Background: PRC is a transcriptional coactivator involved in respiratory chain expression and cell growth.
Results: PRC protein levels are induced in response to various forms of metabolic stress leading to the activation of a program of inflammatory gene expression.
Conclusion: PRC can function as a sensor of metabolic stress.
Significance: Elucidating the molecular basis of chronic inflammatory responses may contribute to our understanding of cancer and degenerative diseases.
Keywords: Coactivator Transcription, Gene Expression, Inflammation, Metabolic Regulation, Signaling, Stress Response
Abstract
PGC-1-related coactivator (PRC) is a growth-regulated transcriptional cofactor that activates many nuclear genes specifying mitochondrial respiratory function. Stable PRC silencing in U2OS cells results in a complex phenotype typical of mitochondrial dysfunction including abundant abnormal mitochondria, reduced respiratory subunit expression, diminished respiratory enzymes and ATP levels, and elevated lactate production. The PRC response to metabolic stress was investigated by subjecting cells to metabolic insults including treatment with the uncoupler carbonyl cyanide 3-chlorophenylhydrazone (CCCP), expression of a dominant negative allele of nuclear respiratory factor 1 (NRF-1), and glucose deprivation. These treatments led to constitutively elevated PRC protein levels, a departure from its normal transient expression upon the initiation of cell growth. A microarray screen identified 45 genes that require PRC for their induction by CCCP. A subset of these genes specific to inflammation and cell stress was also induced by dominant negative NRF-1 and by glucose deprivation, suggesting that diverse metabolic insults converge on the same PRC-dependent inflammatory program. The PRC-dependent inflammatory response was inhibited by N-acetylcysteine, suggesting that PRC may contribute to the inflammatory microenvironment linked to oxidant signaling. The induction of this PRC-dependent program may be an early event in adaptations linked to cancer and degenerative diseases.
Introduction
The transcriptional control of respiratory chain expression in mammalian systems is largely directed by a relatively small number of transcription factors that serve as targets for the PGC-12 family of regulated coactivators (1, 2). Initially, the nuclear respiratory factors NRF-1 and NRF-2 were identified through their interactions with cytochrome c, and cytochrome oxidase promoters and were subsequently associated with many nuclear genes that specify mitochondrial respiratory function (1, 2). Subsequently, several other nuclear transcription factors have been linked to mitochondrial biogenesis. Most notably, the orphan nuclear receptor ERRα along with GABP (GA binding protein) (NRF-2) can regulate nuclear genes specifying mitochondrial energy metabolism (3–5).
Members of the PGC-1 coactivator family provide important links between these transcription factors and the physiological signals controlling cellular functions related to energy metabolism. PGC-1α, the founding member of the family, is described as an activator of the program of adaptive thermogenesis in brown fat (6) and is found to control mitochondrial biogenesis, in part, through its specific interaction with NRF-1 and its activation of NRF target genes (7). PGC-1α is also a potent activator of mitochondrial biogenesis through ERRα (4, 5), consistent with its proposed role in orchestrating complex programs of gene expression through the integration of diverse transcription factor targets. PGC-1β, a close relative of PGC-1α, although not associated with adaptive thermogenesis, is similar to PGC-1α in driving NRF-1 target gene expression (8, 9) and NRF-1 and ERRα-dependent mitochondrial biogenesis (10). The importance of these cofactors has been highlighted by the finding that a PGC-1α/β double knock-out mouse is deficient in the post-natal maturation of mitochondria in both the heart and brown fat (11).
A third member of the PGC-1 family was designated PRC (PGC-1-related coactivator) (12). PRC shares sequence motifs with the other family members including an amino-terminal activation domain, an LXXLL coactivator signature, a central proline-rich region, and a carboxyl-terminal RS domain and RNA recognition motif. Importantly, PRC engages in a direct interaction with transcription factors implicated in respiratory chain expression including NRF-1, CREB (cAMP-response element-binding protein), and ERRα (13). In addition, PRC interacts indirectly with the NRF-2β subunit through host cell factor, a large chromatin-associated coactivator that binds a number of transcription factors and histone modifying activities (14). Like PGC-1α and β, PRC can trans-activate a number of NRF-dependent nuclear genes that are required for mitochondrial respiratory function, including those encoding cytochrome c, 5-aminolevulinate synthase, Tfam, and TFB1M, and TFB2M (12, 15).
PRC differs from the other PGC-1 family members in that its expression pattern is characteristic of regulators of the cell growth program. PRC mRNA and protein are elevated in proliferating cells compared with those that are growth-arrested as a result of serum withdrawal or contact inhibition. PRC mRNA also has a relatively short half-life and is abundantly up-regulated upon serum treatment of quiescent cells in the absence of de novo protein synthesis (12, 13). These properties define the immediate early genes that are expressed early in the cell growth program and the members of which encode chemokines, growth factors, proto-oncogenes, serine-threonine kinases, enzymes of nucleic acid metabolism, and transcription factors among others (16). Serum induction of PRC accompanies a respiratory gene expression profile that is similar to that observed in response to elevated levels of PGC-1α (15, 17). As expected for a cell growth regulator, PRC silencing in two independent lentiviral transductants results in a diminished growth rate on glucose. One transductant, exhibiting nearly complete PRC silencing, also had a severely diminished growth rate under conditions that require mitochondrial respiration. This was accompanied by abundant abnormal mitochondria with defective cristae, reduced expression of respiratory subunits and complexes, and decreased oligomycin-sensitive ATP production (18). Similar phenotypic features have been associated with the loss of function of a number of key nucleus-encoded factors that are essential for mitochondrial respiratory function including Tfam (19), Mterf3 (20), and Tfb1m (21). These observations are consistent with the conclusion that PRC can function as a positive regulator of the mitochondrial respiratory apparatus.
Here, we investigated the response of PRC to a number of metabolic insults including chemical uncoupling, dominant negative inhibition of nuclear respiratory genes, and glucose deprivation. All of these result in a constitutive up-regulation of PRC protein levels leading to a PRC-dependent induction of a set of genes involved in inflammation and cell growth. We postulate that this novel retrograde pathway may represent an early response to diverse metabolic stresses leading to the activation of a chronic inflammatory program.
EXPERIMENTAL PROCEDURES
Cell Culture
U2OS and HEK293 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Invitrogen). Three lentiviral shRNA U2OS cell transductants designated as control shRNA, PRC shRNA1, and PRC shRNA4, as described previously (18), were grown in the same medium with the inclusion of blasticidin to maintain selection. For treatment with uncoupler, log phase cells were plated at a density of 1 × 106 cells/10-cm dish, grown for 12 h, and then treated with either 40 μm carbonyl cyanide 3-chlorophenylhydrazone (CCCP); Sigma) in DMSO or 1 mm 2,4-dinitrophenol (Sigma) in methanol for various times. Vehicle controls were treated with 40 μl of either DMSO or MeOH as appropriate.
Transfections
Transient transfections of U2OS cells were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were plated at a density of 170,000 cells/35-mm-diameter well in 6-well culture dishes and transfected with 1 μg of hCOX17/pGL3Basic reporter and 50 ng of pRL-null control vector (Promega) containing a SpeI and NheI promoter deletion. Trans-activations by PRC were carried out by co-transfecting 2 μg of PRC/pSV-Sport expressing full-length PRC or the pSV-Sport control as described previously (15).
Expression of dominant negative NRF-1 was performed by transfecting pSG5/dnNRF-1 (7) into U2OS cells. This plasmid has a carboxyl-terminal deletion between NRF-1 residues 305 and 503 containing the transcription activation domain (22). Cells were plated at a density of 1 × 106 cells/10-cm-diameter plate, grown for 2 days, and transfected with 15 μg of either empty pSG5 expression vector or pSG5/dnNRF-1 using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's protocol. After 6 h the cells were washed twice with Dulbecco's phosphate-buffered saline (Invitrogen) and grown for an additional 18 h in fresh medium. Cells were harvested for the preparation of whole cell extract and RNA as described (18).
