Abstract
Biological utilization of cellulose is a complex process involving the coordinated expression of different cellulases, often in a synergistic manner. One possible means of inducing an organism-level change in cellulase activity is to use laboratory adaptive evolution. In this study, evolved strains of the cellulolytic actinobacterium, Thermobifida fusca, were generated for two different scenarios: continuous exposure to cellobiose (strain muC) or alternating exposure to cellobiose and glucose (strain muS). These environmental conditions produced a phenotype specialized for growth on cellobiose (muC) and an adaptable, generalist phenotype (muS). Characterization of cellular phenotypes and whole genome re-sequencing were conducted for both the muC and muS strains. Phenotypically, the muC strain showed decreased cell yield over the course of evolution concurrent with decreased cellulase activity, increased intracellular ATP concentrations, and higher end-product secretions. The muS strain increased its cell yield for growth on glucose and exhibited a more generalist phenotype with higher cellulase activity and growth capabilities on different substrates. Whole genome re-sequencing identified 48 errors in the reference genome and 18 and 14 point mutations in the muC and muS strains, respectively. Among these mutations, the site mutation of Tfu_1867 was found to contribute the specialist phenotype and the site mutation of Tfu_0423 was found to contribute the generalist phenotype. By conducting and characterizing evolution experiments on Thermobifida fusca, we were able to show that evolutionary changes balance ATP energetic considerations with cellulase activity. Increased cellulase activity is achieved in stress environments (switching carbon sources), otherwise cellulase activity is minimized to conserve ATP.
Keywords: Bacterial Genetics, Bioenergetics, DNA Sequencing, Energy Metabolism, Evolution, Genome Re-sequencing, Thermobifida fusca, Cellulase, Mutation
Introduction
Cellulose is the most common organic compound on earth and can be potentially used as an inexpensive and renewable raw material for many applications. To more fully utilize cellulose, efforts are being made to identify (1), characterize (2), and utilize (3) cellulolytic organisms. Collectively, cellulolytic microorganisms are a small fraction of all known species with different species found in many different branches of life (4)such as Clostridium thermocellum (5), Humicola insolens (6), Aspergillus niger (7). While generally poorly characterized, cellulolytic microbes possess great physiological and biochemical diversity making them interesting organisms to study from both a basic and applied perspective. Thermobifida fusca is a particularly interesting cellulolytic actinobacterium that is aerobic, thermophilic (8), and is more efficient at producing cellulases than most anaerobic cellulolytic bacteria (5).
One of the main challenges to utilizing cellulose is that cellulose degradation/hydrolysis is a slow and difficult process. The roles, activity, and regulatory mechanisms controlling expression of different cellulases are all being studied with a goal of being able to understand and increase the rate of cellulose hydrolysis. Another potential approach to increase the rate of cellulose hydrolysis is to conduct adaptive evolution experiments with a cellulolytic organism. Laboratory adaptive evolution has been conducted with a number of different microorganisms (9–13) as a means for inducing systemic changes (12–16). In cases where selection is based upon growth rate, it has generally been found in non-cellulolytic microorganisms that adaptive evolution leads to an increased growth rate that is related to increased consumption of the limiting substrate (17–20). Thus, laboratory evolution may be a means for increasing the rate of cellulose hydrolysis in a cellulolytic organism.
In T. fusca, there are six different cellulases that have been purified and identified. Of the identified cellulases, three are endocellulases, Cel9B, Cel6A, and Cel5A (21–25), two of them are exocellulases, Cel6B and Cel48A (24, 25), and one is a novel processive endocellulase, Cel9A (26). Importantly, CelR (Tfu_0938) has previously been identified as a regulator of several cellulase genes, is a member of the lactose repressor family, and binds cellobiose with a mm Kd (27–29). It has been found that physiological concentrations of cellobiose can activate cellulase-related gene transcription (30). Glucose has been found to repress transcription of cellulase-related genes (30) and impair growth of T. fusca (31, 32).
In this study, we conducted laboratory adaptive evolution experiments using T. fusca for growth in two different environmental conditions: 1) evolution of T. fusca on cellobiose and 2) evolution of T. fusca by alternating carbon sources between cellobiose and glucose daily to produce an evolved specialist strain and generalist strain (33), respectively. The evolved T. fusca strains were characterized for phenotypic changes during evolution and full genome re-sequencing using two different high-throughput sequencing platforms was conducted to identify genetic mutations that arose during evolution. Phenotypic effects of identified point mutations were tested by isolating and characterizing different subpopulations of T. fusca in the middle of the evolutionary trajectory.
EXPERIMENTAL PROCEDURES
Adaptive Evolution of T. fusca
Starting with the wild-type strain of T. fusca whose genome has recently been sequenced (8), laboratory evolution experiments were conducted for growth in two conditions (both at 55 °C): 40 days on Hagerdahl medium with 0.1% cellobiose (31) or 40 days on Hagerdahl medium where the carbon source was alternately switched every day between 0.1% cellobiose or 0.1% glucose. Two different evolved strains of T. fusca (mutant grown on cellobiose, muC2 and mutant grown by switching sugars, muS) were generated by serial passage of cells into fresh medium (9) while cells remained in exponential growth. The phenotypes (cell growth and specific cellulase activity) were measured every day and the amount of dilution at each passage was adjusted daily to account for changes in growth rate. After 40 days of adaptive evolution, the cell growth and cellulase activity were stable and the evolution experiment was stopped. The muC strain was gained by propagating for nearly 284 generations and the muS strain was gained by propagating for ∼220 generations. For subsequent testing, isogenic colonies were selected by diluting T. fusca spores on agar plates and picking single colonies for culturing.
Grow T. fusca on 96-Well Micro-plate with Different Substrates
T. fusca from preculture was harvested and washed three times using purified water. To scatter filamentous clumps, filaments were treated using a sonicator (Branson Sonifier 450) at 10% strength for 2 min. The treated cells were then added to 300 μl of Hagerdahl medium with xylose, mannose, sucrose, acetate, or citrate, respectively, on 96-well micro-plate with 5% inoculum. The microplate was cultured in a VersaMax EXT microplate reader (Molecular Devices) at 45 °C for 24 h. The optical density (OD) was measured as absorbance at 600 nm every 10 min.
