Background: mTORC2 is a major regulatory kinase of Akt, and its regulation remains poorly characterized.
Results: The reconstitution of mTORC2 indicates that mTOR as the functional kinase of the complex phosphorylates SIN1 and maintains its stability.
Conclusion: The integrity of mTORC2 depends on the kinase activity of mTOR.
Significance: mTOR is a critical component of mTORC2 that carries its functional activity and controls integrity of the complex.
Keywords: Akt PKB, Lysosomes, Phosphorylation, Protein Degradation, TOR Complex (TORC)
Abstract
In higher eukaryotes, growth factors promote anabolic processes and stimulate cell growth, proliferation, and survival by activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway. Deregulation of PI3K/Akt signaling is linked to human diseases, including cancer and metabolic disorders. The PI3K-dependent signaling kinase complex mTORC2 (mammalian target of rapamycin complex 2) has been defined as the regulatory Ser-473 kinase of Akt. The regulation of mTORC2 remains very poorly characterized. We have reconstituted mTORC2 by its assembly in vitro or by co-expression its four essential components (rictor, SIN1, mTOR, mLST8). We show that the functional mTOR kinase domain is required for the mTORC2 activity as the Ser-473 kinase of Akt. We also found that mTOR by phosphorylation of SIN1 prevents its lysosomal degradation. Thus, the kinase domain of mTOR is required for the functional activity of mTORC2, and it controls integrity of mTORC2 by maintaining the protein stability of SIN1.
Introduction
Growth factor signaling coordinates a variety of cellular processes, including proliferation, metabolism, and survival (1). The serine/threonine kinase Akt/PKB, a member of the AGC (protein kinase A, G, and C) kinase family, is a critical downstream effector of the growth factor-dependent PI3K pathway. Akt comprises three mammalian isoforms (Akt1/PKBα, Akt2/PKBβ, Akt3/PKBγ) that are derived from distinct genes and share a conserved structure, which includes three functional domains: an N-terminal pleckstrin homology domain, a central kinase domain, and a C-terminal regulatory domain containing the hydrophobic motif (HM)2 phosphorylation site (FXXF(S/T)Y) (2). Through the phosphorylation of a diverse set of substrates, Akt regulates cell proliferation, survival, and metabolism. Following activation of PI3K and accumulation of phosphatidylinositol 3,4,5-trisphosphates, the Akt kinase is translocated to the plasma membrane by binding to the phosphatidylinositol 3,4,5-trisphosphates via its N-terminal pleckstrin homology domain. In addition to the pleckstrin homology domain, it has been reported recently that ubiquitination of Akt takes place as a step facilitating its membrane translocation (3). At the plasma membrane, Akt is phosphorylated on two sites required for its full activation. The Thr-308 site of Akt that resides on its activation loop is phosphorylated by the phosphoinositide-dependent kinase (PDK1), and this phosphorylation is required for kinase activity of Akt (4, 5). The second Ser-473 site is located within HM at the C terminus of Akt and regulates the kinase activity of Akt. The structural analysis of Akt provides an insight into how phosphorylation of Thr-308 and Ser-473 is engaged in activation of the kinase domain of Akt (6).
mTORC2 (mammalian target of rapamycin complex 2) has been identified as the main Ser-473 kinase of Akt and other related AGC kinase members carrying the HM domain at the C terminus such as PKCα (protein kinase Cα) and SGK1 (serum-and glucocorticoid-induced protein kinase 1) (7–11). The functional role of mTORC2 as the regulatory kinase of Akt has been supported by mouse genetic studies (12–15). The mTOR kinase is a central component of the essential and highly conserved signaling pathway. Biochemical studies reveal that mTOR and its interacting proteins mLST8, also known as GβL, and DEPTOR exist at least in two distinct complexes. The binding of raptor to mTOR defines a nutrient-sensitive mTORC1 that regulates protein synthesis by phosphorylating its substrates S6K1 and 4EBP1. Rapamycin in a complex with its intracellular receptor FKBP12 specifically binds the FKBP12/rapamycin binding domain on mTOR and inhibits the mTORC1 function (16). The second mTOR complex, mTORC2, is assembled by binding of rictor (rapamycin-insensitive companion of mTOR) and SIN1 (stress-associated protein kinase-interacting protein 1) to mTOR and has been initially identified as a regulator of PKCα and cytoskeleton (14, 17, 18). mLST8, a small adaptor protein containing seven WD40 repeats, binds to the kinase domain of mTOR (17, 18) and has been shown to regulate the kinase activity of mTOR (19). Genetic studies identified that mLST8 is essential for the mTORC2 function (15). The other two components, rictor and SIN1, define mTORC2 and most likely carry the regulatory functions of this kinase complex. In addition to the essential components of mTORC2 (rictor, SIN1, mTOR, and mLST8), PROTOR has been identified as a rictor-interacting protein and a component of mTORC2. The functional study of PROTOR indicates that it is not required for mTORC2 to phosphorylate Akt on Ser-473 (20, 21), but it is critical in the phosphorylation of another mTORC2 substrate SGK1 (22).
