Background: Gle1 is a regulator of DEAD-box RNA helicases with roles in mRNA export and translation.
Results: Gle1 functionally interacts with the DEAD-box protein Ded1 in vitro and in vivo.
Conclusion: Gle1 functions in translation initiation as a negative regulator of Ded1.
Significance: Gle1 is poised to coordinate different steps of gene expression through control of multiple DEAD-box helicases.
Keywords: Inositol Phosphates, mRNA, Nuclear Transport, RNA Helicase, Translation Initiation Factors, Dbp5, mRNA Export, Start Site Selection
Abstract
DEAD-box protein (Dbp) family members are essential for gene expression; however, their precise roles and regulation are not fully defined. During messenger (m)RNA export, Gle1 bound to inositol hexakisphosphate (IP6) acts via Dbp5 to facilitate remodeling of mRNA-protein complexes. In contrast, here we define a novel Gle1 role in translation initiation through regulation of a different DEAD-box protein, the initiation factor Ded1. We find that Gle1 physically and genetically interacts with Ded1. Surprisingly, whereas Gle1 stimulates Dbp5, it inhibits Ded1 ATPase activity in vitro, and IP6 does not affect this inhibition. Functionally, a gle1–4 mutant specifically suppresses initiation defects in a ded1–120 mutant, and ded1 and gle1 mutants have complementary perturbations in AUG start site recognition. Consistent with this role in initiation, Gle1 inhibits translation in vitro in competent extracts. These results indicate that Gle1 has a direct role in initiation and negatively regulates Ded1. Together, the differential regulation of two distinct DEAD-box proteins by a common factor (Gle1) establishes a new paradigm for controlling gene expression and coupling translation with mRNA export.
Introduction
In eukaryotic cells, nuclear export of messenger (m)RNA is required to link the transcription, processing, and translation mechanisms. Recent studies have revealed several independent mechanisms by which these steps can be functionally coupled to one another. On the one hand, factors can be co-transcriptionally recruited to the mRNA for subsequent action. For example, during transcription and splicing, mRNA export factors are incorporated in the maturing mRNA-protein complex (mRNP)3 to allow targeting to and translocation through the nuclear pore complex (NPC) (1). Likewise, in Saccharomyces cerevisiae, specific RNA polymerase II subunits (Rpb4 and 7) are loaded into the mRNP and function during translation initiation (2). Alternatively, such factors can be multi-functional and execute several distinct steps in the gene expression pathway. This includes the cap-binding translation initiation factor eIF4E, which also has a role in nuclear export of a subset of cell cycle-related mRNAs (3). Furthermore, the DEAD-box protein Dbp5 is required for both mRNP export through NPCs and for translation termination (4–6). Investigating the mechanisms by which these factors are recruited and activated should reveal how gene expression is controlled between the nuclear and cytoplasmic compartments.
Previously, we and others have shown that Gle1 and a small molecule, inositol hexakisphosphate (IP6), are required for Dbp5 function (7–10). Gle1 is encoded by an essential gene in S. cerevisiae (11). It is conserved in metazoans (12), and mutation of human GLE1 is linked to a fatal developmental disorder termed lethal congenital contracture syndrome 1 (LCCS1) (13). Gle1 was first identified as an mRNA export factor, binding to the NPC through nucleoporin-42 (Nup42) in yeast and hCG1 and Nup155 in human cells (11, 14–16). Yeast Gle1 directly binds IP6 (8–10, 17), and Gle1:IP6 serves to enhance binding of ATP by Dbp5, thus allowing efficient RNA-stimulated ATPase activity (8, 9, 18). The conformational change to the ADP-bound state allows Dbp5 to promote changes in mRNP composition during mRNA export (18, 19). When Dbp5 binds directly to Nup159 on the cytoplasmic NPC face, ADP is released and Dbp5 is recycled for another round of Gle1-IP6 interaction and mRNP remodeling (18). The docking sites for both Dbp5 and Gle1:IP6 at the NPC are positioned such that mRNP remodeling is restricted to the NPC exit step. This is thought to confer directionality to the transport mechanism (4, 19–21).
Intriguingly, in a manner that is distinct from their roles in mRNA export at the NPC, Gle1, Dbp5, and IP6 are also all required for efficient translation (6, 10, 22). During translation termination, Gle1:IP6 and Dbp5 interact with eukaryotic release factor-1 (eRF1, Sup45 in yeast) and affect eRF3 (Sup35) association with the termination complex (6, 22). Remarkably, we also found that Gle1 plays a separate, uncharacterized role in translation initiation that is independent of both Dbp5 and IP6 (22). Translation initiation, defined as the assembly of translation-competent ribosomes on mRNA, requires a number of steps. Preinitiation complex (PIC) formation by the eIFs, the 40 S ribosomal subunit, and the mRNA precedes scanning by the PIC for the AUG start codon in a 5′ to 3′ direction (23). Once the PIC reaches an appropriate AUG, start site recognition takes place, resulting in a conformational change to a “closed,” scanning-incompetent form (for review, see Ref. 24). Dissociation of eIF1 then allows phosphate release by eIF2-GDP, committing the PIC to initiation at that site. Most of the eIFs dissociate, and eIF5B stimulates subunit joining, forming a competent 80 S ribosome. The mechanism of Gle1 function during initiation is unknown.
Several members of the ubiquitous DEAD-box protein family have been implicated in translation initiation (25–27). Ded1, a conserved, essential DEAD-box protein, has been proposed to function by unwinding structured RNA in the 5′ untranslated region to facilitate start site scanning by the 43S PIC (for review see Ref. 28). In this regard, the Ded1 role is similar to that proposed for eIF4A, although some data suggest that Ded1 is the more critical factor for this activity (29, 30). However, the exact role of Ded1 in initiation has remained unclear, and no regulatory factors have been identified. In this study, we define a novel functional interaction between Gle1 and Ded1. We show that Gle1 negatively regulates Ded1 in an IP6-independent manner and that this modulation is crucial for proper translation initiation. Further, we present data suggesting that the two proteins may play roles in AUG start site recognition. By demonstrating that a single factor, Gle1, can differentially modulate the activity of more than one DEAD-box protein (Dbp5 and Ded1), this work reveals a potential new mechanism for coupling the function of multiple DEAD-box proteins during the life cycle of an mRNA.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains and plasmids used are listed in supplemental Tables S1 and S2. SWY4092 (ded1::KAN +pDED1) was constructed by integration of a Kanamycin resistance cassette at the DED1 locus in a W303 strain background. Double mutants were generated by crossing ded1::KAN +pDED1 to other mutant strains via standard methods. Mutant ded1–120, DED1-GFP, and control strains were generated by plasmid shuffle. All strains are isogenic and generated from psi− parental strains, including gle1–4 (10). Yeast growth assays were performed by serial dilution as previously described (19). For Gal-induction growth assays, transformed strains were initially cultured in selective media with 2% glucose, then plated on media containing glucose or 2% galactose. Plasmids were constructed by conventional methods, and mutants were generated via site-directed mutagenesis.