Cellulase Activity Assay
Two assays were used to measure the overall cellulase and endoglucanase activity of culture supernatants. Filter paper was used as the starting material to measure overall cellulase activity, and endoglucanase activity was assayed by the measurement of reducing sugars generated from 0.5% medium-viscosity carboxymethylcellulose (CMC). All the enzyme activity assays were measured by a microplate-based assay (34). Assays were conducted in a 60-μl volume as follows. A 20-μl aliquot of raw enzyme was added into the wells of PCR plates containing 40 μl of 50 mm NaAc buffer and a filter paper disk (7 mm diameter) for cellulase activity or 40 μl of 50 mm NaAc buffer with 0.5% CMC for endoglucanase activity. After 60 min of incubation at 50 °C, 120 μl of 3,5-dinitrosalicylic acid (DNS) solution was added into each reaction and incubated at 95 °C in a thermal cycler (iCycler® Thermocycler, Bio-Rad) for 5 min. Finally, a 36-μl aliquot of each sample was transferred to the wells of a flat-bottom plate containing 160 μl of H2O, and the absorbance at 540 nm was measured on VersaMax EXT microplate reader (Molecular Devices). One enzyme unit (unit) is defined as an average of one μmole of cellobiose equivalent released per min in the assay reaction. All the enzyme activity values presented were averages obtained from triplicate measurements.
Cell Density and End-product Measurement
Because of the growth physiology of T. fusca (filamentous cells that aggregate), the culture density of T. fusca was determined by measuring cytoplasmic protein content. 1 ml culture was centrifuged at 10,000 × g for 5 min. The pellets were resuspended in fresh media and centrifuged at 10,000 × g for 5 min again. Sediments were dissolved in 200 μl 50 mm Tris-HCl buffer (pH 6.8) containing 2% SDS, 0.1 m DTT, and 50% glycerol. Samples were then pulsar sonicated at 70% strength for 10 min in an ice bath. After centrifuging at 10,000 × g for 5 min, the proteins in the supernatant were measured by the Bradford protein assay (35) The dry cell weight (DCW) is proportionally related to the overall protein content. Metabolic end-products were measured using an HPLC system (Dionex Ultimate3000) equipped with Bio-Rad HPX-87H ion exclusion column. The mobile phase was 0.005 mol/liter H2SO4 at the rate of 0.6 ml/min and RI, and UV detectors were used.
ATP Determination
ATP was measured by the ATP determination kit from Invitrogen using the protocol suggested by the manufacturer. Specific ATP levels were defined as the amount of ATP divided by the amount of cytoplasmic proteins in T. fusca. For ATP assays, cells were sampled during mid-log phase growth. Experiments were conducted to assay for ATP levels for growth in a bioreactor at 55 °C with the 0.375 vvm aeration and a stir speed of 250 rpm and in 500 ml flasks at 55 °C that were shaken at 250 rpm. 1 ml culture was centrifuged at 10,000 × g for 10 min. The pellets were resuspended in the purified water and centrifuged at 10,000 × g for 5 min again. Samples were then sonicated (Branson Sonifier 450) at 10% strength for 5 min in the ice bath to release the ATP. After centrifuging at 10,000 × g for 5 min, 10 μl supernatant was added into the wells of PCR plates containing 90 μl reaction solutions. After incubating in the VersaMax EXT microplate reader (Molecular Devices) for 30 s at room temperature, the absorbance was measured at 560 nm.
DNA Sequencing
Genomic DNA was isolated using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer's instructions. All the genomic sequencing libraries were prepared according to the manufacturer's instructions. We sequenced the isolates from muS and muC using 454 GS-FLX sequencer and then these two isolates were also sequenced by Illumina Genome Sequencer. In all cases, we generated single-end reads. We used Sanger sequencing of PCR products to confirm mutations. Primers used for PCR were designed by Primer5.0. PCR was done in a 25 μl volume using Pfu DNA polymerase (Stratagene) and cycled on an MJ Research thermal cycler. Products were checked on an agarose gel and purified using a QIAquick PCR purification kit (Qiagen).
SNP Detection from Sequence Data
Sequencing reads obtained using the Roche 454 FLX platform were analyzed by manufacturer-supplied software and processed by a core facility at Virginia Commonwealth University. Illumina reads were mapped directly to the T. fusca reference sequence using the MOM algorithm (36). During the alignment of the Illumina data, sequence fragments (between 27 bp and 38 bp in length) were called to a unique genome position and only fragments with a perfect sequence match or a single base mismatch were used. The set of filtered 454 SNP calls that overlapped with filtered Illumina SNP calls (excluding any calls in repetitive sequences) was used to approximate the “true” set of SNPs for each of the two isolates.
Testing Intermediate Evolutionary Time Points (supplemental Fig. S1)
To test for heterogeneity in the evolving T. fusca strains, subpopulations were tested by isolating individual colonies by serial dilution of T. fusca spores onto agar plates. For each intermediate evolutionary time point tested, 10 single colonies were selected from agar plates and regrown.
For growth characterization on non-evolutionary subtrates (xylose, mannose, sucrose, acetate, and citrate), colonies selected from plates were grown as a preculture for 1 day. Cells from precultures were centrifuged, washed three times with the water, and used to inoculate a 96-well plate to test for growth on xylose, mannose, sucrose, acetate, and citrate. Cell growth was monitored by measuring optical density (aborbance at 600 nm) in 10 min increments for 24 h using an incubator/shaker microplate reader (VersaMax EXT).
For testing of the day 20 time points, 10 colonies were selected and tested for growth on non-evolutionary substrates. Based upon the results of the growth testing, single colonies were identified and tested for the presence of point mutations by DNA sequencing. Genomic DNA from the selected colonies was isolated and sequencing was conducted by Sanger sequencing to test for mutations in the 18 genes that had mutations in the end point strains.
