Abstract
Although esterification of free cholesterol to cholesteryl ester in the liver is known to be catalyzed by the enzyme acyl-coenzyme A:cholesterol acyltransferase, ACAT, the neutral cholesteryl ester hydrolase (nCEH) that catalyzes the reverse reaction has remained elusive. Because cholesterol undergoes continuous cycling between free and esterified forms, the steady-state concentrations in the liver of the two species and their metabolic availability for pathways, such as lipoprotein assembly and bile acid synthesis, depend upon nCEH activity. On the basis of the general characteristics of the family of rat carboxylesterases, we hypothesized that one member, ES-4, was a promising candidate as a hepatic nCEH. Using under- and overexpression approaches, we provide multiple lines of evidence that establish ES-4 as a bona fide endogenous nCEH that can account for the majority of cholesteryl ester hydrolysis in transformed rat hepatic cells and primary rat hepatocytes.
Keywords: Cholesterol Metabolism, Hepatocyte, Hydrolases, Liver, Rat
Introduction
Cytosolic cholesterol pools are in a dynamic state of turnover between the free (FC)2 and esterified forms (CE) (1–3). The identification of the enzyme(s) in the liver that catalyzes the hydrolysis of CE at neutral pH has been elusive. In contrast, the CE that enters cells as part of apoB-containing lipoproteins through receptor- or heparan sulfate proteoglycan-mediated uptake is known to be hydrolyzed by lysosomal acid lipase, with the ensuing FC available to enter various cellular pools. A fraction of the FC trafficked to the endoplasmic reticulum (ER) is esterified by isoforms of acyl-coenzyme A:cholesterol acyltransferase (ACAT). When potentially cytotoxic levels of FC are reached, there is a significant increase in the formation of CE, which is then stored in cytosolic lipid droplets (4, 5).
In the liver, CE is a component of very low density lipoproteins (VLDL). The source of CE can be either direct (i.e. synthesized completely in the ER membrane) or indirect through hydrolysis of cytosolic CE and re-esterification of FC by ACAT (6, 7). FC can also become part of VLDL, and, as in all cells, FC resulting from CE hydrolysis can be effluxed to HDL or used in the synthesis of other molecules. In liver, a major fate of FC is its conversion to bile acids and oxysterols. The interconversion between the non-lysosomal cellular pools of FC and CE in the liver is under metabolic regulation, with the key enzymatic components attributed to ACAT and neutral cholesteryl ester hydrolases (nCEHs) (8–12).
The explicit identification of nCEHs in the liver has been elusive. Ghosh and colleagues (13) have recently reported a human macrophage nCEH active in foam cells in atherosclerotic plaques that was related to a previously studied enzyme proposed as a rat hepatic nCEH (14). The latter was cloned from rat liver, and, when overexpressed in COS cells, the lysates exhibited nCEH activity against exogenously provided CE (14). However, whether this candidate operates as a nCEH under physiological conditions in hepatic cells has never been established. Another candidate that has been recently proposed in the liver is hormone-sensitive lipase (15), which had been studied mainly as a triglyceride lipase in adipose tissue and as a nCEH in macrophages (16). In HSL−/− mice, liver CE content increased by 47% over that in wild-type mice, although CE hydrolysis in intact cells was not measured directly (15).
This study was designed to identify a potential nCEH by employing multiple types of assays, including functional measurements of CE hydrolysis in intact primary rat hepatic cells and hepatic cell lines (Fu5AH and McArdle RH7777). Rat hepatic cells were chosen given the historical relevance of rat liver as a model of human hepatic lipid and lipoprotein metabolism. The candidate enzymes studied belong to the rat carboxylesterase family. They are expressed in hepatocytes and hydrolyze a wide variety of lipid ester substrates in vitro. The carboxylesterase family was thought to include over 30 rat liver “carboxylesterases,” but biochemical genetic studies have shown that these enzymes are the products of five major loci in linkage group V (17). These enzymes are designated as ES-2, ES-3, ES-4, ES-10, and ES-15, of which ES-3, ES-4, and ES-10 account for 95% of rat liver microsomal carboxylesterase activity (18). The function of ES-3 has not been clearly determined, but it can hydrolyze retinyl palmitate in vitro (18). ES-4 was shown to hydrolyze palmitoyl-CoA (19) and other lipid esters (20) in vitro. ES-10 was shown to hydrolyze retinol esters in vitro, but not CE (21).
Here we present multiple independent lines of evidence that strongly support a role for ES-4 as a potent nCEH in rat liver, with 55% of CE turnover attributable to this single enzyme in primary rat hepatic cells.
EXPERIMENTAL PROCEDURES
Cell Lines
The rat hepatoma cell line McArdle RH7777 (McA) was purchased from the ATCC. McA cells were maintained in DMEM (Invitrogen) with 10% fetal bovine serum, 10% horse serum, 1% streptomycin/penicillin, and 1% l-glutamine in a 37 °C incubator with 5% CO2 atmosphere. Fu5AH rat hepatoma cells and RAW macrophage cell were maintained in DMEM (+10% FBS, 1% streptomycin/penicillin, and 1% l-glutamine). The stellate cell line (HSC-T6) was a gift from Dr. Scott L. Friedman (The Mount Sinai School of Medicine, New York) and was maintained according to the published protocol (22). Cells were maintained on tissue culture plates (VWR, West Chester, PA) coated with type I collagen from rat tail (Sigma).
Primary Rat and Mouse Hepatocytes
All animal procedures were approved by the New York University School of Medicine Institutional Animal Care and Usage Committee. Male Sprague-Dawley Rats 4 to 6 weeks old (Taconic, Germantown, NY) were treated with a combination of ketamine (5 mg/kg) and xylazine (2 mg/kg) following the Institutional Animal Care and Usage Committee guidelines. The livers were perfused using Hanks' balanced salt solution without CaCl2, MgSO4, or phenol red and with 10 mm HEPES at pH7.4 (buffer 1) saturated with 5% CO2 and 95% O2 for 5 min, followed by buffer 1 with added collagenase type I (30 mg/100 ml, Worthington) for 25 min. Cells were then filtered through a 70-μ mesh and subjected to a 40% Percoll gradient to isolate viable cells. Hepatocytes were then washed twice in PBS followed by maintenance media (McA media with 10 nm insulin) and plated on collagen-coated plates. To isolate rat primary stellate cells, collagenase-treated liver was filtered through a 70-μ mesh and gently centrifuged (50 × g) to remove hepatocytes. The remaining cells in the supernatant were subject to further centrifugation steps and pelleted at (200 × g) (23). Cells were then plated on collagen-coated plates.
siRNA
McA cells were plated at a concentration of 2 × 105 per well in collagen-coated 6-well plates. The following day, cells were transfected using siRNA (ES-4 X81825, ES-3 NM_031565, or ES-10 NM_133295) smart pool (Dharmacon, Lafayette, CO) at a final concentration of 50–100 nm, in combination with transfection reagent (siQuest/Mirus, Madison, WI) according to the manufacturer's protocol. To account for the effects of the transfection procedure, controls were separately transfected with nonspecific siRNA (Dharmacon). After 24 h of transfection, the cells were replated on new collagen-coated plates. 48 h post-transfection, cells were washed twice in PBS and harvested for RNA, protein, lipid mass, or CE turnover studies.
