Background: The dual mTORC1/mTORC2 inhibitor AZD8055 can induce autophagy.
Results: Autophagy induction by AZD8055 antagonizes chemotherapy-induced cell death in association with inhibition of ULK1 phosphorylation at Ser757 and down-regulation of p62/sequestosome 1.
Conclusion: AZD8055 induces pro-survival autophagy that attenuates cell death by down-regulating p62.
Significance: mTOR kinase inhibition may reduce the efficacy of cytotoxic chemotherapy.
Keywords: Apoptosis, Autophagy, Cell Death, DNA Damage, mTOR, AZD8055, ULK1, p62/Sequestosome 1
Abstract
AZD8055 is an ATP-competitive inhibitor of mammalian target of rapamycin (mTOR) that forms two multiprotein complexes, mTORC1 and mTORC2, and negatively regulates autophagy. We demonstrate that AZD8055 stimulates and potentiates chemotherapy-mediated autophagy, as shown by LC3I-II conversion and down-regulation of the ubiquitin-binding protein p62/sequestosome 1. AZD8055-induced autophagy was pro-survival as shown by its ability to attenuate cell death and DNA damage (p-H2AX), and to enhance clonogenic survival by cytotoxic chemotherapy. Autophagy inhibition by siRNA against Beclin 1 or LC3B, or by chloroquine, partially reversed the cytoprotective effect of AZD8055 that was independent of cell cycle inhibition. The pro-survival role of autophagy was confirmed using ectopic expression of Beclin 1 that conferred cytoprotection. To determine whether autophagy-mediated down-regulation of p62/sequestosome 1 contributes to its pro-survival role, we generated p62 knockdown cells using shRNA that showed protection from chemotherapy-induced cell death and DNA damage. We also overexpressed wild-type (wt) p62 that promoted chemotherapy-induced cell death, whereas mutated p62 at functional domains (PB1, UBA) failed to do so. The ability of ectopic wt p62 to promote cell death was blocked by AZD8055. AZD8055 was shown to inhibit phosphorylation of the autophagy-initiating kinase ULK1 at Ser757 and inhibited known targets of mTORC1 (p-mTOR Ser2448, p70S6K, p-S6, p4EBP1) and mTORC2 (p-mTOR Ser2481, p-AKT Ser473). Knockdown of mTOR, but not Raptor or Rictor, reduced p-ULK1 at Ser757 and enhanced chemotherapy-induced autophagy that resulted in a similar cytoprotective effect as shown for AZD8055. In conclusion, AZD8055 inhibits mTOR kinase and ULK1 phosphorylation to induce autophagy whose pro-survival effect is due, in part, to down-regulation of p62.
Introduction
Mammalian target of rapamycin (mTOR)2 kinase is a downstream effector of phosphoinositide 3-kinase (PI3K/Akt) signaling that has been shown to regulate colorectal tumorigenesis (1). Furthermore, targeted inhibition of mTORC1 and mTORC2 was shown to prevent the establishment of colorectal cancer metastases in vivo (2), suggesting the rationale for evaluating inhibitors of mTOR kinase for the therapy of this malignancy. AZD8055 is an ATP-competitive inhibitor of mTOR kinase activity. mTOR is a serine/threonine kinase and therapeutic target in cancer cells that serves as a sensor of mitogenic signals, nutrient availablility, and energy levels and regulates cell growth (3) (4). Mitogenic signals are transduced to mTOR via upstream PI3K/AKT (5, 6). mTOR kinase forms two distinct multiprotein complexes known as mTORC1 (contains Raptor, mLST8, and PRAS40) and mTORC2 (contains Rictor, mLST8, and Protor). mTORC1 phosphorylates p70S6K and 4E-BP1 that regulate protein translation (7), whereas mTORC2 phosphorylates AKT on Ser473 that enhances its enzymatic activity (8, 9). Raptor is essential for the efficient phosphorylation of mTORC1 downstream targets whereas Rictor is critical for mTORC2 function(10). In contrast to AZD8055 and other mTOR kinase inhibitors, rapamycin is an allosteric inhibitor of mTOR that, in association with FKBP12, binds mTOR adjacent to its catalytic domain and therefore, leads to incomplete inhibition of mTOR activity. Furthermore, rapamycin-resistant functions of mTORC1 can lead to increased AKT signaling that may be overcome by dual mTORC1/mTORC2 inhibition (11–13). In this regard, AZD8055 inhibits the phosphorylation of mTORC1 substrates p70S6K and 4E-BP1 as well as phosphorylation of the mTORC2 substrate AKT and downstream proteins (13), thereby blocking the negative feedback that can activate AKT (12). AZD8055 is a more potent inhibitor of cell proliferation than is rapamycin and has also been shown to inhibit the growth of xenografts representing a broad range of human tumor types (13). AZD8055 is currently being studied in phase I clinical trials, and its combination with cytotoxic chemotherapy or targeted agents is a promising therapeutic strategy.
mTOR has been shown to negatively regulate autophagy (14–16), which involves the direct phosphorylation of the autophagy-initiating serine/threonine kinase ULK1 (Unc51-like kinase; hATG1) on Ser757 (17) to prevent formation of its complex with Atg13, FIP200, and Atg101 (18). AZD8055 has been shown to be a more potent inducer of autophagy compared with rapamycin (13) and its analogs (7, 19) that may be due to its more complete inhibition of mTORC1 and/or inhibition of mTORC2. Under conditions of cellular stress (nutrient deprivation, hypoxia, or cytotoxic stress), tumor cells induce autophagy to maintain energy metabolism and to enable cell survival. Therefore, autophagy serves as a mechanism of stress tolerance that can protect tumor cells from anti-cancer therapy (20) (21). In some cellular contexts, sustained or prolonged autophagy may trigger cell death although its relevance in tumors remains unknown (22). Autophagy is regulated by Beclin 1 that forms a complex with a class III PI3 kinase, Vps34, that is involved in autophagosome formation (19) during which the cytosolic microtubule-associated protein light chain 3 (LC3-I) is converted to the membrane-bound LC3-II form (23). LC3-II is known to bind to the ubiquitin-binding protein p62/sequestosome 1 (SQSTM 1) that transports protein aggregates to the lysosome for their degradation by autophagy (24). Autophagy induction is associated with down-regulation of p62, whereas autophagy-deficient mice show p62 accumulation (25). Evidence suggests that p62 acts as a signaling hub through its ability to recruit and oligomerize important signaling molecules into cytosolic aggregates to control cell survival and apoptosis (26) (27).