Plasmids
hCOX17/pGL3Basic was constructed by cloning an Acc65I/HindIII PCR product (sense 5′-AAAAAAGGTACCGTTACGAGATCTCTCAGTTGTC-3′ and antisense 5′-AAAAAAAAGCTTGCCGATTCGTCCGCAGTCACTTC-3′ primers) containing 262 nucleotides of the human COX17 proximal promoter along with 26 nucleotides of 5′-untranslated region into Acc65I/HindIII-digested pGL3Basic (Promega). Site-directed mutagenesis of the NRF-1 recognition sites within the promoter was performed by PCR utilizing hCOX17/pGL3 as a template as described (15). Pairs of internal overlapping oligonucleotides with the desired base changes along with flanking hCOX17/pGL3 primers were used to generate mutations. The mutagenized promoter fragments were subcloned as Acc65I/HindIII fragments into pGL3Basic and their sequences verified. Sense (S) and antisense (AS) mutagenesis primers with mutated nucleotides underlined were as follows: hCOX17/NRF-1mutA(S), GCCTCCTTGATCATGCATTGAAGG; hCOX17/NRF-1mutA(AS), CCTTCAATGCATGATCAAGGAGGC; hCOX17/NRF-1mutB(S), GCCTCTTCTGCATACGATCTCCTT; and hCOX17/NRF-1mutB(AS), AAGGAGATCGTATGCAGAAGAGGC. The construction of FL PRC/pSV Sport (12) and pSG5/dnNRF-1 (22) have been described.
Immunoblotting
Whole cell lysates were prepared in Nonidet P-40 lysis buffer as described previously (12). Extracts were subjected to denaturing gel electrophoresis and the proteins transferred to nitrocellulose membranes (Schleicher & Schuell) as described (18). Primary antibodies were: rabbit anti-PRC-(400–467) and rabbit anti-PRC-(1047–1379) (13), used interchangeably; rabbit anti-Sp1 (Santa Cruz Biotechnology); mouse anti-c-Myc (Roche Applied Science); rabbit anti-NRF-1 (23); rabbit anti-NRF2α and anti-NRF2β (14); and rabbit anti-Tfam (a gift from David A. Clayton, Howard Hughes Medical Institute). The relative levels of the five human OXPHOS complexes were determined using the MitoProfile human total OXPHOS complexes detection kit (MitoSciences) as described (18).
Quantitative Real Time PCR
Total RNA was purified using TRIzol reagent (Invitrogen) and quantitative real time PCR carried out as described (18). The sequences of new primers designed for this study are shown in supplemental Table 1S.
Lactate Assay
U2OS WT and the lentiviral transductants expressing the control shRNA or PRC shRNA1 (18) were grown in DMEM for 24 h followed by treatment of either 40 μl of DMSO in 10 ml of medium (vehicle) or 40 μm CCCP for 16 h. The concentration of l-lactate released to the culture medium was measured using a lactate assay kit (Biomedical Research Service Center, State University of New York at Buffalo).
Complex I Enzyme Activity Assay
U2OS cells and the lentiviral transductants expressing the control shRNA or PRC shRNA1 (18) were treated with CCCP as described above and assayed for the activity of human complex I (NADH-ubiquinone oxidoreductase) using a complex I enzyme activity assay kit (MitoSciences) according to the manufacturer's protocol. Samples were processed and the relative activities determined as described (18).
Microarray Analysis
Total RNA was isolated from control shRNA and PRC shRNA1 transductants that had been untreated or treated with CCCP for 16 h as described above. Microarray analysis was performed on quadruplicate samples using an Illumina human HT12 bead chip as described (18).
RESULTS
Constitutively High PRC Protein Expression in Response to Respiratory Chain Uncouplers
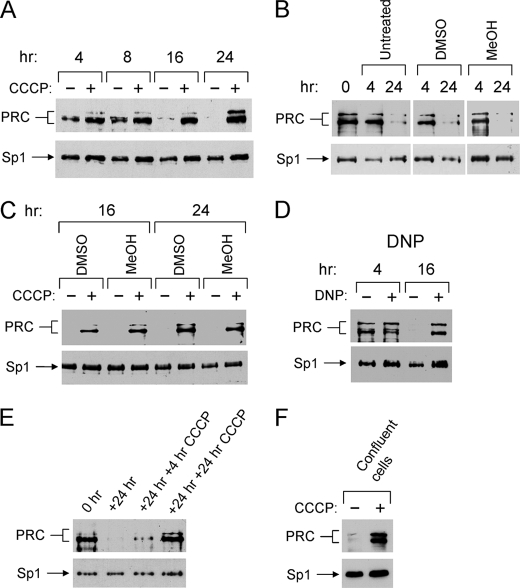
To test the hypothesis that PRC might be under retrograde regulation in response to metabolic stress, human U2OS cells were treated with the uncoupler CCCP. Cells were plated and subjected to treatment with 40 μm CCCP for various times. This concentration was compatible with maintaining cell viability throughout the course of the experiment. As shown in Fig. 1A, PRC protein levels drop precipitously in the untreated cells at about 24–36 h following the initial plating. This is consistent with the fact that PRC is an immediate early gene product that is transiently expressed when cells begin to proliferate (12, 13). In contrast, increased PRC levels were observed within 4 h of CCCP treatment and remain constitutively elevated throughout the 24 h time course (Fig. 1A). The decline in PRC levels in the untreated cells was not the result of an inhibitory effect of the vehicle control. As shown in Fig. 1B, untreated cells behaved identically to those treated with DMSO or MeOH in the amounts used in the treated samples. The identical PRC response to CCCP was also observed with either DMSO or MeOH as vehicle, arguing that the increased PRC levels are CCCP-dependent and not the result of the solvent used (Fig. 1C). Similar results were obtained when 2,4-dinitrophenol was used as an uncoupler, demonstrating that the PRC response was to respiratory chain uncoupling and not to some off-target effect of CCCP (Fig. 1D).
FIGURE 1.
Induction of PRC by respiratory chain uncouplers. A, human log phase U2OS cells were plated and 12 h later (T = 0 h) were either vehicle-treated (−) or treated with CCCP (+) for the indicated times. Total cell extracts from subconfluent cells were subjected to immunoblotting using affinity-purified rabbit anti-PRC or rabbit anti-Sp1 antibodies. B, U2OS cells were plated, and 12 h later PRC protein levels were measured by immunoblotting, as described in A, for the indicated times. To test the effects of solvents used for the dissolution of CCCP, cells were either untreated (−) or treated (+) for the indicated times with either DMSO or methanol. C, the 16- and 24-h time points for CCCP treatment as measured in A were compared using either DMSO or methanol as solvents for the dissolution of CCCP. D, cells were plated as described in A and treated with either vehicle (−) or dinitrophenol (DNP +) for the indicated times. E, cells were plated and 12 h later were harvested for protein extracts (T = 0 h). Cells were grown for an additional 24 h (T = 24 h) and then either untreated or treated for 4 or 24 h with CCCP. F, cells were grown to confluence and either untreated or treated with CCCP for 16 h. B–F, PRC and the Sp1 control were detected by immunoblotting as described in A.
Because CCCP was present throughout the time course of the experiment, it was of interest to determine whether CCCP was acting to maintain the initial levels of PRC or whether it could induce PRC de novo in cells where it had normally declined. CCCP was therefore added to cultures following the normal decline in PRC (Fig. 1E) or to confluent cells where PRC levels were low (Fig. 1F). In both cases, CCCP treatment resulted in a substantial induction of PRC. Thus, PRC was markedly up-regulated in response to uncoupling agents that are known to dissipate the electrochemical proton gradient required for ATP synthesis.
Diminished Respiratory Chain Expression and Function in Response to CCCP Induction of PRC
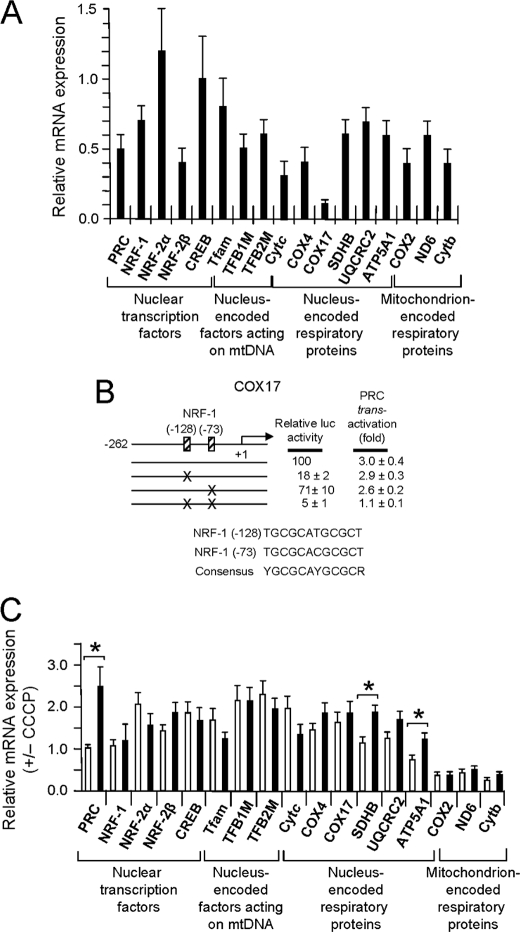
One explanation for the constitutive expression of PRC in response to uncoupling is that PRC is part of a retrograde response to mitochondrial stress. Because PRC silencing leads to severe mitochondrial dysfunction (18), one might expect that the up-regulation of PRC would enhance mitochondrial function in response to uncoupling. This possibility was tested by measuring the effect of CCCP on the expression of a battery of both regulatory and structural genes required for respiratory chain expression and function. PRC dependence was assessed by comparing the gene expression profiles in control lentiviral transductants that express PRC (control shRNA) to those where PRC is silenced (PRC shRNA1). As demonstrated previously (18), the PRC shRNA1 transductant exhibits diminished expression of many nuclear and mitochondrial genes required for the transcriptional expression of respiratory chain constituents (Fig. 2A). This transductant displayed a 50% reduction of PRC mRNA expression but nearly complete loss of PRC protein (Ref. 18 and Fig. 3B). The down-regulated genes include those for nucleus-encoded mitochondrial transcription factors (Tfam and TFB2M) as well as nucleus-encoded (Cytc, COX4, SDHB, UQCRC2, and ATP5A1) and mitochondrion-encoded (COX2, ND6, and Cytb) respiratory subunits. Of particular note is COX17, a putative cytochrome oxidase assembly factor in which expression is greatly diminished upon PRC silencing. Cloning and analysis of the human COX17 promoter region showed a nearly complete dependence on tandem NRF-1 recognition sites for both promoter activity and trans-activation by PRC (Fig. 2B). This is consistent with gene arrays showing that both mitochondrial import and respiratory chain assembly factors are diminished in the PRC shRNA1 transductant (2, 18).