Protein Purification and in Vitro Binding Reactions
Recombinant His-Ded1 and His-ded1-R/G were generated essentially as in Ref. 31. A pET28a plasmid encoding the protein was transformed into BL21 (Rosetta) cells and induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside overnight at 23 °C. Purified protein fractions were diluted 1:1 in 50 mm HEPES pH7.5 and 30% glycerol, and concentration was determined by Bradford assay (Pierce). His-Nup159 (N terminus) was purified similarly. Gle1, gle1-VAI/DDD, and other proteins were purified as in (Ref. 19). For binding reactions, 200 pmol (0.5 μm) of indicated proteins were incubated for 45 min at room temperature with 20 μl of Ni-agarose in 400 μl of modified His lysis (MHL) buffer (50 mm NaHPO4 pH7.4, 150 mm NaCl, 10% glycerol, 10 mm imidazole). For RNase-treated samples (supplemental Fig. S1B), 100 μg/ml RNase A (Qiagen) was added to the incubation buffer. Samples were transferred to new tubes, washed 4× with MHL buffer, and resuspended in 2× SDS sample buffer for SDS-PAGE and Coomassie staining.
Immunoprecipitations
As described in Ref. 22, bound proteins were washed with a high stringency buffer (50 mm Tris-HCl pH 8.0, 250 mm NaCl, 2% Triton X-100, 5 mm MgCl2), eluted with SDS sample buffer, resolved by SDS-PAGE and immunoblotted. Ded1 was detected using rabbit polyclonal antibodies raised against full-length recombinant His-Ded1 (VU318) (supplemental Fig. S1A). Protein-A-tagged Gle1 was detected by anti-Ded1 and/or secondary antibody on the same blot. Secondary antibodies were HRP-conjugated (Jackson), and blots were developed via SuperSignal West Pico ECL (Pierce).
ATPase Assays
Coupled colorimetric reactions were conducted essentially as described in Ref. 10. 100 nm His-Ded1 and/or His-ded1-R/G, 100 to 400 nm Gle1 or gle1-VAI/DDD, and 200 nm IP6 were used as indicated, with 0.3 μg/ml RNA (0.3 μg/ml, unless designated otherwise, isolated from yeast by hot phenol extraction). Ded1 was preincubated with Gle1 for 15 min prior to addition of other components. Significance was determined by Student's t test. Prism 4 software (Graphpad) was used for ATP and RNA concentration-dependent calculations. ATP-dependence values were fitted to a hyperbolic one-site binding equation and RNA-dependence values to a sigmoidal dose-response curve with variable slope.
In Situ Hybridization for Poly(A)+ RNA Localization
Cells were fixed in formaldehyde/methanol, processed as described in Ref. 32, incubated overnight with a digoxigenin-oligo(dT)30 probe, and detected with fluorescein-labeled antidigoxigenin. DNA was stained with 0.1 μg/ml DAPI. Cells were observed using a BX50 Olympus microscope, using an Uplan 100×/1.3 objective. Images were taken using a Photometrics Cool Snap HQ camera with ImagePro Express 6.0 software, and images were processed in Adobe Photoshop CS3.
Translation Analysis
Polysome analysis was performed as described in Ref. 22 with log-phase cultures of indicated strains grown at 27.5 °C and shifted to 20 °C for 60 min. Scanning reporter assays were performed similarly to those described in Ref. 29. Cell pellets were processed for luciferase assays as described in Ref. 22. Start site selection assays were conducted identically to GCN4 reporter assays as in Ref. 22. For in vitro translation assays, extracts were prepared as described in Ref. 33. Capped luciferase RNA was prepared by in vitro transcription of DraI-linearized pLucA50 using an Ampliscribe T7-Flash kit (Epicenter) and purified by RNeasy (Qiagen). RNA was added to translation reactions at 0.9 ng/μl along with Gle1, gle1-VAI/DDD, or Dbp5, and samples were incubated for 45 min at 25 °C. Luciferase assays were performed as above to determine the extent of translation.
RESULTS
Gle1 Interacts with and Inhibits the DEAD-box Protein Ded1
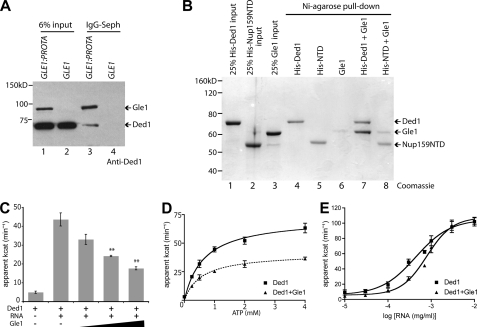
Because Gle1 regulates a DEAD-box protein (Dbp5) in mRNA export and translation termination (8, 9, 22), we speculated that Gle1 might act with a different DEAD-box protein during translation initiation. To address this, co-immunoprecipitation experiments were conducted with yeast lysates from cells expressing either protein-A tagged (Gle1-ProtA) or untagged Gle1. Bound fractions were immunoblotted using a Ded1-specific antibody (Fig. 1A and supplemental Fig. S1A). Ded1 was only co-isolated with Gle1-ProtA (Fig. 1A, lane 3). To test for a direct Gle1:Ded1 physical interaction, in vitro soluble binding assays were conducted with purified recombinant His-tagged Ded1 and Gle1 (Fig. 1B, lanes 1 and 3). The His-tagged N-terminal domain (NTD) of Nup159 was used as a control (lane 2). His-Ded1 or His-NTD were incubated with purified Gle1 and bound to Ni-agarose. Bound fractions were eluted and evaluated by SDS-PAGE. Near-stoichiometric levels of Gle1 were isolated with His-Ded1 (lane 7), revealing a direct interaction. Minimal Gle1 binding was detected with His-NTD (lane 8) or with Ni-agarose alone (lane 6). The Gle1:Ded1 interaction was maintained after RNase treatment, indicating it is not mediated through potentially co-purified RNA contaminants (supplemental Fig. S1B). Thus, Gle1 interacts with Ded1 both in vivo and in vitro.
FIGURE 1.
Gle1 interacts with and inhibits Ded1. A, lysates from protein A-tagged Gle1 (GLE1:PROTA) and wild-type (GLE1) control were subjected to immunoprecipitation with IgG-Sepharose. Immunoprecipitate and 6% input samples were run on SDS-PAGE and blotted using a specific anti-Ded1 antibody (supplemental Fig. S1). Gle1-ProtA is also recognized by the primary and/or secondary antibody. B, recombinant purified Gle1, His-Ded1, and His-Nup159 (N-terminal domain) were incubated for 45 min at room temperature and pull-downs were performed with Ni-agarose. Pull-down and 25% input samples were subjected to SDS-PAGE and Coomassie stained. C, PK/LDH-coupled ATPase assays were performed with recombinant Ded1 (100 nm) in the presence of 0.3 μg/ml total cellular RNA and 1 mm ATP. Purified Gle1 was added at 100, 200, and 400 nm. **, p < 0.01 versus Ded1 with no Gle1. n = 3 to 7 for mean and standard error of means (S.E.) calculations. D, ATPase assays were performed with 100 nm Ded1, 0.3 μg/ml RNA, and 400 nm Gle1. ATP concentration was varied from 0 to 4 mm. E, ATPase assays were performed with 100 nm Ded1 and 1 mm ATP, and 400 nm Gle1. Concentration of total cellular RNA was varied from 10 ng/ml to 10 μg/ml.