In previous studies several phosphorylation sites of rictor have been identified. Among several sites the Thr-1135 site attracted attention as the growth factor-dependent site. Functional characterization determined that S6K1 is the kinase of Thr-1135 on rictor. This phosphorylation was not essential in regulation of the mTORC2 kinase activity (23–26). Recently, the inhibition of mTORC2 by endoplasmic reticulum (ER) stress has been linked to the phosphorylation of rictor on Ser-1235 mediated by the glycogen synthase kinase-3β (27). This phosphorylation event causes inhibition of mTORC2 kinase activity by interfering with its substrate binding. A role of the endoplasmic reticulum in regulation of mTORC2 is becoming more evident. A recent study has indicated the predominant localization of mTORC2 in the endoplasmic reticulum (28). Importantly, the functional regulation and association of mTORC2 with ribosomes have been reported (29, 30).
mTORC2 carries an important regulatory role in growth factor signaling by mediating the regulation between PI3K and its crucial downstream effector Akt. The mTORC2 kinase complex remains poorly characterized. In our study, we have addressed the functional role of the mTOR kinase domain by developing the mTORC2 reconstitution system.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following sources: DMEM/F12 from Invitrogen; FBS from Hyclone, FuGENE 6 transfection reagent and Complete protease inhibitor mixture from Roche Applied Science; protein G-Sepharose from Pierce; and insulin-like growth factor I (IGF-1) from Peprotech. Antibodies to Xpress, myc, and V5 tag were from Invitrogen; antibodies to mTOR, rictor, S6K1, p27, Akt, Ser(P)-2481 mTOR, phospho-S6K1 Thr-389, Thr(P)-450, and Ser(P)-473 Akt were from Cell Signaling Technologies. Antibodies to rictor, HRP-labeled anti-rabbit, anti-mouse, anti-goat secondary antibodies, and tubulin were from Santa Cruz Biotechnology. Lentiviral shRNAs targeting human mTOR were generated and applied as described previously (7). Plasmids pRK5-myc-GCP2, -myc-rictor, myc-raptor, myc-mTOR-WT, myc-mTOR-KD, HA-mLST8, and HA-tubulin were kindly provided by David M. Sabatini (Whitehead Institute for Biomedical Research, Cambridge, MA). pcDNA3.1-Sin1.1-V5 and pcDNA4-Xp-rictor were generated by the TOPO cloning system (Invitrogen).
Cell Lines and Culture
HEK-293T, HeLa, MDA-MB-435, and COS-7 cells were obtained from American Type Culture Collection and cultured in DMEM/F12 with 10% FBS, penicillin/streptomycin in 5% CO2 at 37 °C. All of the above cell lines were cultured at a density that allowed cell division throughout the course of the experiment.
Cell Lysis and Immunoblotting
All cells were rinsed with ice-cold PBS before lysis in buffer containing (40 mm HEPES, pH 7.5, 120 mm NaCl, 1 mm EDTA, 10 mm sodium pyrophosphate, 10 mm sodium glycerophosphate, 50 mm NaF, 1% Triton X-100, and protease inhibitor mixture (Roche Applied Science)). The scraped lysates were incubated for 20 min at 4 °C for complete lysis. The soluble fractions of cellular lysates were isolated by centrifugation at 13,000 rpm at 4 °C for 12 min. Samples of the cellular lysates containing the equal amount of proteins were resolved by SDS-PAGE and transferred to PVDF membrane. Proteins were then visualized by immunoblotting and detected with enhanced chemoluminescence (ECL) from the Immobilion Western kit (Millipore).
Immunoprecipitations and Kinase Assays
For immunoprecipitation experiments the lysis buffer contained 0.3% CHAPS to preserve the integrity of the mTOR complexes as described previously (31). One microgram of rictor antibody was added to the cleared cellular lysates (1 mg of protein content in 700 μl) and incubated with rotation at 4 °C for 90 min. Followed by a 1-h incubation with 40 μl of the 50% slurry of protein G-agarose, immunoprecipitates captured by protein G-agarose were washed four times with the CHAPS-containing lysis buffer and once with the rictor-mTOR kinase buffer (25 mm HEPES, pH 7.5, 100 mm potassium acetate, 2 mm MgCl2). For kinase reaction, immunoprecipitates were incubated in a final volume of 15 μl at 37 °C for 20 min in the rictor-mTOR kinase buffer containing 500 ng of inactive Akt1-GST and 1 mm ATP. The reaction was stopped by the addition of 235 μl of ice-cold dilution buffer (20 mm MOPS, pH 7.0, 1 mm EDTA, 0.3% CHAPS, 5% glycerol, 0.1% 2-mercaptoethanol, 1 mg/ml BSA). After a quick spin, the supernatant was removed from the protein G-agarose, and a 15-μl portion was analyzed by immunoblotting for Ser(P)473-Akt and total Akt detection. The pelleted protein G-agarose beads were also analyzed by immunoblotting to determine the levels of rictor, mTOR, and SIN1 in the immunoprecipitates.
Purification of the Soluble FLAG-mLST8/myc-mTOR Heterodimer
FLAG-mLST8 was co-transfected with myc-mTOR cDNA in HEK-293T cells. After a 48-h transfection, cells were washed with cold PBS, lysed with 0.3% CHAPS buffer, and incubated with mild agitation for 20 min at 4 °C. The lysate was transferred to the spinning columns for centrifugation for 15 min at 10,000 rpm. After centrifugation, the supernatant was used for the FLAG affinity purification as described previously (32). FLAG M2 affinity resins were washed three times in 0.3% CHAPS lysis buffer and packed into a 10-ml Bio-Rad column. The cellular lysates were applied through the column seven times. After running the lysates, the column was washed with 20 ml of lysis buffer and 10 ml of elution buffer (40 mm HEPES, pH 7.4, 500 mm NaCl, 0.1% CHAPS). The FLAG-mLST8/myc-mTOR heterodimer was eluted by the elution buffer with 0.5 mg/ml of the FLAG peptide. The eluted fractions from number 2 to 6 were combined, dialyzed with mTORC2 kinase buffer, and concentrated with Millipore Amicon Ultra-15 centrifugal filter units. The purified FLAG-mLST8/myc-mTOR heterodimer was analyzed by immunoblotting and applied for the in vitro mTORC2 assembly.
