Background: Antibiotic tagetitoxin inhibits bacterial RNA polymerases (RNAPs) and RNAP III from eukaryotes.
Results: We constructed a structural model of tagetitoxin bound to the transcription elongation complex.
Conclusion: Tagetitoxin interacts directly with the β′ subunit trigger loop, stabilizing it in an inactive conformation.
Significance: Results have implications for designing new antibiotics and understanding principles of RNAP functioning and regulation.
Keywords: Antibiotic Action, Molecular Modeling, RNA Polymerase, Transcription, Transcription Elongation Factors
Abstract
Tagetitoxin (Tgt) inhibits multisubunit chloroplast, bacterial, and some eukaryotic RNA polymerases (RNAPs). A crystallographic structure of Tgt bound to bacterial RNAP apoenzyme shows that Tgt binds near the active site but does not explain why Tgt acts only at certain sites. To understand the Tgt mechanism, we constructed a structural model of Tgt bound to the transcription elongation complex. In this model, Tgt interacts with the β′ subunit trigger loop (TL), stabilizing it in an inactive conformation. We show that (i) substitutions of the Arg residue of TL contacted by Tgt confer resistance to inhibitor; (ii) Tgt inhibits RNAP translocation, which requires TL movements; and (iii) paused complexes and a “slow” enzyme, in which the TL likely folds into an altered conformation, are resistant to Tgt. Our studies highlight the role of TL as a target through which accessory proteins and antibiotics can alter the elongation complex dynamics.
Introduction
RNA polymerases (RNAPs)3 undergo complex conformational transitions at all stages of transcription. These transitions frequently serve as targets for antibiotics and regulatory cellular proteins. The β′ trigger loop (TL), a highly flexible element of the enzyme that is required for catalysis and substrate selection, also serves as a target for RNAP inhibitors. A structure of the Thermus thermophilus (Tth) transcription elongation complex (EC) bound to the antibiotic streptolydigin (Stl) revealed a “preinsertion” intermediate, as well as dramatic structural transitions of the β′ TL (1). Structures of the yeast RNAP II elongation complex (2) and core enzyme (3) bound to a fungal toxin α-amanitin suggest that the inhibitor acts through restricting the TL mobility. These and other crystallographic data led to the widely accepted model of the nucleotide addition cycle, common for all multisubunit RNAPs, wherein the metamorphic TL undergoes a sequence of structural transitions, most notably between a substrate-free partially unstructured random coil state and the NTP-bound α-helical hairpin state (trigger helices, THs), with every nucleotide added to the nascent RNA (1, 4–8). The central role postulated for the TL in this model makes it a natural target for regulatory inputs, including those by small molecules.
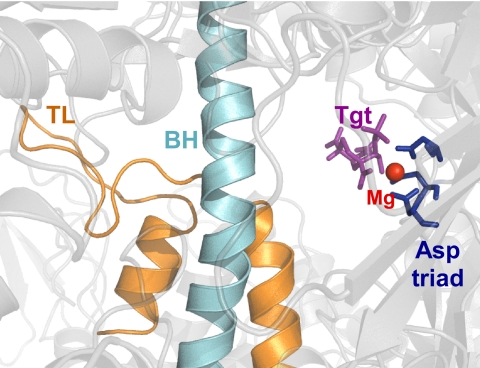
Tagetitoxin (Tgt), 4-O-acetyl-3-amino-1,6-anhydro-3-deoxy-d-glucose 2-phosphate, a phytotoxin produced by Pseudomonas syringae pv. tagetis, causes apical chlorosis in infected plants. This effect has been attributed to inhibition of the bacterial type chloroplast RNAP (9). Tgt also efficiently inhibits bacterial RNAPs and RNAP III from yeast, insects, and vertebrates in vitro but not single-subunit phage RNAPs and nuclear RNAPs I and II (9, 10). We have reported a structure of Tgt bound within the secondary channel of Tth RNAP holoenzyme near the active site of the enzyme (Fig. 1). Tgt makes contact with several residues in the β and β′ subunits of the core RNAP (α2ββ′ω subunit composition); we showed that substitutions of these residues in the Escherichia coli (Eco) RNAP confer resistance to Tgt in vitro (11).
FIGURE 1.
