancer and also identify a novel mechanism for the loss of Merlin expression in breast cancer.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA138850 (to L. A. S.) and CA140472 (to R. S. S.). This work was also supported by the Mayer Mitchell Award (to L. A. S.) and a grant from the University of South Alabama (USA) Mitchell Cancer Institute.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental table and three figures.
- OPN
- osteopontin
- ROC
- receiver operating characteristic.
REFERENCES
- 1. Lau Y. K., Murray L. B., Houshmandi S. S., Xu Y., Gutmann D. H., Yu Q. (2008) Cancer Res. 68, 5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rushing E. J., Cooper P. B., Quezado M., Begnami M., Crespo A., Smirniotopoulos J. G., Ecklund J., Olsen C., Santi M. (2007) J. Neurooncol. 85, 297–305 [DOI] [PubMed] [Google Scholar]
- 3. Muranen T., Grönholm M., Lampin A., Lallemand D., Zhao F., Giovannini M., Carpén O. (2007) Hum. Mol. Genet. 16, 1742–1751 [DOI] [PubMed] [Google Scholar]
- 4. Begnami M. D., Palau M., Rushing E. J., Santi M., Quezado M. (2007) Hum. Pathol. 38, 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fouladi M., Helton K., Dalton J., Gilger E., Gajjar A., Merchant T., Kun L., Newsham I., Burger P., Fuller C. (2003) Cancer 98, 2232–2244 [DOI] [PubMed] [Google Scholar]
- 6. James M. F., Han S., Polizzano C., Plotkin S. R., Manning B. D., Stemmer-Rachamimov A. O., Gusella J. F., Ramesh V. (2009) Mol. Cell. Biol. 29, 4250–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sainio M., Zhao F., Heiska L., Turunen O., den Bakker M., Zwarthoff E., Lutchman M., Rouleau G. A., Jääskeläinen J., Vaheri A., Carpén O. (1997) J. Cell Sci. 110, 2249–2260 [DOI] [PubMed] [Google Scholar]
- 8. Poulikakos P. I., Xiao G. H., Gallagher R., Jablonski S., Jhanwar S. C., Testa J. R. (2006) Oncogene 25, 5960–5968 [DOI] [PubMed] [Google Scholar]
- 9. Kim H., Lim J. Y., Kim Y. H., Kim H., Park S. H., Lee K. H., Han H., Jeun S. S., Lee J. H., Rha H. K. (2002) Mol. Cells 14, 108–114 [PubMed] [Google Scholar]
- 10. Morrison H., Sherman L. S., Legg J., Banine F., Isacke C., Haipek C. A., Gutmann D. H., Ponta H., Herrlich P. (2001) Genes Dev. 15, 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morrison H., Sperka T., Manent J., Giovannini M., Ponta H., Herrlich P. (2007) Cancer Res. 67, 520–527 [DOI] [PubMed] [Google Scholar]
- 12. Xiao G. H., Gallagher R., Shetler J., Skele K., Altomare D. A., Pestell R. G., Jhanwar S., Testa J. R. (2005) Mol. Cell. Biol. 25, 2384–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kissil J. L., Johnson K. C., Eckman M. S., Jacks T. (2002) J. Biol. Chem. 277, 10394–10399 [DOI] [PubMed] [Google Scholar]
- 14. Bai Y., Liu Y. J., Wang H., Xu Y., Stamenkovic I., Yu Q. (2007) Oncogene 26, 836–850 [DOI] [PubMed] [Google Scholar]
- 15. Tang X., Jang S. W., Wang X., Liu Z., Bahr S. M., Sun S. Y., Brat D., Gutmann D. H., Ye K. (2007) Nat. Cell Biol. 9, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 16. Nunes F., Shen Y., Niida Y., Beauchamp R., Stemmer-Rachamimov A. O., Ramesh V., Gusella J., MacCollin M. (2005) Cancer Genet. Cytogenet. 162, 135–139 [DOI] [PubMed] [Google Scholar]
- 17. Wallace A. J., Watson C. J., Oward E., Evans D. G., Elles R. G. (2004) Genet. Test 8, 368–380 [DOI] [PubMed] [Google Scholar]
- 18. Lasota J., Wasag B., Dansonka-Mieszkowska A., Karcz D., Millward C. L., Ry J., Stachura J., Sobin L. H., Miettinen M. (2003) Lab. Invest. 83, 1361–1371 [DOI] [PubMed] [Google Scholar]
- 19. Arakawa H., Hayashi N., Nagase H., Ogawa M., Nakamura Y. (1994) Hum. Mol. Genet. 3, 565–568 [DOI] [PubMed] [Google Scholar]