We studied the ability of AZD8055-induced autophagy to modulate chemotherapy-induced DNA damage and cell death, and determined the role of autophagy-mediated down-regulation of p62 in this process. AZD8055 was shown to significantly attenuate chemotherapy-induced DNA damage and cell death that was independent of cell cycle inhibition and was associated with induction of pro-survival autophagy. Genetic or pharmacological inhibition of autophagy partially reversed AZD8055-mediated cytoprotection. Autophagy induction by AZD8055 was due to direct inhibition of mTOR kinase and the dephosphorylation of ULK1 at Ser757. Furthermore, autophagy-mediated down-regulation of p62/sequestosome 1 was shown to contribute to the pro-survival effect of autophagy.
EXPERIMENTAL PROCEDURES
Cell Culture and Drugs
Colon carcinoma cell lines (HT-29, DLD-1) were cultured as monolayers in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) FBS and 2 mmol/L of l-glutamine. Cells were treated with AZD8055 [5-{2,4-Bis[(3S)-3-methylmorpholin-4-yl]pyrido[2,3-d]pyrimidin-7-yl}B-2-methoxyphenyl)methanol], obtained from Dr. C. Yu (Roswell Park, Buffalo, NY), or rapamycin (LC Lab., Woburn, MA), alone and in combination with 5-fluorouracil or CPT-11 (both from Sigma) in the presence or absence of chloroquine (Sigma). AZD8055 and rapamycin were prepared as 10 mmol/liter stock solutions in DMSO and stored at −20 °C.
Immunoblotting
Protein samples were prepared in a lysis buffer (5 mmol/liter MgCl2, 137 mmol/liter KCl, 1 mmol/liter EDTA, 1 mmol/liter EGTA, 1% CHAPS, 10 mmol/liter HEPES (pH 7.5)) containing a protease inhibitor mixture (Sigma), and were normalized using nanodrop measurement (Thermo Scientific, Franklin, MA) or a protein assay kit (Bio-Rad). The samples were boiled in LDS sample buffer (Invitrogen) and loaded onto a 14% SDS-PAGE gel for protein separation with electrophoretic transfer onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Membranes were blocked in 0.2% I-Block (Applied Biosystems, Foster City, CA) in 1× PBS/0.1% Tween 20 and then probed with the respective primary antibodies overnight at 4 °C. After washing and incubation with secondary antibodies, the immunoblotted proteins were visualized using the CDP-star alkaline phosphatase chemiluminescence substrate (Applied Biosystems). All primary antibodies were obtained from Cell Signaling (Boston, MA) with the exception of antibodies against p62/SQSTM 1 (MBL, Woburn, MA) and tubulin (Sigma). Experiments were repeated and results from representative immunoblots are shown.
Lentiviral Expression
The lentiviral shRNA for mTOR (plasmid 1855 and 1856) and a scramble shRNA (plasmid 1864) were obtained from Addgene (Cambridge, MA). For p62 knockdown, a shRNA targeting sequence (GCATTGAAGTTGATATCGAT) was cloned into pSIH1-puro, as previously described (28). A non-targeting control sequence (CAACAAGATGAAGAGCACCAA; Sigma) was also cloned into pSIH1-puro. The p27 cDNA (Origene, Rockville, MD) was cloned into a lentiviral cDNA expression vector pCDH1-puro (System Biosciences, Mountain View, CA), as previously described (29). Each of the lentiviral expression constructs was then mixed with two packaging plasmids (Addgene plasmids 12259 and 12260) and transfected into HEK293T cells using Lipofactamine/PLUS reagent (Invitrogen) to produce lentivirus. The resultant lentivirus-containing medium was concentrated by incubating with 10% PEG-8000 (Sigma) and centrifuged at 1,500 × g for 10 min. The virus-containing pellet was then resuspended in a small volume of OPTI-MEM medium (Invitrogen) and directly added to the target cells in the presence of 8 μg/ml polybrene (Sigma).
Transfection of siRNA
siRNA oligonucleotides for mTOR, Beclin 1, and LC3B (all from Cell Signaling) and Raptor or Rictor siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) were transfected into target cells using Lipofectamine™ RNAiMAX Reagent (Invitrogen), per the manufacture's protocol. Briefly, cells were seeded onto 6-well plates at ∼50% confluence in 2.5 ml complete medium without antibiotics and incubated overnight. A total of 75–300 nmol siRNA was diluted into 0.5 ml of OPTI-MEM medium (Invitrogen) containing 7.5 μl of RNAiMAX reagent and incubated at room temperature for 20 min. The siRNA/RNAiMAX complex formed was directly added into the 6-well plates to produce a final concentration of 25–100 nm siRNA. Cells were then returned to incubation and knockdown efficiency was determined at least 72 h post-transfection by immunoblotting.
Retroviral Overexpression of Beclin 1 and p62/Sequestosome 1
Beclin 1 (Origene) or p62 (gift of Dr. S. V. Reddy, Medical University of South Carolina, Charleston, SC) cDNA was cloned into a retroviral expression vector containing an N-terminal 3-tandem tag (s-tag, 2X FLAG and streptavidin-binding protein tag; 3-tag) that was derived from a parent empty vector (30). Using site-directed mutagenesis, p62 mutants were generated at UBA or PB1 domains containing mutations of I431A or D69A sites, respectively, that have been shown to impair the ability of p62 to be sequestered into aggregates (31) (32). The resultant retroviral vectors encoding Beclin 1, p62 (WT or mutant), or empty vector were individually mixed with packaging plasmids and retrovirus was generated, as previously described (30). Retrovirus-containing supernatant was concentrated and utilized to transduce the target cells, as previously described (30). Ectopic expression of the 3-tag proteins was detected using an anti-FLAG antibody or by a size shift from endogenous proteins.
Confocal Immunofluorescence
Cells were plated onto Lab-Tek® II CC2TM chamber slides, incubated overnight in media, and then fixed with 4% paraformaldehyde for 10 min at room temperature. Cells were rinsed with PBS ×2 and then permeabilized with 0.5% Triton X-100 in PBS at room temperature for 10 min. Cells were incubated with the primary antibody (anti-FLAG, Cell Signaling) diluted in 1% BSA in PBS at room temperature for 1 h. After rinsing in PBS ×3, a fluorescence dye-conjugated secondary antibody (AF594, Invitrogen) diluted in 1% BSA in PBS was added and incubated at room temperature for 30 min. Slides were rinsed with PBS and mounted. Immunofluorescence was visualized and captured using an inverted LSM 5 Pascal Laser Scanning Microscope (Carl Zeiss) with appropriate filter and detector combinations according to the spectrum of the fluorochrome used.