Tgt in a co-crystal with Tth RNAP holoenzyme (Protein Data Bank code 2BE5). Tgt is shown as purple sticks, the β′ BH and TL as teal and orange schemes, respectively. The catalytic Mg2+ ion is indicated by a red sphere, the catalytic Asp triad by dark blue sticks.
Both structural modeling and biochemical analysis suggested that Tgt, which is an uncompetitive inhibitor of RNAP (10, 11), can bind to the EC together with an NTP substrate. We speculated that Tgt stabilizes a catalytically inactive enzyme intermediate through a Tgt-bound “inhibitory” Mg2+ ion observed in the crystal (11). Nucleotide addition by all RNAPs requires two Mg2+ ions: a catalytic ion (Mg1), which coordinates the α-phosphate of NTP and the 3′-O of the RNA, and the NTP-binding ion (Mg2), which coordinates the α-, β-, and γ-phosphates of the incoming NTP (12). The precise positioning of these ions through interactions with the active site carboxylates is required to facilitate an in-line nucleophilic attack of 3′-OH on NTPs Pα and to stabilize the pentavalent transition state. The Tgt-bound Mg2+ ion is coordinated by the active site β′Asp-460 (the residue numbers correspond to the Eco RNAP) and may compromise binding of the Mg2 ion. Our observations that Tgt inhibits all catalytic reactions of RNAP (11) are consistent with this model.
However, this mechanism does not explain why Tgt inhibits RNA chain elongation only at some sites. Analysis of transcription by the yeast RNAP III led to a hypothesis that Tgt acts to increase RNAP pausing at sites at which enzyme is delayed even in the absence of the inhibitor (13). These sites occur between a pyrimidine and a purine residue, an arrangement that is characteristic of pausing. Thus, it appears that Tgt targets RNAP only at pause-inducing sites and may thus act as a conformation-specific inhibitor. Presently, there is no structure of the paused bacterial EC, whereas the available co-crystal structure of Tgt and holoenzyme (11) offers no feasible explanation for these preferences. Careful examination of the co-crystal structure also revealed a substantial number of Tgt interactions with the crystallographic solvent, indicating that the Tgt binding pose in solution can differ in some details from the reported one. Additionally, Tgt binding valences in the EC could digress from the ones observed in the co-crystal due to presence of nucleic acids, substrate, and conformational changes accompanying transcription.
During structural studies of a small molecule binding to the bacterial RNAPs, the target is often a reflection of convenience or happenstance rather than of biological relevance. In addition, alternate structures have been reported for apparently the same ligand-target combinations (14–17), likely reflecting idiosyncrasies of the crystallization and structure-solution procedures. This could be potentially ameliorated by subjecting the crystallographic structures to molecular dynamic simulations of their behavior in solution, taking into account conformational flexibility, entropic effects of solvation/water exclusion, etc., as singularly different crystallographic structures might converge on a single lowest energy-solvated binding pose. To the best of our knowledge, none of the reported inhibitor-bacterial RNAP structures had been subjected to such simulations.
The pattern of site-specific effects of, and differential responses of RNAP locked in “slow” and “fast” states to, the inhibitor suggest that Tgt fails to act on the ECs paused at strong regulatory sites. To understand the molecular basis of this specificity, we constructed a structural model of Tgt bound to the Tth EC to determine whether a compatible site existed for Tgt in the Tth EC. A compatible site was found contacting the folded TL, but requiring a change in its position; this repositioning readily explains the dramatic effect of Tgt on all types of catalysis. The model predicts that β′Arg-933 is required for Tgt function; indeed, resistance conferred by a substitution of Arg-933 for Asn (present in RNAP II) explains why RNAP II is insensitive. We propose that paused ECs, in which the TL is believed to be mobile/unfolded as a result of a formation of a pause RNA hairpin (18), backtracking (19), or fraying of the 3′ end of RNA (20), cannot bind to Tgt and are thus resistant to the action of the inhibitor.
EXPERIMENTAL PROCEDURES
Proteins and Reagents
All general reagents were obtained from Sigma and Fisher. NTPs were from GE Healthcare. PCR reagents were from Eppendorf. Restriction and modification enzymes were from NEB. [α-32P]NTPs were from PerkinElmer Life Sciences. Oligonucleotides were obtained from Integrated DNA Technologies. DNA purification kits were from Qiagen. Tgt was from Epicenter Technologies. Core wild-type and mutationally altered RNAPs were purified as described previously (21). Overexpression plasmids for β′Q504A (pIA847), β′R933A (pIA846), β′H936A (pIA853), and β′R933N (pIA940) were constructed in pVS10 (21) by site-directed mutagenesis.