Test Cell Growth in Oleic Acid and Glutamine
For growth characterization on oleic acid and glutamine, colonies selected from plates were grown as a preculture for 1 day. Cells from precultures were centrifuged, washed three times with the water, and used to inoculate a 96-well plate to test for growth on xylose, mannose, sucrose, acetate, and citrate with addition of 0.1% oleic acid or 0.1% glutamine. Cell growth was monitored by measuring optical density (aborbance at 600 nm) in 10 min increments for 24 h using an incubator/shaker microplate reader.
RESULTS
Adaptive Evolution of T. fusca
Two different evolved strains of T. fusca (mutant grown on cellobiose: muC and mutant grown by switching cellobiose and glucose daily: muS) were generated by serial passage of cells into fresh medium while cells remained in exponential growth. The muC strain was propagated for ∼284 generations and the muS strain was propagated for ∼220 generations. The two experimental conditions for evolution were anticipated to produce a specialized phenotype adapted to growth on cellobiose (muC) or a generalist phenotype (muS).
Phenotypes of the Evolved T. fusca Strains
The muC strain was obtained by evolving T. fusca on limiting amounts of cellobiose as the sole carbon source. The muS strain was obtained by culturing T. fusca alternately on glucose and cellobiose which were switched as the sole carbon source daily. Both evolved stains were spread on the agar plates with Avicel as the carbon source and only those with large transparent circles were selected as an indication that the colonies were cellulolytic T. fusca and not a contaminant.
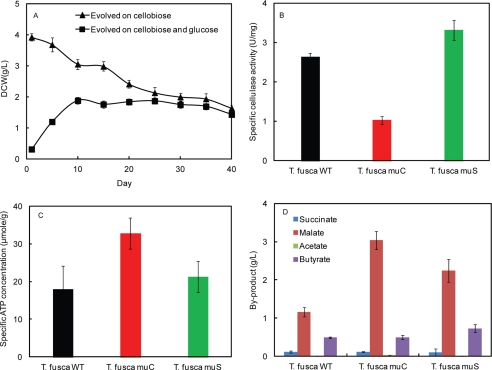
The starting point for the two evolution strains were growth on cellobiose for the muC strain and growth on glucose for the muS strain. It has been previously observed that glucose acts as an inhibitor to growth of T. fusca (37), so the muC and muS strains initially had different cell yields. Over the course of evolution, the muC strain decreased in the maximum cell concentration from 3.92 ± 0.12 g/liter to 1.63 ± 0.10 g/liter, whereas the maximum cell concentration of muS increased from 0.31 ± 0.02 g/liter to 1.43 ± 0.10 g/liter (the starting point and the end point of evolution were conducted on glucose)(Fig. 1A). Because glucose works as a repressor of cellulase production and cell growth in T. fusca, the above phenomenon indicates that the repression of cell growth by glucose in the muS was alleviated by evolving the strain in a condition where sugars were continually switched.
FIGURE 1.
The phenotypes of different T. fusca strains. A, cell growth of T. fusca on cellobiose and on switching cellobiose and glucose daily, whose start and the end point carbon source was glucose. B, specific cellulase activity of T. fusca muS, muC, and WT. C, cytoplasmic ATP concentration of T. fusca muS, muC, and WT. specific ATP levels were defined as the amount of ATP divided by the amount of cytoplasmic proteins in T. fusca. D, secretion of by-products in T. fusca muS, muC, and WT
Concurrent measurement of specific cellulase activity (cellulase activity per dry cell weight) was also conducted using a microtiter plate-based enzyme assay (34) (Fig. 1B). The specific cellulase activity of muC (1.03 ± 0.01 units/mg) was significantly less than the unevolved wild-type (WT) strain (2.63 ± 0.09 units/mg), but the specific cellulase activity of muS (3.31 ± 0.16 units/mg) was higher than both the unevolved wild-type and muC strain.
Additionally, the main metabolic end-products were measured (Fig. 1D) by growing the WT, muC, and muS on cellobiose. Differences in malate and acetate secretion were observed among the three tested strains, with only the muC strain secreting detectable amounts of acetate. Meanwhile, secretion of butyrate and succinate remained largely unchanged in all tested conditions.
In cellulolytic organisms, the metabolic cost of production cellulases can often be very high (38), and it has also been shown that protein expression during adaptation may abide by a metabolic cost-benefit scheme (39). Thus, intracellular ATP levels were also measured (40). It was found that the intracellular ATP levels in the muC strain were higher than those of muS and WT strain (Fig. 1C).
Growth Phenotypes on Various Non-evolutionary Carbon Sources
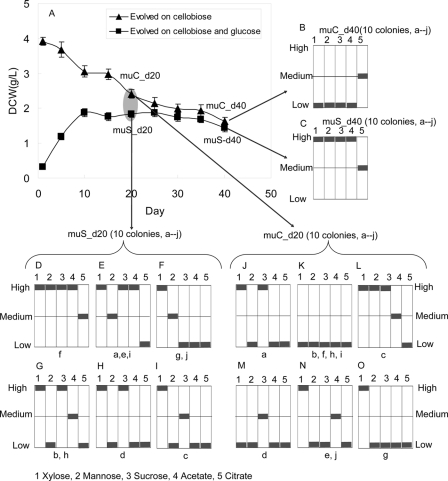
To more thoroughly characterize the phenotypes of the muC and muS strains, the evolutionary endpoints of each strain were grown on a variety of carbon substrates (xylose, mannose, sucrose, acetate, and citrate) that were not used for the evolution experiments. For both the muC and muS strains, 10 colonies (colonies a–j) of the end point strain were selected for testing on the non-evolutionary substrates. For each strain, the growth characteristics of all colonies selected for testing were the same suggesting that the evolutionary end point was relatively homogeneous. The end point of the muC strain (muC_d40) exhibited poor growth on non-evolutionary substrates (low cell yields on xylose, mannose, sucrose, and acetate, Fig. 2B). The end point of the muS strain (muS_d40) exhibited a generalist phenotype with good growth capabilities on xylose, mannose, sucrose, acetate, and citrate (Fig. 2C).
FIGURE 2.
