Abstract
Steroid hormone receptors (SHRs) and nuclear receptors (NRs) in general are flexible, allosterically regulated transcription factors. The classic model is inadequate to explain all their behavior. Keys to function are their regions of intrinsic disorder (ID). Data show the dynamic structure and allosteric interactions of the three classic SHR domains: ligand-binding (LBD), DNA-binding (DBD), and N-terminal (NTD). Each responds to its ligands by stabilizing its structure. The LBD responds to classic steroidal and nonsteroidal small ligands; both may selectively modify SHR activity. The DBD responds differentially to the DNA sequences of its response elements. The NTD, with its high ID content and AF1, interacts allosterically with the LBD and DBD. Each domain binds heterologous proteins, potential allosteric ligands. An ensemble framework improves the classic model, shows how ID regions poise the SHR/NR family for optimal allosteric response, and provides a basis for quantitative evaluation of SHR/NR actions.
Keywords: Allosteric Regulation, Intrinsically Disordered Proteins, Nuclear Receptors, Steroid Hormone Receptor, Transcription Factors, SERM, TMAO, Glucocorticoid Receptor, Protein Ensembles
Introduction
The classic model for nuclear receptors (NRs)2 and the subfamily of steroid hormone receptors (SHRs) has difficulty explaining important aspects of SHR/NR function. The classic model divides SHRs into three domains, ligand-binding (LBD), DNA-binding (DBD), and N-terminal (NTD), and has served well (Fig. 1), but some of its premises are limiting its usefulness. Phenomena for which the classic model is inadequate include tissue- and cell-specific responses to individual ligands, differing transcriptional responses imparted by DNA sequences of response elements (REs), selective effects of isoform-specific activation function 1 (AF1) regions in the NTD, the widely differing size and sequence of NTDs, and the lack of stable tertiary structure in NTDs. Theory and data suggest that such limitations stem from two conceptual weaknesses in the classic model: 1) the presumption that the major NR domains consist of structurally stable peptides and 2) the failure of the model to account for the essential allosteric nature of SHRs/NRs. Herein, we discuss each of the classic domains, citing examples that indicate their structurally dynamic nature. We then show how the intrinsic disorder (ID) found in regions of SHRs/NRs can optimize allosteric responses.
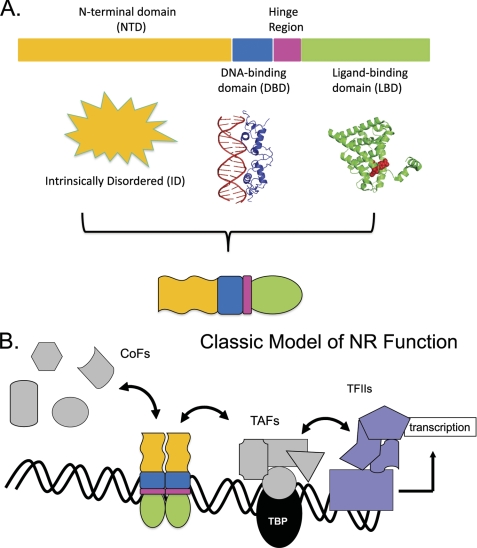
FIGURE 1.
Classic model of SHR function. A, structural architecture: SHRs are composed of three domains with segregated functional roles. The globular C-terminal LBD (green) binds steroidal or other small ligands. This alters the position of the LBD 12th helix to configure the AF2 surface so as to bind “ligand-dependent” CoFs containing LXXLL motifs. The globular DBD (blue) recognizes specific DNA sequences. The hinge region (pink) connects the DBD and LBD. The NTD (yellow; structure undetermined) is of varying length and sequence in the SHR family of TFs. In some NRs, it is absent altogether. When present, the NTD may bind CoFs without requirement for an LBD ligand (“ligand-independent”). The AF1 regions of the NTD vary in potency for regulating transcription. B, functional scheme: binding of steroid localizes the SHR to the nucleus. SHRs homodimerize and other NRs heterodimerize and associate with their RE DNA sequences. The NR-DNA complex attracts CoFs and other proteins of the transcriptional and chromatin-modifying apparatus. TBP, TATA box-binding protein; TAFs, TBP-binding factors; RNA Pol II, RNA polymerase II.
Dynamic Nature of the LBD
In the classic model, the LBD is a stable, globular, 12-helix protein (11 in SHRs) with a hinged final helix. LBD function is defined by its activation function 2 (AF2) subdomain, a surface controlled by the ligand-driven positioning of the final helix. This surface consequently displays differential binding to protein cofactors (CoFs). These interact with SHRs/NRs to collaborate in modulating transcription. Hundreds of such CoFs (Nuclear Receptor Signaling Atlas (NURSA)) (1–3), which serve as chromatin modifiers, form transient complex protein clusters with the primary transcription complex (4). The varied properties of CoFs result in activation or repression of transcription of the relevant gene. Probably more CoFs, particularly those that are specific to particular cell types or tissues, are yet to be found. Success in solving the crystal structures of many DBDs and LBDs as independent proteins has shown their crystallized forms to be globular (www.ncbi.nlm.nih.gov/sites/entrez), but in fact, without ligand, the LBD is rather disordered; it is the binding of ligand that gives it structure (5–8). SHR LBDs interact with steroid hormones, and more generally, most NR LBDs interact with a variety of natural and man-made steroidal and nonsteroidal molecules. The astounding fact that structurally similar ligands, acting on a single receptor isoform, provoke differing tissue- and cell type-specific biological effects to regulate overlapping but also unique sets of genes (9–13) challenges the classic receptor model. Particularly compelling are the effects of tamoxifen, developed as a competitive LBD-binding antagonist to hormonal estrogens. In breast cancer cells, tamoxifen behaves as expected, but it acts as an agonist in bone and uterus, an outcome not easy to explain by simple application of the classic model (14, 15). If, as antagonist, tamoxifen works by repositioning the 12th helix so as to allow AF2 to bind CoFs with co-repressor functions, how can the same ligand act as an agonist in certain tissues, where the same estrogen receptor (ER) AF2 surface presumably must bind CoFs with activator function? Such “mixed function” or “selective response-modifying” LBD ligands have been described for several major classes of SHRs. The dogma that all agonists differ only in potency has been discarded. Although the great bulk of genes are regulated similarly by SHR ligands, data suggest that unique gene sets of varying size are regulated by each natural or xenobiotic LBD ligand. Ligands bound in the LBD exert significant allosteric effects (16–21) locally and throughout their SHR. Selective response-modifying ligands for SHRs are being studied assiduously in hopes of providing just the desired, tissue-specific effects. For success, these studies must consider how the allosteric reach of the LBD extends to other domains.