FIGURE 2.
Expression of regulatory and structural genes required for the biogenesis of the mitochondrial respiratory apparatus. A, the expression of genes from the categories indicated below the x axis included those for nuclear transcription factors (PRC, NRF-1, NRF-2α, NRF-2β, and CREB), nucleus-encoded factors acting in mitochondria (Tfam, TFB1M, and TFB2M), and nucleus-encoded (COX4, COX17, SDHB, UQCRC2, and ATP5A1) and mitochondrion-encoded (COX2, ND6, and Cytb) respiratory subunits. Relative steady-state mRNA levels from the PRC shRNA1 and control shRNA transductants were normalized to 18 S rRNA as an internal control; values are expressed relative to the shRNA control, which was assigned a value of 1. Values are the averages ± S.E. for at least three separate determinations. B, dependence of the human COX17 promoter on NRF-1 recognition sites for both promoter activity and trans-activation by PRC. The 288-nucleotide proximal promoter from the human COX17 gene was cloned into a luciferase reporter plasmid, and the NRF-1 consensus sites were mutated either individually or in combination by site-directed mutagenesis as described under “Experimental Procedures.” Relative promoter activities were measured following transfection of U2OS cells either in the presence or absence of a PRC expression vector. C, the CCCP induction of genes listed under A was measured in both control shRNA (open bars) and PRC shRNA1 (closed bars) transductants. Values were normalized to 18 S rRNA as an internal control and expressed relative to untreated cells which were assigned a value of 1. A significant difference in CCCP induction between the two transductants is indicated by an asterisk denoting a p value of <0.05. Values are the averages ± S.E. for at least three separate determinations.
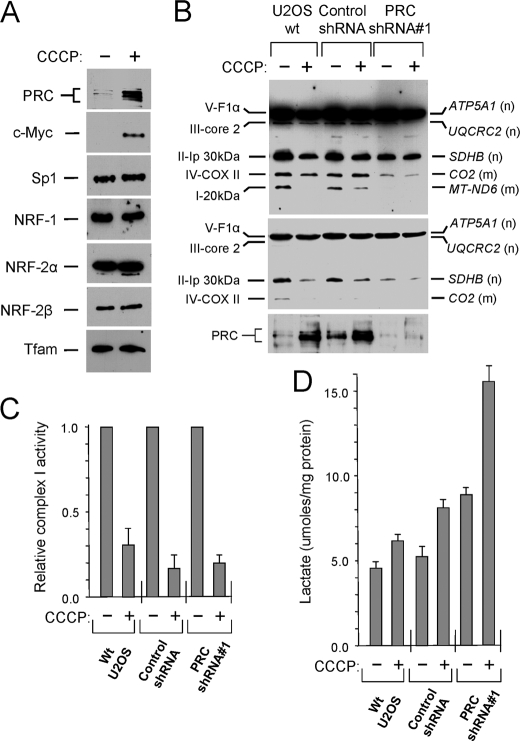
FIGURE 3.
Effect of CCCP treatment on mitochondrial respiratory chain expression and function. A, U2OS cells were allowed to grow for 24 h and then either untreated (−) or treated (+) with CCCP for 16 h. The effect of CCCP treatment on the steady-state levels of key transcription factors associated with the biogenesis of mitochondria and the expression of the respiratory chain was measured by immunoblotting. B, wild-type U2OS cells, along with the control shRNA and PRC shRNA1 lentiviral transductants, were plated and treated with CCCP as in A. The indicated subunits from each of the five respiratory complexes were detected by immunoblotting using a mixture of mouse monoclonal antibodies directed against each subunit. Subunit designations for the respective complexes (I–V) are indicated at the left, and gene names along with their nuclear (n) or mitochondrial (m) assignment are indicated at the right. The middle panel is a lower exposure of the upper panel, and the bottom panel shows PRC expression. C, the activity of respiratory complex I was measured in cell extracts using a dipstick assay (MitoSciences) as described under “Experimental Procedures.” Membranes were scanned for densitometric analysis, and the relative enzyme activity normalized to the untreated control is expressed as the averages ± S.E. for at least three separate determinations. D, lactate in the culture medium was measured enzymatically using an NADH-coupled reaction.
Surprisingly, there was little or no significant difference between the control shRNA and the PRC shRNA1 transductants in the response of this gene battery to CCCP treatment (Fig. 2C). Although several genes were induced significantly in the PRC shRNA1 transductant compared with the control (PRC, SDHB, and ATP5A1), the majority exhibited no PRC-dependent difference in CCCP induction. Even those that were induced in the PRC shRNA1 transductant were not induced beyond control levels because they were initially expressed at reduced levels in this transductant (Fig. 2A). Most notable are the mitochondrion-encoded respiratory subunits (COX2, ND6, and Cytb) in which expression in response to CCCP is greatly decreased.
The effects on mRNA expression were investigated at the protein level by measuring the steady-state amounts of several regulatory factors that are known to affect respiratory chain expression (Fig. 3A). Although PRC and c-Myc were markedly elevated in response to CCCP, the other regulatory factors controlling both nuclear (NRF-1, NRF-2α, and NRF-2β) and mitochondrial (Tfam)α respiratory genes remained unchanged. The steady-state expression of respiratory protein subunits from all five respiratory complexes was also determined (Fig. 3B). In both wild-type cells and the shRNA control, where PRC expression was induced by CCCP treatment, respiratory subunit expression was reduced, particularly for ATP5A1, SDHB, COXII, and ND6. For the latter two, this corresponds to the mRNA levels that were also diminished by CCCP treatment (Fig. 2C). As shown previously, the PRC shRNA1 transductant exhibited reduced expression of several respiratory subunits, which was not diminished further by CCCP. The observed reductions in respiratory subunit expression were also reflected in respiratory function. Complex I activity was decreased by CCCP in all three cell lines (Fig. 3C) and lactate production was increased (Fig. 3D), particularly in the PRC shRNA1 transductant, where mitochondrial structure and function was already severely compromised by PRC silencing. Decreased respiratory enzyme activity and elevated lactate levels are consistent with a respiratory chain deficiency.
Identification of PRC-dependent Inflammatory and Growth-regulatory Genes Up-regulated by CCCP-induced Metabolic Stress
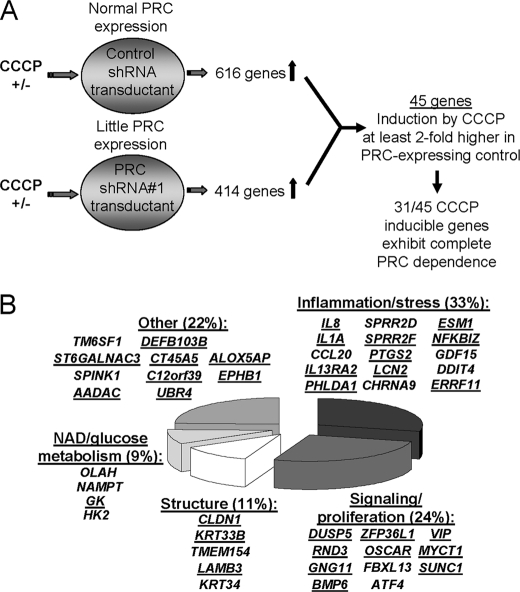
Loss of PRC function through stable gene silencing results in defects in both the respiratory apparatus and mitochondrial structure (18). However, as shown here, PRC induction by the respiratory uncoupler CCCP, rather than enhancing the expression and function of the mitochondrial respiratory chain, had a negative effect on both. These results suggest that the constitutive increase in PRC expression in response to mitochondrial stress may elicit a biological response that is unrelated to its role in maintaining respiratory function. As outlined in Fig. 4A, the target genes associated with this retrograde induction of PRC were investigated by treating both control shRNA and PRC shRNA1 transductants with or without CCCP and comparing the differential effects on global gene expression between the two transductant lines using a gene array.