Transcription Assays
Templates for in vitro transcription were generated by PCR amplification. For steady-state abortive initiation assays, linear T7A1 promoter pIA171 (22) template (2 nm), holo-RNAP (50 nm), ApU (100 μm), and [α32P]CTP were mixed on ice in 50 μl of TGA2 (20 mm Tris acetate, 20 mm sodium acetate, 2 mm magnesium acetate, 5% glycerol, 1 mm DTT, 0.1 mm EDTA, pH 7.9). Tgt was added at the indicated concentrations; reactions were transferred to 37 °C for 20 min and then quenched by the addition of an equal volume of 10 m urea in 45 mm Tris borate, pH 8.3, 50 mm EDTA. For pause assays, halted ECs were formed on a linear pIA171 or pIA349 (21) template (30 nm) with holo-RNAP (40 nm), ApU (100 μm), and starting NTPs (10 μm CTP, ATP, and CTP, 10 μCi if [α-32P]CTP, 3000 Ci/mmol) in TGA2 for 15 min at 37 °C. Transcription was restarted at 37 °C by addition of 150 μm ATP, UTP, CTP, 15 μm GTP, and 10 μg/ml heparin. Reactions were quenched at the indicated times as above. Products were analyzed on 7 m urea, 10% (w/v) acrylamide:bisacrylamide (19:1) denaturing gels, and RNA quantities were determined from PhosphorImager scans of the gels. The assay was repeated at least three times for each variant tested.
Exonuclease Assays
His6-tagged Eco RNAP was immobilized on Co-nitrilotriacetic acid beads (Clontech) and walked along the linear DNA template labeled on the nontemplate strand to a desired position, as described previously (23). The halted ECs were probed with ExoIII (New England Biolabs) as described in Ref. 24 and the supplemental Fig. S4 legend.
RESULTS AND DISCUSSION
Tgt Does Not Accentuate Long Lived Pauses
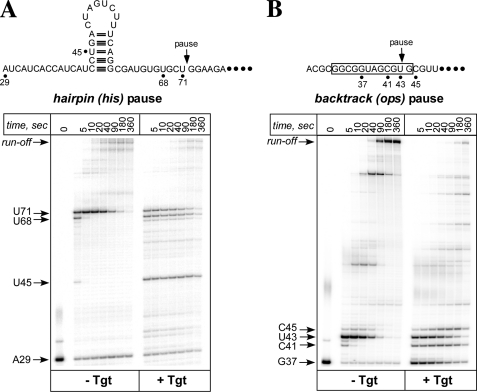
To test whether Tgt preferentially affects the paused RNAP, we used templates that encode the well characterized hairpin-dependent his and a backtracked ops pause sites (22) (Fig. 2). We performed single-round elongation assays by first forming the halted ECs using limiting subsets of initiating NTPs, then adding all four NTPs together with heparin (which inhibits reinitiation) and analyzing RNA products at different time points (from 5 to 360 s). On both templates, addition of Tgt induced pauses at many sites (e.g. C41 and C45 on the ops template and U45 and U68 on the his template) and caused a delay in accumulation of the run-off transcript. Surprisingly, however, transcription at either of the strong, long-lived ops and his pause sites was largely unaffected. These results suggest that although Tgt may act on a distinct state of the EC, the stabilized paused complexes are resistant to inhibition. Instead, Tgt may be specific for an actively transcribing RNAP.
FIGURE 2.
Paused transcription complexes are resistant to Tgt. Radiolabeled halted G37 and A29 complexes were formed on pIA171 (A) and pIA349 (B) templates. These template encode the his and the ops pause signals (shown below). Halted were incubated with Tgt (5 μm) for 2 min at 37 °C and challenged with NTPs and heparin. Aliquots were withdrawn at the indicated times and separated on a 10% denaturing gel. Positions of the halted RNAs, the paused RNA species (U43 at ops, U71 at his site), the run-off transcripts and several other RNA products are indicated with arrows.