The growth of evolved T. fusca strains: muS_d40, muS_d20, muC_d40 and muC_d20 on five different substrates. A, cell growth of T. fusca on cellobiose and on switching cellobiose and glucose daily. B, growth fitness of muC_d40 on five different substrates (end point of adaptive evolution, 10 colonies a–j), C, growth fitness of muS_d40 on five different substrates (end point of adaptive evolution, 10 colonies a–j), D –I, growth fitness of muS_d20 on five different substrates (intermediate strain, 10 colonies a–j); J–O, growth fitness of muC_d20 on five different substrates (intermediate strain, 10 colonies a–j)
Multiplatform Re-sequencing
To study the genetic changes occurring in the evolved strains muC and muS, high-throughput genome re-sequencing of the evolutionary endpoints was conducted using both the Illumina Genome Analyzer and the Roche 454 FLX platforms. Sequence reads generated by the Illumina system were aligned to the wild-type reference genome of T. fusca (8)using the MOM algorithm (36) and 454 pyrosequencing was completed using the manufacturer-supplied base-calling software. For T. fusca muC, we mapped 180,554,318 (98.94% of total) single-end reads with an average depth of 56× coverage by Illumina sequencing, and 159,169 reads (99.48% of total) with an average depth of 17.2× by 454 FLX sequencing. For T. fusca muS we mapped 180,703,707 (98.81% of total) single-end reads with an average depth of 56× coverage by Illumina sequence and 218,595 reads (99.49% of total) with an average depth of 22× coverage by 454 FLX sequencing. Data from the two re-sequencing platforms were analyzed independently and compared with each other to increase confidence in mutation calls.
All generated re-sequencing data were analyzed for point mutations (SNPs) and other mutational events. During the alignment of the Illumina data, sequence fragments (between 27 bp to 38 bp in length) were called to a unique genome position and only fragments with a perfect sequence match or a single base mismatch were used. By these criteria 135 and 144 SNPs with at least 1× coverage were found in T. fusca muC and T. fusca muS, respectively. To increase confidence in the base calling, an arbitrary stringent coverage criteria of 30× was set as the cutoff and resulting in 73 and 70 SNPs being identified in T. fusca muC and T. fusca muS, respectively.
Genome re-sequencing using moderate read-length 454 pyrosequencing was used as a secondary, independent means to evaluate mutations. Using the manufacturer-supplied alignment software, 123 mutations were identified with high quality reads in T. fusca muC. In strain muS, 126 mutations were found with high quality reads.
By identifying mutations that were called by both sequencing methods, 66 mutations (supplemental Table S1) were identified in T. fusca muC and 62 mutations (supplemental Table S2) were found in T. fusca muS. All mutations were further verified by Sanger sequencing. Among them, 48 point mutations (supplemental Table S1 and Table S2) were common to both T. fusca muC (48 of the 66 mutations) and T. fusca muS (48 of the 62 mutations) and were found to be errors in the reference genome sequence of wild-type T. fusca. Finally, 18 out of 66 SNPs in T. fusca muC and 14 out of 62 SNPs in T. fusca muS were found to be confirmed point mutations occurring as a result of the adaptive evolution process (Table 1 and Table 2).
TABLE 1.
Genetic variations detected in T. fusca muC and T. fusca muS
|
T. fusca muC | ||||
|---|---|---|---|---|
| Reference gene | Mutation position | Nucleotide change | Annotation | Amino acid change |
| Tfu_0132 | 155128 | T = >C | Putative hydrolase | C(68) = >R |
| Tfu_0417 | 470157 | T = >C | Peptidyl-tRNA hydrolase | stop(213) = >Q |
| Tfu_0555 | 631259 | T = >C | Signal transduction histidine kinase | NA |
| Tfu_0577 | 658876 | A = >G | HAD-superfamily hydrolase, subfamily IA, variant 1 | NA |
| Tfu_0665 | 771381 | T = >C | tRNA (guanine-N1-)-methyltransferase | NA |
| Tfu_0778 | 919790 | T = >C | Initiation factor 2:Small GTP-binding protein domain | NA |
| Tfu_0913 | 1075652 | A = >G | Oligopeptide/dipeptide ABC transporter, ATP-binding protein | S(224) = >G |
| Tfu_1031 | 1209775 | T = >C | Long-chain fatty-acid-CoA ligase | F(570) = >L |
| Tfu_1108 | 1292195 | A = >G | UDP-N-acetylmuramoylalanine-d-glutamate ligase | A(614) = >G |
| Tfu_1374 | 1592611 | A = >G | Regulatory protein GntR, HTH | W(100) = >R |
| Tfu_1458 | 1685076 | A = >G | Conserved hypothetical protein | T(191) = >A |
| Tfu_1621 | 1880712 | A = >G | Similar to xylanase/chitin deacetylase | NA |
| Tfu_1688 | 1962424 | T = >C | Aldehyde dehydrogenase family protein | I(291) = >V |
| Tfu_1781 | 2082958 | A = >G | Possible ABC transporter, permease protein | NA |
| Tfu_1867 | 2184179 | A = >G | Non-ribosomal peptide synthase:Amino acid adenylation | F(2238) = >L |
| Tfu_2363 | 2780975 | C = >T | Putative zinc proteinase | W(121) = >stop |
| Tfu_2670 | 3143192 | A = >G | Conserved hypothetical protein | NA |
| Tfu_2877 | 3388783 | C = >T | Putative Lsr2-like protein | G(91) = >S |
| T. fuscamuS | ||||
| Tfu_0195 | 221012 | T = >C | Putative iron sulfur-binding reductase | I(33) = >V |
| Tfu_0323 | 365661 | A = >G | Polyphosphate kinase | NA |
| Tfu_0403 | 451972 | T = >C | Hypothetical protein | NA |
| Tfu_0423 | 475686 | A = >G | Flavohemoprotein | T(367) = >A |
| Tfu_0425 | 480685 | A = >G | Adenylyl-sulfate kinase | NA |
| Tfu_0677 | 781810 | A = >G | Elongation factor Ts | K(212) = >E |
| Tfu_0750 | 885777 | T = >C | Putative secreted peptidase | F(446) = >L |
| Tfu_1087 | 1269440 | T = >C | Regulatory protein, MarR | NA |
| Tfu_1426 | 1652369 | A = >G | Hypothetical protein | H(440) = >R |
| Tfu_1472 | 1701773 | T = >C | Short chain dehydrogenase/reductase (SDR) family protein | NA |
| Tfu_1516 | 1748226 | T = >C | Hypothetical protein | H(12) = >R |
| Tfu_2031 | 2375712 | A = >G | Conserved hypothetical protein | Stop(156) = >Q |
| Tfu_2363 | 2780975 | C = >T | Putative zinc proteinase | W(121) = >stop |
| Tfu_2728 | 3207758 | A = >G | Putative DNA methylase | NA |
TABLE 2.