In mRNA export, Gle1:IP6 stimulates the RNA-dependent ATPase activity of Dbp5 (8, 9). To investigate whether Gle1 also alters Ded1 activity, ATPase assays were conducted with purified recombinant proteins at different molar ratios. As expected, we observed an RNA-dependent increase in the ATPase activity of Ded1 (Fig. 1, C and E). However, when Gle1 was added, Ded1 ATPase activity was reduced in a dose-dependent manner (Fig. 1C). This is in direct contrast to Gle1:IP6 activation of Dbp5 ATPase activity. Furthermore, addition of IP6 did not change Ded1 ATPase activity or the Gle1 inhibition (supplemental Fig. S2A). As a control, addition of glutathione S-transferase (GST) or an unrelated RNA-binding protein (Hrp1) had no effect (supplemental Fig. S2B). To examine the ATP and RNA dependence of Ded1 activity with Gle1, ATP, and RNA concentrations were varied in ATPase assays. In altering ATP concentration, the Vmax of Ded1 activity was reduced with Gle1 whereas the EC50 was not significantly altered (Fig. 1D). However, with Gle1, an ∼2-fold increase in total RNA concentration (from 0.4 to 0.8 μg/ml) was required for half-maximal Ded1 ATPase activity (Fig. 1E). Again, this was opposite of the Gle:IP6 effect on Dbp5 wherein ATPase activation is coincident with a decrease in the required RNA concentration (8, 9). The direct binding of Gle1 and inhibition of Ded1 ATPase activity revealed a potential direct role for Gle1 in translation initiation.
Gle1 Inhibition of Ded1 Is Distinct from Activation of Dbp5
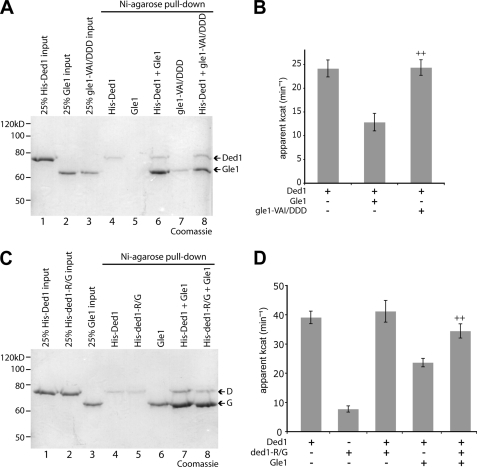
We hypothesized that the Gle1:Ded1 interaction might be mediated in a manner similar to the Gle1:Dbp5 interaction. Extensive structural analysis of Dbp5 has been reported, and very recently an ADP-bound Δ90dbp5L327V:Δ243gle1H337R:IP6 complex was characterized (17). To gain further insight into Gle1:Ded1 binding, we generated altered forms of Gle1 and Ded1 based on the Gle1-Dbp5 findings and assayed for effects on protein interaction and Ded1 activity. First, we utilized a Gle1 protein in which three residues (Val-513, Ala-516, and Ile-520) that are proposed to facilitate binding of the Dbp5 N-terminal RecA domain are changed to aspartic acid (17). The altered gle1-VAI/DDD protein still binds to Dbp5 but is defective in ATPase activation, and the corresponding yeast gle1-VAI/DDD mutant strain is inviable (17). We performed in vitro pull-downs with His-Ded1 and gle1-VAI/DDD and found it bound at levels similar to wild-type Gle1 (Fig. 2A). This indicates that the gle1-VAI/DDD protein is not perturbed for interaction with either Dbp5 or Ded1. However, the gle1-VAI/DDD protein, despite binding to Ded1, was completely deficient in inhibiting the ATPase activity of Ded1 (Fig. 2B). This result is similar, though correspondingly opposite in effect, to ATPase assays performed with Dbp5 and gle1-VAI/DDD (17). Thus, gle1-VAI/DDD is defective for modulating both Dbp5 and Ded1, indicating a potential shared biochemical mechanism of action on the two different DEAD-box proteins.
FIGURE 2.
gle1-VAI/DDD and ded1-R438G affect Gle1-mediated inhibition of Ded1. A, pull-downs with Ni-agarose were performed as in Fig. 1 with purified gle1-VAI/DDD and Ded1. B, ATPase assays were performed with 100 nm Ded1 and 400 nm of Gle1 or aberrant gle1-VAI/DDD protein. ++, p < 0.01 versus Ded1+Gle1. C, pull-downs with Ni-agarose were performed as in Fig. 1 with Gle1 and ded1-R438G. D, ATPase assays performed with 100 nm Ded1 and/or aberrant ded1-R438G protein and 400 nm Gle1. ++, p < 0.01 versus Ded1+Gle1. For B and D, n = 3 to 7 for mean and standard error of means (S.E.) calculations.
Second, a recent report discovered a DBP5 dominant negative mutant (DBP5-R369G), which generates a dbp5-R369G protein that is deficient in RNA binding and sequesters Gle1 (34). The dbp5-R369G protein, which has reduced ATPase activity compared with wild-type due to the lack of RNA binding, inhibits activation of wild-type Dbp5 in vitro by titering away Gle1:IP6 (34). As the changed arginine-369 residue is conserved in multiple DEAD-box proteins (data not shown), we generated an equivalent DED1 mutant (DED1-R438G). We found that the altered ded1-R438G protein bound to Gle1 in pull-down assays (Fig. 2C). Similarly to dbp5-R369G, the ded1-R438G protein had little ATPase activity, and in fact no additive effect compared with wild-type Ded1 was seen when both proteins are added together (Fig. 2D). However, Gle1-mediated inhibition of Ded1 ATPase activity was significantly reversed by the addition of ded1-R438G protein, consistent with the ability of ded1-R438G to still bind Gle1. Thus, both the ded1-R438G and gle1-VAI/DDD proteins affected Ded1 ATPase activity in ways similar to their effects on Dbp5 activity, though reversed in orientation. Taken together, these biochemical data support direct inhibition of Ded1 by Gle1. Moreover, this indicates that Gle1 potentially binds Ded1 in a manner similar to its interaction with Dbp5. However, the interactions are not completely equivalent. We also generated a ded1 mutant (D392K) similar to a dbp5-E323K mutant that reportedly ablates Gle1 binding and activation (35). The ded1-D392K protein bound Gle1, and its activity in the presence of Gle1 was not significantly different from wild-type Ded1 (data not shown). Furthermore, a ded1-D392K mutant strain also had no observable growth phenotypes (data not shown). Overall, Ded1:Gle1 has both shared and distinct mechanistic determinants compared with Dbp5:Gle1:IP6.