Tgt Preferentially Inhibits Fast RNAPs
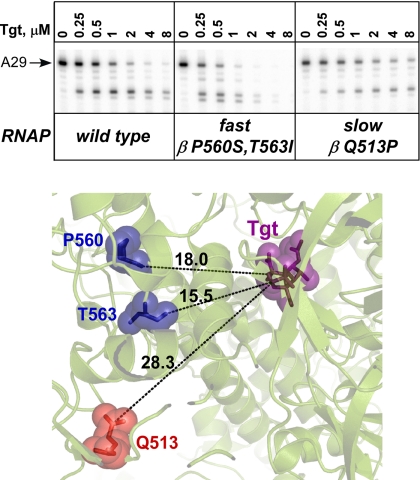
To test this hypothesis, we compared the effect of Tgt on transcription by fast and slow RNAPs. Over the years, many amino acid substitutions in the β and β′ subunits that confer altered responses to regulatory signals during elongation have been isolated (25–29). The fast RNAPs readily transcribe through pause and termination signals whereas the slow variants terminate and pause more efficiently. We compared the effect of Tgt on the wild type, the fast (RpoB5101; βP560S,T563I), and the slow (RpoB8; βQ513P) enzymes to Tgt during formation of halted A29 ECs (Fig. 3). The altered enzymes contain substitutions 15+ Å away from the Tgt binding site (Fig. 3), indicating that they are unlikely to affect the antibiotic binding directly. We found that the fast RNAP was hypersensitive to inhibition, whereas the slow enzyme was more resistant than the wild type. This result is consistent with the failure of Tgt to affect RNAP at pause sites (Fig. 2) because fast and slow RNAPs would be expected to spend less and more time, respectively, off the active elongation pathway. The simplest explanation of these data is that a particular RNAP state may be required for the Tgt binding and action.
FIGURE 3.
Fast and slow enzymes respond differently to Tgt. Top, radiolabeled A29 complexes formed on pIA171 template in the presence of increased concentrations of Tgt (indicated above each lane) with the wild-type (left), RpoB5101 (center), or RpoB8 (right) RNAPs during a 15-min incubation at 37 °C. Reactions were quenched and analyzed on a 10% denaturing gel. Bottom, Distances (in Å) between crystallographic (Protein Data Bank code 2BE5) Tgt (shown as purple sticks and semitransparent spheres) binding site and the positions of fast (blue) and slow (red) amino acid substitutions in RNAP. The residues are numbered as in E. coli.
In Silico Evidence Indicates TL Role in Inhibition by Tgt
We hypothesized that the TL/TH may play a role in Tgt binding. The TH forms a triple-helical bundle with the catalytic β′ bridge helix (BH) and interacts directly with the substrate NTP to “close” the active site and create a perfect catalytic alignment of the active site residues, the 3′-OH, and the NTP phosphates (1). Pausing involves changes in the TL conformation (18–20) that prevent the TH formation and thus the active site closure. In the Tgt-RNAP binary complex, the TL is largely disordered (see supplemental Fig. S1), and Tgt would clash with the TH if modeled into the NTP-bound EC in the orientation captured in the holoenzyme.
Thus, we hypothesized that Tgt may adopt a different docked conformation in the NTP-bound EC. To test this hypothesis in the setting of the low resolution of the target crystallographic structures of RNAP, we first posed the null hypothesis that there would be no structurally compatible site for Tgt in the EC. We reasoned that predicting the actual induced-fit conformation of Tgt bound to the EC starting from a low resolution structure of a slightly distorted conformation of the EC would be nearly impossible, but proving that there is at least one compatible site and then assessing whether it was compatible with the experimental data would not be. Testing the null hypothesis required a computational molecular docking protocol that could take small induced-fit changes and alternative conformations of the TL into account. To accomplish this, we used four-dimensional docking of Tgt to a grid representation of the EC (see supplemental Methods). The grid representation “softens” potentially obstructing full atom hard spheres such as distal side chain atoms, so that slightly distorted Tgt binding pockets in the structure can still be evaluated relative to other molecular surface sites, and the four-dimensional approach can accommodate induced-fit in the TL conformation. Absence of a structurally compatible site for Tgt (e.g. lack of a large enough pocket) can accurately be estimated by docking, and, if the null hypothesis is rejected, a list of preferred contact sites can be identified.