FIGURE 4.
Microarray screen for PRC-dependent, CCCP-inducible genes. A, the strategy for identifying genes that depend upon PRC for their CCCP inducibility is outlined. Lentiviral transductants having either normal PRC expression (control shRNA) or severely reduced PRC expression (PRC shRNA1) were either untreated (−) or treated (+) with CCCP for 16 h in quadruplicate plates. RNA was isolated and subjected to microarray analysis, and for each transductant the -fold CCCP induction was normalized to the untreated control. Of the subset of genes showing a differential response to CCCP between the two transductants, 45 genes had a response to CCCP that was at least 2-fold greater in the shRNA control than in the PRC shRNA1 transductant. The quantitative basis for selecting these genes is shown in Table 1. Of these 45 genes, 31 showed no significant induction by CCCP in the PRC shRNA1 transductant, consistent with complete PRC dependence. B, genes identified in the screen were assigned to the indicated functional categories. Underlined are 31 genes that show complete PRC dependence for their response to CCCP. The quantitative data are in Table 1.
The results of the screen revealed a set of 45 genes in which induction by CCCP in the control shRNA transductant was at least 2-fold greater than that observed in the PRC shRNA1 transductant (Table 1 and Fig. 4A). Of these 45 PRC-dependent CCCP-inducible genes, 31 showed no significant response to CCCP in the shRNA1 transductant, indicating that their induction by CCCP in the control was completely dependent upon PRC (Table 1). The genes were assigned to several broad functional categories (Table 1 and Fig. 4B). The largest of these categories contained genes involved in inflammation/stress (33%) followed by genes associated with signaling and proliferation (24%), cell structure (11%), and NAD and glucose metabolism (9%), with the remainder not unified by any particular function (22%). It is notable that the screen did not identify any PRC-dependent CCCP-inducible genes involved in respiratory chain expression or mitochondrial biogenesis. This is consistent with the data presented in Figs. 2 and 3 showing respiratory genes as either reduced or unchanged along with diminished mitochondrial respiratory subunit expression and function.
TABLE 1.
List of genes in which induction by CCCP is PRC-dependent
| Gene symbol | Gene name | Control shRNA +/− CCCPa |
PRC shRNA#1 +/− CCCPb |
Control/sh1e | ||
|---|---|---|---|---|---|---|
| -Fold inductionc,d | Adjusted p value | -Fold inductionc,d | Adjusted p value | |||
| Inflammation/stress | ||||||
| SPRR2D | Small proline-rich protein 2D | 43.3 (1206.6) | 1.36E-15 | 1.6 (34.2) | 6.80E-03 | 27.1 (35.3) |
| IL8 | Interleukin 8 | 6.5 (2.7) | 2.65E-09 | −2.0 (0.8) | 1.32E-03 | 13.0 (3.4) |
| SPRR2F | Small proline-rich protein 2F | 9.0 (890.7) | 3.10E-15 | 1.1 (17.8) | 5.83E-01 | 9.0 (50.0) |
| IL1A | Interleukin 1α | 5.7 (170.0) | 2.30E-13 | 1.1 (9.5) | 4.62E-01 | 5.7 (17.9) |
| CCL20 | Chemokine (C-C motif) ligand 20 | 10.0 (29.1) | 5.80E-14 | 2.9 (5.6) | 4.57E-08 | 3.5 (5.2) |
| CHRNA9 | Cholinergic receptor, nicotinic, α9 | 3.5 (41.6) | 4.95E-09 | 1.4 (3.3) | 1.46E-02 | 3.5 (12.6) |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 3.1 | 1.88E-12 | 1.1 | 4.87E-01 | 3.2 |
| LCN2 | Lipocalin 2 | 3.1 (9.5) | 2.09E-06 | 1.2 (4.6) | 5.15E-01 | 3.1 (2.1) |
| PHLDA1 | Pleckstrin homology-like domain, family A, member 1 | 3.0 | 4.67E-03 | 3.0 | ||
| ESM1 | Endothelial cell-specific molecule 1 | 2.9 (23.1) | 4.01E-04 | 1.2 (5.3) | 6.03E-01 | 2.9 (4.4) |
| NFKBIZ | Nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor, ζ | 2.4 | 9.34E-06 | 1.1 | 7.08E-01 | 2.4 |
| ERRFI1 | ERBB receptor feedback inhibitor 1 | 2.4 (4.1) | 6.95E-06 | −1.0 (1.2) | 9.25E-01 | 2.4 (3.4) |
| IL13RA2 | Interleukin 13 receptor, α2 | 2.3 | 9.94E-11 | 2.3 | ||
| GDF15 | Growth differentiation factor 15 | 15.5 | 5.67E-14 | 7.4 | 6.53E-11 | 2.1 |
| DDIT4 | DNA damage-inducible transcript 4 | 8.0 | 2.01E-09 | 4.0 | 1.51E-06 | 2.0 |
| Signaling/proliferation | ||||||
| DUSP5 | Dual specificity phosphatase 5 | 5.9 | 3.36E-10 | 5.9 | ||
| RND3 | Rho family GTPase 3 | 3.5 | 8.46E-06 | 1.6 | 6.11E-02 | 3.5 |
| GNG11 | Guanine nucleotide-binding protein (G protein), γ 11 | 3.0 (5.0) | 5.08E-03 | 2.0 (2.5) | 9.37E-02 | 3.0 (2.0) |
| BMP6 | Bone morphogenetic protein 6 | 2.7 (2.9) | 1.72E-07 | −1.1 (0.8) | 6.80E-01 | 2.7 (3.6) |
| ZFP36L1 | Zinc finger protein 36, C3H type-like 1 | 2.4 | 2.07E-04 | 2.5 | ||
| OSCAR | Osteoclast-associated, immunoglobulin-like receptor | 2.3 | 2.57E-08 | 1.1 | 4.02E-01 | 2.3 |
| FBXL13 | F-box and leucine-rich repeat protein 13 | 3.6 | 6.31E-12 | 1.6 | 3.69E-05 | 2.3 |
| VIP | Vasoactive intestinal peptide | 2.2 | 2.73E-07 | 1.1 | 6.08E-01 | 2.3 |
| MYCT1 | Myc target 1 | 2.2 | 1.61E-07 | 2.2 | ||
| SUNC1 | Sad1 and UNC84 domain-containing 1 | 3.6 | 1.48E-05 | 1.8 | 2.50E-02 | 2.0 |
| ATF4 | Activating transcription factor 4 (tax-responsive enhancer element B67) | 4.1 | 5.73E-08 | 2.1 | 4.10E-04 | 2.0 |
| NAD/glucose metabolism | ||||||
| OLAH | Oleoyl-ACP hydrolase | 4.9 | 1.31E-12 | 1.7 | 5.00E-05 | 2.9 |
| NAMPT | Nicotinamide phosphoribosyltransferase | 2.6 | 3.03E-08 | 1.7 | 2.52E-04 | 2.6 |
| GK | Glycerol kinase | 2.4 | 1.15E-07 | 1.2 | 1.91E-01 | 2.4 |
| HK2 | Hexokinase 2 | 3.7 | 6.15E-10 | 1.9 | 2.43E-05 | 2.0 |
| Cell structure | ||||||
| CLDN1 | Claudin 1 | 3.5 | 1.35E-07 | 1.4 | 4.74E-02 | 3.5 |
| KRT33B | Keratin 33B | 2.5 | 1.30E-08 | 2.5 | ||
| TMEM154 | Transmembrane protein 154 | 4.7 | 1.14E-09 | 2.0 | 1.60E-04 | 2.4 |
| LAMB3 | Laminin, β3 | 2.2 | 7.50E-05 | 2.2 | ||
| KRT34 | Keratin 34 | 4.9 | 1.10E-08 | 2.4 | 6.03E-05 | 2.0 |
| Other | ||||||
| ST6GALNAC3 | ST6 (α-N-acetyl-neuraminyl-2,3-β-galactosyl-1,3)-N-acetylgalactosaminide α-2,6-sialyltransferase 3 | 3.3 (6.0) | 4.88E-11 | 1.0 (0.9) | 9.40E-01 | 3.3 (6.7) |
| SPINK1 | Serine peptidase inhibitor, Kazal type 1 | 7.1 | 8.94E-09 | 2.4 | 5.15E-04 | 3.0 |
| AADAC | Arylacetamide deacetylase (esterase) | 2.8 (13.3) | 1.65E-08 | 1.1 (4.7) | 4.30E-01 | 2.9 (2.8) |
| TM6SF1 | Transmembrane 6 superfamily member 1 | 2.7 | 1.36E-11 | 1.2 | 2.54E-02 | 2.3 |
| CT45A5 | Cancer/testis antigen family 45, member A5 | 2.2 | 1.24E-07 | 1.2 | 8.25E-02 | 2.3 |
| C12orf39 | Chromosome 12, open reading frame 39 | 2.2 | 2.65E-09 | 1.2 | 5.77E-02 | 2.2 |
| UBR4 | Ubiquitin-protein ligase E3 component n-recognin 4 | 2.1 | 1.29E-02 | 2.1 | ||
| DEFB103B | Defensin, β 103B | 2.0 | 3.93E-03 | 2.0 | ||
| ALOX5AP | Arachidonate 5-lipoxygenase-activating protein | 2.0 | 1.33E-03 | 2.0 | ||
| EPHB1 | EPH receptor B1 | 2.0 | 9.14E-03 | 1.5 | 1.49E-01 | 2.0 |
| Tfam | Mitochondrial transcription factor A | (1.7) | (1.2) | (1.4) | ||
a Total RNA was isolated from CCCP-treated and untreated U2OS lentiviral transductants expressing the control shRNA. Fold induction values were derived by normalizing the expression of the indicated gene in CCCP-treated cells to that in the untreated cells (+/−).
