Background: It is not known if ubiquitination controls endocytic trafficking of μ-opioid receptors.
Results: Endocytic sorting to the proteolytic compartment is ubiquitination-independent. Ubiquitination specifically in the first cytoplasmic loop promotes multivesicular body sorting required for subsequent destruction of the ligand binding site.
Conclusion: The ubiquitinated first cytoplasmic loop represents a discrete downstream sorting determinant.
Implications: The down-regulation mechanism is hierarchical.
Keywords: 7-Helix Receptor, Intracellular Trafficking, Lysosomes, Opiate Opioid, Ubiquitination
Abstract
μ-Type opioid receptors (MORs) are members of the large seven-transmembrane receptor family which transduce the effects of both endogenous neuropeptides and clinically important opioid drugs. Prolonged activation of MORs promotes their proteolytic degradation by endocytic trafficking to lysosomes. This down-regulation process is known to contribute to homeostatic regulation of cellular opioid responsiveness, but mechanisms that mediate and control MOR down-regulation have not been defined. We show here that lysosomal down-regulation of MORs is ESCRT (endosomal sorting complex required for transport)-dependent and involves ubiquitin-promoted transfer of internalized MORs from the limiting endosome membrane to lumen. We also show that MOR down-regulation measured by conventional radioligand binding assay is determined specifically by ubiquitination in the first cytoplasmic loop. Surprisingly, we were unable to find any role of ubiquitination in determining whether internalized receptors recycle or are delivered to lysosomes. Instead, this decision is dictated specifically by the MOR C-tail and occurs irrespectively of the presence or absence of receptor ubiquitination. Our results support a hierarchical organization of discrete ubiquitin-independent and -dependent sorting operations, which function non-redundantly in the conserved down-regulation pathway to mediate precise endocytic control. Furthermore, they show that this hierarchical mechanism discriminates the endocytic regulation of naturally occurring MOR isoforms. Moreover, they are the first to reveal, we believe, for any seven-transmembrane receptor, a functional role of ubiquitination in the first cytoplasmic loop.
Introduction
μ-Opioid receptors (MOP-Rs or MORs)3 are members of the large family of seven-transmembrane signaling receptors (7TMRs) and mediate the physiological actions of endogenous opioid neuropeptides as well as clinically important opiate drugs (1, 2). MORs are extensively regulated after ligand-induced activation, producing long term changes in cellular opioid responsiveness that influence tolerance and dependence at the level of tissues and whole animals (3–5). The number and functional activity of surface-accessible MORs are regulated by ligand-induced endocytosis via clathrin-coated pits (6), but the functional consequences of MOR endocytosis depend critically on the molecular sorting of receptors after endocytosis. Receptor sorting into the recycling pathway restores the surface-accessible receptor pool and contributes to rapid recovery of cellular opioid responsiveness. Sorting of internalized receptors to lysosomes promotes proteolytic destruction of receptors, a process that is traditionally measured pharmacologically by a time-dependent down-regulation of the total number of ligand binding sites measured in cell or tissue extracts. This down-regulation process is associated with a prolonged attenuation, rather than recovery, of signaling. MOR trafficking in the recycling pathway has been studied in some detail (7), but essentially nothing is known about sorting operations mediating down-regulation of this 7TMR.
The endocytic trafficking itinerary of many signaling receptors is determined by ubiquitin-dependent sorting from the endosome-limiting membrane to the lumenal membrane compartment of late endosomes/multivesicular bodies (8–10). Ubiquitin-directed sorting to the intralumenal compartment is mediated by the endosomal sorting complex required for transport (ESCRT), and ubiquitination is thought to primarily determine whether many signaling receptors recycle or degrade after endocytosis (11–13). It is not known if ubiquitination plays any role in the endocytic sorting or down-regulation of MORs. Furthermore, ubiquitination plays a minor and dispensable role in down-regulating a close MOR paralogue, the δ-opioid receptor (DOR), even though these receptors traverse the canonical multivesicular body pathway (14–17). MORs have the particularly interesting feature that their endocytic trafficking between recycling and lysosomal pathways is regulated by a modular MOR regulatory sequence (MRS) (18, 19), which is present in the receptor distal C-terminal tail and differs across MOR isoforms due to alternate RNA splicing (20–22). The MRS present in the MOR1 variant specifically confers rapid recycling on internalized receptors and effectively inhibits receptor trafficking to lysosomes. It is not known if ubiquitination or the ESCRT machinery plays any role in determining the down-regulation of MORs, nor is it known what functional relationship might exist between these mechanisms and sorting controlled by the MRS.
The presently described study addressed these questions. We show that down-regulation of MORs indeed occurs by the canonical ESCRT-dependent pathway, and that MORs undergo agonist-stimulated ubiquitination which promotes sorting of receptors from the endosome limiting membrane to the lumenal compartment. We also establish the MOR as the first example of a 7TMR whose topological sorting by this mechanism is controlled specifically by ubiquitination in the first cytoplasmic loop. Interestingly, in contrast to the prevailing view, the ubiquitin-directed sorting step does not control the essential trafficking fate of internalized MORs. Instead, the ubiquitin-dependent step functions independently and effectively downstream of a primary “recycling versus degradation” decision that does not require receptor ubiquitination and is determined by the C-terminal MRS. Furthermore, although the ubiquitin-dependent step is not required to direct internalized receptors to lysosomes or initiate their proteolysis, it plays a specific role in promoting subsequent destruction of the receptor opioid binding site. Accordingly, MORs have the remarkable ability to undergo endocytic delivery to lysosomes and incur substantial proteolytic fragmentation without losing the ability to specifically bind an opioid radioligand. This separation of sorting steps is biologically relevant because only the first step distinguishes the down-regulation properties of naturally occurring MOR isoforms. It may also impact the interpretation of studies of MOR regulation in vivo because the second step is rate-limiting for loss of radioligand binding sites as estimated by a traditional pharmacological assay of down-regulation.
EXPERIMENTAL PROCEDURES
Cell Culture, cDNA Constructs, and Transfection
GFP-tagged Rab5 cDNA was a gift from Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics), and the Q79L mutation was generated by site-directed mutagenesis (Stratagene), with the respective coding sequences cloned into pECFP-N1. The MycHRS expression plasmid was a gift from Harold Stenmark (Norwegian Radium Hospital). The N-terminal FLAG-tagged murine MOR1 μ-opioid receptor construct (F-MOR), recycling impaired C-terminal truncation (F-MORΔ17), and MOR1B splice variant (F-MOR1B) have been previously described (18, 23). Lysine-mutant versions (F-MOR-0cK and the F-MORΔ17–0cK) and subsequent single loop mutants as indicated were generated using QuikChange site-directed mutagenesis (Stratagene). Mutated cDNAs were cloned into pcDNA3 (Invitrogen), and sequences were verified by dideoxynucleotide sequencing (ElimBio). F-MORΔ17-GFP, F-MORΔ17–0cK-GFP, F-MORΔ17-K94R,K96R-GFP, and F-MORΔ17–1st loop K-GFP fusion proteins were constructed by amplifying the respective coding sequence by polymerase chain reaction (PCR), with an AgeI restriction site incorporated into the reverse primer. PCR products were then ligated in-frame in pEGFP-N1 (Clontech). All cDNA constructs were verified by sequencing (ElimBio). Stably transfected cells expressing epitope tagged receptors were generated by selection by neomycin resistance using 500 μg/ml G418 (Geneticin, Invitrogen). Resistant colonies were clonally isolated and selected for further study based on comparable levels of receptor expression as assessed by fluorescence microscopy and saturation binding analysis (supplemental Table 1). HEK293 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (University of California, San Francisco, Cell Culture Facility). For transient expression, cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer instructions. Cells expressing FLAG-tagged receptors were harvested by washing with EDTA and plated in 60-mm dishes at 80% confluency before transfection with plasmid DNA. Cells were reseeded into poly-lysine-coated 6- or 12-well plates and cultured for a further 24 h before experimentation. Transfection of cells with RNAi duplexes to Tsg101 (15) was achieved using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions. Cells were transfected at 50% confluency, and experiments were performed 72 h post-transfection.