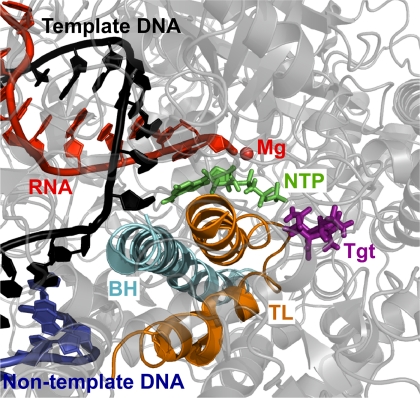
The results show a structurally compatible docking site for Tgt that is dependent on an altered TL conformation (Fig. 4), thereby rejecting the null hypothesis and suggesting that Tgt can interact with the EC. Furthermore, the model is remarkably consistent with the mutational analysis, suggesting that this specific alternative complex conformation may underlie the effect of Tgt on RNA chain elongation. In the final model, Tgt finds a preferred docking site on the Tth EC with the NTP present in the insertion site, but with the Mg2 ion removed and the TL slightly distorted and shifted ∼4 Å toward the catalytic site. In this alternative conformation, the TL bends toward the docked Tgt from its crystallographic position. The NTP remains Watson-Crick paired with the template DNA, but forms a different set of contacts with the RNAP than those seen in the EC, and forms a hydrogen bond with Tgt. Tgt participates in an extensive polar contact and hydrogen bonding network, which includes Lys-780, Arg-783, Lys-908, Met-1238, and Arg-1239 of the β′ subunit, as well as the γ-phosphate of the NTP. Thus, our analysis reveals a structurally compatible site for Tgt in the EC, which is consistent with the mutational data and may explain the effects of Tgt on catalysis.
FIGURE 4.
Tgt interacts with the TL in the Tth EC model. RNA (red scheme), nontemplate DNA (blue scheme), template DNA (black scheme), and substrate NTP (green sticks) are shown; the other colors are as in Fig. 1.
Interactions with the TL Residue Arg-933 Are Critical for Tgt Action
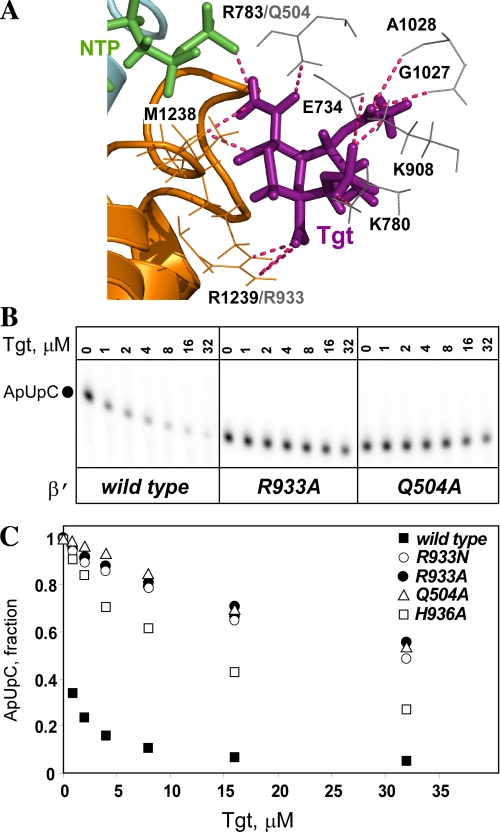
In the model we have generated, Tgt stabilizes the folded TL in a catalytically unproductive configuration through direct contacts with the side chain of β′Arg-1239 and the backbone of β′ Met-1238 (Fig. 4) in the folded TL. β′Arg-1239 interacts with the NTP phosphates in bacterial and yeast ECs (1, 6), and its substitution for Ala confers modest defects on nucleotidyl transfer (8). To test whether this residue is required for Tgt function, we constructed an Ala substitution of the corresponding Eco β′Arg-933. We also constructed Ala substitution of β′His-936, a residue that also interacts with the NTP phosphates in the EC but does not interact directly with Tgt in the model.