b Total RNA was isolated from CCCP-treated and untreated U2OS lentiviral transductants expressing the PRC shRNA-1. Fold induction values were derived by normalizing the expression of the indicated gene in CCCP-treated cells to that in the untreated cells (+/−).
c Numbers are derived from microarray analysis using a Illumina HT12 gene chip.
d Numbers in parentheses are values derived from quantitative real time PCR.
e Numbers represent the ratio of the fold induction of the listed gene in the control shRNA transductant to that in the PRC shRNA-1 transductant derived from either microarray analysis or quantitative real time PCR (in parentheses).
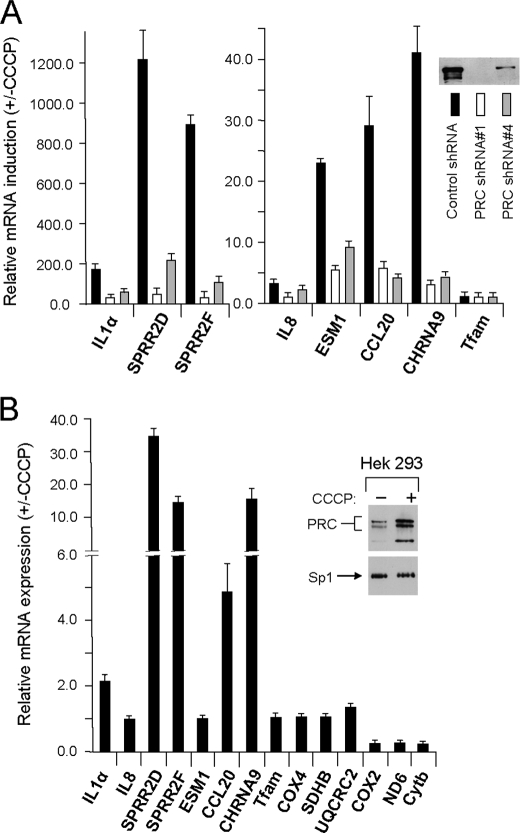
The most abundantly up-regulated category of inflammatory and cell stress genes included an inflammatory cytokine (IL-1α), chemokines (IL-8 and CCL20), the IL-13 receptor (IL13RA2), cytokine- and chemokine-regulated genes (ESM-1 and LCN2), several genes involved in mediating the inflammatory response (PTGS2, NFKBI, and GDF15), and several cell stress-inducible genes (SPRR2D, SPRR2F, and DDIT4). The microarray data were validated by measuring the expression, in both transductant lines, of a subset of the identified genes by quantitative real time PCR (Table 1). The results confirm both the induction of these genes by CCCP and their PRC dependence. In most cases, the differential expression between control and the PRC shRNA1 transductants was in good agreement with the microarray results. In instances where discrepancies occurred, the RT-PCR data usually showed a larger differential effect. Tfam, which was not identified in the array, was used as a negative control for the RT-PCR.
The PRC dependence of several of the genes was confirmed further by measuring their expression in a second lentiviral transductant cell line expressing PRC shRNA4. As shown previously (18) and in Fig. 5A, the PRC shRNA4 transductant, which was derived independently from a different shRNA designed to silence PRC expression, retains ∼15% of the control level of PRC. It also differs from the PRC shRNA1 transductant in having nearly normal mitochondrial structure and respiratory chain expression and function (18). Importantly, the CCCP induction of seven representative PRC-dependent inflammation/stress genes identified in the screen was also markedly reduced in this transductant relative to the control. In all but one case, CCL20, the level of CCCP induction was diminished marginally less than in the shRNA1 transductant, consistent with the less efficient silencing of PRC in the shRNA4 transductant. The negative control, Tfam, was not affected significantly. The results argue that despite the differences in mitochondrial phenotype between the transductants, it is the level of PRC expression that dictates the response of inflammatory/stress genes to CCCP-induced mitochondrial stress.
FIGURE 5.
Validation of a subset of genes assigned to the inflammation/stress category. A, PRC dependence of CCCP induction was validated for a subset of seven genes from the inflammation/stress category by measuring their response to CCCP in a second independent lentiviral transductant (PRC shRNA4) exhibiting ∼85% PRC silencing. The three transductant lines (control shRNA, black bars; PRC shRNA1, open bars; PRC shRNA4, gray bars) were treated for 16 h with CCCP as described under “Experimental Procedures,” and the -fold induction for each gene relative to the untreated control was determined by quantitative real time PCR. The insert shows the relative PRC expression in each transductant line. B, HEK293 cells were either untreated or treated with CCCP for 16 h, and PRC protein was assayed by immunoblotting (insert). The CCCP-dependent expression of the subset of seven inflammation/stress genes was compared with that of the respiratory genes described in the legend to Fig. 2 by quantitative real time PCR. Values are the averages ± S.E. for at least three separate determinations.
It is unlikely that PGC-1α or PGC-1β contributes to the observed changes in gene expression. There was no statistically significant difference in PGC-1α mRNA induction by CCCP in the negative control (5.6 ± 0.8-fold) compared with the PRC shRNA1 transductant (4.6 ± 0.5-fold). In addition, PGC-1β mRNA was induced about 2-fold in the shRNA1 transductant but not at all in the control, where the PRC-dependent genes were markedly induced. Thus, the expression of the other PGC-1 family members seemed not to correlate with the observed changes in PRC protein levels or PRC-dependent gene expression.
Multiple Metabolic Insults Converge on the Induction of the Same PRC-dependent Stress Program
It was of interest to determine whether the results obtained using CCCP in the U2OS cell line were restricted to these cells. To this end, HEK293 cells were treated or untreated with CCCP for 16 h in a manner identical to that performed with the U2OS cells. As shown in Fig. 5B, PRC protein levels were elevated significantly in the CCCP-treated cells relative to the untreated control. An assay of representative inflammatory genes identified by the microarray screen and of a battery of known respiratory genes revealed a differential pattern of expression very similar to that obtained in U2OS cells (Fig. 5B). With two exceptions, IL-8 and CCL20, the inflammatory genes were markedly up-regulated under conditions where the nucleus-encoded respiratory genes were unchanged and the mitochondrion-encoded respiratory genes were diminished. Although the magnitude of inflammatory gene induction was less in the HEK293 cells, the differential between the inflammatory and respiratory genes resembled that obtained with the U2OS cells. The inability of IL-8 and CCL20 to respond likely reflects cell-specific differences.