Biochemical Detection of Receptor Proteolysis and Protein Levels by Immunoblotting
Immunoblotting to assess total cellular receptor levels was carried out as previously described. Briefly, cell monolayers were washed 3 times in ice-cold phosphate-buffered saline (PBS) and lysed in extraction buffer (0.5% Triton X-100, 150 mm NaCl, 25 mm KCl, 25 mm Tris, pH 7.4, 1 mm EDTA) supplemented with a standard protease inhibitor mixture (Roche Applied Science). Extracts were clarified by centrifugation (12,000 × g for 10 min) and then mixed with SDS sample buffer for denaturation. Proteins present in the extracts were resolved by SDS-PAGE using 4–12% Bis-Tris gels (NuPAGE, Invitrogen), transferred to nitrocellulose membranes, and probed for protein by immunoblotting using anti-FLAG-M1 (Sigma) and horseradish peroxidase-conjugated sheep anti-mouse IgG (GE Healthcare Life Sciences) and SuperSignal detection reagent (Pierce). Apparent molecular mass was estimated using commercial protein standards (SeeBlue Plus2, Invitrogen). Band intensities of unsaturated immunoblots were analyzed and quantified by densitometry using FluorChem 2.0 software (AlphaInnotech Corp.).
Biochemical Detection of Receptor Ubiquitination
Cells were plated in 10-cm dishes and then treated before being lysed on ice in 100 μl of 10 mm Tris, pH 7.4, 1% SDS, 10 mm iodoacetamide, and protease inhibitor mixture (Roche Applied Science). The lysate was then diluted with 400 μl of extraction buffer (see above), and samples were sonicated for 5 s at a 30% duty cycle. The extract was then clarified by centrifugation (12,000 × g for 10 min), and the supernatant was passed through a Qia-shredder column (Qiagen) at 3000 rpm for 60 s. Another 500 μl of extraction buffer was then added to give a final volume of 1 ml and then incubated overnight at 4 °C with 2 μg of M2 anti-FLAG antibody (Sigma). 30 μl of protein A/G-agarose (Pierce) was added for 2 h at 4 °C. Immunoprecipitates were collected by centrifugation (3000 rpm, 1 min, 4 °C) and washed 3 times with 500 μl of extraction buffer before the addition of 20 μl of SDS sample buffer (Invitrogen) supplemented with β-mercaptoethanol and analysis by Western blotting using anti-ubiquitin (P4D1, Santa Cruz). Blots were then stripped (Restore Western Blot Stripping Buffer, Pierce) and reprobed with anti-FLAG M2-HRP (Sigma) to verify equal receptor levels.
Analysis of Receptor Levels by Radioligand Binding
Receptor down-regulation was determined by radioligand binding as previously described (16). After transfection, HEK293 cells stably expressing FLAG-tagged receptors were re-plated into 12-well plates. 24 h later 10 μm d-Ala-d-Leu-enkephalin (DADLE) was added to the cells for the indicated time period, cells were washed twice with ice-cold PBS, and 300 μl of PBS was added to the cells, and the plates were frozen. Plates were thawed, and cells were resuspended. Binding assays were performed in triplicate in 96-well plates using 10 nm of the radiolabeled opioid receptor antagonist [3H]diprenorphine (DPN) (Amersham Biosciences, 88 Ci/mmol), a saturating concentration that is sufficient to access both surface and internal receptors under these conditions, and incubated for 1 h at room temperature. Incubations were terminated by vacuum filtration through glass fiber filters (Whatman), and unbound radioligand was removed by repeated washes with TBS. Bound radioactivity was determined by liquid scintillation counting of washed filters. Nonspecific binding, determined by carrying out parallel determinations in the presence of excess unlabeled competitive antagonist (10 μm naloxone), was <10% of specific. In all assays we verified that bound diprenorphine was <10% of the total added to the assay. Data presented represent the specific binding (total minus nonspecific binding) at each time point, expressed as a percentage of specific binding in similarly transfected, but agonist naive cells.
Fluorescence Microscopy
Colocalization of receptors with early endosomes or lysosomes was visualized using HEK293 cells stably expressing the indicated FLAG-tagged receptor constructs plated on poly-lysine-coated glass coverslips (Corning). Cells were incubated in the presence of 10 μm DADLE for 30 or 90 min before fixing with 4% formaldehyde and permeabilizing with 0.1% Triton X-100 in PBS. Cells were labeled using rabbit anti-FLAG (Sigma) and mouse antibodies recognizing either EEA1 (BD Biosciences) or LAMP-1 and -2 (Santa Cruz Biotechnology). Localization was confirmed using a MOR antibody (generated in rabbits against a previously described GST fusion of the MOR C-terminal tail (24)), with injections and bleeds carried out according to approved procedures (Josmine Laboratories, Napa, CA). Immunolabeling was detected using Alexa488-conjugated anti-mouse and Alexa594 anti-rabbit secondary antibodies (Invitrogen). Specimens were imaged by laser-scanning confocal fluorescence microscopy using a Zeiss LSM 510 microscope fitted with a Zeiss 63 × NA1.4 objective, with standard excitation laser lines and emission filter sets verified for lack of detectable cross-channel bleed-through and confocality achieved using a 1 Airy disc pinhole in the emission light path. Acquired optical sections were analyzed with LSM Image Examiner (Zeiss) and rendered with Adobe Photoshop software.
Spinning-disk Confocal Microscopy and Live Image Analysis
The extent of receptor involution was visualized using HEK293 cells transiently transfected with the indicated N-terminal FLAG-tagged, C-terminal GFP-tagged receptor constructs plated onto poly-lysine-coated glass coverslips (Corning Glass). Cells were incubated in the presence of 10 μm DADLE for 90 min before imaging. Cells were imaged in DMEM without Phenol Red supplemented with 1% fetal bovine serum (University of California, San Francisco Cell Culture Facility) and including 30 mm Hepes adjusted to pH 7.4. Live cell imaging was performed using a Yokogawa CSU22 Spinning Disk Confocal (Solamere Technology Group) interfaced to a Nikon TE2000U inverted microscope using a 100 × 1.49 NA TIRF objective and fiber-coupled 488-nm argon laser (Coherent) for excitation. Time-lapse sequences were acquired at a continuous rate of 5 frames/s, and the acquired images were analyzed with Image J software (Wayne Rasband, National Institutes of Health, Bethesda, MD). To quantify the extent of receptor involution, measurements were conducted on raw data of individual endosomes as previously described (17). To minimize the influence of out-of-plane fluorescence from the limiting membrane (in the z axis), only mid-focal plane images of endosomes with a diameter of >3 μm were analyzed. For each optical section, straight-line selections were drawn across the diameter, and pixel intensities across the line were measured. Endosomal diameter was normalized to account for endosomes varying in size. The pixel numbers with the first and second maximum pixel intensities, corresponding to pixels on the limiting membrane of the endosome, were normalized to 0 and 100, respectively. The location across the line of pixel 0 was then subtracted from each pixel situated on the line, and this value was divided by the total diameter (in pixels) of the endosome. This generated normalized pixel distances corresponding to distance across the line occupied by each pixel, expressed as a percentage. Average background fluorescence was subtracted from raw pixel intensity values. The pixel intensities for the pixel numbers normalized to 0 and 100 were also normalized to 0 and 100, respectively, generating normalized fluorescence values. The background-corrected pixel intensity values corresponding to pixels that lay 40–60% across the endosomal diameter were averaged, generating a middle fluorescence value for each endosome. Middle fluorescence values were compiled across multiple cells, and the mean values quantified for each condition are shown. Representative live images shown were rendered using Adobe Photoshop software.