In Vitro mTORC2 Assembly and Kinase Reaction
Myc-rictor and SIN1-V5 cDNAs were co-transfected in HEK-293T cells. After a 48-h transfection, cells were washed with cold PBS, lysed with 0.3% CHAPS buffer, and subjected for centrifugation for 15 min at 10,000 rpm. The supernatant was applied for immunoprecipitation with anti-V5 antibody. The myc-rictor/SIN1-V5 immunoprecipitates were incubated with or without the purified soluble FLAG-mLST/myc-mTOR heterodimer at room temperature for 2 h in mTORC2 kinase buffer with 1 mm ATP. After washing three times with 0.3% CHAPS lysis buffer, the immunoprecipitates were used for in vitro mTORC2 kinase reaction with wild-type Akt as the substrate.
RESULTS AND DISCUSSION
Reconstitution of mTORC2 by Expression of Its Essential Components
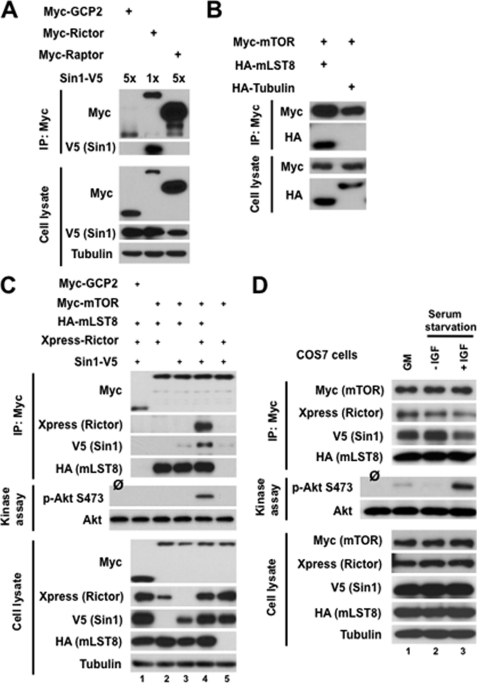
To study a role of the mTOR kinase activity within mTORC2, we reconstituted this signaling complex by co-expression of its four distinctly tagged recombinant components in HEK 293T cells. Previous biochemical studies indicated different binding affinity among mTORC2 components (17, 33). The native mTORC2 complex is preserved in a mild lysis condition containing the CHAPS detergent. This complex dissociates into two heterodimers rictor/SIN1 and mTOR/mLST8 when cells are lysed under stringent buffer containing Triton X-100 detergent. These observations led to an assumption that two heterodimers rictor/SIN1 and mTOR/mLST8 are formed at the initial stage of the mTORC2 assembly, and these heterodimers form a low affinity Triton X-100-sensitive mTORC2 complex. As a first step in the reconstitution of mTORC2, we have studied assembly of the heterodimers by co-expressing their components. We have assembled the rictor/SIN1 heterodimer by expression of rictor and SIN1 (Fig. 1A). The full-length V5-tagged SIN1–1 isoform (33) was co-transfected with the myc-tagged rictor, GCP2, or raptor expression plasmids (Fig. 1A, lower). Immunoprecipitation of the myc-tagged proteins showed the binding of SIN1 only to rictor but not to the control proteins GCP2 and raptor (Fig. 1A, upper). A specific binding between rictor and SIN1 is critical to mutually stabilize each other expression, where SIN1 known to be a highly unstable protein without its binding partner rictor (33). As indicated, to compensate a loss of SIN1 expression when expressed without rictor, we have transfected a 5-fold amount of the cDNA encoding SIN1. In parallel, a similar co-expression experiment indicated a specific assembly of the mTOR/mLST8 heterodimer (Fig. 1B). When the myc-tagged mTOR co-expressed with HA-tagged mLST8 or tubulin, we detected a specific interaction between mTOR and mLST8 but not tubulin in the pulldowns of myc-mTOR. Our initial set of the co-expression studies showed a specific assembly of the rictor/SIN1 and mTOR/mLST8 heterodimers.
FIGURE 1.
Reconstitution of the functional mTORC2 complex by expressing its recombinant components. A, rictor interacts with SIN1. SIN1-V5 plasmid DNA was co-transfected with myc-rictor into HEK-293T cells by Lipofectamine 2000, and cells were analyzed 48 h following transfection. Cell lysates were applied for immunoprecipitation (IP) with anti-myc antibody. The immunoprecipitates and cell lysates were analyzed by immunoblotting with anti-myc and anti-V5 antibodies. First and third lanes, negative controls are presented by expressing myc-GCP2 or myc-raptor with SIN1-V5. B, mTOR interacts with mLST8. Myc-mTOR construct was co-transfected with HA-mLST8 in HEK-293T cells and analyzed as in A. Right 2, a negative control is presented by expressing HA-α-tubulin with myc-mTOR. C, interaction of mTOR with other mTORC2 components. The differently tagged recombinant components of mTORC2 were transiently expressed in HEK-293T cells, and the assembled complexes were purified by the immunoprecipitation of myc-mTOR. Lane 1, a negative control is presented by expressing myc-GCP2 with SIN1-V5, Xp-rictor, and HA-mLST8. D, reconstituted mTORC2 kinase activity was stimulated by IGF-1. The differently tagged recombinant components of mTORC2 were transiently expressed in COS-7 cells, and the assembled complexes were purified by the immunoprecipitation of myc-mTOR. Lane 1, COS-7 cells expressing recombinant mTORC2 were cultured in growth medium containing 10% serum. Lanes 2 and 3, COS-7 cells expressing recombinant mTORC2 were cultured in serum-free medium for 16 h and then treated without or with IGF-I (100 ng/ml) for 30 min, respectively.