For primary hepatocytes, 1 × 106cells/well were plated on collagen-coated plates. After 2 h of incubation, cells were washed once with PBS and transfected with siRNA using the same protocol as above. After 24 h, the media were replaced with fresh media, and cells were harvested 48 h post-transfection.
ES-4 Overexpression
Total RNA from rat liver was isolated using TRIzol (Invitrogen) using the manufacturer's protocol. cDNA was then generated using reverse transcriptase II (Invitrogen). ES-4 cDNA was then amplified using the following primers with HindIII and AflII cloning sites: forward, 5′-TCTCAAGCTTAGATGTGCCTCAGCTTCCTGATCC-3′ and reverse, 5′-GTCCTTAAGATTCACAGCTCGTTGTGGTGTGG-3′. The PCR product was then digested and ligated with digested pcDNA3.1− (Invitrogen) using the rapid ligation kit (Roche) following the manufacturer's protocol. The ligation product was transformed into Oneshot DH5α-competent bacteria (Invitrogen). Colonies were isolated, the plasmids purified, and the presence and correctness of the ES-4 sequence were confirmed by the New York University School of Medicine core DNA sequencing facility. ES-4-GFP fusion protein was generated by subcloning ES-4 into the pGFP-N3 expression vector (Clontech).
mRNA Quantification
RNA was isolated from cells using the RNAeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. RNA was then quantified using the Ribogreen assay kit (Molecular Probes/Invitrogen). Total RNA (10 ng) was then used in a Taqman PCR assay to quantify ES-3 (probe 5′-FAM-ATCCCAATGGACAGGGCCTGCC-3′-BHQ; forward, 5′-CTGGGCAAACTTTGCTAGGAA-3′; reverse, 5′-TTTGGTCATACTCTGGCCAATG-3′), ES-4 (probe 5′-FAM-CTGGATGTGAAACCACCACATCTGCC-3′-BHQ; forward, 5′CAGCCGCTAAGCAAATTGCT-3′; reverse, 5′-ACGAGGCAGTGAACAATGA), ES-10 (probe 5′-FAM-TGGGCTATCCACTCTCCGAAGGCA-3′-BHQ; forward, 5′ AGCAAGAGTTTGGCTGGATCAT-3′; reverse, 5′-AGAGGGATTTGGCTGTTTTCTG-3′), and rat β actin (loading control) (21). Mouse ES-4 (NCBI AU018778), forward, 5′AAGCAAGAATGTGGCTGGCTTCTG-3′; reverse, 5′TCTGGTATGCCACGA-3′. The PCR assays were performed on the 7300 real-time PCR system (Applied Biosystems, Inc., Foster City, CA).
Western Blot Analysis
Cells were lysed in 10 mm PBS (pH 7.4), 125 mm NaCl, 36 mm lithium dodecyl sulfate, 24 mm deoxycholate, 1% Triton X-100, and protease inhibitor mixture (Roche). Protein concentration was determined by the Dc Protein assay (Bio-Rad), and 20 μg of protein was loaded in each well. Proteins were resolved by SDS-PAGE and then transferred to a PVDF membrane (PerkinElmer Life Sciences). The ES-4 primary antibody was used at 1:1000. Anti-ES-4 antibodies were raised in chickens (IgY) against a unique peptide sequence (LAKRQPQPHHN) in the C-terminal end of rat carboxylesterase ES-4 and affinity-purified by Genway (San Diego, CA.) Secondary antibody was used at 1:10,000 (anti-chicken-conjugated HRP, GeneTex, Inc., San Antonio, TX). Goat GAPDH primary antibody (Chemicon, Temecula, CA) was used at a concentration of 1:2000 with secondary goat anti-mouse-conjugated HRP used at (1:10,000) (Calbiochem-Novabiochem). For competition experiments, McA lysates (20 μg/well) were run on SDS-page gel and transferred to PVDF membranes. Three separate sections were cut out and incubated with anti-ES-4 antibody and 0 ng/ml, 1 ng/ml, or 5 ng/ml ES-4 peptide for 1 h. The membranes were washed and then incubated with secondary antibody at 1:10,000 (anti-chicken conjugated HRP). GAPDH was used as a loading control. Signals were displayed on x-ray film by chemiluminescence reagent.
Oil Red O Staining
McA cells were transfected with siControl or siES-4 RNAs, and after 24 h they were replated on collagen-coated Lab-Tek chamber slides (Nunc, Rochester, NY). The cells were then loaded with FC/cyclodextrin (Sigma) (molar ratio, 1:6 cholesterol/MβCD), 20 μg/ml, in serum-free media. After 24 h of loading, cells were equilibrated for 12 h and then fixed in 4% paraformaldehyde in PBS and stained with Oil Red O (Sigma) for neutral lipids (24).
Confocal Fluorescence Microscopy
McA cells were grown on collagen I-coated coverslips (BD Biosciences) to 90% confluence. The cells were fixed with ice-cold methanol for 20 min and incubated with 5% BSA (in phosphate-buffered saline) for 20 min prior to probing with antibodies. Anti-ES-4 chicken-antibody (Genway) was used to probe ES-4, followed by a 1-h incubation with goat anti-chicken IgY conjugated with FITC (Genway) as a secondary antibody. Subcellular compartments were localized with antibodies against lysosomal-associated membrane protein 1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for lysosomal compartments, and GM130 (Santa Cruz Biotechnology, Inc.) for cis Golgi and calnexin for the ER. Additionally, an ER dye (which binds to the sulfonylurea receptors of ATP-sensitive K+ channels that are prominent on the ER, Invitrogen) was also used to localize the endoplasmic reticulum network. For detection of ACAT-2, anti-ACAT-2 antibody made in rabbit was used at a concentration of 1:500 (Dr. Larry Rudel, Wake Forest University). The secondary antibody was either Alexa Fluor 594 conjugated with anti-rabbit IgG-TR (Invitrogen) or goat anti-mouse TR (Santa Cruz Biotechnology, Inc.). In some experiments with cholesteryl ester-loaded cells, Oil Red O staining of the CE droplets was performed along with immunoflourescence for ES-4 localization. All incubations and washes were performed at room temperature. After immunostaining, the coverslips were mounted onto a glass slide using mounting media (Vectashield, Vector Laboratories, Burlingame, CA). Images were captured by a Zeiss LSM 510 Meta confocal microscope.
Immunohistochemistry
Formalin-fixed, paraffin-embedded rat liver tissue sections of 3–5 μm were incubated in an oven at 37 °C for 16 h. After deparaffinizing and rehydration in graded ethanol, sections were pretreated with 3% hydrogen peroxide, blocked with 3% BSA plus 1.28% normal goat serum (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and then incubated with chicken antibodies to rat ES-4 (Genway) overnight at 4 °C. Tissue was then incubated serially with a biotin-conjugated donkey anti-chicken secondary antibody (Jackson ImmunoResearch Laboratories, Inc.), an anti-biotin IgY conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc.), and diaminobenzidine. They were counterstained with hematoxylin (Fisher Scientific, Pittsburgh, PA). Sections were mounted using Vectashield (Vector Laboratories). No immunostaining was observed in the controls (data not shown). Hydrogen peroxide and BSA were obtained from Sigma. Normal goat serum, secondary, and tertiary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. Immunoflourescence competition assays were performed by adding (0.05, 0.5, or 5 ng/ml) of ES-4 peptide in the presence of ES-4 antibody (5 ng/ml) to liver sections. The sections were then washed and incubated with anti-chicken antibody conjugated with Alexa Fluor 488 (1:400, Molecular Probes). Sections were then mounted using Vectashield containing DAPI. Similar experiments were also performed using the Vectastain ABC (peroxidase) system and following the manufacturer's protocol (Vector Laboratories).