Genetic Interactions of gle1 and ded1 Mutants
Next we evaluated the in vivo significance of Ded1 regulation by Gle1. A double mutant strain harboring the temperature-sensitive gle1–4 allele and the cold-sensitive ded1–120 allele was generated and tested for growth at different temperatures (Fig. 3A). Interestingly, the growth defect of the gle1–4 mutant at 30 °C was suppressed in the gle1–4 ded1–120 double mutant strain. This genetic suppression of the growth defect is consistent with the negative biochemical regulation of Ded1 by Gle1 and suggests that Gle1 and Ded1 have opposing actions in vivo. No significant synthetic growth suppression or synthetic lethal effects were observed at other temperatures tested. The lack of suppression of the ded1–120 growth defect at 25 °C and the lack of suppression of the gle1–4 growth defect at 37 °C likely reflect differences in the severity of their defects at different temperatures. In addition, Ded1 and Gle1 each play roles in more than one process and the suppression effect might be linked to only one of these functions (see below) (22). We did not test the gle1-VAI/DDD mutant because it is lethal at all temperatures in vivo and is defective in regulating both Ded1 and Dbp5 (Fig. 2B and Ref. 17).
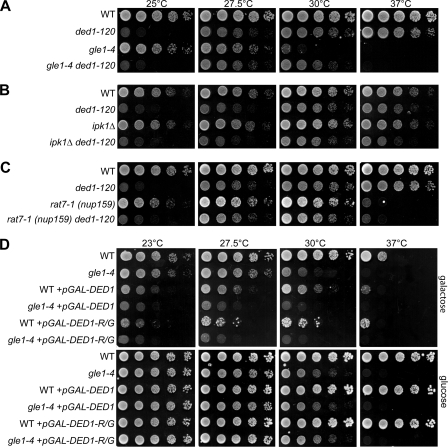
FIGURE 3.
A gle1–4 mutant displays genetic interactions with DED1. A–C, cultures of indicated single and double mutant strains with wild-type controls (WT) were 5-fold serially diluted and spotted on YPD, then incubated 2 days at 25 °C, 27.5, 30, or 37 °C. Strains included cold-sensitive ded1–120 (A–C), gle1–4 (A), ipk1Δ (B), and rat7-1 (nup159) (C). D, wild-type and gle1–4 strains transformed with galactose-inducible DED1 (pGAL-DED1), DED1-R438G (pGAL-DED1-R/G), or control plasmids were incubated at the indicated temperatures for 3 days on selective media containing galactose (top) or glucose (bottom).
In parallel, ipk1Δ and rat7-1 (nup159) mutants were tested as controls. Ipk1 is the 2-kinase responsible for IP6 production (32), and is directly linked to Gle1 function in mRNA export and in translation termination (8–10, 22). However, initiation defects have not been observed in ipk1Δ cells (22). Nup159 is involved in Gle1 function during mRNA export by serving as the NPC docking site for Dbp5, and the rat7-1 (nup159) mutant is specific for defects in mRNA export (20–22). Thus, we predicted that double mutants of ded1–120 with ipk1Δ or rat7-1 (nup159) should not have genetic interactions. Indeed, no growth suppression or synthetic fitness effects were observed in ipk1Δ ded1–120 or rat7-1 (nup159) ded1–120 double mutants (Fig. 3, B and C). This confirmed the specificity of the gle1–4 ded1–120 phenotype and is consistent with the lack of effect of IP6 in Ded1 ATPase assays.
To further strengthen the genetic links between Gle1 and Ded1, we examined the DED1-R438G mutant in vivo. The DED1-R438G did not complement a ded1Δ mutation (data not shown); however, as reported, the corresponding DBP5-R369G mutant is also inviable but acts dominantly to inhibit growth upon overexpression (34). Thus, we constructed plasmids containing Gal-inducible wild-type DED1 and mutant DED1-R438G. Unlike wild-type DBP5, a prior report has shown that overexpression of wild-type DED1 causes growth defects (36). We also observed inhibition of growth for strains harboring the GAL-DED1 plasmid on galactose media at all temperatures (Fig. 3D). However, combining DED1 overexpression with the gle1–4 mutant resulted in a more severe growth inhibition. This effect is most notable at 27.5 °C at which growth of a gle1–4 control strain is not significantly affected but the gle1–4 + pGAL-DED1 strain grows more poorly than wild-type + pGAL-DED1 (Fig. 3D). Furthermore, overexpression of the DED1-R438G mutant resulted in growth defects similar to wild-type DED1 and showed a similar synthetic defect with gle1–4 (Fig. 3D). This is also consistent with overexpression of an ATPase-dead ded1 mutant that was shown by others to inhibit growth (36). On the one hand, this ability of DED1-R438G overexpression to inhibit wild type cell growth indicates that RNA binding is likely not responsible for the growth inhibition by DED1 overexpression. Further, taken in conjunction with the ATPase data (Fig. 2D), these results suggest that overexpression of either DED1 or DED1-R438G could inhibit growth by binding and sequestering of Gle1, resulting in the loss of Gle1 for appropriate Ded1 regulation.
Gle1 Regulation of Ded1 Is Critical for Efficient Translation Initiation
It has been reported previously that ded1 mutants, including the ded1–120 allele, do not have defects in mRNA export (37). However, it was possible that the known defect in mRNA export in the gle1–4 mutant could be affected in the gle1–4 ded1–120 strain (11). To test this, in situ hybridization experiments were performed. Nuclear accumulation of poly(A)+ signal was observed in gle1–4 mutant cells shifted to growth at 30 °C, the temperature at which growth suppression was evident, while no defect was observed in ded1–120 cells (Fig. 4 B, C, E). However, the nuclear accumulation levels in the gle1–4 ded1–120 double mutant cells were similar to that of the gle1–4 single mutant (Fig. 4, D and E). This suggests that the Ded1:Gle1 interaction does not play a role in mRNA export.
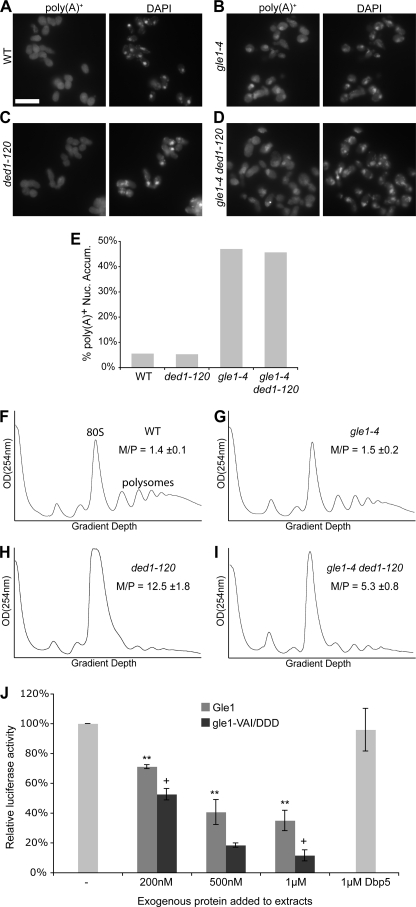
FIGURE 4.