In addition to classic ligands, the various proteins that bind the LBD (CoFs, chaperones, etc.) can be looked upon as an additional ligand class (18, 22, 23). Chaperones and associated proteins that bind to the LBDs of certain SHRs in the absence of small molecule LBD ligands (24, 25) act to preserve the optimal configuration of the small molecule-binding pocket and thus can be thought of as acting locally as classic Koshland allosteric modifiers (26). Some CoFs bind directly to the AF2 surface (and in some instances, other LBD surfaces), where they provide important chromatin-modifying functions and serve as binding platforms for the aggregation of additional co-regulatory proteins that collectively determine the transcriptional response. In binding the LBD, CoFs affect the structural properties of the SHR; thus, CoFs are allosteric effectors/ligands (18, 27, 28). In sum, the LBD is malleable, adopting functionally important, differing structures in response to small molecule and protein ligands. Ligand binding to the LBD results in allosteric effects in remote receptor regions.
Dynamic Nature of the DBD
The DBD classically has been defined as a small globular peptide. In solution, however, the unbound DBD has a flexible structure (29–32). Recently, it has been shown that for optimal DNA binding, an even more flexible C-terminal peptide “extension” of the DBD is required (33, 34). Thus, the complete DBD may be considered to include this extension. The accepted model states that the ligand-activated SHR continually samples DNA across the vast excess of nonspecific weak binding sites until a proper high affinity site is reached (35, 36)). The dynamic nature of these complexes can be studied by newly developed applications of NMR (37, 38). In addition to the SHR itself, other proteins are often involved in the site-specific binding, e.g. HMG proteins (33, 39). This suggests structure-modifying effects of the heterologous proteins on the SHR.
In sum, the processes by which SHR/NR DBDs seek and find their high affinity binding sites strongly suggest flexible dynamic behavior of their DBDs. The stable interaction of the DBD with its cognate RE also results in allosteric effects, as discussed next.
Allosteric Effects of RE-DBD Binding
Contrary to the classic model, the binding interaction between the DBD of an SHR/NR and its RE does more than anchor the receptor to a proper genome site. Evidence shows that the RE is an allosteric ligand, acting through the DBD of SHRs to influence NR structure at the DBD and beyond (40–44). In general, higher SHR-RE affinity correlates with stronger transcriptional activity, but in some cases, the opposite has been found (43). An extreme example was given in the comparison of two ER REs with equal affinity for the ER. One estrogen RE engendered a typical transcriptional response; the other was inactive (45). Such sequence-specific RE effects suggest allosteric effects on receptor structure. The effect of high affinity DBD-DNA binding on protein, e.g. SHR structure, was calculated. The results suggested that binding may involve folding of some part of the protein (46). Consistent with this, predictive algorithms (47) indicate that some disorder exists in the DBD. Globular proteins often contain structurally dynamic regions important for function (48, 49). Indeed, NMR studies of dynamics (29, 50–54) show the DBD in solution to be an ensemble of conformers. The ensemble concept for globular proteins has been validated crystallographically (55). Upon DNA binding, the DBD becomes less flexible, and crystallographic studies provide many valuable snapshots of these structures, stabilized from among the ensemble of conformations. Considering the above, it was reasoned that the specific sequence of a specific binding site for a transcription factor (TF) affected its structure and function (56). Indeed, glucocorticoid receptor (GR) DBD structure is subtly altered according to the specific sequence of the RE to which it is bound. These structural differences correlate with differences in the spectrum and extent of genes regulated (43).
The allosteric influence of SHR-DNA binding is not limited to local DBD effects. In the progesterone receptor, additional NTD structure upon DNA binding was seen in both the A and B isoforms (57, 58). Work with the thyroid receptor/retinoid X receptor heterodimer showed that the sequence of the TR RE to which it bound influenced the structure and function of the heterodimer (59). Extensive work on ERα and ERβ has shown that DNA binding and sequence influence overall receptor structure, binding of CoFs, and transcriptional function (42, 60–62). GR DBD binding to an RE causes acquisition of secondary and tertiary structure in the disordered NTD (63), along with increased binding of several CoFs, the essential function of AF1. It was predicted that binding of DBDs to REs should cause acquisition of structure and function in the unstructured NTDs/AF1s of other SHRs (64, 65) and that the structural changes should affect AF1 binding to and selection of CoFs. Consistent with this, it was shown subsequently that the RE- and AF1-dependent recruitment of TATA box-binding protein in vivo correlated with gene induction (66).
Combining the knowledge that RE-DBD interaction results in altered structure in the DBD and the NTD/AF1 (and the DNA; not reviewed here), it seems plausible that this binding event will also show effects on the regions of the NRs that lie C-terminal to the DBD. The allosteric influences of DNA binding could thus result in selectivity of further SHR-NR interactions with various heterologous proteins. Such structural effects will be important for explaining the cell- and gene-specific effects of SHRs/NRs and their ligands. As different cells expose differing regulatory regions of their genomes to occupancy by these TFs, cell-specific patterns of gene regulation result.