Esterase Assay
The nonspecific esterase assay was adapted from Beaufay et al. (25). Cell lysates from cells transfected with control vector, ES-4 vector, siControl or siES-4 RNAs were diluted in K2PO4, Triton X-100, and EDTA buffer to a final concentration of 10 μg of total protein. The assay was initiated by the addition of 1 mm p-nitrophenyl acetate (Sigma) in cold methanol. The conversion to p-nitrophenol was monitored spectrophometrically at 420 nm at 5-min intervals.
Cholesterol Loading and Cholesterol/Cholesteryl Ester Mass Measurements
McA cells were incubated with FC-cyclodextrin complexes (20 μg/ml of cholesterol) following a published protocol (26). Briefly, cells were incubated with the complexes for 24 h in serum-free media. In experiments with radiolabeled [3H]cholesterol (PerkinElmer Life Sciences), 0.5 μCi/ml (specific activity, 40 Ci/mmol) was also added to the medium. In primary hepatocytes, the cells were not loaded with FC-cyclodextrin complexes because of detectable levels of CE. To determine cholesterol mass content, cells were washed twice in PBS, and the lipids were extracted overnight with isopropanol. Total cholesterol was determined with a cholesterol E kit (Wako, Richmond, VA), and FC was determined with a Free cholesterol kit (Wako). CE was determined by subtracting FC from total cholesterol. Protein content was determined by Dc protein assay (Bio-Rad).
CE Hydrolysis by Cell Lysates
Purified cholesterol droplets containing [3H]CE were prepared as described before (27, 28). Neutral CE hydrolase activity was assessed in a total reaction volume of 0.4 ml. Reaction mixtures contained 0.1 m Tris-maleate buffer (pH 7) and 10 μg of cell homogenate protein from McA cells transfected with vector control or ES-4 expression vector. Assays were initiated within 10 min of homogenization by the addition of radiolabeled CE droplets. The droplets were added at 40,000–80,000 cpm per incubation. One portion of the cell homogenate was boiled for 5 min before addition of the droplets to act as a negative control. Samples were incubated at 37 °C for 4 h. Reactions were terminated by the addition of organic solvents and extraction of the product by the method of Bligh and Dyer (29).
The lipid extract was dried under nitrogen and resuspended in 50 μl of chloroform/methanol. Of this, 20 μl were spotted on instant TLC plates, and the plates were run in 90:10:1 pet ether:ethyl ether:acetic acid. Non-labeled unesterified and esterified cholesterol standards were run in separate lanes on each plate to determine where the lipids migrated to on the plate. After running the plates in solvent, they were exposed to iodine to visualize the lipid on the plates. Spots corresponding to the unesterified and esterified cholesterol were cut and counted to determine percent 3H in each lipid class.
CE Hydrolysis in Intact Cells
McA cells were transfected with the control vector, ES-4 expression vector, control siRNA, or ES-4 siRNA for 24 h. Cells were then incubated with FC-cyclodextrin complexes (20 μg/ml) and 0.5 μCi [H3]cholesterol for 24 h in serum-free medium. In primary hepatocytes, cells were not loaded with cholesterol because in preliminary experiments, rat liver contained enough endogenous CE mass to detect without exogenous cholesterol loading. 0.5 μCi [3H]cholesterol was added 24 h after transfection for 24 h. Cells (McA or primary hepatocytes) were washed three times in PBS, and lipids from time zero plates were isolated using isopropanol. Re-esterification of hydrolyzed CE was prevented by the addition of ACAT inhibitor F-1394 (1 μm) for 12 h as described before (30). After 12 h, samples were washed three times in PBS, and lipids were extracted using isopropanol. The lipids were then dried under nitrogen, and CE and FC was determined using the instant TLC method described above and in Ref. 31. Protein levels were determined by Dc protein assay (Bio-Rad). CE hydrolysis was determined by comparison of the CE content (per mg of cell protein) between the 12 h and zero time points.
Adenovirus Generation and Expression
ES-4 cDNA was subcloned into the DUEL-CCM-EGFP plasmid (Vector Biolabs, Philadelphia, PA). The plasmid was then used by Vector Biolabs to generate an adenovirus expressing ES-4. Control adenovirus containing GFP was also obtained from Vector Biolabs. ApoE-deficient mice (apoE−/−) were given retro-orbital sinus injections of purified recombinant adenovirus (ES-4 or GFP) at a concentration of 109 plaque-forming units (n = 4 per group). Two weeks after the injection, mice were harvested, and livers were collected. Lipid and protein concentrations were determined as described above.
RESULTS
Expression of ES Family Members in Rat Hepatic Cell Types
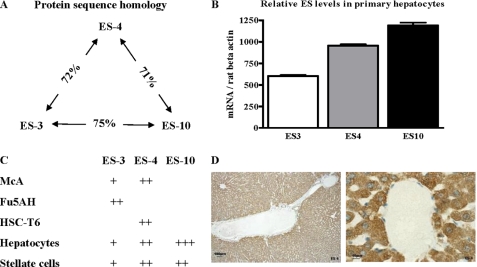
ES-4 and ES-10 have been shown to be expressed at the protein level by Western blotting of rat liver lysates (21, 32). We screened primary hepatocyte and stellate cells isolated from rat liver, as well as corresponding transformed cell lines, for the expression of ES-3, ES-4, and ES-10 mRNAs to help select appropriate models for in vitro studies. All three family members are highly related and were expressed at the RNA level in rat primary hepatocytes (Fig. 1, A–C), consistent with our previous results (21). Immunohistochemistry of rat liver sections showed diffuse cytoplasmic expression of ES-4 (Fig. 1D). In the McA cell line, ES-10 mRNA was not detectable, whereas ES-4 mRNA had a higher level of expression than that of ES-3 (Fig. 1C). In another rat hepatic cell line, Fu5AH, only ES-4 was expressed (Fig. 1C). Stellate cells play important roles in retinyl ester metabolism, therefore we also examined ES expression in primary stellate cells (isolated from rat liver) and in the rat stellate cell line HSC-T6 because some carboxylesterases can hydrolyze both retinyl and cholesteryl esters. As shown in Fig. 1C, the primary stellate cells contained mRNAs for all three esterases, whereas the stellate cell line expressed predominately ES-4.
FIGURE 1.
Carboxylesterase family members and their relative expression in rat liver and rat cell lines. A, protein sequence comparisons of the three major carboxylesterases (ES-3, ES-4, and ES-10) in the rat liver., showing the degrees of homology as percent. B, relative esterase mRNA levels in rat primary hepatocytes as determined by RT-PCR (mean ± S.E.). C, relative ES-3, ES-4, and ES-10 mRNA levels in rat hepatic cell lines (McA, Fu5AH), a rat stellate cell line (HSC-T6), and primary rat hepatocytes and stellate cells. D, rat liver sections were immunostained for ES-4 (brown). Note the diffuse expression in hepatocytes, which is more clearly visible under higher power (right panel).