In our following experiment, we reconstituted the functional mTORC2 complex by co-expressing its four essential components (Fig. 1C). By pulling down myc-mTOR and detecting its interacting proteins, we found that rictor or SIN1 alone does not bind to the mTOR/mLST8 heterodimer. In this setting, rictor and SIN1 show low expression levels because their stability requires heterodimer formation. Importantly, the stabilized rictor and SIN1 proteins by their co-expression did not bind to mTOR, indicating that no formation of a heterotrimer takes place in assembly of mTORC2. Only when all four mTORC2 components were co-expressed did we detect assembly of mTORC2 (Fig. 1C, top). This reconstituted complex is a functional kinase complex as indicated by its activity to phosphorylate Akt on the mTORC2-dependent Ser-473 site. Our mTORC2 reconstitution study is in line with the previous findings that rictor and SIN1 are the essential mTORC2 components and required for the complex formation and its function (14, 33). Besides, mLST8 is a critical factor in mTORC2 function (15) and assembly because we observe that mTOR without mLST8 fails to bind the rictor/SIN1 heterodimer. We find that the transient expression of rictor, SIN1, mTOR, and mLST8 reconstitutes assembly of mTORC2 without interference from the endogenous proteins. Most likely, within a short period of time the protein-protein interactions among the endogenous components are saturated to form a native form of mTORC2, and the stable expression of recombinant mTORC2 component such as rictor is capable of forming the functional complex by assembly with the endogenous proteins (31). Based on our initial study, we interpret this as meaning that mTORC2 is assembled by formation of the complex between the rictor/SIN1 and mTOR/mLST8 heterodimers, and formation of these heterodimers is an initial step in the mTORC2 assembly.
mTORC2 kinase activity is regulated by growth factor signaling (7, 33). To address whether the reconstituted complex is regulated by growth factor signaling as its native form, we studied activity of the reconstituted mTORC2 in growth factor-responsive COS-7 cells (34). Following co-expression of the mTORC2 components in COS-7 cells, we stimulated serum-starved cells by IGF-1, purified the reconstituted mTORC2, and performed an in vitro kinase assay (Fig. 1D). We detected a low basic activity of the complex pulled down by myc-mTOR in cells growing in 10% serum. The similar pulldown of the complex from the serum-starved cells did not show the kinase activity toward Akt. On the contrary, following stimulation of the serum-starved cells by IGF-1, we detected a high kinase activity of the reconstituted complex as shown by a robust phosphorylation of Akt on Ser-473. Therefore, the functional study of the reconstituted mTORC2 assembled by co-expressing its essential components behaves similarly to the endogenous mTORC2 that is also regulated by growth factor signaling.
mTOR Kinase Activity Is Required for mTORC2 Function
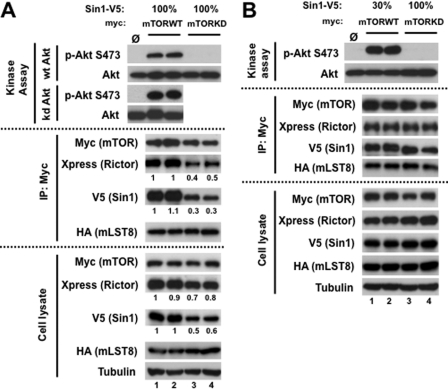
In activation of Akt, the phosphorylation of Ser-473 is a key step in stabilizing the active conformation state of the kinase (6). Therefore, defining the Akt Ser-473 kinase (PDK2) provides a critical point in understanding the regulation of Akt. The identity of PDK2 has been controversial for many years. Although several candidates were proposed earlier as PDK2, the biochemical (7) and mouse genetic studies defined a role of mTORC2 as the major Ser-473 kinase of Akt (12–15). Most likely, mTOR functions as a kinase within mTORC2, but no direct evidence has been presented indicating the function of mTOR as the Ser-473 kinase of Akt. To define a role of the mTOR kinase within mTORC2, we applied our mTORC2 reconstitution system by replacing mTOR with its kinase-dead form. We co-expressed the wild-type or kinase-dead mTOR cDNA with other mTORC2 components and analyzed the functional activity of mTORC2 in the duplicate independent experiments. Our in vitro study did not detect kinase activity of mTORC2 assembled with the kinase-dead form of mTOR toward its substrate Akt (Fig. 2A, top), indicating a critical role of the mTOR kinase domain in the functional activity of mTORC2. Previously, it has been also reported that the autophosphorylation of Akt on Ser-473 site takes place in regulation of Akt (36). To prevent a potential effect of autophosphorylation of Akt, we also carried the kinase assay with the kinase-inactive form of GST-Akt. We found that mTORC2 assembled with the wild-type mTOR shows a high kinase activity toward the inactive Akt kinase, indicating that autophosphorylation of Akt does not occur in the mTORC2-dependent phosphorylation of Akt.
FIGURE 2.