Tabulation of non-synonymous mutations tested in intermediate strains of adaptive evolution
|
T. fusca muC |
T fusca muS |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reference gene | Mutation Position | muC endpoint | muC_d20 colony b | muC_d20 colony c | Reference gene | Mutation Position | muS endpoint | muS_d20 colony c | muS_d20 colony f |
| Tfu_0132 | 155128 | X | X | X | Tfu_0132 | 155128 | X | X | |
| Tfu_0417 | 470157 | X | X | X | Tfu_0417 | 470157 | X | X | |
| Tfu_0913 | 1075652 | X | X | X | Tfu_0913 | 1075652 | X | X | |
| Tfu_1031 | 1209775 | X | X | X | Tfu_1031 | 1209775 | X | X | |
| Tfu_1108 | 1292195 | X | X | X | Tfu_1108 | 1292195 | X | X | |
| Tfu_1374 | 1592611 | X | X | X | Tfu_1374 | 1592611 | X | X | |
| Tfu_1458 | 1685076 | X | X | X | Tfu_1458 | 1685076 | X | X | |
| Tfu_1688 | 1962424 | X | X | X | Tfu_1688 | 1962424 | X | X | |
| Tfu_1867 | 2184179 | X | X | None | Tfu_1867 | 2184179 | X | 2184174 | |
| Tfu_2363 | 2780975 | X | None | None | Tfu_2363 | 2780975 | X | None | None |
| Tfu_2877 | 3388783 | X | X | X | Tfu_2877 | 3388783 | X | X | X |
| Tfu_0195 | 221012 | X | X | Tfu_0195 | 221012 | X | X | X | |
| Tfu_0423 | 475686 | 475670 | X | Tfu_0423 | 475686 | X | 475670 | X | |
| Tfu_0677 | 781810 | X | X | Tfu_0677 | 781810 | X | X | X | |
| Tfu_0750 | 885777 | X | X | Tfu_0750 | 885777 | X | X | X | |
| Tfu_1426 | 1652369 | X | X | Tfu_1426 | 1652369 | X | X | X | |
| Tfu_1516 | 1748226 | X | X | Tfu_1516 | 1748226 | X | X | X | |
| Tfu_2031 | 2375712 | X | X | Tfu_2031 | 2375712 | X | X | X | |
In the muC strain, 11 non-synonymous mutations were found. The mutation of Tfu_0417, which encodes for a peptidyl-tRNA hydrolase, changed a stop code to a glutamine in T. fusca muC, adding 24 amino acids to this protein. Peptidyl-tRNAs are recycled by peptidyl-tRNA hydrolase, which hydrolyzes the ester link between the tRNA and the peptide (41). If the addition of 24 amino acids to peptidyl-tRNA hydrolase (213 amino acids on the backbone) disrupted the function of this protein, the mutation could lead to the accumulation of peptidyl-tRNAs, which can cause cellular toxicity (42)in the T. fusca muC strain.
Tfu_1688 encodes an aldehyde dehydrogenase (8) that catalyzes acetate to acetaldehyde. The mutation of Tfu_1688 caused a change from isoleucine to valine at amino acid position 291. Of the T. fusca strains studied here, only the muC strain produced acetate as a metabolic end-product when grown on cellobiose (Fig. 1D), which indicates that the mutation of Tfu_1688 may have affected the conversion between acetate and acetaldehyde leading to the accumulation and secretion of acetate.
Another mutation found in the muC strain was found in the gene Tfu_1374 that encodes a repressor of the gluconate operon (GntR) in T. fusca. Gluconate is an important intermediate in the pentose phosphate pathway where xylose and mannose enter central metabolism. In the cases where growth was studied on non-evolutionary substrates, the growth rate of the muC strain was significantly lower than muS on xylose and mannose (Fig. 2, B and C). This is potentially a reflection of the affect of the mutation in Tfu_1374 and the role of gluconate as a metabolic intermediate for growth on xylose and mannose.
Fitness Contribution of Acquired Mutations
After characterizing the phenotypic and genetic characteristics of the evolutionary endpoints for the muC and muS strains, the day 20 timepoints of each strain (muC_d20 and muS_d20) were studied to better understand the evolutionary dynamics of each strain.
As previously described, the muS and muC strains had the significantly different growth fitness on the five different non-evolutionary substrates (the muS is generalist but the muC strain is specialist). Based upon the evolutionary paradigm of random mutation occurrence, selection based upon fitness, and fixation in the population (11), evolving populations should be more heterogeneous at earlier points of evolution rather than the end point where dominant genotype-phenotype could overtake a population. Thus, phenotypic and genetic characterization of the intermediate, day 20 time point of evolution was conducted to study the occurrence and eventual fixation of specific characteristics in the evolved populations.
Cells from day 20 (midpoint) of evolution for both the muC and muS strains were cultured on agar plates to allow for selection of single colonies. Ten colonies of each sample were then cultured on the five non-evolutionary substrates to test growth characteristics. As expected, there was a greater diversity of phenotypes found at the day 20 point than at the end point of evolution. Of the muS_d20 colonies tested, colony f (Fig. 2D) exhibited the best growth characteristics on non-evolutionary substrates and phenotypically was similar to the muS end point. Colonies c, g, and j exhibited poor growth on non-evolutionary substrates. Of the muC_d20 colonies that were tested, colonies b, f, h, i exhibited no appreciable growth on the non-evolutionary substrates tested. Colony c had the best growth characteristics. Overall, growth characterization of colonies taken at day 20 of evolution showed greater diversity in phenotype, but still followed the generalist of specialist phenotype observed at the end point (i.e. muS_d20 colonies tended to grow better on non-evolutionary substrates similar to the muS end point and muC_d20 colonies showed poor growth similar to the muC end point).