With our main interest in hepatic parenchymal CE metabolism, and because ES-4 is highly expressed in all three hepatic cell types as well as in hepatocytes in intact rat liver (Fig. 1D), we initially focused on the effects of ES-4 in McA cells (derived from a rat hepatoma), a commonly used and easily transfected model for rat hepatic lipid and lipoprotein metabolism.
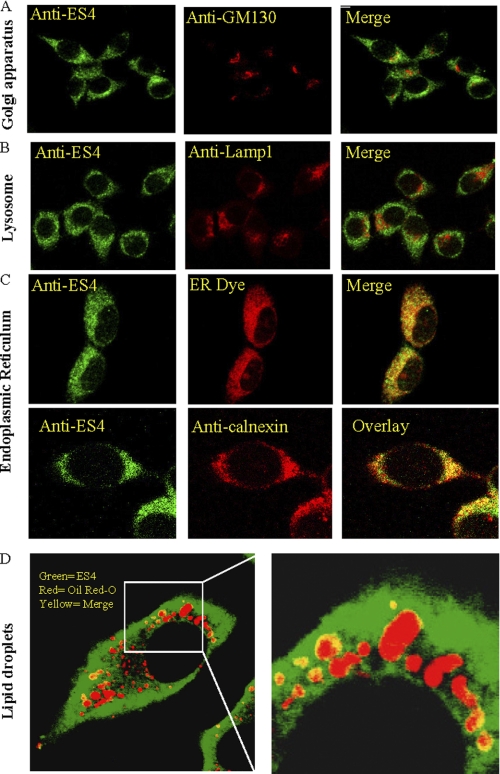
Subcellular Localization of ES-4 in McA Cells
ES-4 antibody specificity was tested in a number of ways (supplemental Fig. 1). In one approach, the ES-4 peptide used to generate the anti-ES-4 antibody competed for signal in Western blot analyses of McA cell lysates and in immunofluorescence and immunohistochemical experiments in rat liver. In another experiment, RAW macrophages, which do not express ES-4, had no detectable immunofluorescent signal until they were transfected with an ES-4 expression vector. We also examined the specificity of ES-4 antibody in McA cells. These cells already express ES-4, and when they were transfected with ES-4-GFP, the distribution of the immunofluorescent signal resembled that of GFP fluorescence.
With the specificity for the ES-4 antibody for immunohistochemistry established, we then performed additional confocal microscopic studies of McA cells (Fig. 2). As shown, there was little, if any, ES-4 present in the Golgi or lysosomal compartments, as ES-4 was not colocalized with GM130, a cis-Golgi matrix protein, or with glycosylated type 1 lysosome-associated membrane proteins (Lamp1) (Fig. 2, A and B). In contrast, ES-4 immunostaining was found to be almost exclusively associated with the ER (Fig. 2C), as judged by its colocalization with an ER tracker dye and with calnexin (as well as with ACAT2, a known ER protein, supplemental Fig. 2). Additional experiments were performed in which Golgi and the ER fraction of cell homogenates were separated using sucrose gradients (33). Probing for ES-4 showed that it was restricted to the ER fractions (data not shown).
FIGURE 2.
Subcellular localization of ES-4 in McA cells. McA cells were plated on collagen-coated coverslips. A–D, the cells were fixed, and ES-4 was probed with anti-ES-4 antibody followed by anti-rabbit FITC-conjugated secondary antibody (green). Subcellular compartments were identified using an antibody to cis-Golgi, GM130 (A) or to the lysosome (anti-Lamp1) (B). C, an ER-specific dye or anti-calnexin antibody was used to identify the endoplasmic reticulum. The secondary antibodies (either Alexa Fluor 594 conjugated with anti-rabbit IgG (Invitrogen) or goat anti-mouse TR) are shown in red. Colocalization of ES-4 with a subcellular compartment is indicated in yellow. D, to determine whether ES-4 colocalizes with lipid droplets, McA cells were cholesterol-loaded for 48 h (see “Experimental Procedures”) and fixed and stained with Oil Red O and with FITC-labeled anti-ES-4 antibody (green). As seen in the low- and higher-magnification micrographs, there are areas (yellow) in which cholesteryl ester droplets (red) show colocalization with ES-4 (green). All images were captured using a Zeiss LSM 510 Meta confocal microscope.
Overall, the data suggest that ES-4 is associated with the ER, a known location of cholesterol, CE, and triglyceride metabolism. In addition, when the McA cells were loaded with cholesterol, CE droplets accumulated in the cytoplasm (Fig. 2D), and some were contiguous with the ER, as shown by their proximity to the ES-4 staining. Significantly, there were numerous, discrete areas where we observed colocalization of ES-4 and the lipid droplet surface, as shown in the enlarged image where the two signals are merged (Fig. 2D).
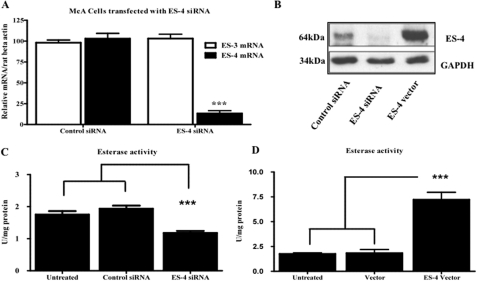
Effects of ES-4 Under- and Overexpression on Esterase Activity in McA Cells
McA cells were transfected with either control siRNA or an ES-4 siRNA “smart pool,” and 48 h following transfection, RNA was purified and quantified by Taqman PCR. As shown in Fig. 3A, in ES-4 siRNA-treated cells there was a 90% reduction in ES-4 mRNA compared with the control. We also tested the ability of ES-4 siRNA to modulate ES-3, a closely related family member also expressed in McA cells. As shown in Fig. 3A, there was little to no change in ES-3 mRNA abundance. Treatment of McA cells with ES-4 siRNA also led to a significant reduction (by approximately 90%) of ES-4 protein levels (Fig. 3B). We also tested the effects of the rat ES-4 siRNA on the expression of its mouse homologue, which has a 90% sequence homology to the rat gene. The results showed that rat ES-4 siRNA is extremely species-specific (supplemental Fig. 3).
FIGURE 3.
Altering ES-4 levels changes total esterase activity in McA cells. McA cells were transfected for 72 h with control siRNA or ES-4 siRNA. A, RNA was isolated, and mRNA levels for ES-4 and ES-3 were quantified using Taqman PCR. B, Western blot analyses were performed on lysates from cells transfected with control siRNA, ES-4 siRNA, and ES-4 expression vector and probed for ES-4 and GAPDH (control). Shown is an example representative of three separate blotting experiments. C, lysates from untreated cells or cells transfected with control or ES-4 siRNAs were assayed for general esterase activity using p-nitrophenyl acetate as substrate (see “Experimental Procedures”). D, lysates from untreated cells or cells transfected with empty vector (Vector) or ES-4 expression vector were assayed for general esterase activity. The results for A, C, and D are from experiments done twice in triplicate wells and are expressed as mean ± S.E. Statistical analysis was by two-tailed Student's t test (A) or one-way analysis of variance followed by Bonferroni's multiple comparison test (C and D). ***, p < 0.001.