Flow Cytometric Analysis of Internal and Recycled Receptors
Fluorescence flow cytometry was used to quantify internalization and recycling of receptors by measuring cell surface fluorescence, as previously described (25). Briefly, stably transfected cells were treated for 30 min with 10 μm DADLE (internalization) before placing on ice or washing and returning to 37 °C for 45 min in the presence of Naloxone (recycling). Cells were then washed and lifted with ice-cold PBS and incubated with Alexa647-conjugated M1 anti-FLAG antibody (Molecular Probes). Fluorescence intensity was measured using a FACSCalibur (BD Biosciences), counting 10,000 cells/sample in duplicate. Recycling was determined from surface fluorescence (F) as follows (Fnaloxone − FDADLE)/(Funtreated − FDADLE) × 100.
Statistical Analysis
Quantitative data were averaged across multiple independent experiments, with the number of experiments specified in the corresponding figure legend. Unless indicated otherwise, error bars represent the S.E. determined after compiling mean determinations across experiments. The statistical significance of the indicated differences was analyzed using the appropriate variations of one or two-way ANOVA and post-test or Student's t test as specified in the figure legends, calculated using Prism 4.0 software (GraphPad Software, Inc.). The relative significance of each of the reported differences is specified by calculated p values that are also listed in the figure legends and annotated graphically in the figures.
RESULTS
MOR Down-regulates by Lysosomal Proteolysis Requiring the ESCRT
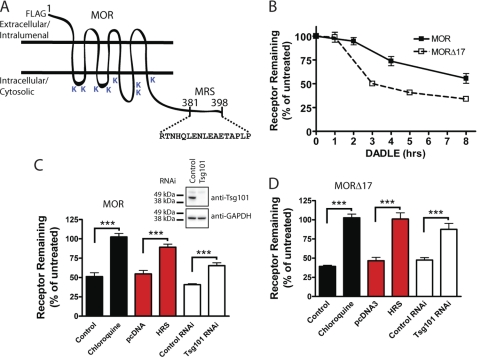
We generated stably transfected cell lines expressing a FLAG epitope-tagged receptor construct (F-MOR, Fig. 1A) at moderate levels (supplemental Table 1). Agonist-induced down-regulation was assessed by radioligand binding assay using [3H]DPN, which binds to MORs with high affinity and does not detect any endogenous receptors in untransfected HEK293 cells, by the standard method of determining specific binding at saturating concentration to estimate Bmax. There was little change in net cellular F-MOR binding for ∼2 h after the addition to the culture medium of 10 μm DADLE, an opioid peptide agonist that robustly promotes MOR endocytosis. However, a significant reduction of net binding sites was observed after longer agonist exposure (Fig. 1B, filled squares). Truncation of the distal 17 residues from the MOR cytoplasmic tail (F-MORΔ17), which removes the previously defined MRS promoting receptor recycling after short term agonist exposure (18), markedly accelerated agonist-induced down-regulation observed in the prolonged presence of DADLE (Fig. 1B, open squares). Thus, although MORs predominantly recycle after brief agonist-induced activation and endocytosis, they undergo significant down-regulation after prolonged stimulation, and the degree to which they do so is controlled by the previously described C-terminal MRS.
FIGURE 1.
Both MOR and MORΔ17 undergo ESCRT-dependent down-regulation by lysosomes. A, shown is a diagrammatic representation of the F-MOR and F-MORΔ17 constructs, indicating the location of cytoplasmic lysine residues and the previously identified MRS. B, shown is the time course of the down-regulation of F-MOR and F-MORΔ17. HEK293 cells stably expressing either receptor were incubated at 37 °C with 10 μm DADLE for the indicated time period, and then radioligand binding assay was carried out using the high affinity radiolabeled opioid antagonist [3H]DPN at 10 nm to estimate Bmax, as described under “Experimental Procedures.” Data points represent specific binding of [3H]DPN measured at each time point, expressed as a percentage of the specific binding in cells not exposed to agonist. Points represent mean determinations from independent experiments, with each time point analyzed in triplicate tubes in each experiment. Error bars represent the S.E. calculated across the experiments (n = 3–5). C and D, shown is the effect of the indicated experimental manipulations on DADLE-induced down-regulation of F-MOR (panel C) and F-MORΔ17 (panel D) in stably transfected HEK293 cells. Cells were transfected with “empty” pcDNA3 or pcDNA3-HRS (red bars) or transfected with control (scrambled) siRNA or siRNA targeting TSG101 (white bars). A final set of cells was pretreated with 200 μm chloroquine for 20 min before the start of the experiment (black bars). Cells were then left untreated or exposed to 10 μm DADLE for 5 h (F-MORΔ17, panel D) or 8 h (F-MOR, panel C), chosen according to the different down-regulation kinetics of these constructs, before carrying out the radioligand binding assay using 10 nm [3H]DPN. Bars represent the specific binding detected after agonist treatment, expressed as a percentage of that detected to identically manipulated cells except not exposed to agonist. In each experiment binding was determined in triplicate tubes. Bars represent average determinations, and error bars S.E., across multiple experiments (n = 3–6; ***, p = <0.001, two way ANOVA). The inset shows correlative immunoblot data verifying Tsg101 depletion by the siRNA.
Agonist-induced down-regulation of F-MOR was inhibited by chloroquine, verifying that MORs indeed down-regulate by lysosomal proteolysis. Down-regulation was also inhibited by HRS overexpression, which disrupts ESCRT0 function, as well as by siRNA-mediated knockdown of the essential ESCRT1 component TSG101/hVPS23 (Fig. 1C and inset). Down-regulation of F-MORΔ17, assessed using the same manipulations (except at 5 rather than 8 h after agonist addition due to its substantially faster kinetics), exhibited the same dependence (Fig. 1D). Thus, pharmacological down-regulation of MORs requires the ESCRT machinery, and the C-terminal MRS controls the extent to which internalized MORs traverse the ESCRT-dependent down-regulation pathway after prolonged activation.