We introduced Ala substitutions into the Eco rpoC gene by site-directed mutagenesis, purified the altered RNAPs, and tested their response to Tgt using a steady-state transcription assay in vitro (Fig. 5B). As a control, we used RNAP with β′Gln-504 substituted with Ala; Gln-504 (Arg-783 in Tth) makes direct contact with the acetyl group of Tgt in the binary complex (supplemental Fig. S1) and in the model (Fig. 5A) and is essential for inhibition of Eco RNAP by Tgt (11). An increase in Tgt concentration to 32 μm led to a >90% decline in the synthesis of the ApUpC transcript by the wild-type Eco RNAP. In contrast, β′Q504A and β′R933A RNAPs retained 55–60% activity even at 32 μm Tgt. The β′H936A substitution conferred weaker resistance (supplemental Figs. S2 and S3); we hypothesize that the effect of this substitution is exerted through its impact on the TL structure/deformability and/or creating a pocket next to the Tgt binding site. These results support the importance of the Tgt/β′Arg-933 contacts inferred from the model.
FIGURE 5.
Substitutions of Eco β′Arg-933 (Tth Arg-1239) abolish inhibition by Tgt. A, polar contacts between Tgt (purple sticks) and amino acids (orange and gray sticks) in the model of Tgt/EC (shown in Fig. 4; the color scheme is preserved). B and C, inhibition of abortive transcription on the T7A1 promoter by Tgt. Formation of the radiolabeled ApUpC RNA was followed as a function of Tgt concentration (from 0 to 32 μm) with the wild-type or altered Eco RNAPs. The key is shown in the figure.
The observed contacts between Tgt and β′Arg-933 could also explain the differential effect of Tgt on eukaryotic enzymes. Indeed, in the Tgt-sensitive RNAP III β′Arg-1239 is conservatively substituted for a Lys, whereas the Tgt-resistant RNAP II contains a structurally and chemically distinct Asn at this position (Fig. 5A). In support of this idea, we found that Eco RNAP with β′Arg-933 substituted for Asn was highly resistant to Tgt (Fig. 5C).
Tgt May Stabilize the Folded TL to Inhibit Translocation
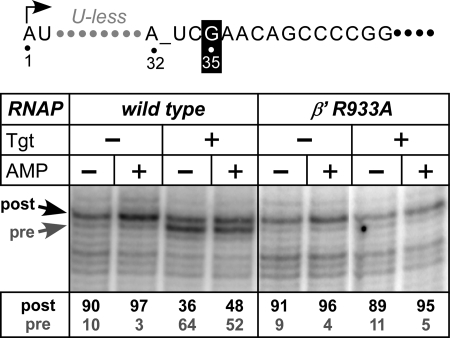
Following nucleotide addition, RNAP releases PPi and moves 1 nucleotide downstream. At some point during this transition, the TH unfolds, and the incoming NTP binds to a posttranslocated inducing the TL→TH transition. Upon misincorporation, RNAP backtracks to thread the nascent RNA through the active site; backtracking is incompatible with the folded TL. By stabilizing the folded TL, Tgt would be expected to inhibit RNAP translocation in either forward or reverse direction. To test this prediction, we used exonuclease footprinting to determine the translocation register of the RNAP (Fig. 6).
FIGURE 6.
Tgt inhibits RNAP translocation. The translocation registers of the EC35 formed on a template shown on top with the wild-type or β′R933A Eco RNAPs determined by ExoIII footprinting; Tgt (60 μm) and AMP (1 mm) were added where indicated. The stop positions that correspond to the post- and pretranslocated ECs are shown by arrows, and their ratios are shown below each lane. See also supplemental Fig. S4.
In EC35 formed by the wild-type RNAP (Fig. 6, left), the enzyme occupied the post- and pretranslocation states at a ratio of 90:10. As expected, addition of a substrate analog AMP stabilized the posttranslocated state (93:7). Tgt shifted equilibrium to the pretranslocated state both in the absence and in the presence of the substrate analog (36:64 and 48:52, respectively). The β′R933A RNAP was mostly in a posttranslocated state, which was further stabilized by AMP, both in the absence and in the presence of Tgt (Fig. 6, right). Tgt also stabilized the pretranslocated state in a backtrack-prone EC33 (supplemental Fig. S4), blocking reverse translocation. Thus, Tgt inhibits RNAP translocation in either direction.
These observations are consistent with a model in which structural transitions of the TL control equilibrium between the posttranslocated (TH) and pretranslocated (TL) states; the bound Tgt will stabilize the TH and inhibit its unfolding into the TL. However, this effect is unlikely to play a major role in the Tgt inhibitory mechanism, as the translocation is preceded by the chemistry (NTP condensation) step, also inhibited by Tgt.