Overexpression of ES-4 was accomplished by transfection with a plasmid containing rat hepatic ES-4 cDNA. McA cells were also transfected with an empty vector (control). After 48 h, lysates were analyzed by Western blot analysis using the ES-4 antibody. The results showed a 5-fold increase in ES-4 protein levels compared with control transfection (Fig. 3B), although this was still below that found in wild-type rat primary hepatocytes (data not shown).
We next evaluated the effects of changing ES-4 protein levels on total esterase activity. ES-4 catalyzes hydrolysis in vitro of p-nitrophenyl, as demonstrated previously in transfected COS cells (19, 34). Lysates from McA cells transfected with siRNA (control or siES-4) or with control or ES-4 expression vector, were used as enzyme sources for the “nonspecific” esterase assay (25). As shown in Fig. 3C, cells transfected with control siRNA showed no change in esterase activity. In contrast, cells treated with ES-4 siRNA exhibited a 40% decrease (p < 0.001) in esterase activity compared with control (1.18 ± 0.15 versus 1.94 ± 0.23 units/mg, respectively). When ES-4 protein was overexpressed, the esterase activity was increased (p < 0.001) approximately 3-fold compared with the control (7.215 ± 1.82 versus 1.83 ± 0.89 units/mg, Fig. 3D), similar to a previous report (19). These experiments show that changes in the ES-4 expression levels in McA cells lead to corresponding changes in esterase activity. The siRNA results in particular indicate that this single enzyme makes a major contribution to the carboxylesterase activity in McA cells.
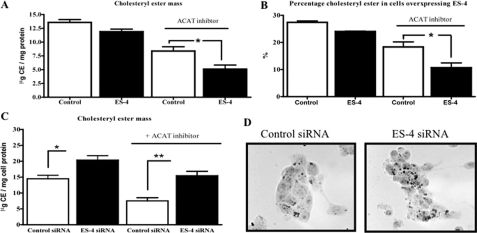
ES-4 Expression Levels Determine CE Mass in Cholesterol-loaded McA Cells
With the ability to manipulate functional ES-4 levels (Fig. 3), we now turned to the metabolic effects of the enzyme on cellular cholesterol metabolism. McA cells under typical culture conditions, like most cells, have little CE. Thus, 24 h after first transfecting with siRNA (control or siES-4) or with control or ES-4 expression vector, the cells were loaded with FC-cyclodextrin complexes for 24 h (26) to promote CE formation. Compared with the unloaded cells, the cellular pools of cholesterol species in the loaded cells were approximately 215% (total cholesterol), approximately 170% (FC), and approximately 525% (CE) (data not shown).
After cholesterol loading, the cells were incubated for 12 h without or, to prevent re-esterification of FC (30, 35), with an ACAT inhibitor (see “Experimental Procedures”), and the amount of CE mass was measured. As shown in Fig. 4A, in cells in which the CE cycle (hydrolysis and re-esterification) was ongoing, when ES-4 expression was increased, there was a slight but non-significant decrease in CE content (13.57 ± 0.93 versus 12.13 ± 0.93 μg/mg protein). In contrast, when re-esterification was blocked by the ACAT inhibitor, in cells transfected with either the ES-4 expression or control vector, as expected, there were decreases in CE content (Fig. 4A). Notably, there was a greater decrease (p < 0.05) in CE mass in ES-4-transfected compared with control-transfected cells (5.09 ± 1.27 versus 8.36 ± 1.35 μg/mg protein, respectively). When CE content was calculated as a percentage of total cellular cholesterol, there was still a similar quantitative impact of ES-4 expression with a significant 40% reduction of CE in the ES-4 overexpressing versus control cells (Fig. 4B). Overall, the data demonstrate that the increased expression of ES-4 not only increases esterase activity (Fig. 3C) but also decreases the net amount of CE in cholesterol-loaded cells (Fig. 4A). Although these data suggest that ES-4 is a bona fide hepatic nCEH, the data are also consistent with results that could be obtained from the increased expression of a non-physiologically relevant esterase.
FIGURE 4.
ES-4 levels regulate cholesteryl ester mass in McA cells. A, McA cells were transfected with control vector (Control) or ES-4 expression vector, and cholesterol-loaded (see “Experimental Procedures”) with or without ACAT inhibitor. CE mass was measured in the cell lysates. B, the data from A are expressed as the percent of total cellular cholesterol that was CE. C, McA cells were transfected with either control or ES-4 siRNA, and an experiment similar to the one in A with and without ACAT inhibitor was performed. D, Oil Red O staining of CE droplets in McA cells treated with control or ES-4 siRNAs as in C. For A--C, the results are representative of two experiments performed in triplicate wells (mean ± S.E.). Statistical analysis was determined by two-tailed Student's t test. *, p < 0.05, **, p < 0.01.
Thus, to obtain more rigorous support for ES-4 as an endogenous hepatic nCEH, a set of underexpression studies was conducted. In the absence of the ACAT inhibitor, McA cells treated with ES-4 siRNA accumulated significantly more CE after FC loading than the cells transfected with the control vector (Fig. 4C, 20.3 ± 4.2 versus 14.0 ± 2.4 μg/mg protein, or 144% of the control). These cholesterol-loaded cells were also stained with Oil Red O, which binds to CE, and those transfected with ES-4 siRNA had more stained lipid droplets compared with control vector-transfected cells (Fig. 4D). Increased CE content was also observed when cells were treated with ES-4 siRNA in the presence of the ACAT inhibitor (Fig. 4C), although the contents in both the control- and ES-4-treated cells were lower under the corresponding conditions but with the inhibitor absent (from the left, compare the first bar to the third bar and the second bar to the fourth bar). This difference most likely reflects the ongoing hydrolysis of CE by non-ES-4 CEH(s) and the inability to re-esterify the FC so generated to replenish the pool. Taken together, these experiments clearly show that increases or decreases in ES-4 expression translate into reciprocal changes in cellular CE content.
CE Mass Is Regulated by ES-4 at the Level of Turnover
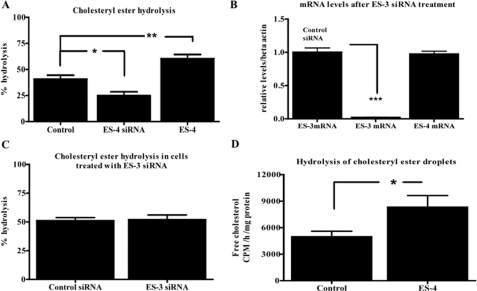
Given its esterase activity, the simplest explanation for the CE mass modulation is that ES-4 is a nCEH. To directly demonstrate this in living cells, we performed CE turnover studies by following protocols we have used previously (22, 36–38). ES-4 levels were modulated by transfecting either control vectors, ES-4 siRNA, or the ES-4 expression vector, and FC loading was done as described above. After the loading period, cellular pools of cholesterol were equilibrated in 0.2% BSA for 8 h. After equilibration, a set of wells was extracted, and the lipid fraction was reserved for baseline assays of CE. ACAT inhibitor (F-1394) was added for an additional 12 h to prevent re-esterification. The difference between CE before and after ACAT treatment in each group, therefore, represents CE hydrolysis.