The Gle1:Ded1 interaction is linked to translation initiation. A–D, poly(A)+ RNA was localized by in situ hybridization with oligo-dT probes in wild-type (WT), gle1–4, ded1–120, and double mutant strains after shifting to 30 °C for two hours. Representative images are shown. Size bar in A corresponds to 10 μm. E, quantification of in situ results as the percentage of cells displaying nuclear accumulation of poly(A)+ signal. Cells were scored as positive if the nuclear poly(A)+ signal substantially exceeded cytoplasmic signal. Counts represent n >100 cells for each sample from at least five fields. F–I, cultures of indicated strains were shifted to 20 °C for 1 h, and polysome profiles were generated by subjecting cell lysates to 7–47% sucrose density centrifugation and OD254 nm analysis of the gradient. Monosome/polysome (M/P) ratio was determined by comparing the area of the monosome peak to the sum of the areas of the first four polysome peaks. Error represents S.E. from n = 3. The monosome (80 S) and polysome peaks are labeled in F for reference. J, for in vitro translation assays, competent extracts from wild-type cells were incubated with in vitro-transcribed luciferase mRNA, and luciferase reactions were performed. Purified Gle1, gle1-VAI/DDD, and Dbp5 were added to the reactions as indicated.
Rather, we hypothesized that the Gle1-Ded1 interaction is important for translation initiation. To examine this, we performed sucrose density sedimentation of total yeast cell lysates and quantified the relative 80 S monosome to polysome levels (M/P ratio) (Fig. 4, F–I). We previously showed that the gle1–4 strain displayed a defect in the M/P ratio when shifted to 37 °C (22). However, the ded1–120 strain displayed a more severe defect than gle1–4 at elevated temperatures (data not shown), thus we could not test whether there was rescue of the gle1–4 phenotype in the double mutant. Alternatively, we examined whether gle1–4 could rescue the defect observed in the ded1–120 strain. After growth at 20 °C, a permissive temperature for the gle1–4 strain, the polysome profile was similar to that from wild-type cells (Fig. 4, F and G). However, the polysome profile of the cold-sensitive ded1–120 strain was severely affected (Fig. 4H), with a greatly increased 80 S monosome peak and decreased polysome peaks (M/P = 12.5). This is characteristic of an initiation defect (e.g. Refs. 22 and 38). Of note, this defect was reduced in the gle1–4 ded1–120 mutant (Fig. 4I), which had a comparatively smaller monosome/polysome ratio (M/P = 5.3). Thus, the gle1 mutant partially rescued the altered polysome phenotype in the ded1–120 mutant. This supports the hypothesis that Gle1 and Ded1 have opposing cellular actions and are both functionally linked to translation initiation.
To test whether the effects of Gle1 on translation initiation are direct, we performed in vitro translation assays with ribosome-enriched extracts from wild-type cells and in vitro-transcribed luciferase mRNA. When Gle1 protein was co-incubated with the translation reactions, luciferase activity was markedly reduced in a dose-dependent manner, indicating that Gle1 is acting to inhibit translation (Fig. 4J). Interestingly, when gle1-VAI/DDD protein was added to the translation reactions, it reduced activity to a greater extent than wild-type Gle1. These results suggest that Gle1 might not be a translation inhibitor per se; rather, the proper level of functional Gle1 is critical for efficient translation. Thus, adding excess wild-type Gle1 reduces translation, whereas adding gle1-VAI/DDD might replace functional Gle1 and reduce translation efficiency. This conclusion is also consistent with the complementary suppression observed below in Fig. 5, B and C. Furthermore, as a reflection of the specificity of the effect of Gle1, addition of 1 μm Dbp5 did not significantly alter the translation activity (Fig. 4J).
FIGURE 5.
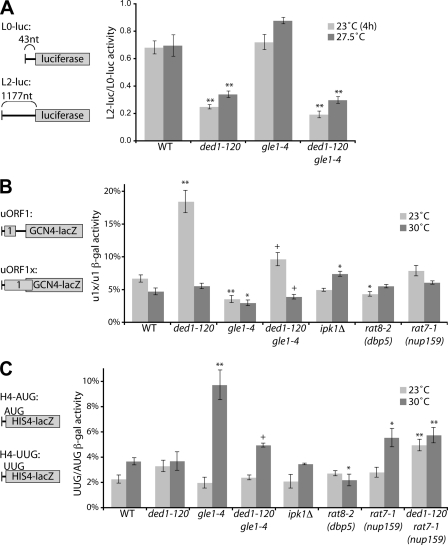
Gle1 regulates Ded1 in AUG start site selection. A, wild-type (WT), gle1–4, ded1–120, and gle1–4 ded1–120 strains were transformed with L0- and L2-luciferase reporters (reporter mRNAs diagrammed at left). Strains were grown overnight at 27.5 °C in selective media, then shifted to 23 °C for 4 h or not. Luciferase assays were performed on cell lysates, and scanning efficiency was calculated by the ratio of activity from L2-LUC to L0-LUC normalized for cell number. B, indicated strains were transformed with uORF1- and uORF1x-GCN4-lacZ reporters (diagram at left), and cultures were diluted in selective media. Following growth overnight at 23 °C or 30 °C, β-galactosidase assays were performed. Leaky scanning activity was determined as a percent ratio of uORF1x to uORF1 β-galactosidase activity. *, p < 0.05 versus WT; **, p < 0.01 versus WT; +, p < 0.05 versus ded1–120 strain. C, indicated strains were transformed with HIS4-lacZ-AUG and -UUG reporters (diagram at left), and cultures were diluted in selective media. Following growth overnight at 23 °C or 30 °C, β-galactosidase assays were performed. Start site fidelity was determined as a percent ratio of HIS4-UUG to HIS4-AUG β-galactosidase activity. *, p < 0.05 versus WT; **, p < 0.01 versus WT; +, p < 0.05 versus gle1–4 strain. For all panels, n = 3 to 10 for mean and S. E.
Ded1 and Gle1 Play Roles in AUG Start Site Recognition
A set of in vivo reporter-based assays was used to assess the efficiency of specific translation initiation steps in gle1–4 and ded1–120 mutant cells. Each set of reporters consisted of both a test reporter and a control. We then took a ratio of the activity of the test reporter to the control construct, thus normalizing the results for differences due to mRNA export defects and prior effects on translation as well. Ded1 has been proposed to facilitate the efficiency of AUG start codon scanning by the 43 S pre-initiation complex (PIC) through unwinding local RNA structure and/or remodeling the mRNP in the 5′-UTR (28). To measure scanning efficiency, the ratio of luciferase activity produced was measured from two reporters with 5′-UTRs of either 43 (L0) or 1117 (L2) nucleotides (Fig. 5A). This method was previously used to document scanning defects in a ded1–57 temperature-sensitive mutant strain (29). Consistent with that report, we observed a 63% reduction in the L2/L0 ratio in the ded1–120 mutant at 23 °C, a non-permissive temperature for this strain (Fig. 5A). We also observed a 51% reduction at 27.5 °C, a semi-permissive temperature for growth of the ded1–120 mutant. In contrast, there was no defect in gle1–4 mutant cells at any temperature tested (Fig. 5A and supplemental Fig. S3). Furthermore, the gle1–4 ded1–120 double mutant strain had scanning defects similar to those of the ded1–120 single mutant (Fig. 5A). These results are consistent with the hypothesis that Ded1 has a role in scanning; however, in this assay, Gle1 does not inhibit this Ded1 function, suggesting that different roles ascribed to Ded1 could be distinct.