Control of Transcription by Motions of the TL
Multisubunit RNAPs carry out a complex reiterative process of mechano-chemical work, which requires a certain degree of conformational mobility of their subunits and/or domains, characteristic of all macromolecular motors (30). Range of motions associated with transcription varies greatly from small oscillations of a catalytic loop (31) to large scale ratcheting of the entire RNAP (32). Intermediate scale transitions of the metamorphic TL are thought to play crucial roles in transcription mechanism and regulation. NTP-dependent refolding of the TL into TH allows the RNAP to sample substrate concentrations, read out the state of the EC, and link catalytic and mechanical steps in a robust fashion still amenable to regulation. Folded TH completes the substrate binding site and appears to play a role in the catalytic step as well, after which it (together with BH) acts as a molecular pawl in RNAP translocation along the nucleic acid scaffold (32). Conformation of the TL may also restrict action of some transcription factors, such as Gre (5), to particular ECs.
The TL was crystallized in a number of different states, some apparently mobile or statistically disordered, and coarse-grained analysis of Tth RNAP structures indicated that the TL, together with the BH, possesses a number of amino acid residues with increased flexibility/deformability (33). Taking advantage of this pliability, Tgt binds to the TH concurrently with the substrate and, by inducing deformation of the TH tip, disrupts chemical (NTP condensation) and subsequent mechanical (translocation) steps. An unrelated RNAP inhibitor Stl binds to, and in the path of a proposed movement of, the unfolded TL in apo-RNAP, thereby blocking transcription prior to binding of the substrate (16, 17). In a context of the EC, Stl binds together with the substrate, forming contacts with the downstream DNA and displacing a partially folded TH (1). The variable mode of Stl binding, invariably trapping the TL, is consistent with its inhibitory effects on initiation, elongation, and pyrophosphorolysis. Consistent with their ability to trap the TL, Tgt and Stl have similar effects on RNAP translocation (supplemental Fig. S5) and fail to affect the his paused complexes (Fig. 2A and Ref. 8), suggesting that an altered state of the TL that is thought to exist in a paused EC is incompatible with binding of either antibiotic.
The proposed mechanism of Tgt underscores the key role of the TL in modulation of RNAP activity by antibiotics and cellular accessory proteins. Those regulators that bind in the secondary channel could interact with the TL directly whereas others may alter the TL folding indirectly, through coupled changes in other flexible elements of the RNAP.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health grants (to I. A., E. N., S. M. N., and T. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. S1–S5, and additional references.
- RNAP
- RNA polymerase
- BH
- bridge helix
- EC
- elongation complex
- Eco
- Escherichia coli
- NTP
- nucleotide triphosphate
- Stl
- streptolydigin
- Tgt
- tagetitoxin
- TH
- trigger helice
- TL
- trigger loop
- Tth
- Thermus thermophilus.
REFERENCES
- 1. Vassylyev D. G., Vassylyeva M. N., Zhang J., Palangat M., Artsimovitch I., Landick R. (2007) Nature 448, 163–168 [DOI] [PubMed] [Google Scholar]
- 2. Brueckner F., Cramer P. (2008) Nat. Struct. Mol. Biol. 15, 811–818 [DOI] [PubMed] [Google Scholar]
- 3. Kaplan C. D., Larsson K. M., Kornberg R. D. (2008) Mol. Cell 30, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nudler E. (2009) Annu. Rev. Biochem. 78, 335–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roghanian M., Yuzenkova Y., Zenkin N. (2011) Nucleic Acids Res. 39, 4352–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., Kornberg R. D. (2006) Cell 127, 941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuzenkova Y., Zenkin N. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10878–10883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J., Palangat M., Landick R. (2010) Nat. Struct. Mol. Biol. 17, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathews D. E., Durbin R. D. (1990) J. Biol. Chem. 265, 493–498 [PubMed] [Google Scholar]