Down-regulation Assessed Pharmacologically by Agonist-induced Loss of Radioligand Binding Requires MOR Ubiquitination
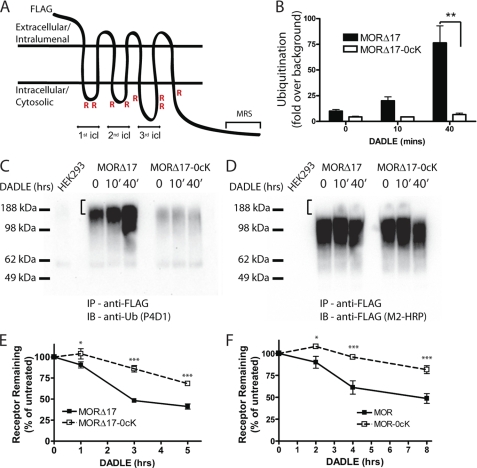
To focus on whether down-regulation of MORs also involves receptor ubiquitination, we examined the effect of mutating all eight intracellular lysine residues in the background of mutant receptor lacking the C-terminal motif (F-MORΔ17–0cK, Fig. 2A). The F-MORΔ17 receptor construct was clearly ubiquitinated, and ubiquitin incorporation increased in an agonist-stimulated manner (Fig. 2B). Lysyl mutation prevented this ubiquitination, as verified by comparison of anti-ubiquitin and anti-receptor blots prepared from the corresponding anti-FLAG immunoprecipitates (Fig. 2, C and D, respectively). Interestingly, the most highly ubiquitinated MOR species were not detectable in the corresponding anti-FLAG (receptor) blot when exposed appropriately to resolve the major species of epitope-tagged receptor protein (indicated by the bracket in Fig. 2, C and D). Nevertheless, specific detection of these ubiquitinated species was verified by the lack of signal in the control (HEK293) lysate (left lane in each panel). This suggests that a small fraction of the total cellular receptor pool is highly ubiquitinated at any one time in intact cells. A similar observation has been made for other 7TMRS, including the DOR and β2 adrenergic receptor (16, 26, 27). In these experiments relatively harsh extraction and washing conditions were used to assure dissociation of residual non-receptor ubiquitinated proteins (see “Experimental Procedures”). This caused immunopurified receptors to resolve by SDS-PAGE as higher order (dimeric and multimeric) species rather than as the primarily monomeric species detected in cell extracts prepared under more gentle conditions (compare Figs. 2D and 3A). Interestingly, radioligand binding assay indicated that preventing ubiquitination in this manner effectively blocked agonist-induced down-regulation measured by diprenorphine binding for both the MRS-truncated and full-length receptor constructs (F-MORΔ17–0cK and F-MOR-0cK, Figs. 2, E and F, respectively).
FIGURE 2.
MOR down-regulation measured radioligand binding requires receptor ubiquitination. A, shown is a diagram of the F-MOR-0cK construct indicating cytoplasmic domains containing the Lys → Arg mutation (R) as well as the MRS that is devoid of lysine residues. icl, cytoplasmic loop. B, shown is a densitometric analysis of ubiquitin incorporation into the F-MORΔ17 and the lysyl-mutant F-MORΔ17–0cK, assessed after immunopurification of receptors in the presence of SDS. Data shown represent the mean and S.E. of densitometry from anti-ubiquitin immunoblot analysis carried out in n = 3 independent experiments. Scanning densitometry was carried out in the linear range and is expressed as -fold over background (defined by nonspecific signal detected in parental HEK293 cells not expressing the indicated FLAG-tagged receptor). C, a representative anti-ubiquitin immunoblot (IB) from the analysis summarized in panel B; IP, immunoprecipitate. D, shown is the same blot as in panel C, except it was stripped and reprobed with anti-FLAG to verify comparable loading and transfer of immunopurified receptors. E and F, shown is the effect of the indicated lysyl mutations on DADLE-induced down-regulation. HEK293 cells stably expressing F-MORΔ17 or F-MORΔ17–0cK (panel E) or F-MOR or F-MOR-0cK (panel F) were exposed to 10 μm DADLE for the indicated time period before estimating Bmax by radioligand binding to [3H]DPN. Points represent specific binding at each time point, expressed as a percentage of the specific binding in cells not exposed to agonist. In each experiment triplicate tubes were analyzed; points represent averages and error bars S.E. across experiments (n = 4; *, p < 0.05; ***, p < 0.001, two way ANOVA).
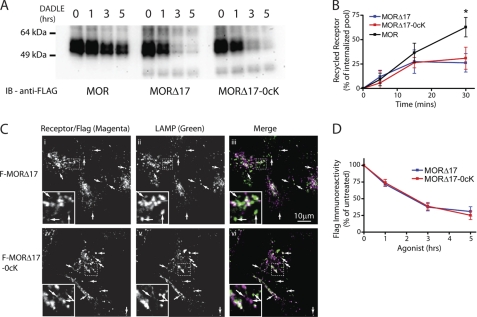
FIGURE 3.
MOR trafficking to lysosomes, assessed biochemically or immunochemically, does not require receptor ubiquitination and is dictated by the C-tail. A, representative anti-FLAG immunoblots (from three-six independent experiments) show the effects of exposing cells to 10 μm DADLE for the indicated time period on FLAG-tagged receptor signal detected in cell extracts. Stably transfected HEK293 cells initially expressing similar levels of F-MOR (left), F-MORΔ17 (middle), or F-MORΔ17–0cK (right) were analyzed. Numbers above each lane indicate the time period of DADLE incubation in hours. B, shown is a comparison of recycling between F-MOR (black) F-MORΔ17 (blue) and F-MORΔ17–0cK (red). Stably transfected cells expressing the indicated receptor construct were incubated for 30 min in the presence of 10 μm DADLE to drive endocytosis and then washed and incubated at 37 °C in the presence of 10 μm naloxone for the indicated times before surface labeling of Alexa647-conjugated M1 anti-FLAG and quantifying surface receptor immunoreactivity by flow cytometry. Displayed are the proportion of internalized receptors that recovered to the plasma membrane at the indicated time point after agonist washout, calculated as described under “Experimental Procedures” (mean ± S.E., n = 3–4; *, p < 0.05, two-way ANOVA, Bonferroni post-test). C, shown are representative confocal micrographs showing the localization of F-MORΔ17 (i–iii) or F-MORΔ17–0cK (iv–vi) relative to LAMP1/2 immunoreactivity in stably transfected HEK293 cells fixed after exposure to 10 μm DADLE for 90 min. Merged images (iii and vii) display receptor and LAMP channels pseudocolored in green and magenta, respectively, with areas of colocalization appearing white. Insets show a magnified region of the image as illustrated by the dotted box. Arrows indicate puncta that appear to be only single colors due to differences in relative intensity of one or the other label but are in colocalized when examined at the level of individual images. D, quantification of agonist-induced proteolysis of F-MORΔ17 and F-MORΔ17–0cK, derived from multiple experiments corresponding to the example shown in panel A, were determined by exposure of anti-FLAG immunoblots in the linear detection range and scanning densitometry. Points indicate mean and error bars S.E. (n = 6 experiments).
Down-regulation Assessed Biochemically by Destruction of the Receptor Ectodomain Does Not Require MOR Ubiquitination
Although loss of diprenorphine binding sites represents the traditional method for assaying MOR down-regulation, we sought to verify mutational effects on proteolytic destruction of receptors biochemically. To do so we assessed receptor proteolysis by loss of receptor protein detected in anti-FLAG immunoblots. F-MORs present in freshly prepared cell extracts resolved by SDS-PAGE with an apparent molecular mass ranging from ∼50 to 60 kDa (Fig. 3A), consistent with the complex-glycosylated, monomeric receptor form shown previously to predominate in cell extracts freshly prepared in isoionic buffer and in the absence of ionic detergents (18). Only a small decrease of wild type receptor immunoreactivity was detected over a 5-h time course of DADLE exposure (F-MOR, Fig. 3A, left panel), consistent with the slow rate of down-regulation measured by radioligand binding (Fig. 1B). Truncating the C-terminal MRS markedly accelerated receptor proteolysis assessed biochemically (F-MORΔ17, Fig. 3A, middle panel), also as expected from previous work (18) and consistent with the radioligand binding data (Fig. 1B). Surprisingly, a marked disconnect between pharmacological and biochemical measures was revealed when the effects of preventing receptor ubiquitination were examined. Whereas lysyl mutation strongly inhibited pharmacological down-regulation of the lysosome-targeted truncated mutant receptor, as assessed by radioligand binding (F-MORΔ17–0cK, Fig. 2E), it did not prevent proteolysis of receptors measured biochemically in parallel studies of the same receptor construct and the same stably transfected cell clones (Fig. 3A, the right panel shows a representative example). Quantification of multiple immunoblots further verified this observation (Fig. 3D, compare with Fig. 2E).
