Background: MID1, a regulator of PP2A, forms part of a messenger ribonucleoprotein complex.
Results: MID1 complex binds a purine-rich sequence motif in mRNAs and regulates their translation.
Conclusion: The translation of mRNAs bound by the MID1 complex is impaired in patients with Opitz syndrome.
Significance: This study shows how coordinated translation of specific mRNAs can be regulated and provides new strategies for therapies and biotechnology.
Keywords: mTOR, PP2A, RNA, Translation, Ubiquitin Ligase, MID1
Abstract
We have shown previously that the ubiquitin ligase MID1, mutations of which cause the midline malformation Opitz BBB/G syndrome (OS), serves as scaffold for a microtubule-associated protein complex that regulates protein phosphatase 2A (PP2A) activity in a ubiquitin-dependent manner. Here, we show that the MID1 protein complex associates with mRNAs via a purine-rich sequence motif called MIDAS (MID1 association sequence) and thereby increases stability and translational efficiency of these mRNAs. Strikingly, inclusion of multiple copies of the MIDAS motif into mammalian mRNAs increases production of the encoded proteins up to 20-fold. Mutated MID1, as found in OS patients, loses its influence on MIDAS-containing mRNAs, suggesting that the malformations in OS patients could be caused by failures in the regulation of cytoskeleton-bound protein translation. This is supported by the observation that the majority of mRNAs that carry MIDAS motifs is involved in developmental processes and/or energy homeostasis. Further analysis of one of the proteins encoded by a MIDAS-containing mRNA, namely PDPK-1 (3-phosphoinositide dependent protein kinase-1), which is an important regulator of mammalian target of rapamycin/PP2A signaling, showed that PDPK-1 protein synthesis is significantly reduced in cells from an OS patient compared with an age-matched control and can be rescued by functional MID1. Together, our data uncover a novel messenger ribonucleoprotein complex that regulates microtubule-associated protein translation. They suggest a novel mechanism underlying OS and point at an enormous potential of the MIDAS motif to increase the efficiency of biotechnological protein production in mammalian cells.
Introduction
Mutations in the RING finger protein MID1 cause the X-linked form of the Opitz BBB/G syndrome (OS),5 a malformation syndrome that is characterized by defective ventral midline development (1). MID1 associates with microtubules and interacts with the α4 protein, a regulatory subunit of PP2A (2). Its ubiquitin ligase activity regulates the modification and degradation of the microtubule-associated catalytic subunit of protein phosphatase 2A (PP2Ac) (3), an important component of the mTOR pathway. PP2A opposes mTOR kinase by dephosphorylating 4EBP1 and p70S6K, thereby down-regulating translation of mRNAs containing oligopyrimidine tracts at their 5′-transcription start sites (5′-terminal oligopyrimidine sequences; reviewed in Ref. 4), most of which code for components of the translational apparatus. In OS patients, MID1-mediated regulation of PP2Ac is disrupted, leading to accumulation of PP2A and to hypophosphorylation of its targets and resultant protein defects (3, 5). Additionally, we could very recently show that the increased levels of PP2A in cells of OS patients also control mTOR signaling by attenuating the formation of mTORC1 resulting in reduced p70S6K phosphorylation, cell size, and cap-dependent translation (6).
Previously, we found that MID1 together with the PP2A subunit α4 form the core of a large microtubule-bound multiprotein complex that associates with active polyribosomes and mRNAs without 5′-terminal oligopyrimidine sequences (7), suggesting a novel and 5′-terminal oligopyrimidine sequence-independent role of the MID1-α4 protein complex and mTOR/PP2A signaling in protein translation. We had furthermore discovered that association of RNA to the MID1-α4 protein complex is mediated by guanine-rich (G-rich) sequences. Here, we report that the MID1 complex associates with mRNAs via a sequence motif, called MIDAS (MID1 association sequence), which includes a purine-rich sequence motif with the ability to form a secondary structure. This association enhances the translation efficiency of these mRNAs severalfold. Our data suggest that MID1 targets specific mRNAs to the microtubule network and regulates their efficient translation, functions that would suggest loss of cytoskeleton-bound protein translation as a novel mechanism underlying the developmental defects observed in Opitz syndrome patients (8). Involvement of not only MID1 but also the α4 protein, which links PP2A with the mTOR kinase, suggests a PP2A/mTOR-dependent mechanism. Interestingly, PDPK-1, an important regulator of mTOR/PP2A signaling, is among the MIDAS-containing mRNAs, and we have found that PDPK-1 protein synthesis is significantly reduced in cells from an OS patient, which further potentiates mTOR dysfunction in MID1 mutation carriers.
Furthermore, our data point out a huge potential of the MIDAS sequence to be used to improve the production of proteins in eukaryotic cell systems for a wide range of applications in modern biotechnology, including research into the biological function of proteins, their production as biopharmaceuticals and diagnostic reagents, and the development of transgenic animals and plants. The typically low productivity of eukaryotic cell systems despite heavily optimized gene transcription in cells used for large scale protein production is still a major problem for many of these processes. We show here that, by targeting protein translation, inclusion of one or more MIDAS sequences into the 3′UTR of a gene of interest can significantly increase productivity of the respective proteins in eukaryotic cells and can also be used to fine-tune protein expression in eukaryotic cell systems.
EXPERIMENTAL PROCEDURES
Constructs
pGL3-CMV, pGL2-CMV, and pRL-CMV vectors were purchased from Promega. To create pEGFP-MIDAS, a 60-bp fragment of pGL3 containing the MIDAS motif was cloned between XhoI and HindIII in the pEGFP-C1 vector (Clontech). In vitro mutagenesis was performed using the QuikChange® site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions. To produce pGL3m-MIDAS and pGL3m-MIDASm, a 175-bp DNA-fragment, including the MIDAS or MIDASm motifs, was cloned into the XbaI restriction site of pGL3m. MID1 cDNA was excised with HindIII and SalI from pEGFP-C1 and ligated into pCMV-Tag2C (Stratagene). Mutant MID1 clones have been established using in vitro mutagenesis (Stratagene) according to mutations that have been identified in OS patients previously (9, 10). pEGFP-2×MIDAS was created by inserting annealed oligonucleotides with HindIII and EcoRI overhangs into the HindIII and EcoRI restriction sites of GFP-MIDAS. pEGFP-3×MIDAS was created by analogously inserting annealing oligonucleotides ES with EcoRI and SalI overhangs into the EcoRI and SalI restriction sites of pEGFP-2×MIDAS. To create GFPi-MIDAS, GFP together with the MIDAS motif was excised with NheI and EcoRI from GFP-MIDAS and cloned in the MCS of pIRES-DsRed-Express (Clontech). Sequence of all constructs was confirmed by direct sequencing. Primer sequences are summarized in supplemental Table 2.
G-quartet Extraction
Based on G-quartet sequences described in the literature (11, 12) combined with re-analysis of known target genes of fragile X mental retardation protein (11), a consensus sequence for the prediction of G-quartet structures was extracted (WGGN1–4WGGN1–4WGGN1–4WGG). Based on Bioperl regular expressions, an algorithm for the prediction of such mRNA structures was made. Following this algorithm, the luciferase genes in both vectors, pGL3 and pGL2, were scanned for the presence of putative G-quartets.
RNA-Protein Pulldown Assay
Firefly and Renilla cDNAs were amplified by PCR with primers, including the T7 promoter sequence from the different vectors. In vitro transcriptions were performed using the RiboMAXTM large scale RNA production system T7 (Promega), following the manufacturer's instructions with some modifications. Briefly, 2–5 μg of purified PCR product was transcribed in the presence of 0.75 mm biotin-16-uridine-5′-triphosphate (Roche Applied Science) and 5 mm UTP. Biotinylated RNAs were purified by phenol/chloroform extraction and kept in nuclease-free water after ethanol precipitation.
3 × 107 HeLa cells were lysed in TKM buffer (20 mm Tris, 50 or 75 mm KCl, 5 mm MgCl2) supplemented with proteinase inhibitors (complete mini, Roche Applied Science), RNase inhibitor (Promega), 1% Nonidet P-40, and 1 mm DTT on ice, using a 27-gauge needle. The lysate was cleared by centrifugation for 15 min at 12,000 × g.