Following scanning, the PIC must properly recognize the AUG start codon, which is linked to a series of conformational changes that ultimately lead to initiation (24). To test for a defect in start site recognition, we employed two different sets of reporters. First, we assayed for leaky scanning, which occurs when the PIC fails to recognize a start codon and continues scanning (39). For this, we utilized a GCN4-based reporter that lacked the inhibitory uORF2, 3, and 4 and retained only the stimulatory uORF1 (40). This reporter's mRNA will normally be translationally active and was used as a control. A second reporter extended the reading frame of uORF1 past the start site of the GCN4-lacZ coding region (40). This version will only translate the fusion protein if the start site in uORF1 is bypassed through leaky scanning. Interestingly, we observed a striking increase in leaky scanning activity in ded1–120 cells at a non-permissive temperature of 23 °C (Fig. 5B), over 18% compared with less than 6% in wild type cells. Normalized activity in gle1–4 cells was reduced by a modest but significant amount compared with wild-type at either 23 °C or 30 °C (Fig. 5B). Importantly, however, the gle1–4 ded1–120 double mutant showed a clearly reduced leaky scanning defect at 23 °C compared with ded1–120 alone. This correlates with the rescued polysome defect in ded1–120 gle1–4 cells (Fig. 4I) and with Gle1 inhibition of Ded1 activity. No substantial changes in leaky scanning were observed in ipk1Δ, rat8-2 (dbp5), or rat7-1 (nup159) mutants, indicating that the defects are not due to general effects on mRNA export or translation termination and could reflect Gle1 and Ded1 functional effects on start site recognition.
Defects in start site recognition can also lead to changes in start site fidelity. Such fidelity perturbations are characterized by an increase in initiation at near-cognate start codons. To test for fidelity defects, we employed a third set of reporters coding for HIS4-lacZ fusion proteins (Fig. 5C). One version of the reporter contained a consensus AUG start codon, whereas the other had a mutated UUG start site (41). An increase in the ratio of activity of UUG to AUG indicates defects in start site fidelity. With these reporters, the ded1–120 mutant was not significantly different from wild-type at permissive or non-permissive temperatures (Fig. 5C). However, gle1–4 cells had an increase in the UUG/AUG ratio at a semi-permissive temperature of 30 °C, indicating translation fidelity defects. Importantly, this defect was again largely rescued in gle1–4 ded1–120 cells, correlating with the rescue of the gle1–4 growth defect by ded1–120 (Fig. 3A). Neither ipk1Δ nor rat8-2 (dbp5) mutants showed fidelity defects (Fig. 5C). Although a small defect was observed in the rat7-1 (nup159) mutant, this was not rescued by ded1–120. Overall, the reporter assays indicate that the negative regulation of Ded1 by Gle1 has specific consequences in translation initiation and reveal a potential role for the Gle1:Ded1 interaction in determining proper start site recognition.
DISCUSSION
In this study, we have defined the target for Gle1 action during translation initiation. We find that Gle1 negatively regulates the DEAD-box protein Ded1, inhibiting its ATPase activity in vitro and affecting the function of Ded1 in translation in vivo. This is a strikingly different and surprising role for Gle1 compared with its established function as an activator of the DEAD-box protein Dbp5 in mRNA export and translation termination. Moreover, Gle1 inhibition of Ded1 does not require IP6 binding, unlike the IP6 enhancement of Dbp5 activation by Gle1. As such, Gle1 is uniquely positioned to differentially control distinct steps in the gene expression pathway.
Complementary genetic and biochemical evidence support a model for Gle1 control of Ded1 in translation initiation. In accordance with inhibition of Ded1 ATPase activity by Gle1, Gle1 interacts with Ded1 in vitro and in vivo (Fig. 1). Furthermore, the gle1–4, ded1–120, and gle1–4 ded1–120 mutant phenotypes correlate well with these biochemical results. The suppression of gle1–4 temperature-sensitive growth by the ded1–120 mutant and the synthetic growth defect in DED1-overexpressing gle1–4 cells agree with the in vitro biochemical inhibition of Ded1 by Gle1 and the suppression of translation fidelity defects in gle1–4 ded1–120 (Figs. 2, 3, A and D, and 5C). Likewise, the severe polysome defect of the ded1–120 mutant was partially suppressed by gle1–4 at 20 °C (Fig. 4, H and I), which correlates with the rescue of the ded1–120 leaky scanning defect in the double mutant strain (Fig. 5B). We did not observe suppression of the ded1–120 cold-sensitive growth defect by gle1–4 (Fig. 3A); however, the ded1–120 mutant has perturbations in at least one other function, promotion of scanning efficiency, which is not affected by Gle1 (Fig. 5A). Thus, multiple effects in ded-120 cells may contribute to growth defects in this strain. On the other hand, we suggest that at least part of the growth sensitivity of the gle1–4 strain is due to the inability of the aberrant gle1 protein to inhibit Ded1 in initiation. This interpretation means that overly active Ded1 is detrimental for cell growth, which is supported by the inhibition of cell growth upon DED1 overexpression (Ref. 36 and Fig. 3D). Alternatively, DED1 overexpression could be sequestering essential Gle1 activity, as overexpression of either DED1-R438D (Fig. 3D) or ATPase-dead ded1 (36) also inhibits cell growth. Thus, in the gle1–4 ded1–120 double mutant strain, the defect in the ability of gle1–4 protein to inhibit Ded1 is partially counterbalanced by attenuated ded1–120 protein activity at 30 °C. Taken together, these results firmly establish a link between Gle1, Ded1, and translation initiation.
It is not unexpected that different phenotypes of gle1–4 and ded1–120 mutants are impacted in different ways. Although the severity of the defects in these mutants varies with temperature, both mutants are partially compromised at all temperatures. Therefore, suppressive effects can be evident even if a particular phenotype is not observed at a specific temperature in one of the single mutants. For example, gle1–4 suppresses the ded1–120 defect in leaky scanning at 23 °C (Fig. 5B), even though the gle1–4 single mutant does not display a significant fidelity defect at that temperature (Fig. 5C). This is likely due to the partial reduction in the gle1–4 protein activity even at 23 °C, as well as reflecting differences in the sensitivities and thresholds of these two distinct assays (in this case, gle1–4 also shows a slight reduction in leaky scanning at 23 °C (Fig. 5B)). Conceptually, the same phenomenon is observed in synthetic fitness/lethal defects, which are frequently manifested at temperatures at which neither of the individual mutants has an observable phenotype. Therefore, the genetic suppression we have observed is consistent with the in vitro data indicating that Gle1 inhibits Ded1 in initiation.