For some 7TMRS, preventing ubiquitination may increase recycling after agonist-induced endocytosis (26, 28, 29). To investigate if this is true for MORs, FLAG-tagged receptors were endocytosed by exposing cells to 10 μm DADLE for 30 min followed by recovery in the absence of agonist and subsequent measurement of surface receptor immunoreactivity. As shown previously, truncation of the MRS markedly inhibited recycling relative to the wild type MOR (compare MORΔ17 to MOR, blue and black lines in Fig. 3B, respectively). Interestingly, mutating all cytoplasmic lysine residues failed to cause any detectable increase of recycling from this inhibited level (MORΔ17–0cK, red line). These observations are fully consistent with the proteolysis data and support the hypothesis that MOR ubiquitination, although specifically important for efficient pharmacological down-regulation of receptors, does not play a major role in determining the overall endocytic itinerary of MORs between recycling or lysosomal fates.
To verify and further investigate this unanticipated observation, we took advantage of the fact that the critical transition of internalized MORs from the early to late endocytic pathway can be monitored immunochemically by colocalization relative to EEA1 (early endosome) and LAMP1 (late endosome/lysosome) (18). We did so by focusing on the F-MORΔ17 construct (lacking the MRS) because this receptor is efficiently sorted for lysosomal down-regulation and restricted analysis to 90 min after DADLE application, before the occurrence of significant cleavage of the epitope tag (90 min, Fig. 3A and Ref. 18). As expected, both the ubiquitinated F-MORΔ17 and ubiquitination-defective F-MORΔ17–0cK constructs imaged by confocal fluorescence microscopy localized to EEA1-marked early endosomes 30 min after DADLE application as determined (supplemental Fig. 1,A and B). Importantly, both constructs clearly localized to LAMP1-positive late endosomes/lysosomes within 90 min (Fig. 3C, iii and vi; colocalized structures appear white in the merged image, and arrows indicate examples of colocalized structures). As another independent verification, we repeated this colocalization experiment using an antibody recognizing the receptor endodomain and obtained the same results (supplemental Figs. 1, C and D). Thus, despite the fact that ubiquitination is required for efficient receptor down-regulation as assessed by loss of radioligand binding, the ubiquitin-independent MRS appears to control the dominant sorting operation determining delivery of internalized receptors to the proteolytic compartment.
Ubiquitination Controls Receptor Distribution between the Limiting Endosome Membrane and Lumen but Is Not Essential for Receptor Delivery to the Proteolytic Compartment
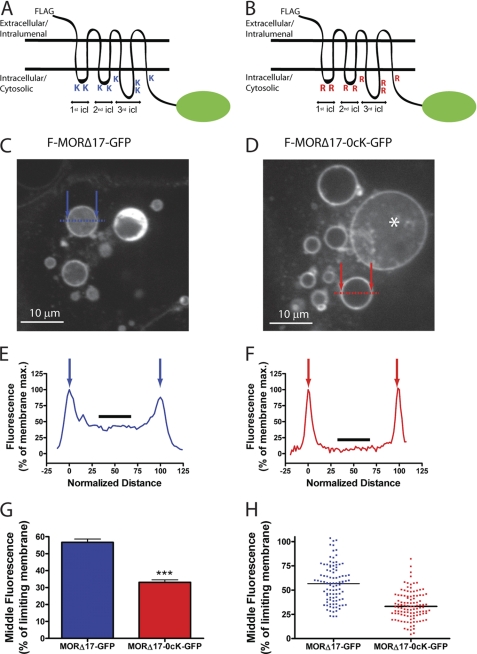
Because MOR ubiquitination plays a specific and relatively late function in the down-regulation pathway, we next asked whether MORs undergo ubiquitin-dependent transfer to intralumenal membranes of endosomes. GFP-tagged versions of F-MORΔ17 and F-MORΔ17–0cK (F-MORΔ17-GFP and F-MORΔ17–0cK-GFP, respectively, Fig. 4, A and B) were co-expressed with constitutively active Rab5 (CFP-Rab5-Q79L) to enlarge endosomes and facilitate optical resolution of lumen from limiting membrane in living cells (17, 30), and cells were imaged live by spinning disc confocal microscopy after agonist treatment for 90 min. F-MORΔ17-GFP was resolved both in the limiting membrane and endosome lumen, and intralumenal localization of F-MORΔ17-GFP was quite uniform across individual endosomes (Fig. 4, C and E, supplemental Fig. 2A). F-MORΔ17–0cK-GFP localized prominently in the limiting endosome membrane, with most endosomes showing little intralumenal fluorescence (Fig. 4, D and F, supplemental Fig. 2B). GFP-tagged receptor distribution across numerous endosomes and experiments, quantified by line scan analysis (Figs. 4, E–H, arrows indicate positions of the limiting membrane), verified inhibited intralumenal localization of lysyl-mutant receptors (Fig. 4, G and H). Thus, despite the fact that MOR ubiquitination has no detectable effect on the primary recycling versus degradation sorting decision, it strongly affects the “limiting membrane versus lumen” localization of receptors. Thus, recycling versus degradation and limiting membrane versus lumen evidently represent discrete sorting operations, which are hierarchically organized and controlled by distinct biochemical determinants (the MRS or ubiquitination, respectively).
FIGURE 4.
Ubiquitination promotes receptor redistribution from the limiting to intralumenal endosome membranes. A and B, shown are schematic representations of the F-MORΔ17-GFP (panel A) and F-MORΔ17–0cK-GFP (panel B) constructs used for live imaging, with the fused GFP indicated as a green oval. icl, cytoplasmic loop. C and D, a representative optical section shows endosomes in HEK293 cells co-transfected with CFP-Rab5Q79L and either F-MORΔ17-GFP (panel C) or F-MORΔ17–0cK-GFP (panel D) after incubation for 90 min at 37 °C with 10 μm DADLE followed by imaging at 37 °C by spinning disc confocal microscopy in the continuous presence of agonist. Essentially all of the enlarged endosomes contained visible F-MOR-GFP fluorescence, whereas intralumenal F-MORΔ17–0cK-GFP was rarely observed. The asterisk in panel D shows an example of such a rare endosome in which detectable intralumenal F-MORΔ17–0cK-GFP fluorescence was detected. Blue and red symbols overlaid on each image indicate the position of line scans used for quantification. Scale bars represent 10 μm. E and F, shown is a representative line scan analysis to quantify intralumenal fluorescence. Normalized distance represents the diameter of the endosome shown, where 0 and 100 correspond to the pixel distances between the first and second maximum pixel intensities measured across the dashed line, respectively (indicated by arrows in the diagrams overlaid on panels C and D). Normalized fluorescence represents the normalized pixel intensity measured across the dashed line, where the maximum pixel intensity across the line is normalized to 100. The graphs shown in panels E and F show line scans of the representative endosomes highlighted in panels A and B, respectively. The black line highlights the normalized fluorescence values of pixels from 40 to 60% of the normalized diameter, used for determining the mean value of intralumenal fluorescence for each endosome, as described under “Experimental Procedures,” G and H, shown are compiled results from the line scan analysis diagrammed in panels E and F. Panel G shows compiled data as the mean and S.E. Panel H shows the distribution of internal fluorescence values from individual analyzed endosomes (n = 98 and 109 endosomes, respectively, each imaged from 12 independent dishes and cells; ***, p < 0.001, Student's t test).