3 μg of in vitro transcribed and biotinylated RNA were then incubated with 200 μg of these lysates in 500 μl of TKM buffer for 1 h at 4 °C and, subsequently, with 40 μl of 50% slurry of M-280 streptavidin-coated magnetic beads (Dynal). Beads were washed three times with TKM buffer for 10 min at 4 °C and boiled off in magic mix (15 mm Tris, 48% urea, 8.7% glycerol, 1% SDS) for 10 min at 95 °C. Bound proteins were analyzed on Western blots with anti-FLAG antibody (Stratagene).
RNA-protective Immunoprecipitation
4 × 106 HeLa cells were transfected with LipofectamineTM 2000 (Invitrogen) and 15 μg of a FLAG-MID1 construct (pCMV-2C-MID1) in 150-cm2 cell culture flasks according to the manufacturer's instructions. After 24 h, cells were lysed in TKM buffer (20 mm Tris, 100 mm KCl, 5 mm MgCl2) supplemented with proteinase inhibitors (complete mini, Roche Applied Science), RNase inhibitor (Promega), 1% Nonidet P-40, and DNase I (Roche Applied Science, 20 units/ml TKM buffer) on ice.
Immunoprecipitation was carried out using either 100 μl of anti-FLAG-M2-agarose (Sigma) or 100 μl of mouse IgG-agarose (Sigma). The co-precipitated RNA was purified by phenol/chloroform extraction followed by ethanol precipitation, and the pellet was resuspended in 10 μl of diethyl pyrocarbonate-treated H2O. cDNA was synthesized using SuperScript VILOTM (Invitrogen) and used for subsequent RT-PCR and quantitative real time PCR.
Cell Culture and Transfection
COS-7, HEK293T, and CHO cells were transfected with LipofectamineTM 2000 (Invitrogen) and HeLa cells with DreamFectTM (OZ Biosciences) and 1.5 μg of DNA in 6-well plates according to the manufacturer's instructions. For the luciferase assay, HeLa cells were seeded on 12-well plates and transfected with 200 ng of Firefly, 10 ng of Renilla, and 400 ng of MID1 construct.
siRNA Transfection
2 × 104 HeLa cells (12-well plate) or 1 × 105 HeLa cells (6-well plate) were transfected with OligofectamineTM reagent (Invitrogen) and siRNA oligonucleotides. 48 h after knockdown cells were transfected with DreamFectTM (OZ Biosciences) according to manufacturer's instructions. Sequences of siRNA oligonucleotide are summarized in supplemental Table 2.
Western Blot
50 μg of proteins were loaded on a 10% SDS-PAGE and incubated with specific anti-α4 (3), anti-MID1 (Sigma), anti-GFP (Roche Applied Science), anti-actin (Sigma), and anti-tubulin antibodies (AbD). For quantification, protein bands were scanned and quantified by densitometry using Aida Image Analyzer software version 3.28.
Luciferase Assay
24 h after transfection, HeLa cells were harvested in 1× luciferase passive lysis buffer (Promega). Firefly and Renilla luciferase activities were measured using a Centro LB 960 luminometer (Berthold Technologies) and the Dual-Luciferase assay system (Promega) according to the manufacturer's instructions. Firefly light units were normalized to Renilla light units.
In Vitro Transcription/Translation
Specific PCR products of the Firefly luciferase cDNAs were transcribed in vitro with the mMessage mMachine® T7 kit (Ambion). Capped transcripts were translated in vitro with the Flexi® rabbit reticulocytes lysates (Promega) according to the manufacturer's instructions. After 90 min, Firefly light units were measured using the luciferase assay reagent (Promega) in a luminometer.
FACS Analysis
24 h after transfection, cells were trypsinized and fixed with ethanol. FACS analysis was performed on a FACSCaliburTM (BD Biosciences). GFP was excited with a 488 nm laser, and emission was collected using a 530-nm emission filter. A total of 10,000 events/experiment were collected for each construct in three independent experiments. Transfection efficiencies with the different constructs were comparable.
Quantitative RT-PCR Analysis
RNA from 6-well dishes was isolated using Qiagen RNeasy kit. cDNA was synthesized using the TaqMan reverse transcription reagents kit (Applied Biosystems) with random primers. Primers for real time PCR analysis were designed with the Primer Express® software version 2.0. For real time PCR analysis, absolute quantification with standard curve using SYBR Green PCR master mix (Applied Biosystems) was performed on an Applied Biosystems ABI Prism 7900HT Sequence detection system equipped with SDS software version 2. Primer sequences used are summarized in supplemental Table 2.
Native Gel Electrophoresis or Anomalous Migration in Native Gels
Oligonucleotides containing the MIDAS core sequence (Fig. 5A) were synthesized using t-butylmethylsilyl phosphoramidite chemistry, deprotected in ammonia/methylamine (1:1) at 60 °C for 10 min, and evaporated to dryness. The t-butylmethylsilyl groups were removed by triethylamine trihydrofluoride (65 °C, 2.5 h), and the oligonucleotides were purified using Glen-Pak cartridges (Glen Research) followed by denaturing PAGE. Oligonucleotides were heated to 95 °C, cooled on ice, and analyzed on native 15% TBE polyacrylamide gels (29:1 polyacrylamide/bisacrylamide) in the presence or absence of 50 mm KCl at room temperature. Electrophoresis buffers were recirculated at >1 liter/h. Gels were run at 3 V/cm until the bromphenol blue marker dyes had migrated 17 cm and stained with SYBR Gold.
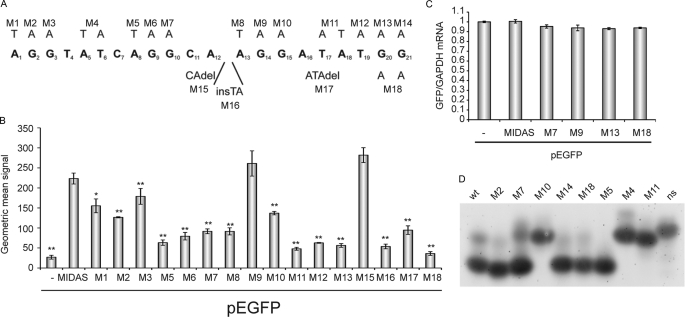
FIGURE 5.
MIDAS motif is a novel RNA protein-binding sequence with the ability to form hairpin structures. A, summary of MIDAS mutations inserted into pEFGP-MIDAS used. B, intensities of green fluorescence recorded from HeLa cells transfected with EGFP-MIDAS mutants, measured by FACS. Standard deviations are calculated from triplicates. t test (two-tailed, homoscedastic): *, p < 0.01; **, p < 0.001. C, quantification of GFP-mRNAs of selected pEGPF-MIDAS mutants after transfection into HeLa cells. GFP-mRNAs were quantified by real time PCR relative to GAPDH. Standard deviations were calculated from triplicates. D, native gel electrophoresis of RNA oligonucleotides carrying selected MIDAS mutations. Salt-free TBE buffer was used, and gels were stained with SYBR Gold.
Cycloheximide Half-life Experiment
150,000 OS fibroblast cells (253/99, control 220/28) per well (6-well) were seeded in DMEM the day before. 80 μg/ml cycloheximide was added, and cells were harvested at each time point (0, 1, 3, 8, 24, and 32 h). PDPK1 protein was detected on Western blot by using anti-PDPK1 (Santa Cruz Biotechnology, sc-17765), and the amount of PDPK1 protein was densitometrically analyzed using anti-GAPDH (Millipore, MAB374) as loading control.
Rescue Experiment and MID1 Overexpression in OS Fibroblast Cells
106 OS fibroblast cells (253/99) per 100-mm dish were seeded in 15 ml of DMEM with 10% FBS and without antibiotics the day before transfection. Either the empty pCMV-Tag2c vector or pCMV-Tag2C-MID1 was transfected using Lipofectamine 2000 (Invitrogen). For each dish, 24 μg of DNA were diluted into 1.5 ml of OptiMEM medium (Invitrogen). In another tube, each 60 μl of Lipofectamine 2000 reagent were diluted in 1.5 ml of OptiMEM. After 5 min of incubation at room temperature, the two solutions were combined, mixed gently, and incubated at room temperature for another 20 min to allow DNA-Lipofectamine 2000 complexes to form. The complexes were then added directly to each dish and mixed gently. After 6 h, complexes were removed from the cells, and fresh medium was added. 48 h after the addition of the DNA-Lipofectamine 2000 complexes, cells were harvested and analyzed by Western blot. MID1 protein was detected using anti-MID1 (ATLAS antibodies, HPA003715). PDPK-1 protein was detected using anti-PDPK-1 (Santa Cruz Biotechnology, sc-17765), and the amount of PDPK-1 protein was densitometrically analyzed using anti-GAPDH (Millipore, MAB374) as loading control.