Cell Cycle and Cell Death Assays
For cell cycle analysis, cells were detached with trypsin, washed once with cold PBS, and then counted. An aliquot of 0.5–1 × 106 cells was centrifuged and pelleted, then resuspended in 0.5 ml of PBS. Cells were fixed by adding 0.5 ml of 100% cold ethanol while vortexing, followed by incubation at −20 °C overnight. Cells were centrifuged, pelleted, and resuspended in 0.5 ml of PI-RNase solution (50 μg/ml propidium iodide (PI) plus 100 μg/ml RNase A (Sigma) in PBS). Cells were stained with PI for at least 20 min at room temperature and then analyzed by flow cytometry. Cell cycle distribution was determined using ModFit LT (Verity Software House, Topsham, ME). Cell death was measured with the annexin V assay (BD Biosciences, San Jose, CA), as previously described (30). After drug treatment, adherent cells were detached from culture dishes by trypsinization for 3 to 5 min and then combined with floating cells. The extent of apoptosis was indicated as the percentage of Annexin V+PI− cells, whereas cells labeled Annexin V+PI+ were interpreted to represent late apoptosis or necrosis.
Colony Formation Assay
Cells were plated in 6-well plates and treated with the study drugs for 3 days. Cells were then lifted using TrypLE™ Express (Invitrogen), a trypsin replacement that is stable at RT, and counted with a cell counter (Bio-Rad). Then, 20 × 104 cells for each condition were plated onto 10-cm dishes and incubated. Cells carrying ectopic Beclin 1 were directly seeded (200 cells/well) onto 6-well plates, allowed to attach overnight, and then treated with the study drugs for 3 days. After incubating in drug-free medium at 37 °C for 7 to 14 days, colonies were fixed and stained with crystal violet. Excess dye was rinsed off with tap water, and the plates were air dried.
Statistical Analysis
The statistical significance of the differences between experimental variables was determined using the Student's t test. The values shown represent the mean ± S.D. for triplicate experiments. p < 0.05 was considered as statistically significant.
RESULTS
AZD8055 Attenuates Chemotherapy-induced Cell Death and DNA Damage
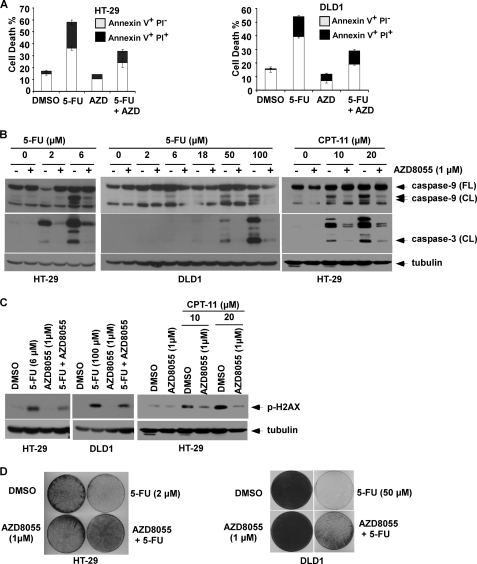
We determined whether AZD8055 can modulate cell death induction by cytotoxic drugs in human colon cancer cell lines (HT-29, DLD1). While AZD8055 alone did not induce cell death, AZD8055 was shown to significantly reduce 5-FU-induced cell death by 41 to 46% as shown by annexin V+ labeling (Fig. 1A), and attenuated 5-FU-induced caspase-9 and -3 cleavage (Fig. 1B) compared with 5-FU alone. AZD8055 treatment was also shown to reduce 5-FU-induced DNA damage in both cell lines, as shown by expression of p-H2AX, a marker of DNA double-strand breaks (Fig. 1C). To demonstrate that the protective effect of AZD8055 was not limited to 5-FU, we utilized the topoisomerase 1 inhibitor CPT-11 (irinotecan) and found that AZD8055 can similarly attenuate CPT-11-induced caspase cleavage and DNA damage (Fig. 1, B and C). We then studied the effect of AZD8055 alone and in combination with 5-FU on long-term cell viability in a clonogenic survival assay. While AZD8055 did not inhibit clonogenic survival in contrast to 5-FU, AZD8055 was shown to significantly attenuate the ability of 5-FU to inhibit colony formation (Fig. 1D).
FIGURE 1.
AZD8055 treatment attenuates 5-FU-induced cell death in human colon cancer cell lines. A, AZD8055 (1 μm) attenuates cell death induced by 5-FU (6 μm: HT-29; 100 μm: DLD1). Cells were incubated with study drugs for 3 days and annexin V/PI labeling was performed. Apoptosis is indicated by annexin V+ PI−-labeled cells and late apoptosis/necrosis is shown by annexin V+PI+ cells. Columns, mean of experiments conducted in triplicate; bars, S.D. B and C, AZD8055 attenuates caspase-9 and -3 cleavage (B) and DNA damage (C) induced by 5-FU (5 days) or CPT-11 (3 days) in HT-29 and/or DLD1 cells. DNA damage was measured by expression of p-H2AX that detects DNA double-stranded breaks. Tubulin served as a control for protein loading. D, suppression of colony formation by 5-FU is reduced by co-treatment with AZD8055 in both cell lines. Cells were incubated with drugs for 3 days and equal numbers of cells were replated and grown for 14 days in drug-free media. Colonies were visualized by staining with crystal violet.
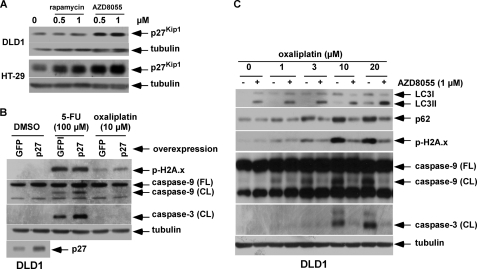
Because AZD8055 is known to potently inhibit cell proliferation (13), we examined the ability of AZD8055 to modulate expression of the cyclin-dependent kinase (cdk) inhibitor, p27Kip1, that inhibits G1 cyclin/cdk complexes to produce cell cycle arrest (33, 34). AZD8055 was shown to induce p27Kip1protein expression in both cell lines (Fig. 2A). To determine whether p27Kip1 can contribute to cytoprotection, we generated DLD1 cells that overexpress a lentiviral p27 construct. Stable overexpression of p27Kip1 increased accumulation of cells in G1 compared with control cells (62.9 ± 0.5% versus 52.9 ± 0.6%). Cells were then treated with 5-FU or oxaliplatin, an alkylating agent whose cytotoxic effect is independent of the cell cycle (35). Overexpression of p27Kip1 failed to attenuate 5-FU- or oxaliplatin-induced caspase cleavage or DNA damage (Fig. 2B). Furthermore, AZD8055 was shown to block oxaliplatin-induced DNA damage and caspase cleavage (Fig. 2C). Together, these data suggest that the cytoprotective effect of AZD8055 is unrelated to cell cycle inhibition.