The Downstream Endocytic Sorting Operation Is Controlled Specifically by Ubiquitination in the First Cytoplasmic Loop
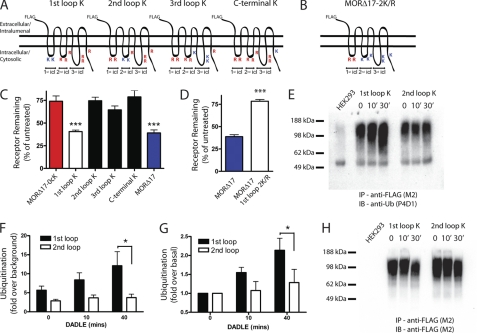
Wild type MORs contain lysine residues in every intracellular domain, and the lysyl-mutant constructs tested so far were devoid of all cytoplasmic lysine residues. Thus, we asked if there is a particular cytoplasmic domain that is the critical site for ubiquitin-dependent control of MOR down-regulation and assessed the ubiquitin-dependent sorting operation quantitatively by monitoring down-regulation by radioligand binding. We first asked if ubiquitination of a particular cytoplasmic domain is sufficient to drive pharmacological down-regulation by systematically reverting arginine to lysine residues in each intracellular domain separately in the F-MORΔ17–0cK background (Fig. 5A). Restoring lysine residues in the first cytoplasmic loop was fully sufficient to produce agonist-induced down-regulation as measured by radioligand binding, whereas restoring lysine residues in any of the other cytoplasmic domains was not (Fig. 5C). To test if ubiquitination of the first cytoplasmic loop is necessary for pharmacological down-regulation, we conversely mutated (to arginine) only this loop in the F-MORΔ17 background (Fig. 5B). This mutation strongly inhibited pharmacological down-regulation of receptors (Fig. 5D) and did so to a degree indistinguishable from that of mutating all cytoplasmic lysine residues (compare the second bar in Fig. 5D to the left bar in Fig. 5B). Thus, the critical location for ubiquitin-dependent control of MOR down-regulation, defined by the location that is both necessary and sufficient for receptor down-regulation measured by radioligand binding, is the first cytoplasmic loop. Further arguing that this represents a bona fide discrete sorting determinant, we also determined by live imaging that the first cytoplasmic loop is necessary and sufficient to promote receptor localization to the intralumenal compartment of endosomes (supplemental Fig. 3).
FIGURE 5.
Ubiquitination specifically of the first cytoplasmic loop is necessary and sufficient for agonist-induced down-regulation measured by radioligand binding. A, shown is a diagram of the series of receptor mutants used to test sufficiency, based on reverting arginine residues to lysine residues within individual cytoplasmic domains of the F-MORΔ17–0cK backbone separately. The positions of lysine (K) or arginine (R) residues in each construct are indicated. B, shown is a diagram of the receptor mutant used to test necessity, based on mutating only the lysine residues present in the first cytoplasmic loop (1st icl) of F-MORΔ17 to arginine, as indicated. C, shown is down-regulation of the receptor constructs diagrammed in panel A, assayed by [3H]DPN binding after incubation of stably transfected cells expressing the indicated receptor construct with 10 μm DADLE for 5 h. In each experiment down-regulation was assessed in triplicate determinations. Bars represent the mean, and error bars are from n = 7 independent experiments (**, p < 0.01; ***, p < 0.001, one way ANOVA, Bonferroni post test). D, shown is down-regulation of the receptor construct diagrammed in panel B. E, shown is a representative anti-ubiquitin immunoblot (IB) used for the densitometry analysis summarized in panels F and G. IP, immunoprecipitate. F and G, shown is quantification of relative ubiquitin incorporation from densitometry of anti-ubiquitin blots expressed as -fold over basal measured in unstimulated cells (panel G) or -fold over background measured in cells not expressing FLAG-tagged receptor (panel F). Bars represent mean and S.E.; error bars, n = 4; * denotes p < 0.05 as determined by two-way ANOVA with the Bonferroni post-test). H, the same blot is shown in panel E except it was stripped and reprobed with anti-FLAG, to verify comparable receptor loading between lanes.
No previous study has mapped a functionally relevant site of ubiquitination to the first cytoplasmic loop of opioid receptors or, we believe, of any 7TMR. Thus, we asked whether this loop is actually ubiquitinated in intact cells and further investigated the specificity of this location by asking if MOR ubiquitination can occur also in other cytoplasmic domain(s). To do so we replaced lysine residues either in the first or second cytoplasmic loops in the F-MORΔ17–0cK background, making the respective loops the only potential ubiquitination sites in the receptor, and investigated ubiquitination of mutant receptors biochemically using immunoblotting. Introducing lysine residues only in the first loop of the F-MORΔ17–0cK background restored both basal receptor ubiquitination and the agonist-dependent increase that is characteristic of the wild type receptor. Introducing lysine residues only in the second cytoplasmic loop restored significant basal receptor ubiquitination but did not restore its agonist-dependent stimulation (Fig. 5, E and H, show examples of anti-ubiquitin and anti-FLAG loading control blots, respectively, and Fig. 5, F and G, show the quantification across multiple experiments). These results, combined with the observation that lysine residues in the first but not second loop are sufficient to confer ubiquitin-dependent control on the downstream sorting event required for pharmacological down-regulation (Fig. 5C), indicate that MOR ubiquitination is not restricted to the first loop but that ubiquitination in this domain is specifically agonist-regulated and critical for controlling receptor sorting in the down-regulation pathway.
Relevance to Naturally Occurring MOR Isoforms
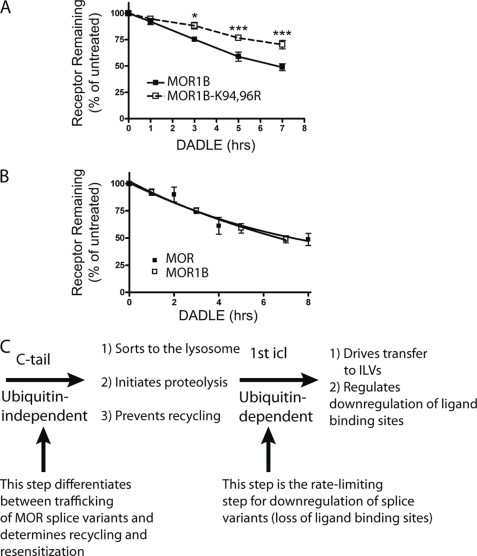
To begin to investigate the physiological significance of MOR ubiquitination in the first loop, we asked whether this post-translational modification controls the down-regulation of naturally occurring receptors. The μ-opioid receptor gene (Oprm1) transcript is subject to alternative splicing that generates structurally distinct MOR variants. The predominant MOR expressed in the rodent brain is MOR1 (corresponding to the MOR construct examined so far in the present study). MOR1B is another variant that is functional and natively expressed (21–23). Interestingly, MOR1B has an identical first cytoplasmic loop as MOR1 but a different C-terminal sequence and, accordingly, lacks the particular MRS present in MOR1. This structural difference was shown previously to selectively drive endocytic delivery of MOR1B to lysosomes (23), and we verified this effect in the present study using the immunoblot assay. Because MOR1B is a naturally expressed receptor for which differential sorting to the lysosome pathway is already known to produce significant functional consequences, we asked if ubiquitination of the first cytoplasmic loop is also relevant to its down-regulation. Indeed, lysyl mutation of only the first cytoplasmic loop in MOR1B clearly inhibited pharmacological down-regulation of receptors measured by radioligand binding assay (Fig. 6A). Interestingly, despite clear biochemical and functional evidence for significant differences in the endocytic sorting of MOR1 relative to MOR1B between recycling and lysosomal fates (as shown previously (23) and verified here), the down-regulation of these isoforms as estimated by radioligand binding was remarkably similar (Fig. 6B). This indicates that ubiquitination of the first cytoplasmic loop is required for (and apparently rate-limiting in) pharmacological down-regulation of both MOR isoforms, as defined by the traditional pharmacological criterion of loss of opiate radioligand binding. Importantly, and as revealed initially by truncation of the MOR1 C-tail, this ubiquitin-dependent sorting operation functions effectively downstream of the (ubiquitin-independent) MRS. Thus, ubiquitination of the first loop plays a shared and highly specific role in pharmacological down-regulation but is not the primary means by which the biochemically and functionally divergent endocytic trafficking itineraries of these naturally occurring receptor isoforms are determined.