Based upon phenotypic similarities to the end point, several of the day 20 colonies were selected for Sanger sequencing to check for genetic mutations. Of the muC_d20 colonies, colony b (poor growth) and colony c (best observed growth) were selected. Of the muS_d20 colonies, colony c (poor growth) and colony f (best growth) were selected. Sanger sequencing was used to sequence the union of all 18 non-synonymous mutations (11 found in muC and 7 in muS) found in the end point strains (Table 1) in each of the four identified day 20 colonies.
Sequencing of muS_d20 colonies c and f identified a number of mutations in common with the end point strain (Table 2). In the day 20 colony c (poorest growth of muS_d20 colonies), 16 genes had the same mutations as found in the end point strains. The only differences found in the muS_d20 colony c sequencing were genes Tfu_2363 and Tfu_0423 where no mutation was found in Tfu_2363 and Tfu_0423 had a mutation in a different position. In the muS_d20 colony f, 16 genes had the same mutations as the end point strains except for Tfu_2363 and Tfu_1867 where no mutation was found in Tfu_2363 and Tfu_1867 had a mutation in a different position.
Sequencing of the muC_d20 colonies b and c also identified a high number of mutations that were also found in the evolutionary endpoints. In day 20 colony b, 16 genes had the same mutations as found in the end point strains except Tfu_2363 and Tfu_0423. There was no mutation found in Tfu_2363 and gene Tfu_0423 had a mutation at a different position than the evolved strain. Interestingly, these pattern of mutations found in muC_d20 colony b were the same as those found in muS_d20 colony c. Even the mutation in gene Tfu_0423 was found in the same position, but the base change in muC_d20 colony b was C = >G whereas the mutation in muS_d20 colony c was C = >T. In the colony c, again 16 genes were found to be the same as the end point strains except for genes Tfu_1867 and Tfu_2363 where no mutations were found
During cellulose degradation, both cellulose-hydrolyzing enzymes and cellobiose dehydrogenase (CDH), flavohemoproteins (6), are typically expressed. There is a flavohemoprotein (Tfu_0423) in T. fusca. The adaptive evolution on switching sugars (the muS strain) leads to change from threonine (position 367) to alanine. One possibility is that this mutation may cause to increase the activity of cellulose dehydrogenase that can lead to improved degradation and consumption of cellobiose by T. fusca (shown in the muS strain).
Influence of Oleic Acid and Glutamine on Intermediate Strain Growth
The gene Tfu_1031 encodes a long-chain fatty-acid-CoA ligase was found to have a site mutation on the position of 1209775 (T = >C) in the muC_d40 strain. To test the influence of the mutation of this gene on the growth of 10 intermediate strains, 0.1% of oleic acid was added to five different substrates (xylose, mannose, sucrose, acetate, citrate). The addition of oleic acid to the growth medium had mixed results with some strains exhibiting increased growth and others showing decreased growth (supplemental Table S3).
The gene Tfu_1108, which encodes UDP-N-acetylmuramoylalanine-d-glutamate ligase was found to have a point mutation at position 1292195 (A = >G) in the muC_d40 strain. To test the influence of the mutation of this gene on the growth of 10 intermediate strains, 0.1% of glutamine was added to five different substrates (xylose, mannose, sucrose, acetate, citrate). As with the testing using oleic acid, the tested strain showed mixed growth results (supplemental Table S3) with no discernable trend related to the mutation results.
DISCUSSION
In this study, the thermophilic, cellulolytic actinobacterium T. fusca was evolved using adaptive laboratory evolution (ALE) in different conditions to generate two different evolved phenotypes (a specialist muC and a generalist phenotype muS). The initial premise of this study was to determine if ALE could be used to increase the growth and cellulolytic activity of T. fusca and to genetically characterize associated adaptive mutations. It was found that ALE of cellulolytic T. fusca did not result in increased growth rates as has been observed for ALE of non-cellulolytic microorganisms (9–13). Thus, analysis of the phenotypic changes and associated genetic changes occurring in T. fusca undergoing ALE cannot be taken to be strictly associated with growth rate changes as has been considered in the past (43).
ALE is fundamentally a dynamic process involving cyclical systemic responses involving selection pressure (environment), adaptation, and fixation into the population. Through the use of high-throughput sequencing techniques, it is possible to study the entire complement of genetic mutations at the end point of evolution and try correlate these to cellular adaptations to match measured or observed phenotypic changes. Here, full genome re-sequencing of both evolved strains using two independent sequencing platforms identified 48 errors in the reference genome of T. fusca and found 18 SNPs in T. fusca muC (evolved on cellobiose) and 14 SNPs in T. fusca muS (evolved on switching between glucose and cellobiose daily). Of the mutations that we positively identified (found both by 454 and Illumina re-sequencing and confirmed by Sanger sequencing), three general categories of mutations arose: 1) mutations correlated to acetate secretion in the muC strain, 2) mutations affecting growth on secondary (non-evolutionary) substrates, and 3) mutations affiliated with transcription/translation.
The evolved muC strain was found to secrete acetate as a metabolic end-product whereas the muS strain did not secrete detectable amounts of acetate. In the initial analysis of the re-sequencing results, a mutation in Tfu_1688, an aldehyde dehydrogenase, was identified that is involved in the conversion of acetate to acetaldehyde. As a follow-up to these results, re-sequencing results from the 454 indicated that the muC strain also contained mutations in Tfu_2248 (fatty acyl-CoA synthetase) and Tfu_2309 (acyl-CoA dehydrogenase). These mutations were not found in the Illumina re-sequencing data and were not considered in our original sequence analysis. Sanger sequencing of these genes confirmed a four base TGTG deletion (2647457–2647461) in Tfu_2248 and a point mutation in position 2716546 (A = >G) in Tfu_2309 in the muC_d40 strain. The confirmed mutations in Tfu_2248 and Tfu_2309 may be correlated to the increase in acetate secretion in the muC strain through the oxidation of fatty acids into acetyl-CoA and also have implications in basal metabolic processes and cellular homeostasis (44, 45, 46).