RESULTS
MID1 Protein Complex Associates with a Specific Sequence Motif
The intriguing composition of the MID1 complex and its association with actively translating polyribosomes and G-rich RNAs prompted us to investigate possible functions of MID1 in the regulation of protein translation. To test if the MID1 complex modulates the efficiency of protein production from its associated mRNAs, we co-expressed either a FLAG-MID1wt construct, two different FLAG-MID1 constructs containing inactive MID1 (A130T, C145S), or an empty pCMV vector together with a Renilla luciferase vector and one or the other of two different Firefly luciferase vectors (Firefly pGL3 promoter-Firefly-wt- or Firefly pGL2 promoter-Firefly2-, respectively) in HeLa cells and analyzed their influence on the expression of Firefly luciferase using a Dual-Luciferase reporter assay. Both MID1 mutations had been identified in OS patients previously and are missense mutations either in the B-Box1 domain (A130T) or in the coiled-coil domain (C145S) and cause loss-of-function of the MID1 protein. Unexpectedly, we observed a >2-fold MID1-dependent induction of Firefly luciferase activity compared with that seen following co-expression with an empty pCMV vector or with either of two different pCMV constructs encoding inactive MID1 proteins, but only in the presence of the pGL3 (Firefly-wt) and not the pGL2 (Firefly2) vector (Fig. 1A). Comparison of the pGL2 and pGL3 vector sequences indicated that all but a few nucleotides within the transcribed sequence of the Firefly luciferase genes were identical (Fig. 1B). Four out of six changed nucleotides in pGL3 reside within a G-rich region comprising a putative protein-RNA-binding motif, which matches with a consensus sequence that we have extracted from genes containing so-called G-quartet structures (Fig. 1D). G-quartets have initially been described as unique structural arrangements intrinsic to guanine-rich DNA, which are able to bind proteins (reviewed in Ref. 13). Later on, similar structures have been discovered in protein-binding RNAs (11, 12, 14). In Firefly2, however, this motif is not present (Fig. 1B). Moreover, in an RNA-protein pulldown assay, we found that FLAG-MID1wt specifically associated with biotin-labeled luciferase mRNA in vitro-transcribed from the Firefly-wt vector but not to mRNAs transcribed from Firefly2 or Renilla (Fig. 1C). Specificity of the reaction was further verified by omission of RNA from the assay. From these data, we have concluded that, although the MID1 protein binds to the Firefly-wt mRNA thereby enhancing its luciferase activity, there is no binding between the MID1 protein and the Firefly2 mRNA and, consequently, no influence of MID1 on the Firefly2 luciferase activity.
FIGURE 1.
MIDAS motif and MID1 expression confer increased protein production from mRNAs that associate with the MID1-PP2A signaling complex. A, Firefly luciferase activity recorded from HeLa cells transfected with vectors either containing (Firefly-wt) or lacking (Firefly2) a functional MIDAS motif and co-transfected with empty pCMV vector (−), wild-type MID1 (wt), or mutated, inactive MID1 (A130T, C145S). Standard deviations were calculated from triplicates. B, sequence comparison of Firefly-wt and Firefly2 vectors. C, RNA-protein pulldown assay. Lysates from FLAG-MID1-overexpressing HeLa cells were incubated with biotinylated RNA transcribed in vitro from either Firefly-wt or Firefly2 vectors or from a Renilla vector without MIDAS motif (Renilla). Binding fractions and lysates were analyzed on a Western blot with an anti-FLAG antibody. D, consensus sequence that has been extracted from mRNAs containing G-quartet like sequence motifs. E, Firefly luciferase activity recorded from HeLa cells transfected solely with Firefly-wt vectors containing either a functional MIDAS motif (Firefly-wt) or a mutated MIDAS motif (Firefly-mut) within the open reading frame, and also co-expressing either empty pCMV vector (−), wild-type MID1 (wt), or mutated, dysfunctional MID1 (A130T, C145S). Standard deviations were calculated from triplicates. F, sequence comparison of the functional (Firefly-wt) and mutated (Firefly-mut) MIDAS motifs of the pGL3 vector. G, RNA-protein pulldown assay. Lysates from FLAG-MID1-overexpressing HeLa cells were incubated alone (No RNA) or with biotinylated RNA transcribed in vitro from a pGL3 vector containing either a functional MIDAS motif (Firefly-wt) or a mutated MIDAS motif (Firefly-mut) or from a Renilla vector lacking the MIDAS motif (Renilla). Binding fractions and lysate were analyzed on a Western blot with an anti-FLAG antibody. H, Firefly luciferase activity recorded from HeLa cells transfected with the Firefly-wt vector and either nonsilencing control siRNA oligonucleotides or α4 or MID1-specific oligonucleotides. Standard deviations were calculated from triplicates. I, real time PCR analysis of the Firefly mRNA relative to Renilla mRNA. All standard deviations were calculated from triplicates. J, knockdown efficiencies were controlled on a Western blot.
G-quartet or G-quartet-like nucleotide motifs are characterized by four guanine residues that are arranged in a planar conformation stabilized by hydrogen bonds. Two to four G quartets can be stacked (12–15). To determine whether the guanines within the motif in Firefly-wt that matches with the G-quartet consensus sequence are required for its ability to bind to the MID1 protein complex and to enhance pGL3-Firefly luciferase activity, one of the core guanines of the motif was mutated to adenine (Firefly-mut, Fig. 1F). This mutation is a silent mutation, and both the mutated and nonmutated codons are common in eukaryotic cells. We found that the mutated vector (Firefly-mut) failed to respond to the induction of luciferase activity caused by co-expression with wild-type MID1, as occurred with the wild-type pGL3 vector. In addition, basal Firefly luciferase activity was lower upon transfection with the mutated pGL3 vector, probably because of the loss of the positive regulatory influence of endogenous MID1 complex upon translation of the mutated mRNA (Fig. 1E). In line with these observations, the ability of the Firefly-mut mRNA to bind to FLAG-MID1 was significantly reduced as compared with that of Firefly-wt mRNA, as shown by RNA-protein pulldown assay (Fig. 1G). Again, Renilla mRNA and an experiment without RNA were used as background controls. Upon these experiments the identified sequence motif was called MIDAS.
MIDAS Effect Depends on MID1 and the α4 Protein, a Rapamycin/mTOR-sensitive Subunit of PP2A
We then verified that the translational efficiency of the RNA transcribed from the MIDAS-containing Firefly-wt vector depends on the MID1 protein. For that, we transfected HeLa with either nonsilencing control or MID1-specific siRNA oligonucleotides and analyzed them in a Dual-Luciferase assay system using the Firefly-wt and a co-transfected Renilla vector. As expected, luciferase activity was significantly reduced after MID1 knockdown with two different sets of oligonucleotides (Fig. 1H and supplemental Fig. 1). The MID1 protein had been shown to be coupled to the catalytic subunit of PP2A via the α4 protein (3), which is the rapamycin- and mTOR-sensitive subunit of PP2A (2), and connects PP2A and MID1 to the mTOR signaling cascade. α4 was originally used to isolate the MID1-mRNP by affinity chromatography (7), suggesting that α4 is an integral part of the MID1-mRNP core, and it had been demonstrated that α4 interacts with poly(A)-binding protein (16) further confirming its role in protein translation. To see if the effect on the translation of a MID1-associated mRNA is PP2A- and mTOR-dependent, effects of knocking down α4 on the activity of the Firefly-wt luciferase were tested. Similar to the knockdown of MID1 itself, knockdown of α4 caused a substantial reduction of Firefly activity produced from the Firefly-wt vector (Fig. 1H). No difference was detected on mRNA levels comparing Firefly relative to Renilla mRNA in all three cell types, transfected with nonsilencing or α4 or MID1-specific siRNA oligonucleotides (Fig. 1I) suggesting that the knockdowns primarily influence protein translation rather than transcription or mRNA stability. Knockdown efficiencies were controlled on Western blots (Fig. 1J).
MIDAS Sequence and Its Influence on Protein Translation Are Transferable to 3′UTR of Any Gene of Choice
To test the transferability of the MIDAS motif and its translation-enhancing properties from the open reading frame into the 3′-untranslated regions of mRNAs, we designed three different vector systems for usage in the following experiments: (i) functional MIDAS motif was cloned 3′ of the open reading frame of GFP in both the pEGFP vector (EGFP-MIDAS) and an pIRES vector (GFPi-MIDAS) and (ii) functional and a nonfunctional (MIDASm) MIDAS motifs were cloned 3′ of Firefly-mut in the pGL3 vector (Firefly-mut-MIDAS and Firefly-mut-MIDASm).