FIGURE 2.
AZD8055-mediated cytoprotection is independent of cell cycle arrest. A, DLD1 and HT-29 cells were treated with rapamycin or AZD8055 at the indicated doses for 48 h and p27Kip1 expression levels were analyzed by immunoblotting. B, DLD1 cells were transduced with a lentivirus encoding GFP or p27Kip1. Cells were then treated with 5-FU or oxaliplatin for 5 days, and expression of p-H2Ax and cleaved caspase-9 and -3 proteins were measured by immunoblotting. C, AZD8055 enhances oxaliplatin-induced autophagy, as shown by LC3I-II conversion and reduced p62/SQSTM 1 expression, which is associated with attenuated DNA damage and caspase cleavage. DLD1 cells were treated with increasing doses of oxaliplatin alone or combined with AZD8055 for 5 days and expression of LC3, p62, p-H2Ax, and cleaved caspase-9 and -3 were analyzed by immunoblotting.
AZD8055 Induces Autophagy to Confer Drug Resistance
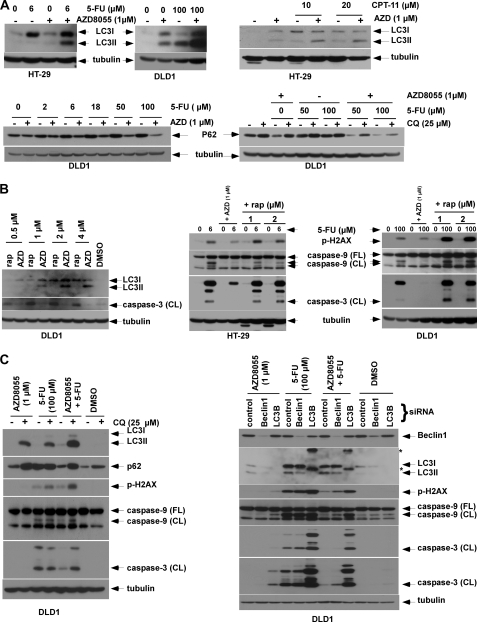
Whereas mTOR is known to negatively regulate autophagy (19), the effect of autophagy induction on chemotherapy-induced cell death is poorly defined. We demonstrate that AZD8055 can induce autophagy and potentiate autophagy induction by 5-FU, CPT-11 (Fig. 3, A and B), or oxaliplatin (Fig. 2C), as shown by LC3I-II conversion and reduced expression of the autophagy substrate, p62/SQSTM1. To confirm that down-regulation of p62 was mediated by autophagy, we utilized the anti-malarial drug CQ that prevents lysosomal acidification to block autophagic catabolism (21). Treatment of cells with CQ or CQ plus AZD8055 and 5-FU was shown to accumulate p62 consistent with inhibition of autophagy (Fig. 3, A and C). AZD8055 was shown to induce autophagy to a greater extent than did rapamycin (Fig. 3B). In contrast to AZD8055 (Fig. 1), rapamycin at equivalent doses induced caspase-3 cleavage (Fig. 3B) and failed to confer cytoprotection against 5-FU (Fig. 3B).
FIGURE 3.
AZD8055 induces autophagy to confer cytoprotection. A, AZD8055 induces autophagy and potentiates autophagy induced by 5-FU (5 d) or CPT-11 (3 days), as shown by conversion of LC3 I to II. Furthermore, treatment with the combination of AZD8055 and 5-FU (5 days) or CPT-11 (3 days) reduce p62/SQSTM 1 expression that is attenuated by the lysomotrophic autophagy inhibitor, chloroquine (CQ). B, DLD1 cells were treated with rapamycin (rap) or AZD8055 (AZD) for 3 days and LC3 and cleaved caspase-3 were measured by immunoblotting. HT-29 and DLD1 cells were treated with 5-FU alone or combined with rapamycin or AZD8055 at the indicated doses for 5 days. Expression of p-H2AX and caspase-9, -3 cleavage were detected using specific antibodies. C, autophagy inhibition by CQ accumulates LC3II and p62 and enhances caspase cleavage and DNA damage induced by 5-FU plus AZD8055 (5 days). Knockdown of Beclin 1 or LC3B using siRNA block LC3I-II conversion and enhance caspase cleavage and DNA damage induced by 5-FU combined with AZD8055 (5 days). *, nonspecific bands.
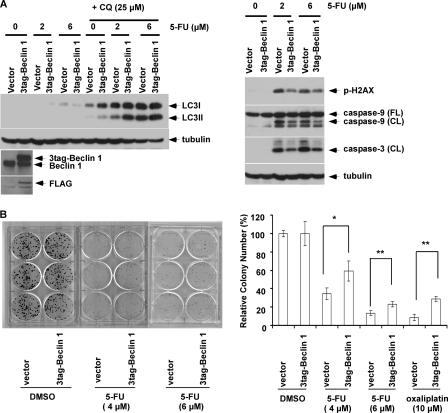
To determine whether pro-survival autophagy can account for the cytoprotective effect of AZD8055, we inhibited autophagy using CQ or by gene knockdown. The addition of CQ to AZD8055 plus 5-FU resulted in the accumulation of LC3II and p62 and enhanced both DNA damage (p-H2AX) and caspase cleavage (Fig. 3C). Furthermore, knockdown of LC3B or Beclin 1 by siRNA attenuated LC3I-II conversion and potentiated caspase cleavage and DNA damage induced by 5-FU alone or combined with AZD8055 (Fig. 3C). To further support autophagy as the mechanism of AZD8055-mediated cytoprotection, we generated HT-29 cells that constitutively express Beclin 1. Overexpression of Beclin 1 (versus vector control) was shown to increase LC3I-II conversion in the presence of CQ or combined with 5-FU (Fig. 4A), indicating the ability of Beclin 1 to increase autophagic flux. Beclin 1 overexpression-protected cells from 5-FU-induced caspase-9 and -3 cleavage and from DNA damage, (p-H2AX) [Fig. 4A]. Furthermore, ectopic Beclin 1 was shown to significantly attenuate the ability of 5-FU or oxaliplatin to reduce colony formation in a clonogenic survival assay (Fig. 4B). Taken together, these data indicate that AZD8055 induces pro-survival autophagy that underlies its ability to attenuate chemotherapy-mediated cell death.