These data collectively urge that any SHR/NR model should include the role of DNA as an allosteric effector, with both local and remote DBD-specific effects. As described next, it appears that the property that mediates the allosteric responses of these proteins is their intrinsic structural disorder.
Disorder Is the Key to NTD Functions
NTDs of SHRs/NRs have been difficult to study structurally; until recently, this limited the understanding of their mechanisms. Mutational mapping and domain swapping studies established the importance of NTDs and their AF1 regions for transcription regulation. Coupled with structural studies, the data show that like AF2 regions, AF1 regions function as allosteric sites that bind various CoFs. The critical property for NTD/AF1 function seems to be ID. To appreciate this property, it is necessary briefly to consider ID proteins in general.
Properties of ID Proteins
ID proteins or regions can be thought of as a set of rapidly interchanging unfolded or only partially folded conformers. These proteins may have evolved so that a compact structure is simply too unstable to be significantly populated under ordinary physiological conditions, or they may never adopt a compact globular structure. It is clear that ID domains can transiently adopt secondary and/or tertiary structure as part of their functional cycle (67). The lack of stable conformation in ID proteins is due to their unique amino acid content: high percentages of certain charged amino acids and low proportions of hydrophobic amino acids (68–72). Although ID proteins lack any signature sequence motif, their special properties permit computer programs to predict their existence (47). These predict that 20–30% of the mammalian proteome exists as ID regions and that 70% of transcription factors (TFs) contain ID regions (71, 73). It appears that the proteins regulating transcription are specifically designed to contain ID regions. SHRs and several other important classes of NRs have ID regions, especially in NTDs and their AF1 subdomains. The enhancement of ID sequences in TFs and the effect of ligand binding on these ID sequences strongly suggest a functional significance for ID in signal transduction.
Role of ID in SHR/NR Function
Isoform comparisons and NTD swapping experiments have shown that differing NTD/AF1s impart uniquely different potencies, gene selectivities, and responses to LBD ligands (74–76). NTDs differ widely in size, sequence, and ID content, and the pattern of ID regions seems specific for each NTD class. The AF1 subregions of NTDs have been shown to have some secondary structure, probably the average of a multitude of distinct states in the ensemble (see Fig. 3). Only detailed studies will reveal their intimate organization. Therefore, hereafter, we refer to NTDs and AF1 regions simply as ID, with the understanding that they consist of a variety of transient conformers, perhaps with some relatively stable regions. Because it is likely that NTDs/AF1s achieve structure(s) as they interact with varying CoFs, it is important to understand the forces controlling the shift from ID to structure and to learn the properties of the structures reached. Several ways of producing structure in AF1 regions of SHRs have been discovered. The first was by use of organic osmolytes, e.g. trimethylamine N-oxide (TMAO). Organic osmolytes of several chemical types, known to induce protein folding, are found in all cells. Such stabilizing osmolytes are well known in nature, where they are produced intracellularly and function to protect cellular proteins against damaging conditions, such as high extracellular osmolarity and extremes of temperature (77–79). Protective osmolytes can induce return of functioning structure to enzymes unable to fold spontaneously (80). TMAO caused a cooperative shift from a disordered to an ordered state in the GR AF1/NTD, and this effect was soon replicated on the androgen receptor (AR) AF1 and the mineralocorticoid receptor (81–83). The folded forms showed both secondary and tertiary structure; these were found to be in a relatively dynamic state, i.e. the conformers with tertiary structure were in rapid exchange with less structured states (84). The stabilized conformers showed increased function by enhanced binding of known CoFs (85).
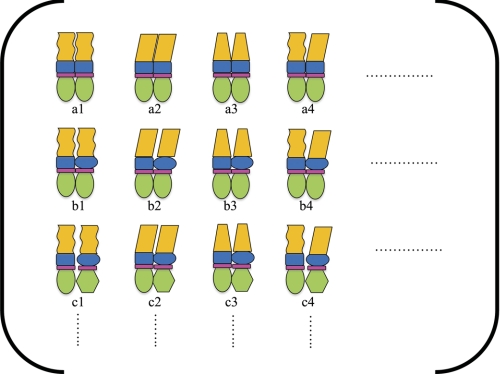
FIGURE 3.
Ensemble model of SHR function. Using the principles derived from the ensemble model depicted in Fig. 2, each domain in the SHR can be depicted as having two or more states (indicated by the different shapes for each domain). A full enumeration of all possible combinations of HA and LA states for different ligands of each domain (by expanding the columns and rows in the ensemble) provides a framework for understanding the potential impact of different ligands on SHR function. By simply modulating the probabilities of different states (c.f. a1 through c4), the effects of the different ligands in Fig. 2B will change. Thus, the ensemble model would suggest that merely asking, “what is stabilized as a result of binding?” is insufficient to understand SHR function. Instead, to what extent something is stabilized and to what degree two domains are coupled are questions that are at the heart of a quantitative and even a qualitative understanding of SHR/NR function.
Because evidence indicates that when TFs are bound to appropriate protein partners, their ID regions are frequently in a relatively stable structured state (67), and because TMAO enhances formation of structure in the GR and AR NTDs, with enhanced binding of certain CoFs, the possibility arises that direct CoF binding could promote structure in SHR NTDs. For the GR, mineralocorticoid receptor, and AR, this was shown to be the case (82, 84, 86).