- 20. Yaegashi S., Sachse R., Ohuchi N., Mori S., Sekiya T. (1995) Jpn. J. Cancer Res. 86, 929–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Craig A. M., Nemir M., Mukherjee B. B., Chambers A. F., Denhardt D. T. (1988) Biochem. Biophys. Res. Commun. 157, 166–173 [DOI] [PubMed] [Google Scholar]
- 22. Shevde L. A., Das S., Clark D. W., Samant R. S. (2010) Curr. Mol. Med. 10, 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shevde L. A., Samant R. S., Paik J. C., Metge B. J., Chambers A. F., Casey G., Frost A. R., Welch D. R. (2006) Clin. Exp. Metastasis 23, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim Y. W., Park Y. K., Lee J., Ko S. W., Yang M. H. (1998) J. Korean Med. Sci. 13, 652–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tuck A. B., Chambers A. F. (2001) J. Mammary Gland Biol. Neoplasia 6, 419–429 [DOI] [PubMed] [Google Scholar]
- 26. Tuck A. B., O'Malley F. P., Singhal H., Harris J. F., Tonkin K. S., Kerkvliet N., Saad Z., Doig G. S., Chambers A. F. (1998) Int. J. Cancer 79, 502–508 [DOI] [PubMed] [Google Scholar]
- 27. Rudland P. S., Platt-Higgins A., El-Tanani M., De Silva Rudland S., Barraclough R., Winstanley J. H., Howitt R., West C. R. (2002) Cancer Res. 62, 3417–3427 [PubMed] [Google Scholar]
- 28. Furger K. A., Menon R. K., Tuck A. B., Bramwell V. H., Chambers A. F. (2001) Curr. Mol. Med. 1, 621–632 [DOI] [PubMed] [Google Scholar]
- 29. Singhal H., Bautista D. S., Tonkin K. S., O'Malley F. P., Tuck A. B., Chambers A. F., Harris J. F. (1997) Clin. Cancer Res. 3, 605–611 [PubMed] [Google Scholar]
- 30. Rodrigues L. R., Teixeira J. A., Schmitt F. L., Paulsson M., Lindmark-Mänsson H. (2007) Cancer Epidemiol. Biomarkers Prev. 16, 1087–1097 [DOI] [PubMed] [Google Scholar]
- 31. Das R., Mahabeleshwar G. H., Kundu G. C. (2003) J. Biol. Chem. 278, 28593–28606 [DOI] [PubMed] [Google Scholar]
- 32. Philip S., Bulbule A., Kundu G. C. (2004) Glycoconj. J. 21, 429–441 [DOI] [PubMed] [Google Scholar]
- 33. Rangaswami H., Bulbule A., Kundu G. C. (2006) Glycoconj. J. 23, 221–232 [DOI] [PubMed] [Google Scholar]
- 34. Metge B. J., Frost A. R., King J. A., Dyess D. L., Welch D. R., Samant R. S., Shevde L. A. (2008) Clin. Exp. Metastasis 25, 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lacroix M. (2009) Cancer Chemother. Pharmacol. 63, 567. [DOI] [PubMed] [Google Scholar]
- 36. Rae J. M., Creighton C. J., Meck J. M., Haddad B. R., Johnson M. D. (2007) Breast Cancer Res. Treat 104, 13–19 [DOI] [PubMed] [Google Scholar]
- 37. Chambers A. F. (2009) Cancer Res. 69, 5292–5293 [DOI] [PubMed] [Google Scholar]
- 38. Robertson B. W., Chellaiah M. A. (2010) Exp. Cell Res. 316, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang G., He B., Weber G. F. (2003) Mol. Cell. Biol. 23, 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okada M., Wang Y., Jang S. W., Tang X., Neri L. M., Ye K. (2009) Cancer Res. 69, 4043–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye K. (2007) Cell Adh. Migr. 1, 196–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai J. L., Wang L., Sahin A. A., Broemeling L. D., Schutte M., Pan Y. (2004) Oncogene 23, 8681–8687 [DOI] [PubMed] [Google Scholar]
- 43. Yang Y., Kitagaki J., Wang H., Hou D. X., Perantoni A. O. (2009) Cancer Sci. 100, 24–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trotman L. C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N. P., Carver B. S., Cordon-Cardo C., Erdjument-Bromage H., Tempst P., Chi S. G., Kim H. J., Misteli T., Jiang X., Pandolfi P. P. (2007) Cell 128, 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao C., Guo H., Mi Z., Grusby M. J., Kuo P. C. (2007) J. Immunol. 178, 1870–1881 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.