Several lines of evidence argue that the effects of Gle1 on Ded1 are specific and not indirect effects from other Gle1 or Ded1 functions. First, the physical interaction and in vitro ATPase assays reflect a direct effect (Fig. 1). This includes assays with gle1-VAI/DDD and ded1-R438G that show specificity of the inhibition (Fig. 2). The inhibition of in vitro translation by relatively low levels of Gle1 also indicates that Gle1 affects translation directly (Fig. 4J). Second, ipk1Δ, rat7-1 (nup159), and rat8-2 (dbp5) mutants do not have genetic interactions with the ded1–120 mutant or start site defects (Figs. 3 and 5). Nup159, Dbp5, and IP6 are all involved with Gle1 in mRNA export (8, 9, 20, 21, 42), and Dbp5 and IP6 also partner with Gle1 in translation termination (6, 10, 22). Therefore, our observations do not appear linked to mRNA export or termination. Third, we observe no effect from the Gle1:Ded1 interaction on bulk mRNA export (Fig. 4, A–E). Fourth, both the leaky scanning and fidelity assays are normalized to the activity from control reporters to eliminate effects from mRNA export perturbations as well as defects in other steps of translation initiation (Fig. 5, B and C). Ded1 has been linked to other processes, including pre-mRNA splicing (although this is primarily genetic and ded1 mutants do not display splicing defects) and processing (P-) body formation (28, 36, 37). It is possible that Gle1 regulates Ded1 in other Ded1 functions, and we cannot rule out that the in vivo results we observe are at least partially due to such regulation. However, the specificity of the reporter assays results argues against solely indirect effects. This evidence, together, highlights the specificity for Gle1 regulation of Ded1.
Our results with the start site recognition reporters suggest that Ded1 and Gle1 affect this step of translation initiation. It is currently unclear whether these are direct functions. However, ded1 mutants have defects in scanning efficiency, while the gle1–4 mutant does not have a perturbation. This observation might indicate that these are distinct functions of Ded1 or that Gle1 is blocked from regulating Ded1 during scanning (Ref. 29 and Fig. 5A). A potential role in start site selection has also recently been proposed for another DEAD-box helicase and translation initiation factor, eIF4A (43); therefore Ded1 and eIF4A might both contribute to start site recognition. The leaky scanning defect that we observed in the ded1–120 mutant suggests that Ded1 promotes start site recognition, whereas the increase in near-cognate recognition in gle1–4 cells implies that Gle1 retards this process, perhaps to ensure fidelity (supplemental Fig. S4). Additionally, the rescue of these effects in the gle1–4 ded1–120 double mutant ties these functions of Ded1 and Gle1 together, suggesting that both are needed for proper AUG recognition. We speculate that Gle1 inhibits Ded1 activity to reduce initiation at improper start sites and that inhibition is relieved by recognition of a correct start codon, possibly through the conformational changes associated with recognition (44, 45). Ded1 might serve to unwind or remodel the mRNA to a particular conformation that aids start site recognition. Alternatively, it could promote one or more of the protein or RNP remodeling steps in the PIC during recognition and subsequent initiation steps. In either case, identifying a target of Ded1 activity in start site recognition would be of great interest and support for this model. A study published during revision of this manuscript has suggested that Ded1 has an ATPase-independent role in translation repression and the formation of stress granules as well as an ATPase-dependent role in promoting 48S PIC assembly through interactions with the eIF4F complex (46). It is thus tempting to speculate that these activities are related to Ded1's role in start site recognition and are also regulated by Gle1 activity.
Taken together, this work reveals multifunctional roles for Gle1 as a highly unique regulator of at least two DEAD-box proteins. Moreover, the regulation by Gle1 is differential: Gle1 activates Dbp5 and inhibits Ded1. Furthermore, unlike Dbp5, addition of IP6 in the presence of Gle1 has no effect on Ded1 activity (supplemental Fig. S2A). It is unlikely that Gle1 is inhibiting Ded1 by competing for binding to RNA. Compared with the known strong affinity of Ded1 for RNA (31), we found that RNA binding by Gle1 was insignificant (data not shown). Moreover, this interpretation is inconsistent with Gle1-mediated activation of Dbp5 wherein further stimulation is achieved by increasing molar ratios of Gle1 to Dbp5 (9). There are at least two other non-exclusive possibilities for the contrasting effects of Gle1 on Dbp5 and Ded1. First, the effects may be due to differences in the binding interfaces for Gle1:IP6 with Dbp5 compared with Gle1 and Ded1. The gle1-VAI/DDD protein, which binds both Dbp5 and Ded1 but is unable to affect the activity of either, suggests that the binding interfaces share at least some commonalities (Ref. 17 and Figs. 1C and 2D). However, the lack of defects in a ded1-D392K mutant, for which the equivalent dbp5-E323K protein has reduced Gle1 binding and activation, argues that the Gle1:Dbp5 and Gle1:Ded1 surfaces are at least partially distinct. A second possibility is that the differential regulation of Dbp5 and Ded1 is a result of the intrinsic activities and cycling of the enzymes. Dbp5 has a poor rate of ATPase hydrolysis in the absence of Gle1, even in the presence of RNA, whereas Ded1 activity (with RNA) is considerably higher (8, 9, 31). The rate-limiting step for Dbp5 might be different than for Ded1, thus leading to different results upon the addition of Gle1. We have begun to investigate these possibilities with the dominant mutants, which further suggest that RNA binding is not required for Gle1 binding to Dbp5 or Ded1 (Ref. 34 and Figs. 1D and 2E).
The specificity of Gle1 function could also be modulated through its targeting to different steps in gene expression. To target its activity, we speculate that Gle1 is directed to specific DEAD-box proteins by unique docking sites, including Nup42 in the NPC, eIF3 in translation initiation, and eRF1 in termination (11, 22). It remains critical to determine whether Gle1 at the NPC, at translation initiation complexes and at translation termination complexes is dynamic. Certainly, human Gle1 has been shown to shuttle between the nucleus and cytoplasm (47). Gle1 might also associate with mRNPs from export through translation and mediate gene expression at multiple steps.
Only a few DEAD-box proteins have known regulatory factors, which include plakophilin-1, PDCD4, and several eukaryotic initiation factors (eIFs) for eIF4A, MAGOH and Y14 for eIF4AIII, Esf2 for Dbp8, and Gle1 for Dbp5 (8, 9, 48–52). The discovery here that a single factor (Gle1) modulates more than one DEAD-box protein (Dbp5 and Ded1) in distinct gene expression steps presents a new paradigm for regulation of the DEAD-box helicase family. This suggests that DEAD-box regulatory factors such as Gle1 could serve as focal points for coordination of gene expression through control of their targets in multiple processes. We predict that further study of Gle1 will shed more light on these paradigms as well as potential functional coupling between late stages of gene expression.
Supplementary Material
Acknowledgments
We thank Drs. T. H. Chang, A. G. Hinnebusch, and J. E. G. McCarthy for plasmids; Y. Zhou, K. N. Noble, and Dr. E. J. Tran for purified proteins; Dr. A. J. Link for equipment use; and Wente laboratory members for discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R37-GM51219 (to S. R. W.) and F32-GM082065 (to T. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- mRNP
- mRNA-protein complex
- Dbp
- DEAD-box protein
- NPC
- nuclear pore complex
- PIC
- preinitiation complex
- NTD
- N-terminal domain.