The third method showed that binding of a GR or progesterone receptor to a palindromic glucocorticoid RE induced structure in the NTD (57, 63). Thus, an RE acts as an allosteric ligand, with structural consequences in the NTD (65). Because we know that the base sequence of an RE affects the choice of genes regulated through transcription and the potency of the regulation, it seems likely that the allosterically induced structure in the NTD/AF1 provides part of the mechanism for that RE-specific regulation. The detailed structures induced in the NTD/AF1 by osmolytes, remote RE-DBD interaction, or direct CoF binding remain unknown. The data clearly show, however, that there is increased tertiary structure and increased helical content. Whatever conformations are formed are still dynamic, an ensemble of interchanging structures that is on average much more stable than the original ID state. Whether this corresponds to an equilibrium between a folded and an unfolded conformation or whether there is an ensemble of partially folded states is unknown. It may be that ID regions fold into more than one structure that can bind to another protein with high affinity (87, 88). Because various cell types contain unique combinations of ubiquitous and cell-specific CoFs with which NRs must interact to regulate transcription, it seems plausible that the ID regions found in the SHR NTDs/AF1s would be able to shift rapidly between conformers. By doing so, NTDs/AF1s may be able to sift through the multiple potential CoFs being encountered.
It may also be the case that a large disordered domain contains multiple CoF-binding regions, such that each ID region adopts a more or less unique conformation when bound to its CoF. Differences in function could result because the various binding sites fold in different combinations in the context of different cellular milieus. We suggest that a productive and more quantitative way to think of these regions is as an ensemble of multiple conformers (89), some of which can be stabilized by allosteric interactions, secondary modifications, and cellular conditions (such as pH, salts, and osmolyte content). The value of this approach to the problem has been demonstrated (90, 91). The properties of such conformers can be examined by quantitative methods. Some in vitro conditions may sufficiently stabilize some conformers so that they can be studied crystallographically. However, this model also predicts that no single static structure will give a complete understanding of the actions of ID regions. For that, methods that follow the dynamic properties of the SHRs and their family will be required.
ID Regions in SHRs, NRs, and TFs Optimize Allosteric Coupling
Because each major domain of an SHR can reversibly bind various ligands, what is the effect of such binding at one domain on the capacity of another domain to bind its own ligands? The fundamental value of the ID regions and how they interact with folded domains can be appreciated in quantitative terms. Consider a two-domain protein, composed of domains I and II, connected through an interface (Fig. 2). This example would be analogous to the observed mutual impact of the DBD and NTD of an SHR wherein the binding to the RE DNA induces structure in the NTD. In Fig. 2, domain I is represented as relatively well folded, like the DBD, and can populate a low (LA) or high (HA) affinity state. Domain II, similar to the NTD, is an ID domain, also with LA and HA states, except that the LA state is disordered and the HA state is structured (similar to the NTD). For this model, domain I can bind ligand A, and domain II can bind ligand B, and the protein can exist in four possible states: 1) both domains I and II in LA states, 2) domain I in an LA state and domain II in an HA state, 3) domain I in an HA state and domain II in a LA state, and 4) both domains I and II in HA states. Classic equilibrium relations allow calculation of the probabilities for the protein to be in any of the four states (91–93), given the intrinsic free energies of domains I and II and the interaction between them. The ability of each domain to “sense” the other domain stems from the fact that the energy of each of the four states, relative to the state in which both domains are in their HA conformations, is determined by the HA/LA energy difference in the relevant domain plus the energy required to break the domain-domain interaction (Δgint). Although the precise magnitude of this coupling will depend on the balance of energies, a necessary condition is that the coupling energy be non-zero (i.e. Δgint ≠ 0). When Δgint is positive, it is energetically unfavorable to break the interaction between the HA forms of each domain, and when Δgint is negative, it is energetically favorable to break the interaction.
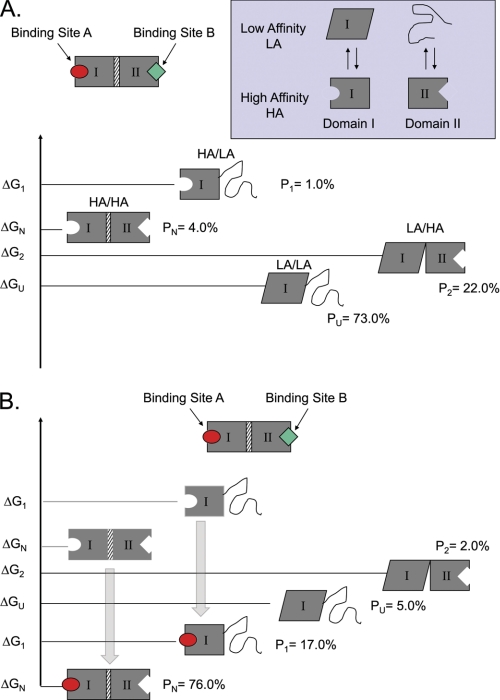
FIGURE 2.
Tuning the protein ensemble can modulate allosteric control. A simple two-domain model can be used to demonstrate how changing the probabilities of states can result in differential allosteric effects. A, a two-domain protein is shown, with the domains connected through an interface. Each domain has an equilibrium between a HA and LA state, which is governed by the intrinsic stability terms (ΔGI and ΔGII). The LA state can be ID (as with the NTDs of the SHRs) or just an alternative conformation (as with the DBDs of SHRs). Coupling between domains is facilitated by an interaction energy (Δgint), which determines the energetic cost of having the HA states populated in each domain simultaneously. B, Le Chatelier's principle dictates that the addition of ligand will stabilize the HA state of a particular domain, causing a ligand-induced redistribution of the ensemble. In this example, the addition of ligand A stabilizes the HA state of domain I, and the resultant redistribution also stabilizes the HA state for domain II. The observed effect is an apparent increase in the affinity of the protein for ligand B (i.e. ligand A is a positive allosteric effector of ligand B). However, the impact that such a redistribution will have on the affinity of the protein for a second ligand will depend on where the equilibrium is poised prior to stimulation with ligand A. The probabilities (P) of each form under specific sets of conditions are shown. When this model was exhaustively explored, it was found that ID could be used to tune the allosteric response of a protein, with some parameter combinations producing positive coupling and others producing negative coupling (91).