The data showed (Fig. 5A) that in ES-4 siRNA-treated cells, CE hydrolysis was decreased by 38% compared with control cells, whereas in the ES-4 overexpressing cells, CE hydrolysis was increased by 48%. As another control for the specificity of the results for ES-4, we also tested the ability of ES-3 to hydrolyze CE by knocking down ES-3 expression (Fig. 5B), but there was no change in CE hydrolysis (C).
FIGURE 5.
ES-4 regulates cholesteryl ester turnover in McA cells. A, McA cells were transfected with either control or ES-4 siRNAs or with ES-4 expression vector (ES-4). After cholesterol loading, re-esterification of FC was prevented by the addition of the ACAT inhibitor, and an isotopic CE hydrolysis assay was performed (see “Experimental Procedures”). B, McA cells were transfected with either control siRNA (left bar) or with ES-3 siRNA. Abundance of ES-3 mRNA (two left bars) or ES-4 mRNA (right bar) was then measured by Taqman PCR. C, a CE hydrolysis assay similar to that in A was performed in McA cells transfected with either control or ES-3 siRNA. D, droplets containing [3H]CE were isolated and used as substrate for a CE hydrolysis assay using lysates from McA cells either untreated (Control) or transfected with ES-4 expression vector (ES-4) as the enzyme source (see “Experimental Procedures”). All experiments were done twice in triplicate (data are mean ± S.E.). Statistical analysis was determined by one-way analysis of variance followed by Bonferroni's multiple comparison test (A) or two-tailed Student's t test (B, C, and D). *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
In addition, given the colocalization of lipid droplets and ES-4 in McA cells (Fig. 2D), we performed experiments using isolated CE lipid droplets in which they were presented in vitro to lysates of McA cells or of ES-4-transfected McA cells. The data showed (Fig. 5D) that cell lysates with increased ES-4 expression had a significant (approximately 67%) increase in CE hydrolysis compared with that of control cell lysate, thereby indirectly linking lipid droplet turnover to ES-4 levels. This is consistent with the data from the intact McA cells. Overall, the results in Fig. 5 establish that the changes in CE mass associated with ES-4 levels are attributable to its activity as a bona fide endogenous nCEH.
ES-4 Regulates CE Metabolism in Rat Primary Hepatocytes
The results so far make a strong case for ES-4 being a nCEH in rat hepatic cells. Although lipid/lipoprotein metabolism in McA cells resembles that in primary hepatocytes, there is still a chance for cell type differences between transformed and non-transformed cells to confound our interpretation. Thus, we investigated the effect of ES-4 on CE metabolism in rat primary hepatocytes.
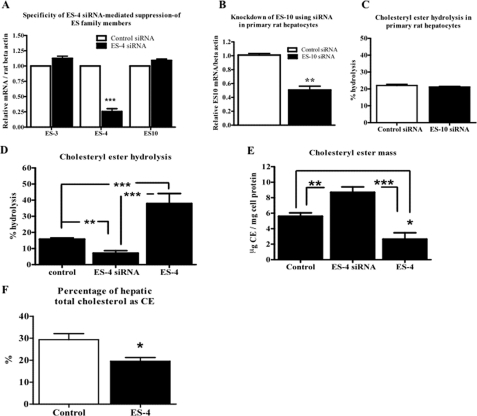
As shown in Fig. 1C, there was no expression of ES-10 in McA cells, but there was detectable mRNA in primary hepatocytes. Because ES-10, or a highly related enzyme, has been proposed by Ghosh and colleagues (14) to be a hepatic nCEH, we first tested whether the siRNA designed to decrease ES-4 expression cross-suppressed ES-10. As shown in Fig. 6A, after 48 h of transfection of rat primary hepatocytes with the siRNAs, there was a 75% decrease in ES-4 mRNA but no change in ES-3 or ES-10 mRNA. Thus, ES-4 expression was specifically inhibited in primary hepatocytes by ES-4 siRNA.
FIGURE 6.
ES-4 regulates cholesteryl ester turnover in primary rat hepatocytes and cholesteryl ester content in vivo. A, primary rat hepatocytes were transfected with either control siRNA (white bars) or ES-4-siRNA (black bars). RNA was isolated 48 h later, and mRNAs for ES-3, ES-4, and ES-10 were quantified and normalized to rat β-actin by Taqman PCR. B, cells were transfected with control siRNA or ES-10 siRNA, and ES-10 mRNA abundance was quantified. C, in parallel plates (after 48 h of transfection with either control or ES10 siRNAs), ACAT inhibitor was added to prevent the re-esterification of FC, and an isotopic CE hydrolysis assay was performed (see “Experimental Procedures”). D, cells were transfected with control vector, ES-4 expression vector, or ES-4 siRNA, and CE hydrolysis assays were performed. E, cells were transfected and treated as in D, and at the end of the experiment, CE mass was measured. F, ES-4 was expressed in apoE-deficient mice using an adenoviral vector (ES-4), with control apoE-deficient mice treated with an adenoviral vector expressing GFP. Two weeks after administration of the viral vectors, liver samples were assayed, and the percentage of the total cholesterol pool that was CE was calculated. A, D, and E summarize results from two experiments performed in triplicate wells. F summarizes results from four mice. Data are displayed as mean ± S.E. Statistical analysis was determined by two-tailed Student's t test (A, B, C, and F) or one-way analysis of variance followed by Bonferroni's multiple comparison test (D and E). *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
To directly explore the contribution of ES-10 to CE hydrolysis, siRNA was used to suppress it (Fig. 6B). The results showed (Fig. 6C) that the suppression of ES-10 did not change CE hydrolysis. In contrast, the suppression of ES-4 led to a significant decrease in CE hydrolysis (55% of control), whereas overexpression of ES-4 led to a significant increase (240% of control) (Fig. 6D). As expected from the hydrolysis data and as shown in Fig. 6E, the suppression of ES-4 resulted in a significant increase in CE mass (155% of control), and overexpression of ES-4 led to a significant decrease in CE mass (53% of control).
We extended the findings in the primary hepatocytes by examining the consequences of expressing ES-4 (by adenoviral vector) in apoE−/− mice, a standard model of atherosclerosis. As shown in Fig. 6F, compared with mice in which a control vector was injected, the percentage of total liver cholesterol that was CE was reduced by 34%, consistent with increased hydrolysis in vivo.
We also examined the correlation in primary hepatocytes between ES-4 protein expression and CE hydrolysis using the data from cells in which ES-4 expression was at wild-type levels, below, or above (the latter as consequences of the siRNA or expression vector treatments, respectively). The relationship between CE hydrolysis and ES-4 expression levels was linear over a wide range (r2 = 0.86), suggesting that if ES-4 is a regulated and not a constitutive enzyme, then variations in its activity would be expected to be reflected in changes in CE hydrolysis.
Overall, the primary hepatocyte data demonstrate that the effects of ES-4 on CE metabolism are not confined to a transformed hepatic cell and that based on the siRNA data, this enzyme can account for up to 55% of CE hydrolysis.
DISCUSSION
The classic observations of Brown and Goldstein (39) in macrophages that the CE pool turns over rapidly stimulated the hunt for the esterifying and hydrolyzing enzymes. The cloning of ACAT (40) and the subsequent identification of its two isoforms, ACAT1, widely expressed (e.g. in macrophages and intestine), and ACAT2, expressed primarily in liver, have led to greater understanding of the esterification reaction. Similar progress has lagged in the understanding of the enzyme(s) mediating hydrolysis. In this study, we have provided multiple lines of evidence demonstrating that ES-4 is a bona fide and potent hepatic nCEH. The results were consistent between the over- and underexpression studies in different transformed and primary hepatic cell types.