FIGURE 4.
Ectopic Beclin 1 expression attenuates 5-FU-induced apoptosis and DNA damage. A, HT-29 cells stably expressing a 3tag-Beclin 1 cDNA or an empty vector were treated with 5-FU in the presence or absence of CQ and protein expression was determined in whole cell lysates. B, clonogenic survival assay was performed using Beclin 1 expressing or control cells that were treated with 5-FU or DMSO for 3 days. Cells were then incubated in drug-free media for 7–14 days, and colonies were developed and stained with crystal violet, counted, and then normalized to vehicle (DMSO)-treated cells. Columns represent mean of experiments that were conducted in triplicate; bars, S.D. *, p < 0.05; **, p < 0.01.
Role of p62/SQSTM 1 Down-regulation in AZD8055-mediated Cytoprotection
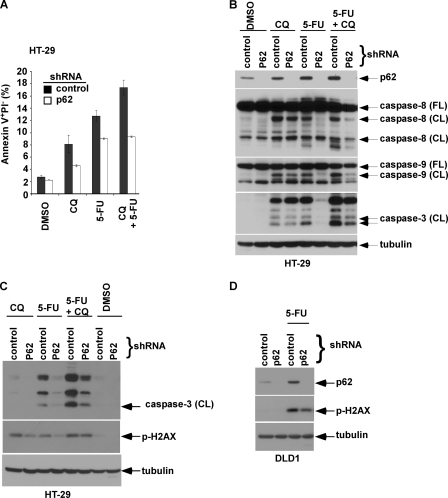
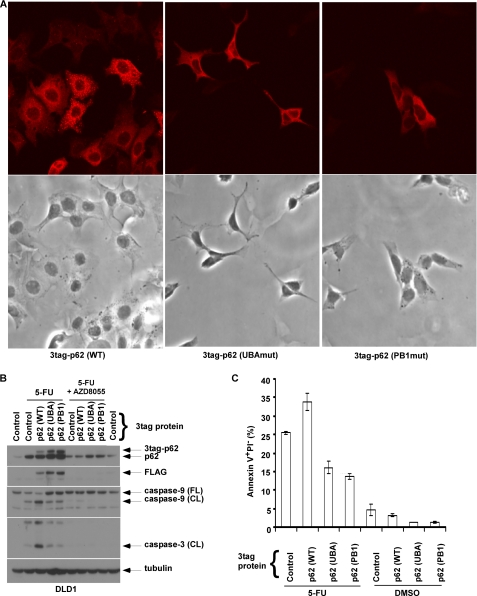
AZD8055 was shown to stimulate autophagy and enhance chemotherapy-induced autophagy that were associated with a reduction in p62/SQSTM 1 expression levels (Fig. 3 A). Evidence suggests that p62 acts as a signaling hub through its ability to recruit and oligomerize important signaling molecules into cytosolic aggregates to regulate cell survival and apoptosis (26, 27). To determine whether autophagy-mediated down-regulation of p62 can contribute to the cytoprotective effect of AZD8055, we generated cells with knockdown of p62 by shRNA. Suppression of p62 was shown to significantly attenuate 5-FU-induced apoptosis, as shown by annexin V+ PI− labeling (Fig. 5A) and caspase-8, -9, -3 cleavage (Fig. 5B). Furthermore, suppression of p62 also attenuated 5-FU-induced DNA damage (Fig. 5, C and D). Despite the ability of the autophagy inhibitor CQ to accumulate p62 (Fig. 3, A and C), attenuation of 5-FU-induced apoptosis by p62 knockdown was maintained in the presence of CQ (Fig. 5, A and B). Next, we generated cells with ectopic wild-type (WT) or mutant p62 proteins that can mimic the accumulation of p62 that occurs when autophagy is disabled. Using site-directed mutagenesis, we generated mutations in p62 at the ubiquitin-associated (UBA: I431A) and PB1 (PB1: D69A) domains (31, 32). To demonstrate loss of function of these p62 mutants, we determined their cellular staining pattern using immunofluorescence microscopy. Cells with ectopic WT p62 exhibited punctate staining indicating the formation of p62 aggregates, whereas cells expressing p62 mutants showed diffuse p62 staining indicating the inability to form aggregates (Fig. 6A). We then determined whether ectopic p62 can promote 5-FU-induced apoptosis and whether p62 mutants retain this ability. Ectopic WT p62 was shown to sensitize DLD1 cells to 5-FU-induced caspase-9, -3 cleavage (Fig. 6B) and apoptosis induction, as shown by annexin V+ PI− labeling compared with control cells (Fig. 6C). In contrast, p62 mutants at UBA and PB1 domains lacked the ability to enhance 5-FU-induced caspase cleavage or apoptosis (Fig. 6, B and C). This finding indicates that both p62 functional domains are required for the apoptosis-promoting effect of p62. Importantly, the addition of AZD8055 was shown to reduce ectopic p62 protein expression (Fig. 6B) consistent with the ability of AZD8055 to down-regulate p62 because of autophagy induction. Moreover, treatment with AZD8055 was shown to abrogate the ability of WT p62 to enhance 5-FU-induced caspase cleavage (Fig. 6B). Taken together, these data suggest that p62 can mediate chemotherapy-induced cell death and DNA damage.
FIGURE 5.
Knockdown of p62/SQSTM 1 by shRNA is shown to attenuate 5-FU-induced apoptosis and DNA damage. A and B, p62 shRNA attenuates apoptosis induced by 5-FU (50 μm) in the presence or absence of CQ, as shown by annexin V+ PI− labeling (36 h) [A] or caspase cleavage (B). Columns represent mean of triplicate experiments; bars, S.D. C and D, knockdown of p62 attenuates DNA damage (p-H2AX) induced by 5-FU alone or combined with CQ in HT-29 (C) and DLD-1 (D) cells.
FIGURE 6.
Ectopic expression of WT p62/SQSTM 1, but not functional p62 mutants, enhance 5-FU-induced apoptosis. A, mutations in p62 at UBA and PB1 domains were generated by site-directed mutagenesis and transduced into DLD1 cells. The staining pattern of ectopic 3 tandem tagged (3tag) p62 was then analyzed by immunofluoresence microscopy using an anti-FLAG antibody. In contrast to WT p62 cells that show punctate p62 staining, mutations at the UBA and PB1 domains resulted in loss of formation of p62 aggregates as shown by a diffuse staining pattern (top). Cellular morphology is shown by bright field images (bottom). B and C, ectopic expression of WT p62 is shown to enhance caspase cleavage (B) and apoptosis indicated by annexin V+PI− labeling (C) in cells treated with 5-FU (100 μm). In contrast to WT p62, p62 mutations were shown to disrupt the ability of p62 to enhance apoptosis induction by 5-FU (B, C). Columns; mean of experiments conducted in triplicate; bars, S.D.