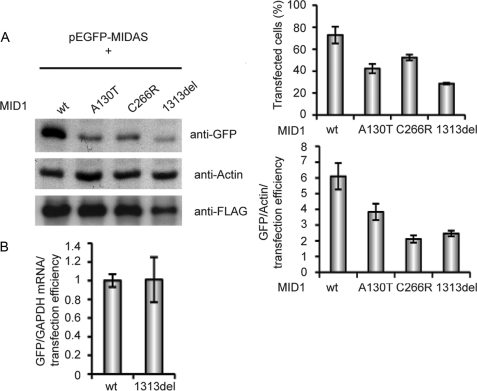
In Dual-Luciferase assays, we had seen that FLAG-MID1 carrying missense mutations as found in OS patients (A130T and C145S, Fig. 1, A and E) and that inactivate the MID1 protein have no influence on the Firefly luciferase activity expressed by Firefly-wt. In a next series of experiments, we co-expressed EGFP-MIDAS with either wild-type MID1 or different MID1 mutants that have been identified in OS patients. The A130T mutant had been analyzed in the previous luciferase experiments (Fig. 1, A and E) and served as negative control. Cell lysates were analyzed by Western blot with an anti-GFP antibody. Densitometric values were normalized to actin and to transfection efficiencies that had been determined by FACS. Results confirmed that an intact MID1 complex is required to augment the synthesis of GFP from a MIDAS sequence containing GFP mRNA (Fig. 2A). Interestingly, OS mutations found in different domains of the MID1 protein, including the B-Box1 (A130T), the coiled-coil domain (C266R), and the C terminus (1313del) destroyed the influence of the MID1 protein on the translation of the MIDAS containing mRNA similarly. No difference was seen on RNA levels, measuring GFP mRNA amounts normalized to GAPDH mRNA and transfection efficiencies in cells co-transfected with EGFP-MIDAS and either wild-type or mutated (1313del) MID1, which again points at an influence of MID1 on the translation efficiency of the MID1 containing EGFP mRNA rather than transcription or mRNA stability (Fig. 2B).
FIGURE 2.
Mutant MID1 loses its influence on MIDAS-containing mRNAs. EGFP-MIDAS was co-transfected with wild-type MID1 (wt) or the A130T mutant as negative control or two other MID1 mutants (C266R and 1313del). A, left panel, lysates were analyzed on a Western blot using specific anti-GFP and anti-actin antibodies. Right upper panel, transfection efficiencies were assessed by FACS. Right lower panel, band intensities were measured, and GFP amounts were adjusted to actin and the number of transfected cells in each sample. Standard deviations were calculated from triplicates. B, real time PCR analysis of the GFP mRNA relative to GAPDH normalized to the transfection control. Standard deviations were calculated from triplicates.
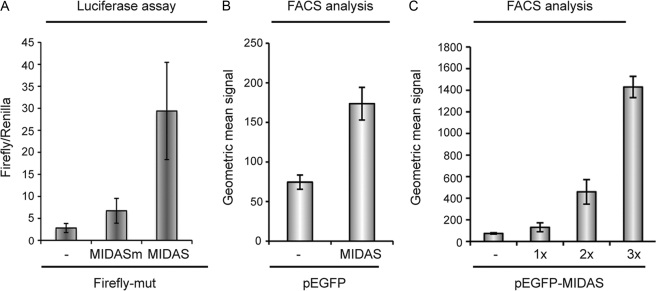
As we have seen in Fig. 1, the endogenous MID1 protein complex has a significant influence on the translation of MIDAS containing mRNAs, making it possible to measure the MIDAS effect without exogenous MID1 expression. In the next series of experiments, we transfected either the Firefly-mut, the Firefly-mut-MIDAS or the Firefly-mut-MIDASm vectors into CHO cells, which are commonly used in biotechnological processes, and we measured the resulting luciferase activities. CHO cells express a reasonable amount of endogenous MID1. The results show a severalfold increase of luciferase activity after transfection of the Firefly-mut vector containing a functional MIDAS motif located 3′ to the luciferase coding sequence, as compared with the same vector carrying a nonfunctional motif (Fig. 3A). A reproducible strong MIDAS effect on the production of GFP was detectable when either an empty pEGFP vector or pEFGP-MIDAS was transfected into CHO cells (Fig. 3B). Amounts of synthesized GFP were quantified by FACS analysis. To validate EGFP quantification by FACS, we had co-expressed EGFP-MIDAS with or without wild-type MID1 and had measured amounts of synthesized GFP either by FACS or by Western blot, previously. Both methods gave similar results (supplemental Fig. 2). Together, these results show that the MIDAS motif can increase cellular production of proteins encoded by transfected genes, independent of the following: (i) its position within the mRNA, (ii) the particular protein produced, and (iii) the vector used for protein expression.
FIGURE 3.
MIDAS motif increases production of proteins. A, luciferase activity in CHO cells expressing a mutated pGL3 vector (Firefly-mut) carrying a functional (Firefly-mut-MIDAS) or a nonfunctional (Firefly-mut-MIDASm) MIDAS motif 3′ to the Firefly luciferase coding region. B, intensities of green fluorescence recorded from CHO cells transfected with GFP-encoding vectors (pEGFP) either containing (MIDAS) or lacking (−) a MIDAS motif 3′ to the coding region for the GFP protein, measured by FACS. C, production of GFP from pEGFP vectors with no (−), single (1×) double (2×) or triple (3×) functional MIDAS motif 3′ of the GFP open reading frame of a pEGFP vector in CHO cells. Fluorescence intensities were measured by FACS. All standard deviations were calculated from triplicates.
Furthermore, we asked whether the MIDAS effect is effectively dose-dependent, such that it can be potentiated by inclusion of multiple MIDAS motifs. To test this, we introduced one and two additional MIDAS sequences 3′ of EGFP in the pEGFP-MIDAS vector. Transfection of CHO cells with these constructs containing multiple MIDAS motifs enhanced the protein expression from these mRNAs up to 20-fold (Fig. 3C), thus showing that increasing the number of MIDAS motifs can lead to a multiplied increase in translation efficiency.
MIDAS Effect Is Cell Type-independent and Can Be Transferred to an in Vitro System
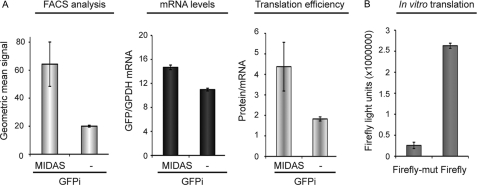
To determine whether the enhancement of protein production by a MIDAS motif is cell type-dependent and also the relative importance of the influence of the MIDAS motif upon mRNA stability and protein translation, we studied the MIDAS effect in the GFP-IRES vector system (GFPi and GFPi-MIDAS) in four different cell lines and simultaneously measured the fluorescence intensity of the proteins produced by FACS and the corresponding mRNA levels by real time PCR. A strong MIDAS effect on the production of GFP was seen in all cell lines tested (HeLa, CHO, COS-7, and HEK293T), whereas its influence on the mRNA levels seemed to play a minor role in the overall effect (Fig. 4A and supplemental Fig. 3). Nevertheless, the strongest MIDAS effect, seen in HEK293T cells (8-fold enhancement), was accompanied by a substantial (50%) increase in GFP mRNA relative to that of GAPDH, suggesting a synergistic effect potentially involving increased mRNA stability as well as translation. Interestingly, the MIDAS effects observed in the different cell lines tend to directly correlate with the relative expression levels of MID1 found in the respective cell lines, being low in HeLa and CHO and high in COS-7 and HEK293T (Fig. 4A and supplemental Fig. 3).
FIGURE 4.
MIDAS motif mainly influences the translation efficiency and to a lesser extent the mRNA stability of MIDAS-containing mRNAs. A, amount of GFP (left panel) and GFP-mRNA (middle panel) produced from a GFP-IRES (GFPi) vector with (MIDAS) or without (−) MIDAS 3′ of the GFP open reading frame in CHO cells. Translation efficiencies are calculated as ratio of protein to mRNA amount (right panel). GFP amounts were measured by FACS, and mRNA amounts were measured by real time PCR. All standard deviations were calculated from triplicates. B, in vitro translation of a Firefly luciferase gene containing either a functional MIDAS (Firefly) or a mutated MIDAS (Firefly-mut) motif. Luciferase activities are shown. All standard deviations were calculated from triplicates.