Upon binding a ligand, ID proteins often fold their disordered domain, and for several SHRs, direct binding of a protein ligand, e.g. a CoF to the disordered NTD/AF1, results in folding of the NTD/AF1. Therefore, in the model, the folding of the NTD can also be allosterically induced by ligand binding in another domain. This can be shown in the model by adding ligand, which redistributes the ensemble probabilities. The impact that the redistribution has on the ensemble properties depends on the probabilities of each state prior to the addition of ligand. Importantly, the response of the system to the addition of ligand depends on where the equilibrium is poised prior to ligand addition, and where it is poised is determined by the magnitudes of the intrinsic stabilities and interaction energies between the domains. For the example shown, the addition of ligand A promotes the transition from the LA state of domain I to the HA state. For the particular parameters chosen for Fig. 2, redistribution to the HA states of domain I also causes a shift to the folded HA states of domain II. Such a scenario is consistent with the impact of RE DNA binding to the DBD of an SHR, where binding of the RE would promote the HA state of the DBD. If the balance between the conformational and interaction coupling energies between the domains is within a specific set of values (see below), redistribution to the HA state of the DBD results in a commensurate redistribution to the HA or folded state of ID domain II. In other words, DNA binding to the DBD could fold the ID NTD and thus increase its affinity for NTD-binding proteins. Is this what is actually happening in SHRs? This simple model has important implications for SHR function. The response of the system depends on the relative probabilities (P) of the different states. Under one particular set of conditions (Fig. 2), the addition of ligand A increases the ability of the system to bind ligand B, making ligand A a positive allosteric effector for ligand B under these conditions. However, because the response is determined by the magnitudes of the probabilities, the same four states can be poised in such a way as to make ligand A a negative allosteric effector for ligand B. Thus, within the context of even the most simple manifestation of the ensemble model, it is straightforward to understand how a single ligand can act as an agonist under one set of conditions and as an antagonist under another set of conditions: the ensemble need only be poised differently prior to activation. In the case of SHRs (Fig. 3), integrating the ensemble model into the classic SHR model provides a new framework within which functional differences can be addressed. For instance, a long-standing problem in explaining all SHR actions is lack of a mechanism as to how these TFs have such tissue and cell specificity of action. The original discovery of SHRs (94) was based on the hypothesis that steroid class-specific receptors would be concentrated in appropriate target tissues, e.g. the ER in the uterus. These SHRs would bind the proper circulating steroid for actions in that tissue. This mechanism may apply sometimes, as in the differential expression of certain SHR isoforms in various tissues or in cell sets within an organ, and at various times during development (95–99). However, it is apparent that many NRs, GRα for example, are abundant in widespread tissues. Thus, the original hypothesis cannot explain the tissue-specific effects of widely expressed SHRs. Many other factors may influence NR behavior, including signaling pathway cross-talk, the metabolic state of each cell, paracrine signaling pathways, and tissue-specific CoFs. We suggest that the ensemble nature of the SHR and, in particular, the ID properties of the NTD/AF1 contribute significantly to the cell and tissue specificity of SHR/NR action. Conformational plasticity provides a wide range of different states that an NR can populate, and the broad conformational repertoire also provides a range of regulatory possibilities. Having multiple conformers in the ensemble allows precise tuning, permitting interactions with differing potential ligands, including proteins, small molecules, and REs. Conformational diversity even allows the same ligand to act as agonist or antagonist depending on how the SHR states are poised prior to activation.
Conclusions
NRs should not be viewed as simple articulated sets of linked globular domains; the allosteric properties of NRs are essential to their function. SHRs (and by extension, all NRs) are structurally much more dynamic proteins than is represented by traditional models, and each of the three classic SHR domains has a dynamic ensemble of structures that respond to a variety of ligands, including small molecules, DNA sequence, and heterologous proteins. The LBD and DBD each undergo ligand-driven structural stabilization that drives remote allosteric responses. LBD ligands include a variety of natural and man-made small molecules (steroids and nonsteroidal xenobiotics), which influence the AF2 surface and other regions in ligand-specific ways. For the DBD, high affinity RE sites, which vary considerably in sequence for each class of SHR, represent ligands that variably affect DBD structure, resulting in differential selection of genes regulated and the extent of that regulation. Besides local allosteric effects, RE-DBD binding causes increased structure in the otherwise ID NTD. This more structured NTD/AF1 shows increased affinity for important known CoFs. It is increasingly clear that ID is important for mediating allosteric control, as demonstrated theoretically and experimentally (91, 92, 100). By examining how the response of the protein ensemble to ligand can drive allosteric coupling, we have shown that the presence of one or more ID regions in a protein can modulate allosteric coupling between domains. Just as important as showing how ID can facilitate allostery is that the ensemble model provides a framework for understanding the broader principles at play in NR function. Because the response of the protein to ligand depends on the relative stabilities of states prior to activation, NRs and indeed most TFs can no longer be viewed as switches with fixed committed responses. To the contrary, the ensemble can be viewed as functionally “pluripotent,” having the capability to up- or down-regulate a specific set of activities depending on the relative stabilities of the various states in the ensemble. It is hoped that this ensemble view of NR function will provoke a re-evaluation of data and provide novel insights from which new experiments will be conducted.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM63747 and National Science Foundation Grant MCB-0446050 (to V. J. H.). This work was also supported by the Sealy Foundation at the University of Texas Medical Branch (to E. B. T.).