AZD8055 Inhibits mTORC1 and mTORC2 Downstream Signaling Events
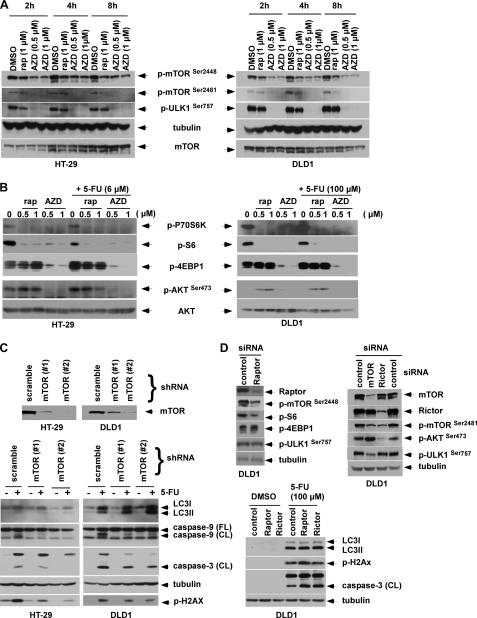
Studies indicate that mTORC1 contains primarily mTOR phosphorylated on Ser2448, whereas mTORC2 contains mTOR phosphorylated on Ser2481 (36). Phosphorylation of mTOR on Ser2481 is a marker for the presence of mTORC2 complexes (36). We detected phosphorylation of mTOR on Ser2448 and Ser2481 in untreated colon cancer cell lines, indicating the presence of both mTORC1 and mTORC2 complexes (Fig. 7A). Treatment of cells with AZD8055 inhibited phosphorylation of mTOR on Ser2448 and Ser2481, consistent with inhibition of both mTORC1 and mTORC2 activity. Rapamycin primarily inhibited p-mTOR on Ser2448 although inhibition of p-mTOR on Ser2481 was also observed at a longer time point (Fig. 7A), as previously described (36). Recent evidence indicates that mTOR kinase can directly phosphorylate the mammalian autophagy-initiating kinase ULK1, a homologue of yeast ATG1, on Ser757 thereby disrupting its association with AMPK and inhibiting autophagy (17). Accordingly, we studied that ability of AZD8055 to inhibit phosphorylation of ULK1 and demonstrate that AZD8055 can potently inhibit p-ULK1 at Ser757 as early as 2 h after treatment (Fig. 7A). This finding provides a mechanism by which AZD8055 can stimulate autophagy. We then confirmed that AZD8055 inhibits both mTORC1 and mTORC2 downstream targets. AZD8055 was shown to potently inhibit the mTORC1 substrate P70S6K and its kinase target ribosome protein S6, as well as 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) at Thr37/46 (Fig. 7B). In contrast to the ATP competitive mTOR kinase inhibitor AZD8055, the allosteric mTOR inhibitor rapamycin failed to inhibit p-4E-BP1 on Thr37/46 consistent with more complete mTORC1 inhibition by AZD8055. Inhibition of mTORC2 by AZD8055 was shown by probing for phosphorylation of AKT on Ser473 that is a known mTORC2 substrate. AZD8055 was found to inhibit p-AKT Ser473 whereas rapamycin did not (Fig. 7B). By suppressing mTORC2, AZD8055 may block the negative feedback resulting from mTORC1 inhibition that can activate AKT (12).
FIGURE 7.
AZD8055 inhibits ULK1 phosphorylation at Ser757 and blocks mTORC1/mTORC2 downstream signaling. A, cells were treated with rapamycin (rap) or AZD8055 (AZD) for the indicated doses and times, and p-mTOR (Ser2448 and Ser2481) and p-ULK1 (Ser757) were analyzed in whole cell lysates by immunoblotting. B, cells were treated with rapamycin or AZD8055 in the presence or absence of 5-FU for 24 h, and analysis of mTORC1 (p-P70S6K, p-S6, p-4E-BP1) and mTORC2 (p-AKT at Ser473) substrates was performed by immunoblotting. C, mTOR knockdown by shRNA is shown to potentiate 5-FU-induced autophagy, indicated by LC3I-II conversion that was associated with protection from 5-FU-induced DNA damage and apoptosis. Suppression of mTOR was achieved by transduction with two lentiviral shRNA sequences targeting mTOR (#1 or #2) compared with a scramble shRNA. Knockdown or control cells were treated with 5-FU (6 μm for HT-29, 100 μm for DLD1) for 5 days and LC3I-II conversion, DNA damage (p-H2AX) and caspase-9, -3 cleavage were probed. D, knockdown of mTOR, but not Raptor or Rictor, confers cytoprotection. DLD1 cells were transfected with mTOR, Raptor or Rictor siRNA for 3 days and phosphorylation of p-mTOR (Ser2448 and Ser2481), p-ULK1 (Ser757), as well as phosphorylation of the substrates of mTORC1 (p-S6, p-4EBP-1) or mTORC2 (p-AKT at Ser473) were analyzed. Raptor, Rictor or control knockdown cells were treated with 5-FU for 5 days, and expression of p-H2AX and caspase-9, -3 cleavage were measured.
To confirm that autophagy induction and cytoprotection by AZD8055 are due to mTOR inhibition, we generated cells with knockdown of mTOR using lentiviral shRNA. Two different shRNA targeting sequences were shown to suppress mTOR expression compared with scramble shRNA (Fig. 7C). Suppression of mTOR was shown to enhance 5-FU-induced LC3I-II conversion (Fig. 7C), consistent with negative regulation of autophagy by mTOR (19). Furthermore, mTOR knockdown was shown to protect cells from 5-FU-induced caspase-9, -3 cleavage and from DNA damage, with the extent of cytoprotection shown to correlate with the relative level of mTOR suppression (Fig. 7C). To determine the contribution of mTORC1 and mTORC2 to the observed cytoprotection, we suppressed Raptor and Rictor that are essential components of mTORC1 and mTORC2 complexes, respectively. Raptor siRNA was shown to inhibit p-mTOR at Ser2448 and p-S6, but not p-4EBP1 (Fig. 7D), that was similar to the allosteric inhibition of these targets by rapamycin (Fig. 7B). Rictor knockdown by siRNA was shown to inhibit p-mTOR on Ser2481 and p-AKT at Ser473, consistent with the known ability of Rictor to modulate mTORC2. However, neither suppression of Raptor nor Rictor in DLD1 cells was able to inhibit p-ULK1 at Ser757, suggesting their inability to directly regulate the initiation of autophagy. In contrast, knockdown of mTOR by siRNA was shown to significantly attenuate p-ULK1 at Ser757 (Fig. 7D), indicating the direct role of mTOR kinase in this autophagy-initiating event. Consistent with this finding, suppression of mTOR (Fig. 7C), but not Raptor nor Rictor, was able to potentiate autophagy and to attenuate 5-FU-induced apoptosis and DNA damage (Fig. 7D).