To confirm that indeed a major part of the MIDAS effect results from an increase in the translation efficiency of a MIDAS-containing mRNA, we performed in vitro transcription/translation from the wild-type and MIDAS-mutated forms of pGL3 (Firefly-wt and Firefly-mut). Using rabbit reticulocyte lysates, which contain endogenous MID1 levels, we observed a 10-fold increase in protein translation from the RNA containing the intact MIDAS motif versus its single-site mutated form (Fig. 4B).
MIDAS Consensus Sequence Contains Several Guanines and Has the Ability to Form a Hairpin
The MIDAS motif and its ability to influence the translational efficiency of its associated mRNAs was further characterized in a systematic in vitro mutagenesis screen of the following: (i) all the Gs within the sequence, (ii) As/Ts before the G-pairs in the sequence, and (iii) some of the nucleotides between the G-pairs (Fig. 5A) in the EGFP-MIDAS construct. Clones were transfected into HeLa cells, analyzed by FACS, and compared with EGFP-MIDAS(wt). Interestingly, although most of the mutations significantly reduced the amount of synthesized GFP, M9 and M15 did not seem to significantly affect GFP signal intensity (Fig. 5B). The observation that the 5th guanine in the MIDAS sequence (M9) did not influence the GFP production significantly suggested that the MIDAS consensus sequence, other than we predicted from the mere spacing of the four G-duplets and the match with the consensus sequences extracted from G-quartet containing genes, is not a G-quartet-like motif but a novel RNA-protein-binding motif that contains several adenines and guanines. Real time PCR analysis of selected mutants confirmed that mRNA levels are not influenced by the MID1 protein complex in HeLa cells (Fig. 5C). In silico analysis of the MIDAS sequence using the RNA secondary structure prediction program Mfold predicted the formation of a hairpin structure between nucleotides 3–7 and 15–19. Analysis of an oligonucleotide comprising the MIDAS sequence and different types of its mutants in a native gel electrophoresis assay showed that indeed the parts between nucleotides 3–7 and 15–19 are necessary for the formation of a secondary RNA structure that leads to a significantly higher mobility of the oligonucleotide through the gel than compared with a nonstructural oligonucleotide. Although most of the guanine exchanges within the MIDAS oligonucleotide did not cause a visible mobility shift, confirming that the MIDAS sequence does not form a G-quartet, mutation of AT (5, 6) into TA (M4), TA (17, 18) into AT (M11), and G (15) into A (M10) leads to a dramatic mobility shift independent of salt conditions (Fig. 5D and supplemental Fig. 4). This is in line with the data obtained before showing that M10 and M11 as well as mutation of T (19) into A (M12) and deletion of nucleotides 16–18 (M17) significantly reduced the ability of the MIDAS motif to drive GFP production (Fig. 5B). Taken together, the results of our in vitro and in vivo mutational analyses of sequence constraints in the MIDAS motif suggested that it is a purine-rich sequence with an intermittent stem-loop structure. Based on these results, we extracted a refined MIDAS consensus sequence (Fig. 6A). In this consensus sequence, all nucleotides that (when mutated) cause a significant decrease of the MIDAS effect appear as set. Nucleotides that do not cause an effect when mutated were set as “N.” Nucleotides 3–7 became the forward (“F”) strand, and nucleotides 15–19 became the reverse (“R”) strand of the stem loop.
FIGURE 6.
mRNAs that are involved in ventral midline development, mTOR signaling, and energy homeostasis carry MIDAS motifs. A, extraction of a MIDAS consensus sequence, F and R are the forward and reverse nucleotides of the stem, respectively. B, binding of a selection of the identified mRNAs to the MID1 protein complex was tested in SHSY cells using FLAG-tagged MID1, RNA-protective immunoprecipitation, and RT-PCR. RT-PCR results were randomly confirmed using real time PCR. C, summary of mRNAs with known functions that were extracted from the data base using the MIDAS consensus sequence. Several mRNAs with related functions were identified.
Most Genes in the Human Genome That Carry MIDAS Motifs Play Important Roles during Development and/or Regulate Energy Homeostasis
An in silico search in current human mRNA databases of the MIDAS consensus sequence using the programs PATSEARCH and RNAMST, accounting for possible GU and UG pairings occurring in RNA, identified a set of 73 MIDAS-containing mRNAs in more than 50,000 sequences (105 megabases). This is a factor of 2.06 above random occurrence of this motif within the calculated sequences. The relatively low number of genes identified gives an indication of the specificity of the MIDAS consensus sequence (supplemental Table 1). We could confirm the specific association of a selection of these mRNAs to the MID1 protein complex using FLAG-MID1 expressing HeLa and SHSY cells by RNA-protective immunoprecipitation, subsequent extraction of the co-precipitated RNA, and RT-PCR as well as quantitative PCR. Although GAPDH does not bind to the complex but is pulled down unspecifically in both experiments, performed with a specific anti-FLAG antibody and unspecific IgGs, all candidate mRNAs showed specific enrichments in the FLAG-containing immunoprecipitate (Fig. 6B), which shows that indeed they are associated with the MID1 protein complex.
Functional hints exist only for 42 of the proteins encoded by the identified mRNAs. Interestingly, grouping the proteins according to their respective functions revealed that 27 of them play important roles in developmental processes involved in ventral midline formation and/or are part of signaling cascades specific to embryogenesis. Another big group includes 10 regulators of energy homeostasis that links them to mTOR, the central controller of the two major cellular anabolic processes, namely translation and lipogenesis (Fig. 6C and supplemental Table 1). Taken together, there seems to be a significant overlap between the OS phenotype, MID1, as a regulatory element in PP2A and mTOR signaling and the function of genes carrying MIDAS motifs.
PDPK-1 Synthesis Is Significantly Reduced in Fibroblasts from an OS Patient
PDPK-1 is a kinase that plays a fundamental role in mTOR signaling. A MIDAS motif has been identified in its 3′-untranslated region (Fig. 7A), and binding to the MID1 protein complex had been demonstrated by RNA-protective immunoprecipitation (Fig. 6B). Control of the PDPK-1 protein synthesis by the MID1 protein would provide a feed-forward regulation loop of MID1 on mTOR activity that is synergistic with its influence on PP2A activity. Regulation of PDPK-1 protein synthesis through the MID1 protein complex would further mean that PDPK-1 protein amounts are significantly reduced in OS patient cells with dysfunctional MID1. To test this hypothesis, fibroblasts from an OS patient carrying a C-terminal loss-of-function mutation in MID1 and age-matched control cells were lysed and analyzed for PDPK-1 protein and mRNA levels, both relative to GAPDH. Although the PDPK-1 mRNA is not reduced in the OS patient cells (Fig. 7C), compared with the control cells, a very clear reduction (by 80%) was observed on the protein level (Fig. 7B) suggesting that the synthesis of the PDPK-1 protein is significantly reduced in cells with dysfunctional MID1. To show that this reduction in PDPK-1 protein in OS cells is due to reduced protein synthesis and not enhanced protein degradation, we determined the half-life of PDPK-1 in control and patient cells (Fig. 7D). The results clearly show that the reduction of PDPK-1 protein in OS patient cells is due to a reduction of PDPK-1 protein translation rather than to decreased protein stability. The determined half-life of PDPK-1 seems even slightly higher in the OS patient than in age-matched control cells. Finally, to confirm that dysfunctional MID1 is responsible for the decrease of PDPK-1 in OS cells, we were able to demonstrate that transformation of OS patient cells with an expression vector encoding functional MID1 can rescue PDPK-1 (Fig. 7E). These data clearly suggest that, as expected from the RNA binding data, MID1 indeed has a regulatory role in the synthesis of the PDPK-1 protein.
FIGURE 7.
PDPK-1 protein amount is significantly reduced in cells from an OS patient (OS) compared with age-matched control cells. A, schematic showing the predicted MIDAS motif within the PDPK-1 mRNA. B, representative Western blot showing the analysis of PDPK-1 and GAPDH and quantification of three independent experiments measuring PDPK-1 protein levels relative to GAPDH. C, RT-PCR analysis of the PDPK-1 mRNA relative to GAPDH. Standard deviations are calculated from triplicates. D, analysis of PDPK-1 protein stability after treatment with cycloheximide in a time course by Western blot and quantification of PDPK-1 protein levels relative to GAPDH. E, rescue of PDPK-1 protein levels by overexpression of wild-type MID1 in OS cells. Representative Western blot showing the analysis of MID1, PDPK-1, and GAPDH and quantification of three independent experiments measuring PDPK-1 protein levels relative to GAPDH (*, p = 0.035).