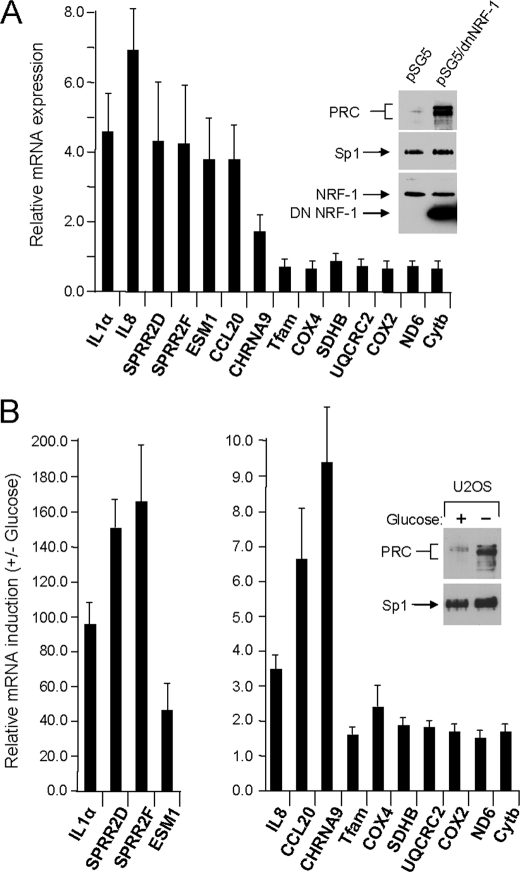
The response to metabolic stress was also examined using the pseudo-genetic approach of overexpressing dominant negative NRF-1. The dominant negative allele binds DNA but is lacking the trans-activation domain and thus acts as a competitive inhibitor of NRF-1 function. Expression of this allele has been shown to exert a negative effect on mitochondrial biogenesis (7). As shown in Fig. 6A, overexpression of dominant negative NRF-1 results in a marked induction of PRC protein under conditions where endogenous NRF-1 and the Sp1 control remain unchanged. The differential in PRC protein is similar to that obtained upon treatment with CCCP. This coincides with a significant induction of all seven of the representative PRC-dependent inflammatory genes. As expected, expression of the nuclear and mitochondrial respiratory genes was either unchanged or diminished. The induction of several of the inflammatory genes (SPRR2D, SPRR2F, CCL20, and CHRNA9) was less robust than with CCCP treatment, possibly reflecting a difference between interfering with nuclear gene expression compared with a more immediate effect induced by chemical uncoupling.
FIGURE 6.
Response of PRC-dependent inflammation/stress genes to diverse metabolic insults. A, a dominant negative allele of NRF-1 (pSG5/dnNRF-1) was expressed in U2OS cells, and the level of PRC protein relative to that in cells transfected with the empty vector control (pSG5) was measured by immunoblotting (insert). Sp1, endogenous NRF-1, and dnNRF-1 were assayed as controls. The expression of the subset of seven inflammation/stress genes was compared with that of the respiratory genes described in the legend to Fig. 2 by quantitative real time PCR (filled bars). Results were normalized to the values obtained from the pSG5-transfected control. B, U2OS cells were plated and after 24 h were fed with glucose-containing (+) or glucose-depleted (−) medium. After 16 h PRC protein was assayed by immunoblotting (insert). The expression of the subset of seven inflammation/stress genes was compared with that of the respiratory genes described in the legend to Fig. 2 by quantitative real time PCR. Results were normalized to the values obtained from the glucose-fed cells. For both panels, values are the averages ± S.E. for at least three separate determinations.
If the induction of the inflammatory genes was truly a consequence of metabolic stress, one might expect that the program would be induced in response to nutrient deprivation. This was tested by starving the cells for glucose for a period equal to the CCCP treatment. As shown in Fig. 6B, glucose deprivation resulted in a similar induction of PRC protein accompanied by a robust induction of the seven representative PRC-dependent inflammatory/stress genes. However, in contrast to CCCP, a modest increase (∼2-fold) was observed for both the nuclear and mitochondrial respiratory genes as well. This may reflect differences in the signaling pathways controlling respiratory gene expression in response to depletion of the major glycolytic substrate. Nevertheless, several different means of inducing metabolic stress all appear to converge on the same or similar PRC-dependent program of inflammatory gene expression.
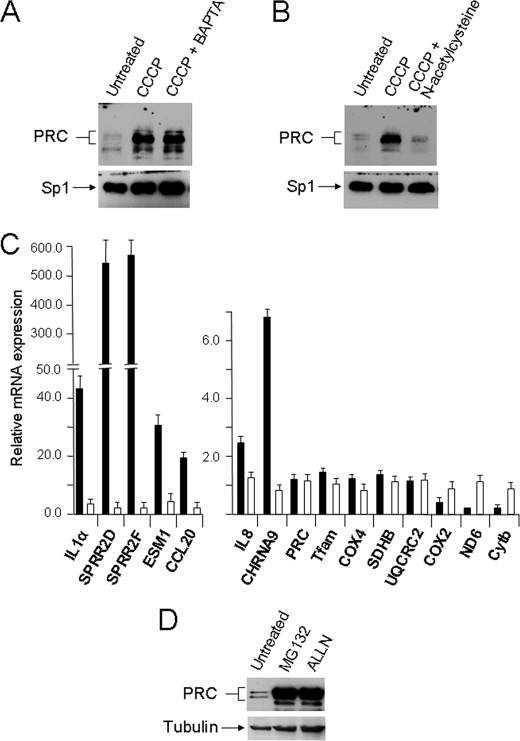
PRC Protein Induction by CCCP Is Inhibited by Antioxidant
Both calcium and reactive oxygen species (ROS) are thought to play an important role in mitochondrial signaling (24, 25). Mitochondrial ROS in particular have been associated with tumor progression (26), and several of the PRC-dependent genes identified here including IL-1α, IL-8, and PTGS2 contribute to the tumor microenvironment in many forms of human cancer (27). The potential role of calcium signaling in the PRC-dependent inflammatory/stress program was tested by treating cells with BAPTA, a potent calcium chelator, prior to induction by CCCP. The results shown in Fig. 7A indicate no significant change in PRC induction associated with BAPTA treatment. Similarly, pretreatment with the anitioxidant N-acetylcysteine was tested for its effect on PRC induction by uncoupler. In contrast to BAPTA, antioxidant treatment resulted in a nearly complete inhibition of PRC induction by CCCP under conditions in which the Sp1 control was unaffected (Fig. 7B). Importantly, expression of the subset of PRC-dependent inflammatory/stress genes is also abrogated by the antioxidant (Fig. 7C). The genes in this subset were induced from severalfold to several hundredfold, and in each case induction was dramatically diminished by N-acetylcysteine pretreatment. This result contrasts sharply with the nucleus-encoded respiratory genes (Tfam, COX4, SDHB, and UQCRC2) where expression was not induced by CCCP, and the N-acetylcysteine had no significant effect. Interestingly, the mitochondrion-encoded respiratory subunit mRNAs (COX2, ND6, and Cytb), which were markedly reduced by CCCP treatment, were restored to normal levels by the antioxidant, indicating that their down-regulation is part of the PRC-dependent program.
FIGURE 7.
Effects of BAPTA and N-acetylcysteine on PRC induction by uncoupler. A, U2OS cells were plated and after 24 h were either untreated, treated with 40 μm CCCP for 16 h (CCCP), or pretreated with 10 μm BAPTA for 1 h following treatment with 40 μm CCCP for 16 h (CCCP + BAPTA). B, U2OS cells were plated and after 24 h were either untreated, treated with 40 μm CCCP for 16 h, or pretreated with 5 mm N-acetylcysteine for 1 h following treatment with 40 μm CCCP for 16 h (CCCP + N-acetylcysteine). Immunoblots in A and B were probed with rabbit anti-PRC or rabbit anti-Sp1. C, RNA was isolated from cells subjected to the treatments described in B, and the expression of the subset of PRC-dependent inflammatory genes was compared with that of the nucleus- and mitochondrion-encoded respiratory genes using quantitative real time PCR. Results obtained with the CCCP-treated (filled bars) or CCCP + N-acetylcysteine-treated (open bars) cells were normalized to those obtained from vehicle-treated controls. Values are the averages ± S.E. for at least three separate determinations. D, U2OS cells were either untreated or treated with 5 μm MG132 or 20 μm N-acetyl-leucinyl-leucinyl-norleucinal (ALLN) for 8 h.
Finally, in contrast to the substantial CCCP induction of PRC protein and its nearly complete inhibition by N-acetylcysteine (Fig. 7B), PRC transcript was unaffected by either treatment (Fig. 7C). Thus, the PRC-dependent program is most likely regulated post-transcriptionally, possibly at the level of protein turnover. This was tested by treating cells with the proteasome inhibitors MG132 and N-acetyl-leucinyl-leucinyl-norleucinal (ALLN). As shown in Fig. 7D, proteasome inhibition with either agent results in a massive increase in PRC protein under conditions in which the tubulin control is only moderately affected. Similar results were obtained using Balb3T3 cells (not shown). Thus, like many cell growth regulators, PRC is likely regulated by protein turnover via the ubiquitin proteasome system. It remains to be determined whether this system is the target for mediating the stress response on PRC expression.