- NR
- nuclear receptor
- SHR
- steroid hormone receptor
- LBD
- ligand-binding domain
- DBD
- DNA-binding domain
- NTD
- N-terminal domain
- RE
- response element
- AF1
- activation function 1
- AF2
- activation function 2
- ID
- intrinsic disorder
- CoF
- cofactor
- ER
- estrogen receptor
- TF
- transcription factor
- GR
- glucocorticoid receptor
- TMAO
- trimethylamine N-oxide
- AR
- androgen receptor
- LA
- low affinity
- HA
- high affinity.
REFERENCES
- 1. York B., O'Malley B. W. (2010) J. Biol. Chem. 285, 38743–38750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hermanson O., Glass C. K., Rosenfeld M. G. (2002) Trends Endocrinol. Metab. 13, 55–60 [DOI] [PubMed] [Google Scholar]
- 3. Thakur M. K., Paramanik V. (2009) Horm. Res. 71, 194–200 [DOI] [PubMed] [Google Scholar]
- 4. Roeder R. G. (2005) FEBS Lett. 579, 909–915 [DOI] [PubMed] [Google Scholar]
- 5. Krasowski M. D., Reschly E. J., Ekins S. (2008) J. Proteome Res. 7, 4359–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cronet P., Petersen J. F., Folmer R., Blomberg N., Sjöblom K., Karlsson U., Lindstedt E. L., Bamberg K. (2001) Structure 9, 699–706 [DOI] [PubMed] [Google Scholar]
- 7. Johnson B. A., Wilson E. M., Li Y., Moller D. E., Smith R. G., Zhou G. (2000) J. Mol. Biol. 298, 187–194 [DOI] [PubMed] [Google Scholar]
- 8. Paramanik V., Thakur M. K. (2011) Mol. Biol. Rep. 38, 4657–4661 [DOI] [PubMed] [Google Scholar]
- 9. Bhasin S., Jasuja R. (2009) Curr. Opin. Clin. Nutr. Metab. Care 12, 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gessi S., Merighi S., Borea P. A. (2010) Curr. Pharm. Des. 16, 3540–3553 [DOI] [PubMed] [Google Scholar]
- 11. Miller A. L., Webb M. S., Thompson E. B. (2007) Steroids 72, 673–681 [DOI] [PubMed] [Google Scholar]
- 12. Pickar J. H., MacNeil T., Ohleth K. (2010) Maturitas 67, 129–138 [DOI] [PubMed] [Google Scholar]
- 13. Reed J. C., Kitada S., Kim Y., Byrd J. (2002) Semin. Oncol. 29, 10–24 [DOI] [PubMed] [Google Scholar]
- 14. Hol T., Cox M. B., Bryant H. U., Draper M. W. (1997) J. Women's Health 6, 523–531 [DOI] [PubMed] [Google Scholar]
- 15. Katzenellenbogen B. S., Montano M. M., Ekena K., Herman M. E., McInerney E. M. (1997) Breast Cancer Res. Treat. 44, 23–38 [DOI] [PubMed] [Google Scholar]
- 16. Allan G. F., Leng X., Tsai S. Y., Weigel N. L., Edwards D. P., Tsai M. J., O'Malley B. W. (1992) J. Biol. Chem. 267, 19513–19520 [PubMed] [Google Scholar]
- 17. De Bosscher K. (2010) J. Steroid Biochem. Mol. Biol. 120, 96–104 [DOI] [PubMed] [Google Scholar]
- 18. Pfaff S. J., Fletterick R. J. (2010) J. Biol. Chem. 285, 15256–15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ricketson D., Hostick U., Fang L., Yamamoto K. R., Darimont B. D. (2007) J. Mol. Biol. 368, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Veleiro A. S., Alvarez L. D., Eduardo S. L., Burton G. (2010) ChemMedChem 5, 649–659 [DOI] [PubMed] [Google Scholar]
- 21. McMaster A., Ray D. W. (2007) Exp. Physiol. 92, 299–309 [DOI] [PubMed] [Google Scholar]
- 22. Joseph J. D., Wittmann B. M., Dwyer M. A., Cui H., Dye D. A., McDonnell D. P., Norris J. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12178–12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kazmin D., Prytkova T., Cook C. E., Wolfinger R., Chu T. M., Beratan D., Norris J. D., Chang C. Y., McDonnell D. P. (2006) Mol. Endocrinol. 20, 1201–1217 [DOI] [PubMed] [Google Scholar]
- 24. Pratt W. B., Galigniana M. D., Morishima Y., Murphy P. J. (2004) Essays Biochem. 40, 41–58 [DOI] [PubMed] [Google Scholar]
- 25. Smith D. F., Toft D. O. (2008) Mol. Endocrinol. 22, 2229–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu E. W., Koshland D. E., Jr. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9517–9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cvoro A., Tzagarakis-Foster C., Tatomer D., Paruthiyil S., Fox M. S., Leitman D. C. (2006) Mol. Cell 21, 555–564 [DOI] [PubMed] [Google Scholar]
- 28. O'Malley B. W., Qin J., Lanz R. B. (2008) Curr. Opin. Cell Biol. 20, 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baumann H., Paulsen K., Kovács H., Berglund H., Wright A. P., Gustafsson J. A., Härd T. (1993) Biochemistry 32, 13463–13471 [DOI] [PubMed] [Google Scholar]
- 30. Berglund H., Kovács H., Dahlman-Wright K., Gustafsson J. A., Härd T. (1992) Biochemistry 31, 12001–12011 [DOI] [PubMed] [Google Scholar]
- 31. Berglund H., Wolf-Watz M., Lundbäck T., van den Berg S., Härd T. (1997) Biochemistry 36, 11188–11197 [DOI] [PubMed] [Google Scholar]