Initially ES-3, ES-4, and ES-10, all of which are members of the rat carboxylesterase family, were considered as candidates. These microsomal enzymes are expressed in liver, and have been shown to hydrolyze a variety of lipid esters in vitro. Initial experiments on expression levels (Fig. 1) and localization of ES-4 to the ER and to the surface of cholesterol lipid droplets (Fig. 2, C and D) implicated ES-4 as a lead candidate among this group. Fortunately, a standard model of rat hepatic lipoprotein and lipid metabolism, McA cells (41–43), has detectable ES-4 mRNA and protein levels, making it a convenient system for performing experiments in which ES-4 levels can be decreased or increased under conditions in which CE content can be easily manipulated.
Although the increased expression data in hepatic cells showing elevated CE hydrolytic activity and, in cells and intact liver, decreased CE content were consistent with ES-4 acting as a nCEH, it did not establish this as its biological function under normal circumstances. More convincing were the data showing that when endogenous ES-4 mRNA and protein expression were inhibited by siRNA, increased CE mass and decreased CE hydrolysis resulted in both McA cells and rat primary hepatocytes. Importantly, CE hydrolysis was measured in living cells using an intact cell assay developed and validated by Rothblat and colleagues (37). Also of note, despite the sequence similarities within the ES family, which could potentially lead to functional redundancies or effects of a siRNA beyond its intended target, the results were specific to ES-4 on the basis of the lack of changes in the levels of ES-3 and ES-10 mRNA in cells treated with ES-4 siRNA (Figs. 3A and 6A), in the level of CE hydrolysis in McA cells (which express ES-3) with ES-3 siRNA treatment (Fig. 5, B and C), and in the level of CE hydrolysis in rat primary hepatocytes (which express ES-10) with ES-10 siRNA treatment (Fig. 6).
Consistent with the CE hydrolysis data are the subcellular localization studies. First, in intact liver, ES-4 was largely expressed in hepatocytes (Fig. 1D). In McA cells it was associated with the ER (Fig. 2). Upon cholesterol loading, the CE droplets were found dispersed in the cytoplasm, contiguous with the reticular network of the ER (Fig. 2), with points of contact with ES-4. These may represent regions of nascent droplet formation resulting from ACAT-catalyzed formation of CE and would suggest that these are sites of both synthesis and hydrolysis of CE.
We noted earlier that there have been a number of failed candidates for the rat hepatic nCEH. Possibly the candidate most studied to date has been the enzyme that Ghosh and colleagues (14) identified. This enzyme is likely to be ES-10 (the reported sequence differs from that of ES-10 by only four amino acids). Only indirect evidence was provided for its ability to hydrolyze CE in liver cells. As noted earlier, the primary supporting data were obtained by overexpressing the cDNA in non-hepatic cells (44). Also, limiting the role of ES-10 in hepatic CE metabolism are our previous demonstration that purified ES-10 is a retinyl ester hydrolase with no nCEH activity in vitro (21) and the data in this study (Fig. 6) showing no effect of ES-10 siRNA on CE hydrolysis in rat primary hepatocytes.
Another major candidate for rat liver nCEH was the pancreatic carboxylester lipase. We have previously excluded it from having a major role in bulk hepatic CE hydrolysis (36), but in more recent studies, Howlett and colleagues (45) have shown that carboxylester lipase may hydrolyze esters derived from selective uptake from HDL.
In a recent study, hormone-sensitive lipase has also emerged as a candidate hepatic nCEH. Hormone-sensitive lipase-deficient mice had higher levels of CE compared with WT mice and had reduced CE hydrolysis in lysate assays (15). The results were weakened by no direct measurements of CE hydrolysis in intact cells and by the mice having increased ACAT activity in both liver lysates and cultured hepatocytes. Nonetheless, even accepting hormone-sensitive lipase as a hepatic nCEH, there was still significant activity unaccounted for in the liver and hepatocyte lysates (66 and 40%, respectively), which would be compatible with the approximately 55% of CE hydrolysis we can explain by ES-4 in intact rat primary hepatocytes.
In future experiments, promising areas to explore are the effects of ES-4 on bile acid production and the CE content of VLDL. Both pathways are relevant to cardiovascular disease risk. For example, the loss of hepatic cholesterol in the form of bile acid would be expected to increase LDL receptor activity and increase LDL clearance from the plasma. Lipid compositional changes in VLDL also may have important consequences, as suggested by the work of Rudel and colleagues. For example, they have shown that ACAT2 regulates the CE content of VLDL (and ultimately, that of LDL) and that this content, in turn, is a determinant of the atherogenicity of LDL. Indeed, in ACAT2 KO/apoE KO mice, atherosclerosis progression was decreased (46).
It will also be interesting to pursue the regulation of ES-4 itself. As for many factors involved in cholesterol metabolism, its expression may be subject to feedback regulation, although in this case of a positive variety on the basis of preliminary observations that after cholesterol-loading, ES-4 mRNA levels were increased in McA and Fu5AH cells by 4.5-fold compared with non-loaded cells (data not shown).
ES-4 may also be an important nCEH in tissues other than liver. Although in some tissues (e.g. adrenal tissue (47) and macrophages (48)) strong candidates have been identified, for the majority of other tissues and cell types the search for the nCEH(s) remains open. Although establishing ES-4 as a major nCEH in liver cells fills a long-standing gap in our understanding of hepatic CE turnover, continued investigation in vitro and in vivo will be needed to fully illuminate its roles in mammalian lipid metabolism.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK44498 (to E. H. H. and E. A. F.), HL58541 (to E. A. F.) and by Postdoctoral Fellowship F32-HL087627–1 (to S. P.). We would also like to thank Lisa Grauer and Courtney Blachford for expert technical assistance.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- FC
- free cholesterol
- CE
- cholesteryl ester
- ER
- endoplasmic reticulum
- ACAT
- acyl-coenzyme A:cholesterol acyltransferase
- VLDL
- very low density lipoprotein
- nCEH
- neutral cholesteryl ester hydrolase
- McA
- McArdle RH7777
- HSL
- hormone-sensitive lipase.