DISCUSSION
PRC is tightly regulated in U2OS cells upon serum stimulation (13), and as confirmed here (Fig. 1), it is dramatically down-regulated following the initiation of cell growth. Here, the results established that PRC is also induced as part of an early response to diverse metabolic insults. This results in the induction of a PRC-dependent inflammatory/stress program in which expression is sensitive to the potent antioxidant N-acetylcysteine. Thus, PRC may serve as a metabolic sensor that can help orchestrate alternative gene expression programs. Under normal conditions, PRC functions as an immediate early gene product in promoting the initiation of cell growth. In this capacity, PRC expression is transient, with the protein markedly down-regulated upon achievement of steady-state growth. The early expression of PRC in response to serum growth factors precedes the induction of a number of genes required for respiratory chain expression and mitochondrial biogenesis (15, 17), and the induction of these genes coincides with increased PRC occupancy of the cytochrome c promoter region (13). However, under conditions of metabolic stress, PRC protein levels depart from their usual transient expression pattern and become constitutively elevated. In this capacity, rather than enhance the expression of respiratory genes, PRC promotes a proinflammatory program. This PRC-dependent inflammatory/stress response may be an early event in triggering adaptive changes in cell structure, metabolism, and growth that enhance survival under adverse conditions.
The screen described here identified 45 genes in which induction in response to a chemical uncoupler was highly PRC-dependent. Thirty-one of these were not induced significantly in a shRNA transductant where PRC expression was efficiently silenced, whereas the remaining 14 showed a markedly reduced response to the uncoupler. The PRC dependence was confirmed by measuring the induction of a subset of genes in a second, independent lentiviral transductant that retained ∼15% of control PRC protein levels (18). In all cases, the response to uncoupler was diminished in this transductant as well. The two transductants were constructed independently from two different PRC shRNAs and differ in the severity of their respiratory phenotypes (18), arguing that it is the PRC levels that determine the response to uncoupler. Genes from this program are also elevated by other metabolic insults including the pseudo-genetic inhibition of respiratory genes by dominant negative NRF-1 or by glucose deprivation. This is suggestive of a novel function for PRC as a sensor of metabolic stress. Many of the inflammatory and growth regulatory genes in this PRC-dependent program are associated with chronic inflammation.
Chronic inflammation is a contributing factor in many human diseases including neurodegenerative disease (28), metabolic diseases (29), and cancer (27, 30). In the latter, inflammatory cells and gene products (cytokines and chemokines) contribute to an inflammatory microenvironment present in most tumors (31). This environment is thought to derive from extrinsic factors or from intrinsic alterations in gene expression. For example, the activation of Ras oncogenes can stimulate cytokine and chemokine expression (32, 33). Such events lead to the activation of key transcription factors NF-κB, STAT3, and HIF1α and the induction of cyclooxygenase 2, an important enzyme in prostaglandin production leading to the inflammatory response. These pathways have a number of important consequences for tumor promotion including the recruitment of inflammatory cells, enhanced cell proliferation, angiogenesis, invasion and metastasis, inhibition of adaptive immunity, and altered response to hormones and chemotherapy (27, 30). Inflammatory pathways are also associated with nutrient sensing through the NLR family, pyrin domain-containing 3 (NLRP3) inflammasome (34). This multiprotein complex facilitates the processing and secretion of proinflammatory cytokines IL-1β and IL-18 in response to pathogens, environmental insults, and metabolites. Recent evidence suggests that reactive oxygen species from damaged mitochondria activate the inflammasome, possibly explaining the association of mitochondrial dysfunction with inflammatory disease (34).
Interestingly, the PRC-dependent inflammatory/stress genes identified here are also activated in other physiological contexts involving chronic inflammation. Some are postulated to promote cell survival under adverse conditions by enhancing cell growth and migration, by conferring resistance to apoptosis, and by stimulating angiogenesis (27). IL-1α, an inflammatory cytokine, IL-8, a chemokine that promotes angiogenesis and tumor promotion, and CCL20, a chemokine that attracts monocytes and dendritic cells, have all been associated with the inflammatory microenvironment in human cancers (27, 31, 35). In particular, IL-8 is a significant regulatory factor promoting angiogenesis, proliferation, and cell migration (36). Notably, PTGS2 encoding cyclooxygenase 2 (COX-2) is induced by proinflammatory cytokines and promotes tumor progression and angiogenesis in many human cancers where it is up-regulated (37). Additional PRC-dependent stress response genes (NFKBIZ, LCN2, ESM-1, ATF4, CHRNA9, GDF15, and IL13RA2) are either cytokine-regulated or associated with inflammation. For example, CHRNA9 encodes a nicotinic receptor that has been linked to neuroinflammation (37).
Also notable are the genes encoding members of the small proline -rich proteins SPRR2D and SPRR2F, which are among the most highly induced PRC-dependent genes. These are part of an 11-member family of small proline rich proteins localized to a 170-kilobase region of human chromosome 1q21 (38, 39). Although associated with the cornified envelope and induced during squamous differentiation, these proteins are present in nonsquamous cell lines and tissues. Only 5% of the total SPRR1 protein is present in the cornified envelope, implying that it has other functions. Interestingly, SPRR1 is elevated by the UV-induced cytokines IL-1 and IL-3 and by proximity to inflammatory cells, providing a link between the SPR proteins and inflammatory mediators (40). Certain family members are induced by tumor promoters as well as by UV irradiation and other DNA-damaging agents. They are also up-regulated upon withdrawal from the cell cycle (38). The two SPRR2 genes identified here are induced by UV irradiation, and the SPRR2 genes in general have been linked to the residual ability for terminal differentiation in cultured cancer cells (41). Thus, in addition to providing barrier function, the SPR protein family represents a class of stress-induced proteins associated with the response to DNA damage and to the cessation of cell proliferation.
Three PRC-dependent genes (PTGS2, PHLDA1, and NAMPT) are thought to be anti-apoptotic. Interestingly, NAMPT, encoding the rate-limiting enzyme in NAD biosynthesis, also functions as a cytokine to regulate cell growth, migration, and apoptosis (42). Thus, resistance to apoptosis may be part of the early stress response. PTGS2 and ESM-1 promote angiogenesis and invasiveness. The screen also revealed several tumor suppressors and growth regulators (LCN2, ATF4, GDF15, OSCAR, FBXL13, DUSP5, ZFP36L1, and RND3). Two genes, HK2 and GK, are modulators of glucose homeostasis. Hexokinase-2, the product of HK2, is localized to the outer mitochondrial membrane and promotes the increased rate of glycolysis observed in many cancer cells (43). As a contributor to the “Warburg effect,” the aerobic glycolysis that is a hallmark of tumor metabolism (44), hexokinase-2 helps generate carbon intermediates that drive malignant growth (45). Interestingly, the up-regulation of c-Myc in tumor cells, in collaboration with HIF-1, can induce hexokinase-2 expression (46). Although a c-Myc transcript was not present in the set of PRC-dependent stress genes, c-Myc protein was elevated in response to uncoupling by CCCP (Fig. 3A). PRC may serve as a cofactor in the c-Myc-dependent induction of hexokinase-2 and possibly other c-Myc target genes possibly accounting for the elevated expression of PRC protein in thyroid oncocytomas (47). Preliminary results suggest that PRC protein levels are markedly elevated in a number of solid tumors (not shown).
Genes involved in the expression of the respiratory chain or mitochondrial biogenesis were notably absent. In fact, the mitochondrial respiratory subunit mRNAs were diminished by CCCP treatment, consistent with the increase in lactate production and the diminished respiratory chain subunits and complex I activity, despite the fact that numerous genes in this category have reduced expression upon PRC silencing in the same cell line (2, 18). PRC control over nuclear respiratory genes can occur through the direct PRC trans-activation of respiratory gene promoters (12–15). As evidenced here for the COX17 gene (Fig. 2B), this trans-activation requires functional NRF-1 sites, or in the case of other related genes, such as TFB1M and TFB2M, a combination of NRF-1 and NRF-2 sites localized near the transcription initiation site (15). The proximal promoters of several of the inflammatory genes identified here (IL-8, 660 bp; SPRR2D, 783 bp; SPRR2F, 765 bp; and ESM1, 853 bp) were tested for PRC trans-activation, and we observed no direct activation of these proximal promoters by PRC. Thus, the stress response elements may not be proximal to the initiation site or PRC may exert indirect control on multiple genes through one or a few regulatory genes that are direct PRC targets. Alternatively, because the PGC-1 coactivators may couple transcription and RNA processing (48), PRC may act post-transcriptionally to modulate RNA levels.