DISCUSSION
Translational control has become a major focus of attention, and research activity in the field of gene regulation and especially developmental biology is a prominent example that has since gained profound insights through studies on translational controls, from bacteriophages (17) to man (18). Here, we introduce the MID1 protein, which is crucial for the development of the ventral midline in the human body, as a highly specific translational regulator. We present several lines of evidence for a novel pronounced effect of this polysome-associated ubiquitin ligase on the translation efficiency of a subset of mRNAs. We show the following: (i) a sequence-dependent association (MIDAS motif) of the MID1 complex with mRNA, which correlates with an enhancement of protein production from this mRNA; (ii) this association is dependent on an intact MID1 protein complex as mutated MID1 variants, like those found in OS patients, do not enhance protein expression of MIDAS-containing mRNAs; (iii) loss of MID1 or α4 function leads to a significant reduction of protein synthesis from MIDAS-containing mRNAs; (iv) the MIDAS effect is also seen in an in vitro translation system, favoring a mechanism involving enhancement of translation rather than transcription; (v) in line with this, comparison of reporter activity with the respective mRNA levels reveals that the effects are caused by enhanced translation rather than mRNA production/stability; (vi) the MIDAS motif originally found in the open reading frame of the Firefly luciferase can be transferred to the 3′UTR of other mRNAs without losing its activity; (vii) introduction of multiple MIDAS motifs results in a dose-dependent enhancement of protein production; (viii) the MIDAS effect can be observed on different vector systems and in different cell lines and is not dependent on ectopic expression of MID1; (ix) the MIDAS consensus sequence can be identified in a small selective subset of mammalian genes, many of which are involved in developmental processes linked to the ventral midline and accordingly to the phenotype seen in OS patients; (x) these MIDAS-containing mRNAs bind the MID1 protein complex; and (xi) finally, further analysis of one of these mRNAs, PDPK-1, proved that PDPK-1 protein levels are significantly reduced in OS patient cells compared with age-matched control cells. This reduction is due to diminished protein synthesis as PDPK-1 mRNA levels and PDPK-1 protein stabilities are not affected. The role of MID1 in PDPK-1 protein synthesis is further corroborated by the finding that PDPK-1 protein levels in OS cells can be rescued by functional MID1.
MID1 Protein Complex Regulates Protein Translation in an mTOR-dependent Manner
These findings clearly show that MID1 not only associates with polyribosomes but also has an important function in the regulation of the translation of specific mRNAs. This is not really surprising, because its ubiquitin ligase function targets the catalytic subunit of PP2A (PP2Ac) for proteasomal degradation, and this phosphatase is the major opponent of the translation master regulator mTOR (4, 19). The degradation of PP2Ac is triggered by the α4 protein, which serves as an adapter between MID1 and PP2Ac (3, 5, 9). It has been reported that the association of α4 with PP2Ac is mTOR-dependent in several cellular contexts (2, 21). Thus, while mTOR phosphorylates key regulators of translation, namely p70S6K and 4EBP1 (19), it can also trigger the association of the PP2Ac-α4-MID1 complex to inhibit PP2A from dephosphorylating p70S6K and 4EBP1. Additionally, α4 is itself part of a positive feedback loop, because its own translation is enhanced in a PI3K- and mTOR-dependent manner (22). These control loops for mTOR signaling are further potentiated by the synergistic influence of the MID1 protein complex on the protein synthesis of PDPK-1. PDPK-1 is an efficient mTOR agonist inducing mTOR activity in two different manners, directly in a leucine-dependent way and indirectly via the activation of Akt (23, 24). By stimulating PDPK-1 protein synthesis, MID1 is therefore further increasing mTOR signaling. Thus, the amount of MID1 associated with a certain mRNA, which is, as we show here, controlled by the number and specific structures of MIDAS motifs integrated into it, could lead to a fine-tuning of mTOR signaling and therefore of protein production from specific mRNAs via control of PP2A activity. The hypothesis of the existence of such a fine-tuned regulation process that involves the ubiquitin ligase MID1, mTOR, PP2A, and PDPK-1 is supported by the observation that knockdown of the rapamycin-sensitive PP2A subunit α4, similar to knockdown of MID1, also leads to a significant reduction of the synthesis of protein from MIDAS-containing mRNAs. Given the complexity of developmental processes, such fine-tuning mechanisms could serve to generate the necessary diversity in developmental gradients and complex compositions (25).
Functions of MIDAS-containing mRNAs Concentrate on Developmental Processes and Energy Control
Scrutinizing the function of the proteins encoded by MIDAS-containing mRNAs (a comprehensive list including accession numbers and explanations of abbreviations are given in supplemental Table 1) strikingly reveals that, in accordance with the OS phenotype, the majority of them is involved in developmental processes and in particular in midline development, like “cleft lip and palate-associated transmembrane protein 1,” SMCA7L, ARS2, INCA-like, and Scribbled (26, 27). Interestingly, SMCA7L maps to exactly the chromosomal region on chromosome 22 where the autosomal gene for OS was linked to previously (28) making SMCA7L an excellent candidate gene for autosomal dominantly inherited OS. Several others of these mRNAs code for developmental regulators of the RhoA/Rac/Rab protein family (e.g. Kalirin, Obscurin PLEKHG5, ARHGAP25, Rabenosyn5/ZFYVE20, and AS160) or for important players in ciliogenesis and cell polarity (e.g. Dynein-DNAH7, CEP131, TNK1, Scribbled, Rabenosyn5/ZFYVE20, and LZTS2). All these molecules are related to known MID1 functions and important processes during the formation of the ventral midline, such as the establishment of left/right symmetry. In addition, ciliogenesis is essential for the function of GLI3 (29), a sonic hedgehog transcription factor, nuclear localization of which is controlled by MID1 (30). Finally, the mRNA encoding HOX11, a homeobox transcription factor and proto-oncogene product, which is expressed in the first branchial arch, which later, among others, forms ventral midline structures like the tongue and the pharynx, also contains a MIDAS motif in its 3′UTR (31).
A second large group of MIDAS-containing mRNAs code for major players in growth factor signaling pathways involved in embryogenesis or are directly linked to the mTOR/PP2A pathway (e.g. GAS6, TrkB, MMP14, Sortilin2, and IRS4), including components of phosphoinositide signaling like the direct Akt activator PDPK1 and PLCδ1 as well as the PLCγ1-interacting kinase TNK1 (32).
A third group consists of proteins involved in basic metabolic processes that are controlled by mTOR to orchestrate intracellular energy homeostasis. These include protein translation (URB1), lipogenesis (Lipin3), mitochondrial biogenesis (COX16 and acyl-CoA-DH), and especially glycolysis (phosphofructokinase) to produce pyruvate, the precursor building block for lipogenesis from glucose. Strikingly, this group also includes basic fuel-providing pathways like the following: (i) the glucose transporter GLUT4 trafficking protein AS160 (33), which is necessary for insulin-dependent activation of glucose transport by Akt, and (ii) the fructose-generating sorbitol dehydrogenase (34).
The finding of a tight connection between MIDAS-containing mRNAs and lipogenesis also sheds further light into recent reports that S14-R (MIG12), a direct binding partner of MID1 (35), is involved in lipogenesis, which is tightly regulated by dietary carbohydrates, glucagon, and thyroid hormones (36) and has recently been found to boost fatty acid synthesis by up-regulating the activity of the key lipogenic enzyme acetyl-CoA carboxylase (37). So far, the biological significance of the binding of such a pro-lipogenesis factor to MID1 was totally unclear. But in light of our results, this basic metabolic function of a MID1-binding partner is clearly in line with a strong feed-forward regulation of mTOR in key metabolic pathways via MIDAS-driven translational enhancement.