DISCUSSION
We studied the ability of AZD8055, an ATP-competitive and dual mTORC1 and mTORC2 inhibitor, to modulate chemotherapy-induced cell death and studied the effect of AZD8055-induced autophagy on this process. Using clinically achievable doses, AZD8055 was shown to significantly attenuate chemotherapy-induced DNA damage and cell death and to promote long-term clonogenic survival. This effect was observed using anti-cancer drugs with diverse mechanisms of action. The cytoprotective effect of AZD8055 was not due to cell cycle inhibition since overexpression of the cdk inhibitor p27, that arrests cells in G1, did not protect cells from 5-FU-induced DNA damage or apoptosis, and AZD8055 conferred similar cytoprotection against the cell cycle-independent drug, oxaliplatin.
We determined whether AZD8055-induced autophagy underlies its cytoprotective effect. AZD8055 was shown to stimulate autophagy and to potentiate autophagy induction by 5-FU, CPT-11, or oxaliplatin. AZD8055 was a more potent inducer of autophagy compared with rapamycin, as has also been shown using other dual mTOR kinase inhibitors (Torin 1 and pp242) (13). Recent data indicate that mTOR kinase can directly phosphorylate the mammalian autophagy-initiating kinase ULK1 on Ser757 to prevent its activation (17), suggesting that mTOR inhibition inhibits ULK1 activation to induce autophagy. We demonstrate for the first time that AZD8055 more potently inhibits ULK1 phosphorylation on Ser757 compared with rapamycin that may contribute to its more potent effect on autophagy stimulation. In this regard, rapamycin failed to protect cells from 5-FU-induced DNA damage and apoptosis compared with AZD8055. Consistent with our data is the finding that the dual mTOR kinase inhibitor OSI-027 can induce autophagy that limited the extent of apoptosis induction in Bcr-Abl expressing leukemic cells and was reversed using chloroquine (37). To demonstrate the potential role of autophagy in conferring cytoprotection by AZD8055, we inhibited autophagy with siRNA knockdown of Beclin 1 or LC3B, or using chloroquine. We found that both strategies were able to partially restore the ability of 5-FU to induce DNA damage and to trigger apoptosis. In contrast, ectopic expression of Beclin 1 was shown to confer cytoprotection as did AZD8055. While AZD8055-mediated cytoprotection was observed in colon cancer cell lines, we cannot exclude the possibility that autophagy-mediated cytoprotection by this drug is tumor cell type-dependent.
AZD8055 induced autophagy and enhanced chemotherapy-induced autophagy that were associated with reduced p62/SQSTM 1 expression. p62 binds ubiquitinated proteins and transports them to lysosomes for autophagic degradation (24). Autophagy is the main mechanism of p62 turnover in tumor cells, and data indicate that p62 may regulate cell death and/or cell survival due to its ability to recruit and oligomerize important signaling molecules into cytosolic aggregates (26) (27). Specifically, p62-mediated aggregation of caspase-8 resulted in its activation and mediated apoptosis induction by a protesome inhibitor (38)and a death receptor agonist(27). We detected p62 aggregates in untreated human colon cancer cells and demonstrate that these aggregates are lost when functional domains of p62 are inactivated by single-site mutations. Mutations within p62 in the UBA domain impair its ability to bind ubiquitin (31) and mutations in PB1 disrupt p62 polymerization (32). We found that ectopic p62 expression increased 5-FU-induced apoptosis indicating a pro-death role for this protein. AZD8055 was also shown to reduce the level of ectopic p62 proteins, compared with untreated cells, that was associated with abrogation of 5-FU-induced apoptosis. Given that mutations at functional domains of p62 resulted in the inability to form p62 aggregates, we determined whether these mutants retain a pro-death function as was seen for WT p62. We found that both p62 mutants can disrupt the ability of p62 to enhance 5-FU-induced apoptosis, indicating that formation of p62 aggregates are critical to the pro-apoptotic function of p62. The mechanism by which p62 can promote cell death is unknown and studies are ongoing in our laboratory to examine the association between the formation of p62 aggregates with their pro-apoptotic function. Taken together, these data suggest that p62 can promote chemotherapy-induced apoptosis, and that autophagy-mediated down-regulation of p62 levels may contribute to the ability of autophagy to attenuate cell death.
AZD8055 is a small molecule that is known to more completely suppress mTORC1 than does allosteric inhibition by rapamycin. AZD8055 inhibited the phosphorylation of the mTORC1 substrates 4E-BP1 at positions 37 and 46 sites which are resistant to rapamycin, in addition to the P70S6K downstream target. Consistent with its dual inhibition of mTORC1/mTORC2, AZD8055 inhibited phosphorylation of the mTORC2 substrate, p-AKT at Ser473. We confirmed that AZD8055-mediated cytoprotection is due to mTOR kinase inhibition and not off-target effects using cells with mTOR knockdown. Suppression of mTOR was shown to potentiate 5-FU-induced autophagy and to attenuate 5-FU-induced DNA damage and apoptosis. Knockdown of mTOR in human glioma cell lines (39) or in TSC-deficient fibroblasts (40) was also shown to reduce cell death that may be related to autophagy induction and related cytoprotection. Activation of mTOR, which forms the catalytic core of multiprotein complexes mTORC1 and mTORC2, is modulated by Raptor and Rictor, respectively. To determine the relative contributions of mTORC1 and mTORC2 inhibition to the cytoprotective effect of AZD8055, we suppressed the expression of Raptor or Rictor that are essential components of mTORC1 and mTORC2, respectively. As expected, suppression of Raptor or Rictor was shown to inhibit mTORC1 and mTORC2 substrates, respectively. However, we found that suppression of mTOR, but not Raptor or Rictor, was able to inhibit p-ULK1 Ser757 indicating the central role of mTOR kinase in the direct regulation of autophagy initiation. Specifically, suppression of mTOR but not Raptor or Rictor increased LC3 conversion and conferred protection against 5-FU-induced DNA damage and apoptosis, suggesting that AZD8055-induced autophagy and cytoprotection result from direct inhibition of mTOR kinase activity, although a potential contribution by downstream targets of mTOR cannot be excluded. In this regard, S6K is a downstream target of mTORC1 that was potently inhibited by AZD8055 in this study and has been shown to negatively regulate autophagy (41).