- 32. Härd T., Kellenbach E., Boelens R., Maler B. A., Dahlman K., Freedman L. P., Carlstedt-Duke J., Yamamoto K. R., Gustafsson J. A., Kaptein R. (1990) Science 249, 157–160 [DOI] [PubMed] [Google Scholar]
- 33. Melvin V. S., Harrell C., Adelman J. S., Kraus W. L., Churchill M., Edwards D. P. (2004) J. Biol. Chem. 279, 14763–14771 [DOI] [PubMed] [Google Scholar]
- 34. Melvin V. S., Roemer S. C., Churchill M. E., Edwards D. P. (2002) J. Biol. Chem. 277, 25115–25124 [DOI] [PubMed] [Google Scholar]
- 35. Lieberman B. A., Nordeen S. K. (1997) J. Biol. Chem. 272, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 36. Yamamoto K. R. (1985) Annu. Rev. Genet. 19, 209–252 [DOI] [PubMed] [Google Scholar]
- 37. Iwahara J., Schwieters C. D., Clore G. M. (2004) J. Am. Chem. Soc. 126, 12800–12808 [DOI] [PubMed] [Google Scholar]
- 38. Takayama Y., Sahu D., Iwahara J. (2010) Biochemistry 49, 7998–8005 [DOI] [PubMed] [Google Scholar]
- 39. O'Brien J. E., Peterson T. J., Tong M. H., Lee E. J., Pfaff L. E., Hewitt S. C., Korach K. S., Weiss J., Jameson J. L. (2006) J. Biol. Chem. 281, 26683–26692 [DOI] [PubMed] [Google Scholar]
- 40. Geserick C., Meyer H. A., Haendler B. (2005) Mol. Cell. Endocrinol. 236, 1–7 [DOI] [PubMed] [Google Scholar]
- 41. Klinge C. M. (2001) Nucleic Acids Res. 29, 2905–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klinge C. M., Jernigan S. C., Mattingly K. A., Risinger K. E., Zhang J. (2004) J. Mol. Endocrinol. 33, 387–410 [DOI] [PubMed] [Google Scholar]
- 43. Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., Yamamoto K. R. (2009) Science 324, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schultz-Norton J. R., Ziegler Y. S., Likhite V. S., Yates J. R., Nardulli A. M. (2008) BMC Mol. Biol. 9, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nardulli A. M., Romine L. E., Carpo C., Greene G. L., Rainish B. (1996) Mol. Endocrinol. 10, 694–704 [DOI] [PubMed] [Google Scholar]
- 46. Spolar R. S., Record M. T., Jr. (1994) Science 263, 777–784 [DOI] [PubMed] [Google Scholar]
- 47. He B., Wang K., Liu Y., Xue B., Uversky V. N., Dunker A. K. (2009) Cell Res. 19, 929–949 [DOI] [PubMed] [Google Scholar]
- 48. Gu J., Hilser V. J. (2008) Structure 16, 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hilser V. J., García-Moreno E. B., Oas T. G., Kapp G., Whitten S. T. (2006) Chem. Rev. 106, 1545–1558 [DOI] [PubMed] [Google Scholar]
- 50. Rastinejad F., Perlmann T., Evans R. M., Sigler P. B. (1995) Nature 375, 203–211 [DOI] [PubMed] [Google Scholar]
- 51. Schwabe J. W., Chapman L., Finch J. T., Rhodes D. (1993) Cell 75, 567–578 [DOI] [PubMed] [Google Scholar]
- 52. Schwabe J. W., Chapman L., Rhodes D. (1995) Structure 3, 201–213 [DOI] [PubMed] [Google Scholar]
- 53. Schwabe J. W., Neuhaus D., Rhodes D. (1990) Nature 348, 458–461 [DOI] [PubMed] [Google Scholar]
- 54. van Tilborg M. A., Bonvin A. M., Hård K., Davis A. L., Maler B., Boelens R., Yamamoto K. R., Kaptein R. (1995) J. Mol. Biol. 247, 689–700 [DOI] [PubMed] [Google Scholar]
- 55. Levin E. J., Kondrashov D. A., Wesenberg G. E., Phillips G. N., Jr. (2007) Structure 15, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lefstin J. A., Yamamoto K. R. (1998) Nature 392, 885–888 [DOI] [PubMed] [Google Scholar]
- 57. Bain D. L., Franden M. A., McManaman J. L., Takimoto G. S., Horwitz K. B. (2000) J. Biol. Chem. 275, 7313–7320 [DOI] [PubMed] [Google Scholar]
- 58. Bain D. L., Franden M. A., McManaman J. L., Takimoto G. S., Horwitz K. B. (2001) J. Biol. Chem. 276, 23825–23831 [DOI] [PubMed] [Google Scholar]
- 59. Ikeda M., Wilcox E. C., Chin W. W. (1996) J. Biol. Chem. 271, 23096–23104 [DOI] [PubMed] [Google Scholar]
- 60. Loven M. A., Likhite V. S., Choi I., Nardulli A. M. (2001) J. Biol. Chem. 276, 45282–45288 [DOI] [PubMed] [Google Scholar]
- 61. Loven M. A., Wood J. R., Nardulli A. M. (2001) Mol. Cell. Endocrinol. 181, 151–163 [DOI] [PubMed] [Google Scholar]
- 62. Wood J. R., Likhite V. S., Loven M. A., Nardulli A. M. (2001) Mol. Endocrinol. 15, 1114–1126 [DOI] [PubMed] [Google Scholar]
- 63. Kumar R., Baskakov I. V., Srinivasan G., Bolen D. W., Lee J. C., Thompson E. B. (1999) J. Biol. Chem. 274, 24737–24741 [DOI] [PubMed] [Google Scholar]
- 64. Kumar R., Thompson E. B. (2003) Mol. Endocrinol. 17, 1–10 [DOI] [PubMed] [Google Scholar]
- 65. Thompson E. B., Kumar R. (2003) Biochem. Biophys. Res. Commun. 306, 1–4 [DOI] [PubMed] [Google Scholar]