The widely used antioxidant N-acetylcysteine completely blocked CCCP induction of PRC under conditions in which the calcium chelator BAPTA had no discernable effect. The antioxidant inhibition of PRC protein levels coincided with the diminished induction of the PRC-dependent inflammatory/stress response under conditions in which nucleus-encoded respiratory genes were unaffected. This provides independent evidence that PRC directs the inflammatory/stress program and argues for a high degree of specificity. The remarkable sensitivity to the redox state of the cell along with enhanced PRC protein expression in several human tumors suggests that ROS signaling may contribute to the PRC-dependent program. Cancer cells produce high levels of ROS, but it is not clear whether ROS signaling is a direct contributor to oncogenic transformation (26). Recent studies indicate that mitochondrial ROS can direct the expression of proinflammatory cytokines through inflammasome-dependent and -independent pathways (25). The increase in PRC levels in response to a proteasome inhibitor suggests that redox signaling to PRC may occur through the ubiquitin proteasome system.
Supplementary Material
Acknowledgments
We thank the Center for Genetic Medicine at Northwestern University Medical School, for processing the Illumina Beadchip and the BioInformatics Core for statistical analysis of the data generated from the microarray.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 32525-29 from NIGMS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1S.
- PGC
- peroxisome proliferator-activated receptor γ coactivator
- PRC
- PGC-1-related coactivator
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- DMSO
- dimethyl sulfoxide
- NRF
- nuclear respiratory factor
- Tfam
- mitochondrial transcription factor A
- TFB
- mitochondrial transcription factor B
- ALLN
- N-acetyl-leucinyl-leucinyl-norleucinal
- ROS
- reactive oxygen species
- BAPTA
- 1,2-bis-(o-aminophenoxy)ethane-N,N,N,N,-tetraacetic acid.
REFERENCES
- 1. Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 2. Scarpulla R. C. (2011) Biochim. Biophys. Acta 1813, 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huss J. M., Kopp R. P., Kelly D. P. (2002) J. Biol. Chem. 277, 40265–40274 [DOI] [PubMed] [Google Scholar]
- 4. Mootha V. K., Handschin C., Arlow D., Xie X. H., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Willy P. J., Schulman I. G., Heyman R. A., Lander E. S., Spiegelman B. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schreiber S. N., Emter R., Hock M. B., Knutti D., Cardenas J., Podvinec M., Oakeley E. J., Kralli A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 7. Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 8. Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B. M. (2002) J. Biol. Chem. 277, 1645–1648 [DOI] [PubMed] [Google Scholar]
- 9. Lin J., Tarr P. T., Yang R., Rhee J., Puigserver P., Newgard C. B., Spiegelman B. M. (2003) J. Biol. Chem. 278, 30843–30848 [DOI] [PubMed] [Google Scholar]
- 10. Shao D., Liu Y., Liu X., Zhu L., Cui Y., Cui A., Qiao A., Kong X., Liu Y., Chen Q., Gupta N., Fang F., Chang Y. (2010) Mitochondrion 10, 516–527 [DOI] [PubMed] [Google Scholar]
- 11. Lai L., Leone T. C., Zechner C., Schaeffer P. J., Kelly S. M., Flanagan D. P., Medeiros D. M., Kovacs A., Kelly D. P. (2008) Genes Dev. 22, 1948–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersson U., Scarpulla R. C. (2001) Mol. Cell. Biol. 21, 3738–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vercauteren K., Pasko R. A., Gleyzer N., Marino V. M., Scarpulla R. C. (2006) Mol. Cell. Biol. 26, 7409–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vercauteren K., Gleyzer N., Scarpulla R. C. (2008) J. Biol. Chem. 283, 12102–12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gleyzer N., Vercauteren K., Scarpulla R. C. (2005) Mol. Cell. Biol. 25, 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winkles J. A. (1998) Prog. Nucleic Acids Res. Mol. Biol. 58, 41–78 [DOI] [PubMed] [Google Scholar]
- 17. Scarpulla R. C. (2008) Ann. N.Y. Acad. Sci. 1147, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vercauteren K., Gleyzer N., Scarpulla R. C. (2009) J. Biol. Chem. 284, 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Wilhelmsson H., Graff C., Li H., Oldfors A., Rustin P., Brüning J. C., Kahn C. R., Clayton D. A., Barsh G. S., Thorén P., Larsson N. G. (1999) Nat. Genet. 21, 133–137 [DOI] [PubMed] [Google Scholar]
- 20. Park C. B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Falkenberg M., Gustafsson C. M., Larsson N. G. (2007) Cell 130, 273–285 [DOI] [PubMed] [Google Scholar]
- 21. Metodiev M. D., Lesko N., Park C. B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M., Larsson N. G. (2009) Cell Metab. 9, 386–397 [DOI] [PubMed] [Google Scholar]
- 22. Virbasius C. A., Virbasius J. V., Scarpulla R. C. (1993) Genes Dev. 7, 2431–2445 [DOI] [PubMed] [Google Scholar]
- 23. Gugneja S., Scarpulla R. C. (1997) J. Biol. Chem. 272, 18732–18739 [DOI] [PubMed] [Google Scholar]
- 24. Mammucari C., Rizzuto R. (2010) Mech. Ageing Dev. 131, 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naik E., Dixit V. M. (2011) J. Exp. Med. 208, 417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schumacker P. T. (2006) Cancer Cell 10, 175–176 [DOI] [PubMed] [Google Scholar]
- 27. Mantovani A., Garlanda C., Allavena P. (2010) Ann. Med. 42, 161–170 [DOI] [PubMed] [Google Scholar]
- 28. Lee Y. J., Han S. B., Nam S. Y., Oh K. W., Hong J. T. (2010) Arch. Pharm. Res. 33, 1539–1556 [DOI] [PubMed] [Google Scholar]
- 29. Baker R. G., Hayden M. S., Ghosh S. (2011) Cell Metab. 13, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grivennikov S. I., Karin M. (2010) Curr. Opin. Genet. Dev. 20, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grivennikov S. I., Greten F. R., Karin M. (2010) Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sparmann A., Bar-Sagi D. (2004) Cancer Cell 6, 447–458 [DOI] [PubMed] [Google Scholar]
- 33. Orjalo A. V., Bhaumik D., Gengler B. K., Scott G. K., Campisi J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17031–17036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tschopp J., Schroder K. (2010) Nat. Rev. Immunol. 10, 210–215 [DOI] [PubMed] [Google Scholar]
- 35. Borrello M. G., Degl'Innocenti D., Pierotti M. A. (2008) Cancer Lett. 267, 262–270 [DOI] [PubMed] [Google Scholar]
- 36. Waugh D. J., Wilson C. (2008) Clin. Cancer Res. 14, 6735–6741 [DOI] [PubMed] [Google Scholar]
- 37. Keibel A., Singh V., Sharma M. C. (2009) Curr. Pharm. Des. 15, 1949–1955 [DOI] [PubMed] [Google Scholar]
- 38. Tesfaigzi J., Carlson D. M. (1999) Cell Biochem. Biophys. 30, 243–265 [DOI] [PubMed] [Google Scholar]
- 39. Cabral A., Voskamp P., Cleton-Jansen A. M., South A., Nizetic D., Backendorf C. (2001) J. Biol. Chem. 276, 19231–19237 [DOI] [PubMed] [Google Scholar]
- 40. Yaar M., Eller M. S., Bhawan J., Harkness D. D., DiBenedetto P. J., Gilchrest B. A. (1995) Exp. Cell Res. 217, 217–226 [DOI] [PubMed] [Google Scholar]
- 41. Lohman F. P., Medema J. K., Gibbs S., Ponec M., van de Putte P., Backendorf C. (1997) Exp. Cell Res. 231, 141–148 [DOI] [PubMed] [Google Scholar]
- 42. Bi T. Q., Che X. M. (2010) Cancer Biol. Ther. 10, 119–125 [DOI] [PubMed] [Google Scholar]
- 43. Mathupala S. P., Ko Y. H., Pedersen P. L. (2009) Semin. Cancer Biol. 19, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gatenby R. A., Gillies R. J. (2004) Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 45. Moreno-Sánchez R., Rodríguez-Enríquez S., Marín-Hernández A., Saavedra E. (2007) FEBS J. 274, 1393–1418 [DOI] [PubMed] [Google Scholar]
- 46. Podar K., Anderson K. C. (2010) Cell Cycle 9, 1722–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Savagner F., Mirebeau D., Jacques C., Guyetant S., Morgan C., Franc B., Reynier P., Malthièry Y. (2003) Biochem. Biophys. Res. Commun. 310, 779–784 [DOI] [PubMed] [Google Scholar]
- 48. Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B. M. (2000) Mol. Cell 6, 307–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.