FIGURE 6.
Ubiquitination in the first cytoplasmic loop is also required for pharmacological down-regulation of the MOR1B isoform. A, HEK293 cells stably expressing FLAG-tagged MOR1B (F-MOR1B) or FLAG-tagged MOR1B in which only the lysine residues present in the first cytoplasmic loop were mutated to arginine (F-MOR1B-K94R,K96R) and treated with 10 μm DADLE for the indicated time period before assessing receptor down-regulation by radioligand binding using [3H]DPN. Points represent mean determinations from independent experiments, with each time point analyzed in triplicate tubes in each experiment. Error bars represent the S.E. calculated across the experiments (n = 4). B, shown is a down-regulation assay comparing the MOR1 and the MOR1B splice variants. Data are replotted from Figs. 1B and 6A to reveal that, in the present experiments, there was no detectable difference in pharmacological down-regulation of the wild type versions of MOR1 (F-MOR) compared with MOR1B (F-MOR1B) isoforms. C, shown is a diagram describing the proposed sequential sorting operations in the hierarchical sorting model. The C-tail (containing the previously described MRS) determines the overall trafficking itinerary of internalized MORs between recycling and lysosomal routes and does not require MOR ubiquitination. Ubiquitination of the first cytoplasmic loop (1st icl) specifically promotes redistribution of receptors from the limiting membrane to lumen of late endosomes/multivesicular bodies. This intra-multivesicular body “topological” sorting operation does not dictate the overall trafficking itinerary of internalized receptors but is required for efficient destruction of the transmembrane helical bundle containing the diprenorphine binding site. This step is effectively rate-limiting for pharmacological down-regulation of both the MOR1 and MOR1B isoforms. Therefore, traditional down-regulation assays based on loss of radioligand binding sites may not be sensitive to functionally significant differences in the regulated endocytic trafficking itinerary of naturally occurring MOR isoforms.
DISCUSSION
Prolonged activation is well known to attenuate cellular opioid responsiveness by inducing proteolytic destruction of opioid receptors through endocytic trafficking to lysosomes. However, mechanisms that mediate and control this fundamental process of homeostatic regulation remain poorly understood. The present results provide several pieces of mechanistic insight. First, the present data establish an essential role of the ubiquitin-ESCRT system in mediating MOR down-regulation and show that this system promotes the efficient transfer of receptors from the limiting to intralumenal membranes of late endosomes. Second, the results show that MOR ubiquitination is specifically required for agonist-induced down-regulation of receptors as determined by loss of radioligand binding. In particular, we show that disrupting MOR ubiquitination has no effect on proteolysis of the N-terminal receptor ectodomain, on delivery of receptors to the late endocytic pathway, or on the specificity of receptor trafficking via the recycling pathway. Together with previous evidence suggesting that the binding site for alkaloid radioligands is located predominantly within the transmembrane helices (31), these observations suggest that MOR ubiquitination, by driving topological sorting to intralumenal membranes, specifically promotes destruction of the transmembrane helical bundle without dictating the overall endocytic fate of receptors. Third, the present results establish a precedent for 7TMR regulation through modification of the first cytoplasmic loop. Studies of other 7TMRs suggest that the location of receptor ubiquitination is not critical for regulation (32) or place the site of ubiquitination within the C-tail (33–35) or third cytoplasmic loop (36, 37). We show that ubiquitination specifically of the first cytoplasmic loop is both necessary and sufficient for pharmacological down-regulation of MORs and for receptor transfer to the endosome lumen even though ubiquitination of receptors is not restricted to this cytoplasmic domain. To our knowledge the present results are the first to establish a specific regulatory function of ubiquitination in the first cytoplasmic loop of a 7TMR.
According to the prevailing view of ESCRT-dependent down-regulation, ubiquitination of signaling receptors mediates the primary sorting operation determining whether they recycle or traffic to lysosomes after endocytosis (8–10). Here we show that MOR down-regulation indeed requires the conserved ESCRT machinery, and that MOR ubiquitination controls topological sorting in endosomes. Interestingly, however, our results clearly show that ubiquitination of MORs is not essential for, and does not determine, the primary sorting decision controlling the delivery of internalized receptors to the proteolytic compartment. Instead, the primary endocytic itinerary of MORs after prolonged agonist exposure is determined by a ubiquitin-independent C-tail sequence (the MRS), which was shown previously to drive MOR recycling after endocytosis induced by short term agonist activation (18, 19). Thus, the mechanism of MOR down-regulation can be understood in terms of hierarchical organization of discrete ubiquitin-dependent and -independent steps, which function in sequence in the same pathway, and which are both required for complete receptor destruction (Fig. 6C). The ubiquitin-independent step is determined by the previously defined C-tail motif, functions effectively upstream, and determines the ultimate trafficking fate of the receptor, either recycling or delivery to the lysosome. The ubiquitin-dependent step operates effectively downstream in the same pathway and determines how efficiently the receptor is transferred to intralumenal membranes of endosomes; this topological sorting operation promotes destruction of the receptor hydrophobic core and, hence, loss of radioligand binding activity.
We note that there is growing evidence that, at least in mammalian cells, topological transfer to intralumenal vesicles is not an absolute requirement either for sorting of signaling receptors to lysosomes or for subsequent proteolytic down-regulation (38, 39). We also note that a similar separation of steps was proposed previously for the related DOR (17). However, for this 7TMR, a structural determinant(s) has not yet been clearly identified for either sorting operation. Furthermore, whereas complete down-regulation of DORs (including destruction of the ligand binding site) can occur with nearly wild type efficiency when DOR ubiquitination is prevented by lysyl mutation (16), proteolysis of MORs is effectively stalled at an intermediate state when receptor ubiquitination is similarly prevented. Accordingly, MORs provide the first example of how receptor sorting between recycling and degradative itineraries and the process of lysosomal destruction itself are separately controlled by discrete yet sequential ubiquitin-independent and -dependent sorting operations.
Together our results support the conclusion that the mechanism directing ESCRT-dependent lysosomal down-regulation of opioid receptors is hierarchical and involves the function of at least two discrete and non-redundant molecular sorting operations. They also emphasize the importance of future work to better define the biochemical machinery mediating the upstream (ubiquitin-independent) sorting operation and to investigate the physiological significance of hierarchical organization as revealed in the present study. In principle, each of the sorting operations in this hierarchy represents a discrete control point, and the present data clearly resolve distinct biochemical determinants mediating each. Thus, we anticipate that selective control of each may occur in vivo, and there may exist different functional consequences of driving one operation relative to the other.