REFERENCES
- 1. Stewart M. (2010) Trends Biochem. Sci. 35, 609–617 [DOI] [PubMed] [Google Scholar]
- 2. Harel-Sharvit L., Eldad N., Haimovich G., Barkai O., Duek L., Choder M. (2010) Cell 143, 552–563 [DOI] [PubMed] [Google Scholar]
- 3. Culjkovic B., Topisirovic I., Skrabanek L., Ruiz-Gutierrez M., Borden K. L. (2006) J. Cell Biol. 175, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Snay-Hodge C. A., Colot H. V., Goldstein A. L., Cole C. N. (1998) EMBO J. 17, 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tseng S. S., Weaver P. L., Liu Y., Hitomi M., Tartakoff A. M., Chang T. H. (1998) EMBO J. 17, 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J. J., Seedorf M., Cole C. N., Krebber H. (2007) Science 315, 646–649 [DOI] [PubMed] [Google Scholar]
- 7. Miller A. L., Suntharalingam M., Johnson S. L., Audhya A., Emr S. D., Wente S. R. (2004) J. Biol. Chem. 279, 51022–51032 [DOI] [PubMed] [Google Scholar]
- 8. Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R. (2006) Nat. Cell Biol. 8, 711–716 [DOI] [PubMed] [Google Scholar]
- 9. Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006) Nat. Cell Biol. 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 10. Alcázar-Román A. R., Bolger T. A., Wente S. R. (2010) J. Biol. Chem. 285, 16683–16692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy R., Wente S. R. (1996) Nature 383, 357–360 [DOI] [PubMed] [Google Scholar]
- 12. Watkins J. L., Murphy R., Emtage J. L., Wente S. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6779–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nousiainen H. O., Kestilä M., Pakkasjärvi N., Honkala H., Kuure S., Tallila J., Vuopala K., Ignatius J., Herva R., Peltonen L. (2008) Nat. Genet. 40, 155–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rayala H. J., Kendirgi F., Barry D. M., Majerus P. W., Wente S. R. (2004) Mol. Cell Proteomics 3, 145–155 [DOI] [PubMed] [Google Scholar]
- 15. Kendirgi F., Rexer D. J., Alcázar-Román A. R., Onishko H. M., Wente S. R. (2005) Mol. Biol. Cell 16, 4304–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strahm Y., Fahrenkrog B., Zenklusen D., Rychner E., Kantor J., Rosbach M., Stutz F. (1999) EMBO J. 18, 5761–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Montpetit B., Thomsen N. D., Helmke K. J., Seeliger M. A., Berger J. M., Weis K. (2011) Nature 472, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noble K. N., Tran E. J., Alcázar-Román A. R., Hodge C. A., Cole C. N., Wente S. R. (2011) Genes Dev. 25, 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tran E. J., Zhou Y., Corbett A. H., Wente S. R. (2007) Mol. Cell 28, 850–859 [DOI] [PubMed] [Google Scholar]
- 20. Schmitt C., von Kobbe C., Bachi A., Panté N., Rodrigues J. P., Boscheron C., Rigaut G., Wilm M., Séraphin B., Carmo-Fonseca M., Izaurralde E. (1999) EMBO J. 18, 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hodge C. A., Colot H. V., Stafford P., Cole C. N. (1999) EMBO J. 18, 5778–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bolger T. A., Folkmann A. W., Tran E. J., Wente S. R. (2008) Cell 134, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kapp L. D., Lorsch J. R. (2004) Annu. Rev. Biochem. 73, 657–704 [DOI] [PubMed] [Google Scholar]
- 24. Mitchell S. F., Lorsch J. R. (2008) J. Biol. Chem. 283, 27345–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jankowsky E., Gross C. H., Shuman S., Pyle A. M. (2001) Science 291, 121–125 [DOI] [PubMed] [Google Scholar]
- 26. Linder P. (2006) Nucleic Acids Res. 34, 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Linder P. (2003) Biol. Cell 95, 157–167 [DOI] [PubMed] [Google Scholar]
- 28. Tarn W. Y., Chang T. H. (2009) RNA Biol. 6, 17–20 [DOI] [PubMed] [Google Scholar]
- 29. Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. (2004) Mol. Microbiol. 51, 987–1001 [DOI] [PubMed] [Google Scholar]
- 30. Marsden S., Nardelli M., Linder P., McCarthy J. E. (2006) J. Mol. Biol. 361, 327–335 [DOI] [PubMed] [Google Scholar]
- 31. Iost I., Dreyfus M., Linder P. (1999) J. Biol. Chem. 274, 17677–17683 [DOI] [PubMed] [Google Scholar]
- 32. York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. (1999) Science 285, 96–100 [DOI] [PubMed] [Google Scholar]
- 33. Balagopal V., Parker R. (2011) RNA 17, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hodge C. A., Tran E. J., Noble K. N., Alcazar-Roman A. R., Ben-Yishay R., Scarcelli J. J., Folkmann A. W., Shav-Tal Y., Wente S. R., Cole C. N. (2011) Genes Dev. 25, 1052–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dossani Z. Y., Weirich C. S., Erzberger J. P., Berger J. M., Weis K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16251–16256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beckham C., Hilliker A., Cziko A. M., Noueiry A., Ramaswami M., Parker R. (2008) Mol. Biol. Cell 19, 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. (1997) Science 275, 1468–1471 [DOI] [PubMed] [Google Scholar]
- 38. Nielsen K. H., Szamecz B., Valásek L., Jivotovskaya A., Shin B. S., Hinnebusch A. G. (2004) EMBO J. 23, 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozak M. (2002) Gene 299, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grant C. M., Miller P. F., Hinnebusch A. G. (1994) Mol. Cell Biol. 14, 2616–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donahue T. F., Cigan A. M. (1988) Mol. Cell Biol. 8, 2955–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Del Priore V., Heath C., Snay C., MacMillan A., Gorsch L., Dagher S., Cole C. (1997) J. Cell Sci. 110, 2987–2999 [DOI] [PubMed] [Google Scholar]
- 43. Watanabe R., Murai M. J., Singh C. R., Fox S., Ii M., Asano K. (2010) J. Biol. Chem. 285, 21922–21933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pestova T. V., Kolupaeva V. G. (2002) Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. (2005) Mol. Cell 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 46. Hilliker A., Gao Z., Jankowsky E., Parker R. (2011) Mol. Cell 43, 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kendirgi F., Barry D. M., Griffis E. R., Powers M. A., Wente S. R. (2003) J. Cell Biol. 160, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grifo J. A., Abramson R. D., Satler C. A., Merrick W. C. (1984) J. Biol. Chem. 259, 8648–8654 [PubMed] [Google Scholar]
- 49. Ballut L., Marchadier B., Baguet A., Tomasetto C., Séraphin B., Le Hir H. (2005) Nat. Struct. Mol. Biol. 12, 861–869 [DOI] [PubMed] [Google Scholar]
- 50. Wolf A., Krause-Gruszczynska M., Birkenmeier O., Ostareck-Lederer A., Hüttelmaier S., Hatzfeld M. (2010) J. Cell Biol. 188, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang H. S., Jansen A. P., Komar A. A., Zheng X., Merrick W. C., Costes S., Lockett S. J., Sonenberg N., Colburn N. H. (2003) Mol. Cell Biol. 23, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Granneman S., Lin C., Champion E. A., Nandineni M. R., Zorca C., Baserga S. J. (2006) Nucleic Acids Res. 34, 3189–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.