Critical role of mTOR kinase activity in mTORC2 function. A, The kinase-dead mTOR forms inactive mTORC2 deficient in its assembly. The study has been performed in duplicate: Xpress-rictor, SIN1-V5, and HA-mLST8 constructs were co-transfected with myc-mTOR wild type (WT) (lanes 1 and 2) or myc-mTOR kinase-dead (KD) (lanes 3 and 4) in HEK-293T cells. Immunoprecipitates (IP) prepared with anti-myc antibody were used for in vitro kinase assay with wild-type or kinase-dead Akt as the substrate. Immunoblotting was used to detect the phosphorylation of Akt and the indicated proteins. B, a study similar to that in A was performed by optimizing expression and assembly of the mTORC2 assembled with the wild-type or the kinase-dead mutant of mTOR. To optimize expression, the myc-mTOR-WT was co-transfected with Xpress-rictor, 30% of SIN1-V5, and HA-mLST8 constructs in HEK-293T cells (lanes 1 and 2).
In a follow-up analysis we found that the kinase-dead form of mTOR did not interfere with expression and binding of its binding partner mLST8, but it caused a substantial decrease in expression of SIN1 and rictor that subsequently translated to a low assembly of mTORC2 (Fig. 2A, middle and bottom). Moreover, the SIN1 protein as a component of mTORC2 shows a slower migratory form only in a complex with mTOR-WT but not with mTOR-KD (Fig. 2A, middle) suggesting that the phosphorylation of SIN1 is dependent on the mTOR kinase activity, and this modification might play a role in regulation of the SIN1 protein half-life. Our initial study in Fig. 2A shows that mTOR-KD is deficient in assembly of mTORC2 because it caused a substantial effect on the rictor and SIN1 expression. To overcome this shortcoming of the mutant form of mTOR, we have optimized expression and assembly of mTORC2 by decreasing assembly of this complex with mTOR-WT by co-expressing only 30% of the cDNA amount encoding SIN1 (Fig. 2B). The immunoprecipitates with the comparable abundance of mTORC2 reconstituted with mTOR-WT or mTOR-KD were analyzed further by performing an in vitro kinase assay. Under this setting, we found that contrary to the wild-type mTOR, its kinase-dead form assembled to mTORC2 again did not show a kinase activity toward Akt. As mentioned above, we also observe that the SIN1 mobility on a gel is consistently altered when it is co-expressed with mTOR-KD, suggesting a role of the mTOR kinase activity in phosphorylation of SIN1. Thus, our data indicate that the mTOR kinase activity is required for the functional activity of mTORC2 as the regulatory Ser-473 kinase of Akt, and it also may play a role in regulation of SIN1 by phosphorylation.
SIN1 Is Phosphorylated When Assembled into mTORC2 Kinase Complex in Vitro
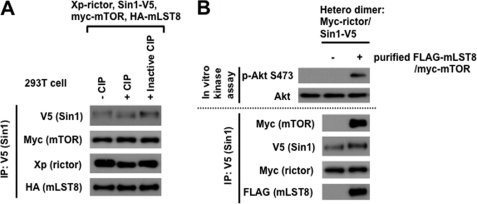
Our experiments shown in Fig. 2 indicate that the SIN1 protein expression and its mobility in a gel are dependent on the kinase activity of mTOR. They suggest that the mTORC2 component SIN1 is a substrate of mTOR. To follow this lead we reconstituted mTORC2 as described in Fig. 2 and following its purification incubated it with phosphatase. We found that only the catalytically active form of phosphatase altered the SIN1 mobility in a gel from a slower to faster migratory form (Fig. 3A), indicating that SIN1 is a phosphorylated protein of mTORC2.
FIGURE 3.
SIN1 is a substrate of mTOR. A, SIN1 as a component of mTORC2 is a phospho-protein. Myc-mTOR-WT cDNA was co-transfected with Xpress-rictor, Sin1-V5, and HA-mLST8 cDNAs in HEK-293T cells. Cell lysates were applied for immunoprecipitation with anti-V5 antibody. The immunoprecipitates were incubated with or without calf intestinal phosphatase (CIP) or inactive CIP (pretreated with 50 mm EDTA) and analyzed by immunoblotting with indicated antibodies. CIP treatment resulted in a faster migratory form of SIN1, suggesting that the mobility shift of SIN1 band is indeed due to phosphorylation/dephosphorylation. B, SIN1 as a component of mTORC2 is a substrate of mTOR. In vitro mTORC2 kinase assay was performed by incubation of two heterodimers, rictor/SIN1 and mTOR/mLST8. First, FLAG purification was conducted from HEK-293T cells with FLAG-mLST/myc-mTOR co-transfection. Then, the purified soluble FLAG-mLST/myc-mTOR heterodimer was incubated with immunopurified myc-rictor/SIN1-V5 by the V5 antibody at room temperature for 2 h in mTORC2 kinase buffer with 1 mm ATP. After washing three times with 0.3% CHAPS lysis buffer, the immunoprecipitates were used for the in vitro mTORC2 kinase reaction by using wild-type Akt as the substrate.