Because of the manner in which the muC and muS strains were generated, testing of growth characteristics on non-evolutionary substrates was conducted to characterize secondary growth phenotypes, producing as expected, a specialist strain (muC) and a generalist strain (muS). Phenotype and genotype analyses for this purpose were conducted on single colonies at both the end point of evolution and at a midpoint of evolution to account for potential heterogeneity of the cell population. Three mutations (Tfu_1374, Tfu_1031, and Tfu_1108) were found in the muC_d40 strain that could potentially affect secondary growth characteristics. The Tfu_1374 gene affects the gluconate operon related to metabolic intermediates in the pentose phosphate pathway in T. fusca and the effects of this mutation were demonstrated by the reduced ability of the muC_d40 strain to grow on xylose or mannose.
Tfu_1031 is a long-chain-fatty-acid-CoA ligase is involved in the breakdown of fatty acids. Growth on oleic acid to test the effect of this mutation on the muC_d40 strain was inconclusive showing no discernable influence on growth properties. Tfu_1108 is a UDP-N-acetylmuramoylalanine-d-glutamate ligase and the mutation in this gene was tested using medium supplemented with glutamine. Again, no discernable change in growth properties was observed so the effect of identified mutations in Tfu_1031 and Tfu_1108 were not able to be identified.
Genes Tfu_1867, Tfu_2363, and Tfu_0423 were the only genes that were found to be different between colonies tested at the midpoint of evolution and colonies from the end point strains. The effect of the mutation in Tfu_1867 at position 2184179 can by hypothesized to be detrimental to cell growth as all three strains possessing this mutation exhibited inhibited growth (muC end point-worse growth on cellobiose and on secondary substrates, muC_d20 colony b-worse growth on secondary substrates, and muS_d20 colony c- worse growth on secondary substrates). The muS end point strain, muC_d20 colony c, and muS_d20 colony f either did not have a mutation in this gene nor had a different mutation (muS_d20 colony f) and all showed good growth on secondary substrates.
The functional consequences of the mutation in Tfu_2363 (a putative zinc proteinase) are not clear. None of the intermediate day 20 colonies had mutations in this gene and both end point strains were found to have the same mutation in this gene.
For gene Tfu_0423 (a flavoprotein), a correlation can be seen between a mutation and growth on secondary substrates. All three populations that possess a mutation at position 475686 (muS end point, muC_d20 colony c and muS_d20 colony f) showed good growth on secondary substrates. The strains that did not have a mutation at that position (muC end point, muC_d20 colony b, and muS_d20 colony c) showed poor growth on secondary substrates.
Outside of the identification and testing of the mutations mentioned above, a general trend was found where many of the mutations appear to be related to basic transcription/translation processes (mutations in Tfu_0417, Tfu_0665, Tfu_0778, Tfu_1867, Tfu_0677, Tfu_2728). While not conclusive, this is potentially analogous to some of the results found in ALE experiments conducted in E. coli where mutations have been found in RNA polymerase subunits such as rpoB, rpoC (43) and rpoS (47, 48). For our study with T. fusca, it appears that the effect is more distributed rather than concentrated on transcriptional subunit that also has regulatory functions, but both cases allude to the broader principle of adaptations aimed at balancing or efficiency of protein expression. This is secondarily supported by the measured changes in available intracellular ATP in the muC and muS strains. In a cellulolytic microorganism such as T. fusca, the expression of cellulolytic genes has a high metabolic cost with the cellulolytic machinery constituting as much as 20% of the total cell mass (38).
Overall, this study describes the phenotypic and genetic changes occurring during adaptive evolution of a cellulolytic microorganism, T. fusca. It appears that even under growth rate selection pressure, T. fusca evolves with a strong consideration of the metabolic cost associated with producing and expressing cellulases. Whereas several mutations were found that are linked to specific primary (secretion of acetate) or secondary (growth on secondary substrates) phenotypes, many of the mutations are more ambiguous in their effect as the primary phenotype change did not result in growth rate increases as have been observed in other microbial evolution experiments.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) 1) T. fusca muC strain by Illumina: SRA023455.3, 2) T. fusca muC strain by 454: SRA023427.6, 3) T. fusca muS train by Illumina: SRA023505.2, 4) T. fusca muS strain by 454: SRA023445.5.
- muC
- mutant grown on cellobiose
- muS
- mutant grown by switching sugars
- CMC
- carboxymethylcellulose
- DNS
- dinitrosalicylic acid
- DCW
- dry cell weight
- CDH
- cellobiose dehydrogenase.