REFERENCES
- 1. Brown M. S., Goldstein J. L. (1983) J. Clin. Invest. 72, 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown M. S., Goldstein J. L. (1984) Sci. Am. 251, 58–66 [DOI] [PubMed] [Google Scholar]
- 3. Brown M. S., Goldstein J. L. (1986) Science 232, 34–47 [DOI] [PubMed] [Google Scholar]
- 4. Tabas I. (2002) J. Clin. Invest. 110, 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parini P., Davis M., Lada A. T., Erickson S. K., Wright T. L., Gustafsson U., Sahlin S., Einarsson C., Eriksson M., Angelin B., Tomoda H., Omura S., Willingham M. C., Rudel L. L. (2004) Circulation 110, 2017–2023 [DOI] [PubMed] [Google Scholar]
- 6. Liang J. J., Oelkers P., Guo C., Chu P. C., Dixon J. L., Ginsberg H. N., Sturley S. L. (2004) J. Biol. Chem. 279, 44938–44944 [DOI] [PubMed] [Google Scholar]
- 7. Brewer H. B., Jr. (2000) J. Clin. Invest. 105, 703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hajjar D. P., Weksler B. B. (1983) J. Lipid Res. 24, 1176–1185 [PubMed] [Google Scholar]
- 9. Hoeg J. M., Demosky S. J., Jr., Brewer H. B., Jr. (1982) Biochim. Biophys. Acta 711, 59–65 [DOI] [PubMed] [Google Scholar]
- 10. Hajjar D. P., Minick C. R., Fowler S. (1983) J. Biol. Chem. 258, 192–198 [PubMed] [Google Scholar]
- 11. Durham L. A., 3rd, Grogan W. M. (1984) J. Biol. Chem. 259, 7433–7438 [PubMed] [Google Scholar]
- 12. Nishide T., Sasaki N., Shirai K., Saito Y., Yoshida S. (1985) Tohoku J. Exp. Med. 146, 123–130 [DOI] [PubMed] [Google Scholar]
- 13. Zhao B., Song J., Chow W. N., St Clair R. W., Rudel L. L., Ghosh S. (2007) J. Clin. Invest. 117, 2983–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghosh S., Mallonee D. H., Hylemon P. B., Grogan W. M. (1995) Biochim. Biophys. Acta 1259, 305–312 [DOI] [PubMed] [Google Scholar]
- 15. Sekiya M., Osuga J., Yahagi N., Okazaki H., Tamura Y., Igarashi M., Takase S., Harada K., Okazaki S., Iizuka Y., Ohashi K., Yagyu H., Okazaki M., Gotoda T., Nagai R., Kadowaki T., Shimano H., Yamada N., Ishibashi S. (2008) J. Lipid Res. 49, 1829–1838 [DOI] [PubMed] [Google Scholar]
- 16. Kraemer F. B., Shen W. J. (2002) J. Lipid Res. 43, 1584–1592 [DOI] [PubMed] [Google Scholar]
- 17. Mentlein R., Ronai A., Robbi M., Heymann E., von Deimling O. (1987) Biochim. Biophys. Acta 913, 27–38 [DOI] [PubMed] [Google Scholar]
- 18. Sanghani S. P., Davis W. I., Dumaual N. G., Mahrenholz A., Bosron W. F. (2002) Eur. J. Biochem. 269, 4387–4398 [DOI] [PubMed] [Google Scholar]
- 19. Robbi M., Van Schaftingen E., Beaufay H. (1996) Biochem. J. 313, 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mentlein R., Rix-Matzen H., Heymann E. (1988) Biochim. Biophys. Acta 964, 319–328 [DOI] [PubMed] [Google Scholar]
- 21. Linke T., Dawson H., Harrison E. H. (2005) J. Biol. Chem. 280, 23287–23294 [DOI] [PubMed] [Google Scholar]
- 22. Vogel S., Piantedosi R., Frank J., Lalazar A., Rockey D. C., Friedman S. L., Blaner W. S. (2000) J. Lipid Res. 41, 882–893 [PubMed] [Google Scholar]
- 23. Riccalton-Banks L., Bhandari R., Fry J., Shakesheff K. M. (2003) Mol. Cell. Biochem. 248, 97–102 [DOI] [PubMed] [Google Scholar]
- 24. Rong J. X., Shapiro M., Trogan E., Fisher E. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13531–13536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beaufay H., Amar-Costesec A., Feytmans E., Thinès-Sempoux D., Wibo M., Robbi M., Berthet J. (1974) J. Cell Biol. 61, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christian A. E., Haynes M. P., Phillips M. C., Rothblat G. H. (1997) J. Lipid Res. 38, 2264–2272 [PubMed] [Google Scholar]
- 27. Minor L. K., Rothblat G. H., Glick J. M. (1989) J. Lipid Res. 30, 189–197 [PubMed] [Google Scholar]
- 28. Kellner-Weibel G., McHendry-Rinde B., Haynes M. P., Adelman S. (2001) J. Lipid Res. 42, 768–777 [PubMed] [Google Scholar]
- 29. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 30. Rong J. X., Kusunoki J., Oelkers P., Sturley S. L., Fisher E. A. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 122–127 [DOI] [PubMed] [Google Scholar]
- 31. Bernard D. W., Rodriguez A., Rothblat G. H., Glick J. M. (1990) Arteriosclerosis 10, 135–144 [DOI] [PubMed] [Google Scholar]
- 32. Poole M., Bridgers K., Alexson S. E., Corton J. C. (2001) Toxicology 165, 109–119 [DOI] [PubMed] [Google Scholar]
- 33. Gusarova V., Brodsky J. L., Fisher E. A. (2003) J. Biol. Chem. 278, 48051–48058 [DOI] [PubMed] [Google Scholar]
- 34. Mukherjee J. J., Jay F. T., Choy P. C. (1993) Biochem. J. 295, 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kusunoki J., Hansoty D. K., Aragane K., Fallon J. T., Badimon J. J., Fisher E. A. (2001) Circulation 103, 2604–2609 [DOI] [PubMed] [Google Scholar]
- 36. Zolfaghari R., Glick J. M., Fisher E. A. (1993) J. Biol. Chem. 268, 13532–13538 [PubMed] [Google Scholar]
- 37. Harrison E. H., Bernard D. W., Scholm P., Quinn D. M., Rothblat GH, Glick J. M. (1990) J. Lipid Res. 31, 2187–2193 [PubMed] [Google Scholar]
- 38. Glick J. M., Adelman S. J., Rothblat G. H. (1987) Atherosclerosis 64, 223–230 [DOI] [PubMed] [Google Scholar]
- 39. Brown M. S., Goldstein J. L., Krieger M., Ho Y. K., Anderson R. G. (1979) J. Cell Biol. 82, 597–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang C. C., Huh H. Y., Cadigan K. M., Chang T. Y. (1993) J. Biol. Chem. 268, 20747–20755 [PubMed] [Google Scholar]
- 41. Blackhart B. D., Yao Z. M., McCarthy B. J. (1990) J. Biol. Chem. 265, 8358–8360 [PubMed] [Google Scholar]
- 42. van Bennekum A. M., Li L., Piantedosi R., Shamir R., Vogel S., Fisher E. A., Blaner W. S., Harrison E. H. (1999) Biochemistry 38, 4150–4156 [DOI] [PubMed] [Google Scholar]
- 43. DeBose-Boyd R. A., Ou J., Goldstein J. L., Brown M. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ghosh S., Natarajan R., Pandak W. M., Hylemon P. B., Grogan W. M. (1998) Am. J. Physiol. 274, G662–668 [DOI] [PubMed] [Google Scholar]
- 45. Camarota L. M., Chapman J. M., Hui D. Y., Howles P. N. (2004) J. Biol. Chem. 279, 27599–27606 [DOI] [PubMed] [Google Scholar]
- 46. Willner E. L., Tow B., Buhman K. K., Wilson M., Sanan D. A., Rudel L. L., Farese R. V., Jr. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kraemer F. B., Shen W. J., Natu V., Patel S., Osuga J., Ishibashi S., Azhar S. (2002) Endocrinology 143, 801–806 [DOI] [PubMed] [Google Scholar]
- 48. Ghosh S. (2000) Physiol. Genomics 2, 1–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.