Next, we studied the mTOR-dependent phosphorylation of SIN1 by reconstitution of mTORC2 in vitro. Our study shows that co-expression of the four components rictor, SIN1, mTOR, and mLST8 is sufficient to assemble the functional mTORC2 kinase complex. The formation of two heterodimers rictor/SIN1 and mTOR/mLST8 takes place as an intermediate step in assembly of mTORC2 as supported by data shown in Fig. 1. We assembled mTORC2 in vitro by co-incubation of the immunoprecipitated myc-rictor/V5-SIN1 heterodimer bound to G protein-agarose beads with the purified soluble FLAG-mLST8/myc-mTOR heterodimer. Following incubation the G proteins beads were washed thoroughly and examined further by analyzing the mTORC2 assembly and its functional activity. We found that after co-incubation the two heterodimers have assembled into mTORC2 as observed by detection of its four components (Fig. 3B). The reconstituted complex of mTORC2 in vitro was functional as indicated by the kinase assay. Besides, we also detected that following assembly of mTORC2 and its kinase assay the SIN1 protein altered its mobility on a gel to the slower migratory form (Fig. 3B), mimicking its phosphorylated form prior the phosphatase treatment (Fig. 3A). Thus, our data show that SIN1 as the component of mTORC2 is a substrate of mTOR.
mTOR Kinase-dependent Stability of SIN1 Is Controlled by Lysosomal Degradation
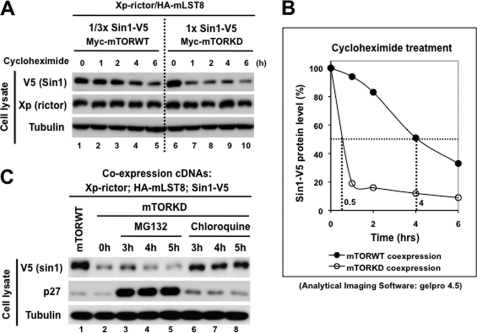
We found that the mTOR kinase activity controls the SIN1 protein expression, which suggests that mTOR by phosphorylation of SIN1 regulates the SIN1 protein stability. To study a role of mTOR in regulation of SIN1, we analyzed its half-life in the reconstituted mTORC2 complex by a transient inhibition of protein synthesis. We normalized the reconstitution of mTORC2 assembled with the mTOR-WT and mTOR-KD by a substantial decreased expression of SIN1 with mTOR-WT as described in Fig. 2B. Following the optimization step, we observed a short half-life of SIN1 when it co-expressed with mTOR-KD (Fig. 4A), indicating that a decrease in the SIN1 protein abundance is linked to accelerated protein degradation. Interestingly, the similar analysis of the SIN1-binding partner rictor did not show dependence of its half-life on the mTOR kinase activity. Quantification of the SIN1 abundance shows a substantial impact of mTOR-KD compared with mTOR-WT by decreasing the SIN1 half-life approximately eight times (Fig. 4B).
FIGURE 4.
mTOR kinase activity stabilizes SIN1 by preventing its lysosomal degradation. A, optimized reconstitution of mTORC2 by co-expression of its components was performed as in Fig. 2B. 48 h following transfection, cells were treated with 20 μg/ml cycloheximide and lysed at the indicated time points. Cell lysates were analyzed by immunoblotting for indicated proteins. B, graphic represents mTOR kinase-dependent SIN1 half-life shown in A. SIN1-V5 signals were quantified by GelPro 4.5 software. The half-life of SIN1 co-expressed with mTOR-WT is shown as filled circles, and its half-life when co-expressed with mTOR-KD is shown as open circles. C, myc-mTOR-KD or its wild-type construct was co-transfected with Xpress-rictor, SIN1-V5, and HA-mLST8 constructs into HEK-293T cells. After 48-h transfection, cells were treated with 30 μm MG132 or 100 μm chloroquine and collected at the indicated time points. The cell lysates were analyzed by immunoblotting as in A.
The ubiquitin-proteasome dependent and lysosomal degradation pathways represent two major and common protein degradation pathways. To identify the degradation pathway controlling the SIN1 protein degradation dependent on the mTOR kinase activity, we treated cells carrying the reconstituted mTORC2 assembled with the kinase-dead form of mTOR with MG132 (proteasome inhibitor) or chloroquine (lysosome inhibitor). The cell cycle regulator p27 protein is known to be degraded via Skp2-mediated ubiquitin-proteasome-dependent pathway (37). Following treatment of cells with MG132 we detected a substantial increase in abundance of p27, indicating that the drug was effective in blocking the proteosomal degradation (Fig. 4C). In this setting, we found that inhibition of the proteosomal degradation did not stabilize expression of SIN1 as detected by its low abundance with or without the MG132 treatment. However, the treatment of cells with the lysosomal inhibitor chloroquine caused an increase of the SIN1 protein abundance similar to the level detected in mTORC2 assembled with the wild-type mTOR (Fig. 4C). Our analysis shows that the mTOR kinase-dependent degradation of SIN1 is linked to the lysosomal but not proteosomal degradation pathway.