REFERENCES
- 1. Himmel M. E., Ruth M. F., Wyman C. E. (1999) Curr. Opin. Biotechnol. 10, 358–364 [DOI] [PubMed] [Google Scholar]
- 2. Jeoh T., Wilson D. B., Walker L. P. (2002) Biotechnol. Prog. 18, 760–769 [DOI] [PubMed] [Google Scholar]
- 3. Lynd L., van Zyl W., McBride J. E., Laser M. (2005) Curr. Opin. Biotechnol. 16, 577–583 [DOI] [PubMed] [Google Scholar]
- 4. Gowen C. M., Fong S. S. (2010) Chem. Biodivers. 7, 1086–1097 [DOI] [PubMed] [Google Scholar]
- 5. Demain A. L., Newcomb M., Wu J. H. D. (2005) Microbiol. Mol. Biol. Rev. 69, 124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Igarashi K., Verhagen M. F., Samejima M., Schülein M., Eriksson K. E., Nishino T. (1999) J. Biol. Chem. 274, 3338–3344 [DOI] [PubMed] [Google Scholar]
- 7. Suto M., Tomita F. (2001) J. Biosci. Bioeng. 92, 305–311 [DOI] [PubMed] [Google Scholar]
- 8. Lykidis A., Mavromatis K., Ivanova N., Anderson I., Land M., DiBartolo G., Martinez M., Lapidus A., Lucas S., Copeland A., Richardson P., Wilson D. B., Kyrpides N. (2007) J. Bacteriol. 189, 2477–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fong S. S., Joyce A. R., Palsson B. (2005) Genome Res. 15, 1365–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Babu M. M., Aravind L. (2006) Trends Microbiol. 14, 11–14 [DOI] [PubMed] [Google Scholar]
- 11. Elena S. F., Lenski R. E. (2003) Nat. Rev. Genet. 4, 457–469 [DOI] [PubMed] [Google Scholar]
- 12. Papadopoulos D., Schneider D., Meier-Eiss J., Arber W., Lenski R. E., Blot M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3807–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wright B. E. (2004) Mol. Microbiol. 52, 643–650 [DOI] [PubMed] [Google Scholar]
- 14. Fong S. S., Burgard A. P., Herring C. D., Knight E. M., Blattner F. R., Maranas C. D., Palsson B. O. (2005) Biotechnol. Bioeng. 91, 643–648 [DOI] [PubMed] [Google Scholar]
- 15. Denamur E., Matic I. (2006) Mol. Microbiol. 60, 820–827 [DOI] [PubMed] [Google Scholar]
- 16. Heidenreich E. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 285–311 [DOI] [PubMed] [Google Scholar]
- 17. Lenski R. E., Travisano M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6808–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fong S. S., Marciniak J. Y., Palsson B. (2003) J. Bacteriol. 185, 6400–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edwards J. S., Ibarra R. U., Palsson B. O. (2001) Nat. Biotechnol. 19, 125–130 [DOI] [PubMed] [Google Scholar]
- 20. Ibarra R. U., Edwards J. S., Palsson B. O. (2002) Nature 420, 186–189 [DOI] [PubMed] [Google Scholar]
- 21. Ai Y. C., Zhang S., Wilson D. B. (2003) Enzyme Microb. Technol. 32, 331–336 [Google Scholar]
- 22. Bevillard E., Goodheart D. B., Karnati S. K., Bayer E. A., Lamed R., Miron J., Nelson K. E., Morrison M. (2004) J. Bacteriol. 186, 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jung H., Wilson D. B., Walker L. P. (2003) Biotechnol. Bioeng. 84, 151–159 [DOI] [PubMed] [Google Scholar]
- 24. Irwin D. C., Zhang S., Wilson D. B. (2000) 267, 4988–4997 [DOI] [PubMed] [Google Scholar]
- 25. Watson D. L., Wilson D. B., Walker L. P. (2002) Appl. Biochem. Biotechnol. 101, 97–111 [DOI] [PubMed] [Google Scholar]
- 26. Zhou W., Irwin D. C., Escovar-Kousen J., Wilson D. B. (2004) Biochemistry 43, 9655–9663 [DOI] [PubMed] [Google Scholar]
- 27. Spiridonov N. A., Wilson D. B. (1999) J. Biol. Chem. 274, 13127–13132 [DOI] [PubMed] [Google Scholar]
- 28. Deng Y., Fong S. S. (2010) Appl. Environ. Microbiol. 76, 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng Y., Fong S. S. (2011) Metab. Eng. 13, 570–577 [DOI] [PubMed] [Google Scholar]
- 30. Chen S., Wilson D. B. (2007) J. Bacteriol. 189, 6260–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferchak J. D., Pye E. K. (1983) Biotechnol. Bioeng. 25, 2855–2864 [DOI] [PubMed] [Google Scholar]
- 32. Ferchak J. D., Pye E. K. (1983) Biotechnol. Bioeng. 25, 2865–2872 [DOI] [PubMed] [Google Scholar]
- 33. Kassen R. (2002) J. Evol. Biol. 15, 173–190 [Google Scholar]
- 34. Xiao Z., Storms R., Tsang A. (2004) Biotechnol. Bioeng, 88, 832–837 [DOI] [PubMed] [Google Scholar]
- 35. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 36. Eaves H. L., Gao Y. (2009) Bioinformatics 25, 969–970 [DOI] [PubMed] [Google Scholar]
- 37. Spiridonov N. A., Wilson D. B. (2001) Curr. Microbiol. 42, 295–301 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y., Lynd L. R. (2003) Anal. Chem. 75, 219–227 [DOI] [PubMed] [Google Scholar]
- 39. Dekel E., Alon U. (2005) Nature 436, 588–592 [DOI] [PubMed] [Google Scholar]
- 40. Deng Y., Fong S. S. (2010) Appl. Microbiol. Biotechnol. 85, 965–974 [DOI] [PubMed] [Google Scholar]
- 41. Menninger J. R. (1979) J. Bacteriol. 137, 694–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh N. S., Varshney U. (2004) Nucleic Acids Res. 32, 6028–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herring C. D., Raghunathan A., Honisch C., Patel T., Applebee M. K., Joyce A. R., Albert T. J., Blattner F. R., van, den, Boom D., Cantor C. R., Palsson B. Ø. (2006) Nat. Genet. 38, 1406–1412 [DOI] [PubMed] [Google Scholar]
- 44. Weimar J. D., DiRusso C. C., Delio R., Black P. N. (2002) J. Biol. Chem. 277, 29369–29376 [DOI] [PubMed] [Google Scholar]
- 45. Black P. N., Zhang Q., Weimar J. D., DiRusso C. C. (1997) J. Biol. Chem. 272, 4896–4903 [DOI] [PubMed] [Google Scholar]
- 46. Spector M. P., DiRusso C. C., Pallen M. J., Garcia del Portillo F., Dougan G., Finlay B. B. (1999) Microbiology 145, 15–31 [DOI] [PubMed] [Google Scholar]
- 47. Notley-McRobb L., King T., Ferenci T. (2002) J. Bacteriol. 184, 806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferenci T. (2008) Heredity 100, 446–452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.