In conclusion, we demonstrate that AZD8055 treatment at clinically relevant doses induces pro-survival autophagy that can antagonize DNA damage and cell death induction by cytotoxic anti-cancer drugs with the potential to reduce treatment efficacy. Induction of autophagy by AZD8055 was associated with dephosphorylation of ULK1 kinase at Ser757 which is known to regulate the initiation of autophagy. Furthermore, the ability of AZD8055 to antagonize chemotherapy-induced cell death involves p62 down-regulation since p62 knockdown conferred chemoresistance, whereas ectopic expression of wild-type, but not mutant, p62 was shown to enhance chemotherapy-induced apoptosis that was abolished by AZD8055 treatment. These novel findings suggest important mechanisms by which AZD8055 can induce autophagy and antagonize cell death which may have important implications in vivo for cancer therapy. Taken together, our findings provide a mechanism by which AZD8055-induced autophagy can regulate chemosensitivity in human cancer cells.
Acknowledgments
We thank David T. Asuzu and Michael R. Bardsley (laboratory of Dr. T. Ordog, Mayo Clinic, Rochester, MN) for assistance with the immunofluorescence experiments and Dr. N. Kang (Mayo Clinic) for confocal microscopy imaging.
This work was supported, in whole or in part, by NCI, National Institutes of Health Grant 1 K05 CA142885-01 (Senior Scientist Award, to F. A. S.), and the Mayo Cancer Center core Grant CA15083.
- mTOR
- mammalian target of rapamycin
- PI3K
- phosphoinositide 3-kinase
- CQ
- chloroquine
- 5-FU
- 5-fluorouracil.
REFERENCES
- 1. Gulhati P., Cai Q., Li J., Liu J., Rychahou P. G., Qiu S., Lee E. Y., Silva S. R., Bowen K. A., Gao T., Evers B. M. (2009) Clin. Cancer Res. 15, 7207–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gulhati P., Bowen K. A., Liu J., Stevens P. D., Rychahou P. G., Chen M., Lee E. Y., Weiss H. L., O'Connor K. L., Gao T., Evers B. M. (2011) Cancer Res. 71, 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindsley J. E., Rutter J. (2004) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139, 543–559 [DOI] [PubMed] [Google Scholar]
- 4. Guertin D. A., Sabatini D. M. (2007) Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 5. Long X., Lin Y., Ortiz-Vega S., Busch S., Avruch J. (2007) J. Biol. Chem. 282, 18542–18551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaw R. J., Kosmatka M., Bardeesy N., Hurley R. L., Witters L. A., DePinho R. A., Cantley L. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Proud C. G. (2009) Biochem. Soc. Trans. 37, 227–231 [DOI] [PubMed] [Google Scholar]
- 8. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 9. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 10. Lee C. H., Inoki K., Guan K. L. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 443–467 [DOI] [PubMed] [Google Scholar]
- 11. Meric-Bernstam F., Gonzalez-Angulo A. M. (2009) J. Clin. Oncol. 27, 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. (2006) Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chresta C. M., Davies B. R., Hickson I., Harding T., Cosulich S., Critchlow S. E., Vincent J. P., Ellston R., Jones D., Sini P., James D., Howard Z., Dudley P., Hughes G., Smith L., Maguire S., Hummersone M., Malagu K., Menear K., Jenkins R., Jacobsen M., Smith G. C., Guichard S., Pass M. (2010) Cancer Res. 70, 288–298 [DOI] [PubMed] [Google Scholar]
- 14. Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. (2010) FEBS Lett. 584, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sudarsanam S., Johnson D. E. (2010) Curr. Opin. Drug Discov. Devel. 13, 31–40 [PubMed] [Google Scholar]
- 16. Chang Y. Y., Juhász G., Goraksha-Hicks P., Arsham A. M., Mallin D. R., Muller L. K., Neufeld T. P. (2009) Biochem. Soc. Trans. 37, 232–236 [DOI] [PubMed] [Google Scholar]
- 17. Kim J., Kundu M., Viollet B., Guan K. L. (2011) Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. (2009) Autophagy 5, 973–979 [DOI] [PubMed] [Google Scholar]
- 19. Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. (2008) Biochimie 90, 313–323 [DOI] [PubMed] [Google Scholar]
- 20. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White E., DiPaola R. S. (2009) Clin. Cancer Res. 15, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kroemer G., Levine B. (2008) Nat. Rev. Mol. Cell Biol. 9, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjorkoy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 25. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 26. Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 28. Huang S., Sinicrope F. A. (2008) Cancer Res. 68, 2944–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang S., Sinicrope F. A. (2010) Autophagy 6, 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang S., Sinicrope F. A. (2010) Mol. Cancer Ther. 9, 742–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donaldson K. M., Li W., Ching K. A., Batalov S., Tsai C. C., Joazeiro C. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8892–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toyoshima H., Hunter T. (1994) Cell 78, 67–74 [DOI] [PubMed] [Google Scholar]
- 34. Polyak K., Kato J. Y., Solomon M. J., Sherr C. J., Massague J., Roberts J. M., Koff A. (1994) Genes Dev. 8, 9–22 [DOI] [PubMed] [Google Scholar]
- 35. Wilkes G. (2003) Clin. J. Oncol. Nursing 7, 353–356 [DOI] [PubMed] [Google Scholar]
- 36. Copp J., Manning G., Hunter T. (2009) Cancer Res. 69, 1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carayol N., Vakana E., Sassano A., Kaur S., Goussetis D. J., Glaser H., Druker B. J., Donato N. J., Altman J. K., Barr S., Platanias L. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12469–12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan J. A., Ullman E., Dou Z., Zong W. X. (2011) Mol. Cell Biol. 31, 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ronellenfitsch M. W., Brucker D. P., Burger M. C., Wolking S., Tritschler F., Rieger J., Wick W., Weller M., Steinbach J. P. (2009) Brain 132, 1509–1522 [DOI] [PubMed] [Google Scholar]
- 40. Lee C. H., Inoki K., Karbowniczek M., Petroulakis E., Sonenberg N., Henske E. P., Guan K. L. (2007) EMBO J. 26, 4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blommaart E. F., Luiken J. J., Blommaart P. J., van Woerkom G. M., Meijer A. J. (1995) J. Biol. Chem. 270, 2320–2326 [DOI] [PubMed] [Google Scholar]