An involvement of the MID1 complex in energy homeostasis is further supported by the fact that several MIDAS-regulated proteins have been implicated in food intake and energy sensing/balance as follows. (i) MMP14 modulates glycolysis and is regulated by leptin, the famous obese gene product (38). (ii) Insulin receptor substrate 4 (IRS4), in addition to its known function in insulin-Akt-mTOR signaling, directly binds to and modulates signaling through the leptin receptor (39). (iii) PDPK-1, CHRNB2, GAS6, and the brain-derived neurotrophic factor receptor TrkB have been implicated in body weight control (40–43). (iv) Anti-lipogenesis transcription factor Nrf2 is negatively modulated by MafK and is a homologue of KEAP1 (44, 45), two proteins also translated from MIDAS-containing mRNAs. Furthermore, the recent finding that the widely used antidiabetic drug Metformin, known to reduce lipogenesis and glucose intolerance (Metformin activates AMP-activated protein kinase, leading to the inhibition of mTOR), also negatively modulates the MID1 complex (46), gives additional evidence toward an important function of MID1 in the orchestration of energy homeostasis downstream of mTOR.
TOR, the target of rapamycin, was originally named due to its inhibition by rapamycin, an effective immunosuppressant, and mTOR is deeply involved in the maintenance of potent immune responses. Interestingly, the fourth group of MIDAS-harboring mRNAs code for components of the immune system, namely MBL2, CD72, and AKNA, a transcriptional regulator of CD40 expression. CD40 and CD72 are important modulatory co-receptors of the B-cell receptor (47, 48), and MBL2 is a basic player in the innate immune response (49), which has recently also been shown to be regulated by mTOR (50). In this respect, it is also intriguing that the MID1-associated α4 protein was originally identified as a “B-cell receptor-associated protein that displays rapamycin-sensitive binding directly to the catalytic subunit of PP2A” (2).
Taken together, our data base screening with the MIDAS consensus sequence resulted in a clearly nonrandom selection of mRNAs, the majority of which not only codes for developmental proteins, as would have been expected from the human phenotype caused by loss of MID1 function, but also for proteins/enzymes that reflect most aspects of the basic mTOR-regulated pathways and are thus in line with an important role of MID1 in post-transcriptional regulation downstream of mTOR. Our model of the MID1 cellular role further extends the recently reported finding of a control function of MID1 on mTORC1 signaling in a PP2A-dependent manner (6) toward more general translational feed-forward mechanisms of MID1 that synergistically enhance processes involving mTORC1 signaling.
In support of our experimental results showing the MID1-dependent regulation of PDPK1, another MIDAS-containing mRNA, namely MMP14, a major regulatory matrix metalloproteinase and proneurotrophin maturation factor, has independently been shown to be reduced in its protein expression after mTOR inhibition with no changes on its mRNA level (51). This supports the hypothesis of a MID1- and mTOR-dependent control mechanism in the translation of MMP14. Furthermore, together with the fact that MMP14 can positively regulate energy production via glycolysis (52), it suggests that, like in the case of PDPK1, mTOR exerts a fine-tuning feed-forward mechanism of regulation via MIDAS-triggered translational enhancement of specific mRNAs involved in body metabolism.
The predominant occurrence of MIDAS motifs in mRNAs coding for groups of proteins involved in basic processes of ventral midline development suggests that MID1 orchestrates complex cellular processes via coordinated translational regulations. A reduction of these proteins in MID1 dysfunctional cells, as we are demonstrating here for PDPK-1, would certainly be a comprehensible factor in the development of an OS phenotype in patients. In the future, it will be interesting to analyze if all the other proteins encoded by MIDAS-containing mRNAs behave in a PDPK-1-similar way in a MID1 dysfunctional context in either patients or in the recently established MID1 knock-out mouse model (20).
Mutant MIDAS Sequences Do Not Necessarily Lose MID1 Binding Ability
From the experiments with the Firefly-wt, Firefly2, and Firefly-mut vectors, we have concluded that some sequence changes in the MIDAS motif significantly reduce its binding activity to the MID1 protein complex, which then loses its influence on the efficiency of protein synthesis. In a series of experiments performing RNA-protein pulldown assays with some of the MIDAS mutants that were generated in Fig. 5, we have seen that, although binding activity of the respective mRNAs to the MID1 protein complex can still be obtained, MIDAS sequences with sequence variations (MIDAS*) have differing potential to stimulate the synthesis of the protein encoded by the respective MIDAS*-containing mRNA (supplemental Fig. 5). This suggests that the sequence of the MIDAS motif not only influences binding to the MID1 protein but also the process of translation induction as such. For example, could the binding of co-factors that enhance the process be reduced in MIDAS* mutants?
Mutant MID1 Loses Its Influence on MIDAS-containing mRNAs
The crucial importance of MID1-regulated translation processes for the pathogenesis of OS is further underscored by the observation that not only mutations in the B-Box1 domain (A130T and C145S) but also a C-terminally truncating OS mutation (1313del) and a point mutation in the coiled-coil domain destroyed the influence of the MID1 protein on the translation of MIDAS-containing mRNA. Loss of its regulatory role on MIDAS-containing mRNAs therefore seems to be a common mechanism for all the mutations found in OS patients, which makes the MID1-mediated translation support an important component for understanding the pathogenesis of OS.
MIDAS Sequences Can Be Used to Enhance Production of Mammalian Proteins
The extent of the MIDAS-dependent enhancement of protein production seen with endogenous levels of MID1 in cell lines commonly used in biopharmaceutical protein production processes suggests an enormous potential for industrial optimization strategies to improve the synthesis of proteins in eukaryotic cell systems. Production of recombinant therapeutic proteins in cultivated mammalian cells has surpassed that in microbial expression systems because of the greater capacity of eukaryotic cells for proper folding, assembly, and post-translational modification of proteins. However, although enhancement of gene transcription has been a focus of biotechnological research for many years and cells that are being used for industrial protein production have been engineered to optimize their gene transcription efficiency, the production of complex proteins in mammalian cells remains inefficient and extremely costly. Consequently, there is high demand for novel approaches to lower costs and enhance protein yields. We anticipate that our present finding of MIDAS motifs, when introduced into mRNAs, markedly enhancing the production of desired proteins will permit considerable improvements in the cost-efficient production of sophisticated biopharmaceuticals. In addition, the observed dose dependence of the MIDAS effect on the number of MIDAS motifs integrated could be exploited in novel vector systems to fine-tune the regulation of protein expression in mouse and tissue culture models.
In summary, our data provide a significant step forward in the understanding of the development of ventral midline structures and its misregulation in patients with Opitz BBB/G syndrome and dysfunctional MID1. In addition, we show that the MIDAS motif is a promising tool to increase production efficiencies of proteins and to allow fine-tuning of protein expression in eukaryotic cell systems.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey B. Tatro for editing the manuscript and Susanne Freier and Ummuhan Demir for tissue culturing.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant SFB 577 (Project A6, to S. S.), Austrian Science Fund Grants FWF-SFB021 (Project 03) and TRP233-B18 (to R. S.), the Tyrolean Future Foundation (to R. S.), Tenovus Scotland (to S. S.), and AtaxiaUK (to S. S.). Patent application pending (EP 07015280.6).
This paper is dedicated to Professor Manfred Schweiger on the occasion of his 75th birthday.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Tables 1 and 2.
- OS
- Opitz BBB/G syndrome;
- p70S6K
- p70 S6-kinase
- mTORC1
- mammalian target of rapamycin complex 1
- MIDAS
- MID1 association sequence
- PDPK1
- phosphoinositide-dependent protein kinase 1
- EGFP
- enhanced GFP.