- 10. Mathews D. E., Durbin R. D. (1994) Biochemistry 33, 11987–11992 [DOI] [PubMed] [Google Scholar]
- 11. Vassylyev D. G., Svetlov V., Vassylyeva M. N., Perederina A., Igarashi N., Matsugaki N., Wakatsuki S., Artsimovitch I. (2005) Nat. Struct. Mol. Biol. 12, 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steitz T. A. (1998) Nature 391, 231–232 [DOI] [PubMed] [Google Scholar]
- 13. Steinberg T. H., Burgess R. R. (1992) J. Biol. Chem. 267, 20204–20211 [PubMed] [Google Scholar]
- 14. Belogurov G. A., Vassylyeva M. N., Sevostyanova A., Appleman J. R., Xiang A. X., Lira R., Webber S. E., Klyuyev S., Nudler E., Artsimovitch I., Vassylyev D. G. (2009) Nature 457, 332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mukhopadhyay J., Das K., Ismail S., Koppstein D., Jang M., Hudson B., Sarafianos S., Tuske S., Patel J., Jansen R., Irschik H., Arnold E., Ebright R. H. (2008) Cell 135, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Temiakov D., Zenkin N., Vassylyeva M. N., Perederina A., Tahirov T. H., Kashkina E., Savkina M., Zorov S., Nikiforov V., Igarashi N., Matsugaki N., Wakatsuki S., Severinov K., Vassylyev D. G. (2005) Mol. Cell 19, 655–666 [DOI] [PubMed] [Google Scholar]
- 17. Tuske S., Sarafianos S. G., Wang X., Hudson B., Sineva E., Mukhopadhyay J., Birktoft J. J., Leroy O., Ismail S., Clark A. D., Jr., Dharia C., Napoli A., Laptenko O., Lee J., Borukhov S., Ebright R. H., Arnold E. (2005) Cell 122, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toulokhonov I., Zhang J., Palangat M., Landick R. (2007) Mol. Cell 27, 406–419 [DOI] [PubMed] [Google Scholar]
- 19. Wang D., Bushnell D. A., Huang X., Westover K. D., Levitt M., Kornberg R. D. (2009) Science 324, 1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sydow J. F., Brueckner F., Cheung A. C., Damsma G. E., Dengl S., Lehmann E., Vassylyev D., Cramer P. (2009) Mol. Cell 34, 710–721 [DOI] [PubMed] [Google Scholar]
- 21. Belogurov G. A., Vassylyeva M. N., Svetlov V., Klyuyev S., Grishin N. V., Vassylyev D. G., Artsimovitch I. (2007) Mol. Cell 26, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Artsimovitch I., Landick R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7090–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nudler E., Gusarov I., Bar-Nahum G. (2003) Methods Enzymol. 371, 160–169 [DOI] [PubMed] [Google Scholar]
- 24. Epshtein V., Cardinale C. J., Ruckenstein A. E., Borukhov S., Nudler E. (2007) Mol. Cell 28, 991–1001 [DOI] [PubMed] [Google Scholar]
- 25. Bar-Nahum G., Epshtein V., Ruckenstein A. E., Rafikov R., Mustaev A., Nudler E. (2005) Cell 120, 183–193 [DOI] [PubMed] [Google Scholar]
- 26. Fisher R. F., Yanofsky C. (1983) J. Biol. Chem. 258, 8146–8150 [PubMed] [Google Scholar]
- 27. Landick R., Stewart J., Lee D. N. (1990) Genes Dev. 4, 1623–1636 [DOI] [PubMed] [Google Scholar]
- 28. McDowell J. C., Roberts J. W., Jin D. J., Gross C. (1994) Science 266, 822–825 [DOI] [PubMed] [Google Scholar]
- 29. Weilbaecher R., Hebron C., Feng G., Landick R. (1994) Genes Dev. 8, 2913–2927 [DOI] [PubMed] [Google Scholar]
- 30. Hwang W., Lang M. J. (2009) Cell Biochem. Biophys. 54, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu T., Janetzko F., Zhang Y., van Duin A. C. T., Goddard W. A. I., Salahub D. R. (2008) Theor. Chem. Accounts 120, 479–489 [Google Scholar]
- 32. Tagami S., Sekine S., Kumarevel T., Hino N., Murayama Y., Kamegamori S., Yamamoto M., Sakamoto K., Yokoyama S. (2010) Nature 468, 978–982 [DOI] [PubMed] [Google Scholar]
- 33. Yildirim Y., Doruker P. (2004) J. Biomol. Struct. Dyn. 22, 267–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.