- 66. Copik A. J., Webb M. S., Miller A. L., Wang Y., Kumar R., Thompson E. B. (2006) Mol. Endocrinol. 20, 1218–1230 [DOI] [PubMed] [Google Scholar]
- 67. Dyson H. J., Wright P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60 [DOI] [PubMed] [Google Scholar]
- 68. Chen J. W., Romero P., Uversky V. N., Dunker A. K. (2006) J. Proteome Res. 5, 879–887, 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fuxreiter M., Tompa P., Simon I. (2007) Bioinformatics 23, 950–956 [DOI] [PubMed] [Google Scholar]
- 70. Le Gall T., Romero P. R., Cortese M. S., Uversky V. N., Dunker A. K. (2007) J. Biomol. Struct. Dyn. 24, 325–342 [DOI] [PubMed] [Google Scholar]
- 71. Liu J., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., Dunker A. K. (2006) Biochemistry 45, 6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tompa P. (2002) Trends Biochem. Sci. 27, 527–533 [DOI] [PubMed] [Google Scholar]
- 73. Fuxreiter M., Tompa P., Simon I., Uversky V. N., Hansen J. C., Asturias F. J. (2008) Nat. Chem. Biol. 4, 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Ström A., Treuter E., Warner M., Gustafsson J. A. (2007) Physiol. Rev. 87, 905–931 [DOI] [PubMed] [Google Scholar]
- 75. Lu N. Z., Cidlowski J. A. (2004) Ann. N.Y. Acad. Sci. 1024, 102–123 [DOI] [PubMed] [Google Scholar]
- 76. Scarpin K. M., Graham J. D., Mote P. A., Clarke C. L. (2009) Nuclear Receptor Signaling 7, e009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bolen D. W. (2001) Methods Mol. Biol. 168, 17–36 [DOI] [PubMed] [Google Scholar]
- 78. Bolen D. W., Rose G. D. (2008) Annu. Rev. Biochem. 77, 339–362 [DOI] [PubMed] [Google Scholar]
- 79. Burg M. B. (1995) Am. J. Physiol. 268, F983–F996 [DOI] [PubMed] [Google Scholar]
- 80. Baskakov I., Bolen D. W. (1998) J. Biol. Chem. 273, 4831–4834 [DOI] [PubMed] [Google Scholar]
- 81. Baskakov I. V., Kumar R., Srinivasan G., Ji Y. S., Bolen D. W., Thompson E. B. (1999) J. Biol. Chem. 274, 10693–10696 [DOI] [PubMed] [Google Scholar]
- 82. Fischer K., Kelly S. M., Watt K., Price N. C., McEwan I. J. (2010) Mol. Endocrinol. 24, 1935–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reid J., Kelly S. M., Watt K., Price N. C., McEwan I. J. (2002) J. Biol. Chem. 277, 20079–20086 [DOI] [PubMed] [Google Scholar]
- 84. Kumar R., Volk D. E., Li J., Lee J. C., Gorenstein D. G., Thompson E. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16425–16430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kumar R., Lee J. C., Bolen D. W., Thompson E. B. (2001) J. Biol. Chem. 276, 18146–18152 [DOI] [PubMed] [Google Scholar]
- 86. McEwan I. J. (2004) Endocr.-Relat. Cancer 11, 281–293 [DOI] [PubMed] [Google Scholar]
- 87. Dastidar S. G., Lane D. P., Verma C. S. (2008) J. Am. Chem. Soc. 130, 13514–13515 [DOI] [PubMed] [Google Scholar]
- 88. Tompa P., Fuxreiter M. (2008) Trends Biochem. Sci. 33, 2–8 [DOI] [PubMed] [Google Scholar]
- 89. Hilser V. J. (2010) Science 327, 653–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bai F., Branch R. W., Nicolau D. V., Jr., Pilizota T., Steel B. C., Maini P. K., Berry R. M. (2010) Science 327, 685–689 [DOI] [PubMed] [Google Scholar]
- 91. Hilser V. J., Thompson E. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8311–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cutler T. A., Loh S. N. (2007) J. Mol. Biol. 371, 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Luque I., Leavitt S. A., Freire E. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 235–256 [DOI] [PubMed] [Google Scholar]
- 94. Gorski J., Toft D., Shyamala G., Smith D., Notides A. (1968) Recent Prog. Horm. Res. 24, 45–80 [DOI] [PubMed] [Google Scholar]
- 95. Cintra A., Zoli M., Rosén L., Agnati L. F., Okret S., Wikström A. C., Gustaffsson J. A., Fuxe K. (1994) Neuroscience 62, 843–897 [DOI] [PubMed] [Google Scholar]
- 96. Pereira F. A., Tsai M. J., Tsai S. Y. (2000) Cell. Mol. Life Sci. 57, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sugiyama N., Andersson S., Lathe R., Fan X., Alonso-Magdalena P., Schwend T., Nalvarte I., Warner M., Gustafsson J. A. (2009) Mol. Psychiatry 14, 223–232, 117 [DOI] [PubMed] [Google Scholar]
- 98. Sugiyama N., Barros R. P., Warner M., Gustafsson J. A. (2010) Trends Endocrinol. Metab. 21, 545–552 [DOI] [PubMed] [Google Scholar]
- 99. Younes M., Honma N. (2011) Arch. Pathol. Lab. Med. 135, 63–66 [DOI] [PubMed] [Google Scholar]
- 100. Freiburger L. A., Baettig O. M., Sprules T., Berghuis A. M., Auclair K., Mittermaier A. K. (2011) Nat. Struct. Mol. Biol. 18, 288–294 [DOI] [PMC free article] [PubMed] [Google Scholar]