REFERENCES
- 1. Quaderi N. A., Schweiger S., Gaudenz K., Franco B., Rugarli E. I., Berger W., Feldman G. J., Volta M., Andolfi G., Gilgenkrantz S., Marion R. W., Hennekam R. C., Opitz J. M., Muenke M., Ropers H. H., Ballabio A. (1997) Nat. Genet. 17, 285–291 [DOI] [PubMed] [Google Scholar]
- 2. Inui S., Sanjo H., Maeda K., Yamamoto H., Miyamoto E., Sakaguchi N. (1998) Blood 92, 539–546 [PubMed] [Google Scholar]
- 3. Trockenbacher A., Suckow V., Foerster J., Winter J., Krauss S., Ropers H. H., Schneider R., Schweiger S. (2001) Nat. Genet. 29, 287–294 [DOI] [PubMed] [Google Scholar]
- 4. Jacinto E., Hall M. N. (2003) Nat. Rev. Mol. Cell Biol. 4, 117–126 [DOI] [PubMed] [Google Scholar]
- 5. Short K. M., Hopwood B., Yi Z., Cox T. C. (2002) BMC Cell Biol. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu E., Knutzen C. A., Krauss S., Schweiger S., Chiang G. G. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 8680–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aranda-Orgillés B., Trockenbacher A., Winter J., Aigner J., Köhler A., Jastrzebska E., Stahl J., Müller E. C., Otto A., Wanker E. E., Schneider R., Schweiger S. (2008) Hum. Genet. 123, 163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schweiger S., Schneider R. (2003) BioEssays 25, 356–366 [DOI] [PubMed] [Google Scholar]
- 9. Liu J., Prickett T. D., Elliott E., Meroni G., Brautigan D. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6650–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. So J., Suckow V., Kijas Z., Kalscheuer V., Moser B., Winter J., Baars M., Firth H., Lunt P., Hamel B., Meinecke P., Moraine C., Odent S., Schinzel A., van der Smagt J. J., Devriendt K., Albrecht B., Gillessen-Kaesbach G., van der Burgt I., Petrij F., Faivre L., McGaughran J., McKenzie F., Opitz J. M., Cox T., Schweiger S. (2005) Am. J. Med. Genet. 132A, 1–7 [DOI] [PubMed] [Google Scholar]
- 11. Brown V., Jin P., Ceman S., Darnell J. C., O'Donnell W. T., Tenenbaum S. A., Jin X., Feng Y., Wilkinson K. D., Keene J. D., Darnell R. B., Warren S. T. (2001) Cell 107, 477–487 [DOI] [PubMed] [Google Scholar]
- 12. Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. (2001) Cell 107, 489–499 [DOI] [PubMed] [Google Scholar]
- 13. Oganesian L., Bryan T. M. (2007) BioEssays 29, 155–165 [DOI] [PubMed] [Google Scholar]
- 14. Schaeffer C., Bardoni B., Mandel J. L., Ehresmann B., Ehresmann C., Moine H. (2001) EMBO J. 20, 4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williamson J. R. (1994) Annu. Rev. Biophys. Biomol. Struct. 23, 703–730 [DOI] [PubMed] [Google Scholar]
- 16. McDonald W. J., Sangster S. M., Moffat L. D., Henderson M. J., Too C. K. (2010) J. Cell. Biochem. 110, 1123–1129 [DOI] [PubMed] [Google Scholar]
- 17. Herrlich P., Rahmsdorf H. J., Pai S. H., Schweiger M. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 1088–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Averous J., Proud C. G. (2006) Oncogene 25, 6423–6435 [DOI] [PubMed] [Google Scholar]
- 20. Lancioni A., Pizzo M., Fontanella B., Ferrentino R., Napolitano L. M., De Leonibus E., Meroni G. (2010) J. Neurosci. 30, 2880–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kong M., Fox C. J., Mu J., Solt L., Xu A., Cinalli R. M., Birnbaum M. J., Lindsten T., Thompson C. B. (2004) Science 306, 695–698 [DOI] [PubMed] [Google Scholar]
- 22. Grech G., Blázquez-Domingo M., Kolbus A., Bakker W. J., Müllner E. W., Beug H., von Lindern M. (2008) Blood 112, 2750–2760 [DOI] [PubMed] [Google Scholar]
- 23. Dowling R. J., Topisirovic I., Fonseca B. D., Sonenberg N. (2010) Biochim. Biophys. Acta 1804, 433–439 [DOI] [PubMed] [Google Scholar]
- 24. Sanchez Canedo C., Demeulder B., Ginion A., Bayascas J. R., Balligand J. L., Alessi D. R., Vanoverschelde J. L., Beauloye C., Hue L., Bertrand L. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E761–E769 [DOI] [PubMed] [Google Scholar]
- 25. Wilhelm J. E., Smibert C. A. (2005) Biol. Cell 97, 235–252 [DOI] [PubMed] [Google Scholar]
- 26. Iden S., Collard J. G. (2008) Nat. Rev. Mol. Cell Biol. 9, 846–859 [DOI] [PubMed] [Google Scholar]
- 27. Luo T., Xu Y., Hoffman T. L., Zhang T., Schilling T., Sargent T. D. (2007) Development 134, 1279–1289 [DOI] [PubMed] [Google Scholar]
- 28. Robin N. H., Feldman G. J., Aronson A. L., Mitchell H. F., Weksberg R., Leonard C. O., Burton B. K., Josephson K. D., Laxová R., Aleck K. A., Allanson J. E., Guion-Almeida M. L., Martin R. A., Leichtman L. G., Price R. A., Opitz J. M., Muenke M. (1995) Nat. Genet. 11, 459–461 [DOI] [PubMed] [Google Scholar]
- 29. Ashique A. M., Choe Y., Karlen M., May S. R., Phamluong K., Solloway M. J., Ericson J., Peterson A. S. (2009) Sci. Signal. 2, ra70. [DOI] [PubMed] [Google Scholar]
- 30. Krauss S., Foerster J., Schneider R., Schweiger S. (2008) Cancer Res. 68, 4658–4665 [DOI] [PubMed] [Google Scholar]
- 31. Lonyai A., Kodama S., Burger D., Faustman D. L. (2008) Immunol. Cell Biol. 86, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Felschow D. M., Civin C. I., Hoehn G. T. (2000) Biochem. Biophys. Res. Commun. 273, 294–301 [DOI] [PubMed] [Google Scholar]
- 33. Chen S., Wasserman D. H., MacKintosh C., Sakamoto K. (2011) Cell Metab. 13, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao W., Aghajanian H. K., Haig-Ladewig L. A., Gerton G. L. (2009) Biol. Reprod. 80, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berti C., Fontanella B., Ferrentino R., Meroni G. (2004) BMC Cell Biol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsatsos N. G., Augustin L. B., Anderson G. W., Towle H. C., Mariash C. N. (2008) Endocrinology 149, 5155–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim C. W., Moon Y. A., Park S. W., Cheng D., Kwon H. J., Horton J. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schram K., Ganguly R., No E. K., Fang X., Thong F. S., Sweeney G. (2011) Endocrinology 152, 2037–2047 [DOI] [PubMed] [Google Scholar]
- 39. Wauman J., De Smet A. S., Catteeuw D., Belsham D., Tavernier J. (2008) Mol. Endocrinol. 22, 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lijnen H. R., Christiaens V., Scroyen L. (2011) J. Pharmacol. Exp. Ther. 337, 457–464 [DOI] [PubMed] [Google Scholar]
- 41. Noble E. E., Billington C. J., Kotz C. M., Wang C. (2011) Am. J. Physiol. Regul. Integr. Comp. Physiol. 300, R1053–R1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iskandar K., Cao Y., Hayashi Y., Nakata M., Takano E., Yada T., Zhang C., Ogawa W., Oki M., Chua S., Jr., Itoh H., Noda T., Kasuga M., Nakae J. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E787–E798 [DOI] [PubMed] [Google Scholar]
- 43. Landgren S., Engel J. A., Andersson M. E., Gonzalez-Quintela A., Campos J., Nilsson S., Zetterberg H., Blennow K., Jerlhag E. (2009) Brain Res. 1305, S72–S79 [DOI] [PubMed] [Google Scholar]
- 44. Huang J., Tabbi-Anneni I., Gunda V., Wang L. (2010) Am. J. Physiol. Gastrointest. Liver Physiol. 299, G1211–G1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dhakshinamoorthy S., Jaiswal A. K. (2000) J. Biol. Chem. 275, 40134–40141 [DOI] [PubMed] [Google Scholar]
- 46. Kickstein E., Krauss S., Thornhill P., Rutschow D., Zeller R., Sharkey J., Williamson R., Fuchs M., Köhler A., Glossmann H., Schneider R., Sutherland C., Schweiger S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21830–21835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clark M. R., Tanaka A., Powers S. E., Veselits M. (2011) Mol. Immunol. 48, 1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu H. J., Bondada S. (2009) J. Clin. Immunol. 29, 12–21 [DOI] [PubMed] [Google Scholar]
- 49. Muller Y. L., Hanson R. L., Bian L., Mack J., Shi X., Pakyz R., Shuldiner A. R., Knowler W. C., Bogardus C., Baier L. J. (2010) Diabetes 59, 2080–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Säemann M. D., Haidinger M., Hecking M., Hörl W. H., Weichhart T. (2009) Am. J. Transplant. 9, 2655–2661 [DOI] [PubMed] [Google Scholar]
- 51. Berthier C. C., Wahl P. R., Le Hir M., Marti H. P., Wagner U., Rehrauer H., Wüthrich R. P., Serra A. L. (2008) Nephrol. Dial. Transplant. 23, 880–889 [DOI] [PubMed] [Google Scholar]
- 52. Sakamoto T., Seiki M. (2010) J. Biol. Chem. 285, 29951–29964 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.