Abundance of Endogenous SIN Is Dependent on mTOR Expression and Its Kinase Activity
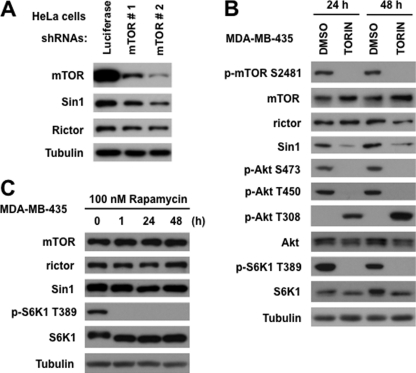
The transient co-expression of the essential components of mTORC2 is sufficient to form the functional kinase complex regulated by growth factor signaling. Our study based on this reconstitution system revealed that the SIN1 protein half-life is dependent on mTOR kinase activity. To validate our finding, we have addressed a role of mTOR in regulation of the endogenous SIN1 protein. We found that the knockdown of mTOR in HeLa cells caused a decrease in abundance of the endogenous SIN1 level (Fig. 5A). This effect correlates well with the efficiency of the mTOR knockdown because the lowest abundance of SIN1 is detected in cells expressing the mTOR shRNA#2 causing the most robust inhibitory impact on the mTOR expression. Similar to our study with the reconstituted mTORC2 complex, the rictor abundance has not been so sensitive to the loss of mTOR as SIN1. Our data show that the SIN1 abundance is dependent on mTOR, and in our next experiment we studied whether inhibition of the mTOR kinase activity is sufficient to mimic the effect of the kinase-dead form of mTOR on SIN1 abundance. To inhibit the mTOR kinase activity in cells, we have applied the recently developed and highly specific mTOR inhibitor TORIN (38, 39). The treatment of MDA-MB-435 cells with TORIN for 24 and 48 h caused a potent inhibition of mTOR as detected by phosphorylation of the mTORC1 substrate S6K1 (Thr-389) and the mTORC2 substrate Akt (Ser-473 and Thr-450) as shown in Fig. 5B. On the contrary, the PDK1-dependent Thr-308 site of Akt has been substantially induced by inhibition of the mTOR kinase activity, which is consistent with the previous study (40). Most likely, this effect is linked to a compensatory mechanism caused by the mTORC2-dependent inhibition of Akt. The autophosphorylation Ser-2481 site on mTOR (41) has been also inhibited by TORIN. Importantly, a prolonged inhibition of the mTOR kinase activity in cells by TORIN has also caused a substantial decrease in abundance of SIN1. The kinase activity of mTOR is critical in regulation of SIN1 because rapamycin, a highly specific compound targeting the FKBP12/rapamycin binding domain of mTOR and inhibiting mTORC1 (35), did not show any substantial impact on abundance of SIN1 (Fig. 5C). Thus, our study indicates that expression of the endogenous SIN1 protein is dependent on mTOR and its kinase activity, which is coherent with our study of the reconstituted mTORC2.
FIGURE 5.
Endogenous SIN1 expression is dependent on mTOR kinase activity. A, SIN1 protein abundance decreased with a loss of mTOR. The mTOR knockdown has been performed by the lentiviral expression of two shRNAs in HeLa cells. A nonspecific luciferase shRNA was used as a control. The abundance of the indicated proteins has been analyzed by immunoblotting. B, mTOR kinase activity controls abundance of SIN1. MDA-MB-435 cells were incubated with specific mTOR inhibitor (0.5 μm TORIN) for 24 or 48 h and analyzed as in A. C, rapamycin does not affect the SIN1 abundance. MDA-MB-435 cells were incubated with 100 nm rapamycin for 1, 24, and 48 h. The abundance of the indicated proteins has been analyzed by immunoblotting.
Our work shows that a transient co-expression of the essential components of mTORC2 (rictor, SIN1, mTOR, and mLST8) is sufficient to form the functional complex with the kinase activity regulated by growth factor signaling. We have applied this mTORC2 reconstitution system to address the functional role of the mTOR kinase domain in the functional activity of the complex to phosphorylate Akt on the HM Ser-473 site. The initial purpose of this study was to exclude that another kinase co-purified with mTORC2 might act as the kinase of Akt. It is unlikely, because mTOR as the kinase of mTORC1 is known to phosphorylate the HM Thr-389 site on its substrate S6K1, and mTOR as the kinase of mTORC2 carries a kinase activity toward a similar HM motif on Akt. As expected, we found that replacement of the wild-type mTOR with its kinase-dead mutant makes mTORC2 deficient in the kinase activity to phosphorylate Akt. The assembly of mTORC2 in vitro also shows that mTOR is required for the kinase activity. Our study clearly indicates that the functional activity of mTORC2 is carried by the mTOR kinase activity required to phosphorylate Akt on its regulatory Ser-473 site. This finding eliminates a role of any other kinase that might associate with mTORC2 and phosphorylate Akt.
Our functional analysis of the mTOR kinase activity also led us to the unexpected finding indicating a critical role of the kinase in controlling integrity of mTORC2. We found that the mTOR kinase activity is required to maintain the SIN1 protein stability by preventing its lysosomal degradation. Assembly of mTORC2 with the kinase-dead form of mTOR causes an accelerated degradation of SIN1 that is linked to its phosphorylation as detected by the phosphatase-dependent SIN1 protein mobility in a gel. The mTOR-dependent phosphorylation of SIN1 does not take place when rictor and SIN1 are co-expressed in cells and form a heterodimer. Importantly, the mTOR-dependent phosphorylation of SIN1 takes place only following assembly of the functional mTORC2 kinase complex, indicating a high kinase specificity of this site on SIN1 toward mTOR.
We believe that within the mTORC2 complex SIN1 is constitutively phosphorylated by mTOR and this modification prevents a rapid turnover of SIN1. The SIN1 phosphorylation by mTOR is not dependent on growth factor signaling because under serum-starved conditions we do not detect any effects on integrity of mTORC2. A basic kinase activity of mTOR within the complex might be sufficient to maintain phosphorylation of SIN1. It will be important to characterize the mTOR-dependent regulation of SIN1 further by identifying the phosphorylation site on SIN1, how this phosphorylation regulates the lysosomal degradation of SIN1, and also the biological role of this regulation.
Acknowledgments
We thank Dr. David M. Sabatini (Whitehead Institute for Biomedical Research, Cambridge, MA) for the mTOR wild type and its kinase-dead mutant plasmids; Dr. Nathanael Gray (Dana-Farber, Cancer Institute, Boston, MA) for the TORIN compound; and our laboratory member Dr. Tattym Shaiken for the GST-Akt substrate.
This work was supported, in whole or in part, by National Institutes of Health Grant CA133522 (to D. D. S.).
- HM
- hydrophobic motif
- IGF
- insulin-like growth factor
- mTOR
- mammalian target of rapamycin
- mTORC2
- mTOR complex 2
- PDK
- phosphoinositide-dependent kinase
- SIN1
- stress-associated protein kinase (SAPK)-interacting protein 1.