Although much of the work leading to the present understanding of hierarchical MOR sorting emerged from the study of truncated mutant receptors lacking the C-tail MRS, the present results also provide evidence that our findings are relevant to the regulation of naturally expressed receptors. This was demonstrated by comparison of two naturally occurring MOR isoforms, MOR1 and MOR1B, which result from alternative splicing of the same receptor transcript. Both of these natural receptor isoforms possess the same first loop sequence but differ in the region of the C-tail controlling the upstream sorting operation. These isoforms are known to differ markedly in their endocytic itinerary, trafficking preferentially through the recycling pathway (MOR1) or to lysosomes (MOR1B), as established previously both by biochemical analysis of receptor proteolysis and by functional assay of cellular opioid responsiveness (23). Nevertheless, when down-regulation was assessed pharmacologically using the conventional radioligand binding assay, distinct regulation of these MOR isoforms was difficult if not impossible to discern (Fig. 6B). This is fully consistent with the hierarchical sorting model and with the ubiquitin-dependent step occurring downstream (and shared by both receptor isoforms) being rate-limiting for destruction of the ligand binding site. Thus, the present results, in addition to providing new insight to the mechanistic basis of MOR down-regulation and identifying the first example of functionally significant regulation mediated by ubiquitination of the first cytoplasmic loop, have potentially important implications for interpreting in vivo studies. In particular, they suggest that the traditional radioligand assay of receptor down-regulation, as widely used in studies of opioid effects in vivo and in various ex vivo preparations, may underestimate the occurrence of lysosomal trafficking as a cellular mechanism of opioid regulation. Moreover, they reveal a previously unappreciated layer of control in MOR endocytic trafficking, whose elucidation may reveal new targets for therapeutic manipulation of the endogenous opioid system.
Supplementary Material
Acknowledgments
We gratefully acknowledge Kurt Thorn and the Nikon Imaging Center at University of California, San Francisco, for advice and access to equipment used for live imaging experiments. We thank Graeme Cottrell for advice on the ubiquitination blots, Ben Lauffer for sharing opioid receptor antibody, Michael Tanowitz for valuable discussions, and Adriano Marchese, Harald Stenmark, and Marino Zerial for essential constructs.
This work was supported, in whole or in part, by National Institutes of Health Grants DA010711 and DA012864 (to M. v. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- MOR
- μ-opioid receptor
- 7TMR
- seven-transmembrane signaling receptor
- DADLE
- 2-d-Ala, 5-d-Leu enkephalin
- DOR
- δ opioid receptor
- DPN
- diprenorphine
- EEA1
- early endosome antigen 1
- ESCRT
- endosomal sorting complex required for transport
- HRS
- hepatocyte growth factor regulated tyrosine kinase substrate
- LAMP1
- lysosome-associated membrane protein 1
- MRS
- MOR regulatory sequence
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ANOVA
- analysis of variance.
REFERENCES
- 1. Walwyn W. M., Miotto K. A., Evans C. J. (2010) Drug Alcohol Depend. 108, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kieffer B. L., Evans C. J. (2009) Neuropharmacology 56, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koch T., Höllt V. (2008) Pharmacol. Ther. 117, 199–206 [DOI] [PubMed] [Google Scholar]
- 4. Martini L., Whistler J. L. (2007) Curr. Opin Neurobiol. 17, 556–564 [DOI] [PubMed] [Google Scholar]
- 5. Williams J. T., Christie M. J., Manzoni O. (2001) Physiol. Rev. 81, 299–343 [DOI] [PubMed] [Google Scholar]
- 6. von Zastrow M., Svingos A., Haberstock-Debic H., Evans C. (2003) Curr. Opin. Neurobiol. 13, 348–353 [DOI] [PubMed] [Google Scholar]
- 7. Hislop J. N., von Zastrow M. (2011) Traffic 12, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roxrud I., Stenmark H., Malerød L. (2010) Biol. Cell 102, 293–318 [DOI] [PubMed] [Google Scholar]
- 9. Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 10. Hurley J. H., Hanson P. I. (2010) Nat. Rev. Mol. Cell Biol. 11, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saksena S., Sun J., Chu T., Emr S. D. (2007) Trends Biochem. Sci. 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 12. Katzmann D. J., Odorizzi G., Emr S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893–905 [DOI] [PubMed] [Google Scholar]
- 13. Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanowitz M., Von Zastrow M. (2002) J. Biol. Chem. 277, 50219–50222 [DOI] [PubMed] [Google Scholar]
- 15. Hislop J. N., Marley A., Von Zastrow M. (2004) J. Biol. Chem. 279, 22522–22531 [DOI] [PubMed] [Google Scholar]
- 16. Hislop J. N., Henry A. G., Marchese A., von Zastrow M. (2009) J. Biol. Chem. 284, 19361–19370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henry A. G., White I. J., Marsh M., von Zastrow M., Hislop J. N. (2011) Traffic 12, 170–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanowitz M., von Zastrow M. (2003) J. Biol. Chem. 278, 45978–45986 [DOI] [PubMed] [Google Scholar]
- 19. Thompson D., Pusch M., Whistler J. L. (2007) J. Biol. Chem. 282, 29178–29185 [DOI] [PubMed] [Google Scholar]
- 20. Pasternak G. W. (2004) Neuropharmacology 47, 312–323 [DOI] [PubMed] [Google Scholar]
- 21. Koch T., Schulz S., Pfeiffer M., Klutzny M., Schröder H., Kahl E., Höllt V. (2001) J. Biol. Chem. 276, 31408–31414 [DOI] [PubMed] [Google Scholar]
- 22. Koch T., Schulz S., Schröder H., Wolf R., Raulf E., Höllt V. (1998) J. Biol. Chem. 273, 13652–13657 [DOI] [PubMed] [Google Scholar]
- 23. Tanowitz M., Hislop J. N., von Zastrow M. (2008) J. Biol. Chem. 283, 35614–35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whistler J. L., Enquist J., Marley A., Fong J., Gladher F., Tsuruda P., Murray S. R., Von Zastrow M. (2002) Science 297, 615–620 [DOI] [PubMed] [Google Scholar]
- 25. Hanyaloglu A. C., McCullagh E., von Zastrow M. (2005) EMBO J. 24, 2265–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacob C., Cottrell G. S., Gehringer D., Schmidlin F., Grady E. F., Bunnett N. W. (2005) J. Biol. Chem. 280, 16076–16087 [DOI] [PubMed] [Google Scholar]
- 27. Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. (2001) Science 294, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 28. Chen L., Davis N. G. (2002) Traffic 3, 110–123 [DOI] [PubMed] [Google Scholar]
- 29. Berthouze M., Venkataramanan V., Li Y., Shenoy S. K. (2009) EMBO J. 28, 1684–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wegner C. S., Wegener C. S., Malerød L., Pedersen N. M., Progida C., Prodiga C., Bakke O., Stenmark H., Brech A. (2010) Histochem. Cell Biol. 133, 41–55 [DOI] [PubMed] [Google Scholar]
- 31. Mansour A., Taylor L. P., Fine J. L., Thompson R. C., Hoversten M. T., Mosberg H. I., Watson S. J., Akil H. (1997) J. Neurochem. 68, 344–353 [DOI] [PubMed] [Google Scholar]
- 32. Cottrell G. S., Padilla B., Pikios S., Roosterman D., Steinhoff M., Gehringer D., Grady E. F., Bunnett N. W. (2006) J. Biol. Chem. 281, 27773–27783 [DOI] [PubMed] [Google Scholar]
- 33. Marchese A., Benovic J. L. (2001) J. Biol. Chem. 276, 45509–45512 [DOI] [PubMed] [Google Scholar]
- 34. Liang W., Hoang Q., Clark R. B., Fishman P. H. (2008) Biochemistry 47, 11750–11762 [DOI] [PubMed] [Google Scholar]
- 35. Hicke L., Riezman H. (1996) Cell 84, 277–287 [DOI] [PubMed] [Google Scholar]
- 36. Martin N. P., Lefkowitz R. J., Shenoy S. K. (2003) J. Biol. Chem. 278, 45954–45959 [DOI] [PubMed] [Google Scholar]
- 37. Xiao K., Shenoy S. K. (2011) J. Biol. Chem. 286, 12785–12795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pons V., Luyet P. P., Morel E., Abrami L., van der Goot F. G., Parton R. G., Gruenberg J. (2008) PLoS Biol. 6, e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Futter C. E., Collinson L. M., Backer J. M., Hopkins C. R. (2001) J. Cell Biol. 155, 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.