REFERENCES
- 1. Shaw R. J., Cantley L. C. (2006) Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 2. Scheid M. P., Woodgett J. R. (2001) Nat. Rev. Mol. Cell Biol. 2, 760–768 [DOI] [PubMed] [Google Scholar]
- 3. Yang W. L., Wang J., Chan C. H., Lee S. W., Campos A. D., Lamothe B., Hur L., Grabiner B. C., Lin X., Darnay B. G., Lin H. K. (2009) Science 325, 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stephens L., Anderson K., Stokoe D., Erdjument-Bromage H., Painter G. F., Holmes A. B., Gaffney P. R., Reese C. B., McCormick F., Tempst P., Coadwell J., Hawkins P. T. (1998) Science 279, 710–714 [DOI] [PubMed] [Google Scholar]
- 5. Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 6. Pearce L. R., Komander D., Alessi D. R. (2010) Nat. Rev. Mol. Cell Biol. 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 7. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 8. Ali S. M., Sabatini D. M. (2005) J. Biol. Chem. 280, 19445–19448 [DOI] [PubMed] [Google Scholar]
- 9. García-Martínez J. M., Alessi D. R. (2008) Biochem. J. 416, 375–385 [DOI] [PubMed] [Google Scholar]
- 10. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Q., Inoki K., Ikenoue T., Guan K. L. (2006) Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shiota C., Woo J. T., Lindner J., Shelton K. D., Magnuson M. A. (2006) Dev. Cell 11, 583–589 [DOI] [PubMed] [Google Scholar]
- 14. Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 15. Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. (2006) Dev. Cell 11, 859–871 [DOI] [PubMed] [Google Scholar]
- 16. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 17. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 18. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 19. Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 20. Pearce L. R., Huang X., Boudeau J., Pawłowski R., Wullschleger S., Deak M., Ibrahim A. F., Gourlay R., Magnuson M. A., Alessi D. R. (2007) Biochem. J. 405, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Woo S. Y., Kim D. H., Jun C. B., Kim Y. M., Haar E. V., Lee S. I., Hegg J. W., Bandhakavi S., Griffin T. J., Kim D. H. (2007) J. Biol. Chem. 282, 25604–25612 [DOI] [PubMed] [Google Scholar]
- 22. Pearce L. R., Sommer E. M., Sakamoto K., Wullschleger S., Alessi D. R. (2011) Biochem. J. 436, 169–179 [DOI] [PubMed] [Google Scholar]
- 23. Treins C., Warne P. H., Magnuson M. A., Pende M., Downward J. (2010) Oncogene 29, 1003–1016 [DOI] [PubMed] [Google Scholar]
- 24. Julien L. A., Carriere A., Moreau J., Roux P. P. (2010) Mol. Cell. Biol. 30, 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boulbes D., Chen C. H., Shaikenov T., Agarwal N. K., Peterson T. R., Addona T. A., Keshishian H., Carr S. A., Magnuson M. A., Sabatini D. M., Sarbassov dos D. (2010) Mol. Cancer Res. 8, 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dibble C. C., Asara J. M., Manning B. D. (2009) Mol. Cell. Biol. 29, 5657–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen C. H., Shaikenov T., Peterson T. R., Aimbetov R., Bissenbaev A. K., Lee S. W., Wu J., Lin H. K., Sarbassov dos D. (2011) Sci. Signal. 4, ra10. [DOI] [PubMed] [Google Scholar]
- 28. Boulbés D. R., Shaiken T., Sarbassov dos D. (2011) Biochem. Biophys. Res. Commun. 413, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oh W. J., Wu C. C., Kim S. J., Facchinetti V., Julien L. A., Finlan M., Roux P. P., Su B., Jacinto E. (2010) EMBO J. 29, 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zinzalla V., Stracka D., Oppliger W., Hall M. N. (2011) Cell 144, 757–768 [DOI] [PubMed] [Google Scholar]
- 31. Chen C. H., Shaikenov T., Peterson T. R., Aimbetov R., Bissenbaev A. K., Lee S. W., Wu J., Lin H. K., Sarbassov dos D. (2011) Sci. Signal. 4, ra10. [DOI] [PubMed] [Google Scholar]
- 32. Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A. (2010) Mol. Cell 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A., Sabatini D. M. (2006) Curr. Biol. 16, 1865–1870 [DOI] [PubMed] [Google Scholar]
- 34. Roudabush F. L., Pierce K. L., Maudsley S., Khan K. D., Luttrell L. M. (2000) J. Biol. Chem. 275, 22583–22589 [DOI] [PubMed] [Google Scholar]
- 35. Chen J., Zheng X. F., Brown E. J., Schreiber S. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4947–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toker A., Newton A. C. (2000) J. Biol. Chem. 275, 8271–8274 [DOI] [PubMed] [Google Scholar]
- 37. Carrano A. C., Eytan E., Hershko A., Pagano M. (1999) Nat. Cell Biol. 1, 193–199 [DOI] [PubMed] [Google Scholar]
- 38. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Q., Chang J. W., Wang J., Kang S. A., Thoreen C. C., Markhard A., Hur W., Zhang J., Sim T., Sabatini D. M., Gray N. S. (2010) J. Med. Chem. 53, 7146–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., Sabatini D. M. (2009) Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peterson R. T., Beal P. A., Comb M. J., Schreiber S. L. (2000) J. Biol. Chem. 275, 7416–7423 [DOI] [PubMed] [Google Scholar]