SUMMARY
RNA-binding proteins coordinate the fates of multiple RNAs, but the principles underlying these global interactions remain poorly understood. We elucidated regulatory mechanisms of the RNA-binding protein HuR, by integrating data from diverse high-throughput targeting technologies, specifically PAR-CLIP, RIP-chip, and whole-transcript expression profiling. The number of binding sites per transcript, degree of HuR-association, and degree of HuR-dependent RNA stabilization were positively correlated. Pre-mRNA and mature mRNA containing both intronic and 3′ UTR binding sites were more highly stabilized than transcripts with only 3′ UTR or only intronic binding sites, suggesting that HuR couples pre-mRNA processing with mature mRNA stability. We also observed HuR-dependent splicing changes and substantial binding of HuR in poly-pyrimidine tracts of pre-mRNAs. Comparison of the spatial patterns surrounding HuR and miRNA binding sites provided functional evidence for HuR-dependent antagonism of proximal miRNA-mediated repression. We conclude that HuR coordinates gene expression outcomes at multiple interconnected steps of RNA processing.
INTRODUCTION
The timing and dosage of gene expression are fundamental determinants of phenotype. Regulation of gene expression is a coordinated multi-layered process involving many trans-acting factors (Maniatis and Reed, 2002; Keene, 2001). Whereas most genomic investigations have focused on transcriptional regulation, the rich and ancient world of post-transcriptional regulation (PTR) has only recently been investigated using high-throughput approaches. RNA-binding proteins (RBPs) and non-coding RNAs (ncRNAs) interact with messenger RNAs (mRNAs) in a combinatorial manner and coordinate PTR to achieve appropriate spatio-temporal expression of the encoded proteins. Therefore, to advance our understanding of gene expression it is necessary to investigate the binding properties of RBPs and ncRNAs at a systems level.
RBPs and ncRNAs interact with mRNAs to form dynamic multi-component ribonucleoprotein complexes (mRNPs). RBPs regulate all aspects of the lives of mRNAs (Moore, 2005). These include early events such as splicing and poly-adenylation, as well as export to and localization in the cytoplasm, where they coordinately regulate the stability and translation of subsets of mRNAs (Keene, 2007). Biochemical, genetic, and computational approaches have been utilized, alone and in combination, to identify and validate RBP binding sites or RNA recognition elements (RREs) and their cognate RBPs or ncRNAs. However, the degenerate nature and/or short length of many RREs, as well as the lack of definitive biochemical techniques to identify precise in vivo occupancy make it challenging to determine the combinatorial regulation underlying an “RNP code”.
The first developed genomic method to identify multiple in vivo targets of a given RBP using cDNA arrays was RNP immunoprecipitation (RIP-chip) under conditions that preserve endogenous RNA-protein interactions, followed by detecting the bound RNAs (Tenenbaum et al., 2000). RIP-chip studies from diverse organisms and biological conditions have uncovered putative regulatory elements, regulatory modules, and RNP remodeling in response to stimuli (reviewed in (Morris et al., 2010; Halbeisen et al., 2008)). These methods provided data for the formalization and subsequent support of the post-transcriptional operon/regulon model, in which RNPs coordinate the expression of transcripts encoding functionally related proteins through combinatorial and dynamic interactions (Keene and Tenenbaum, 2002; Hogan et al., 2008).
The primary deficiency of RIP-chip is the inability to precisely identify the location of the binding site of the RNA-interacting component. A number of subsequent methods utilizing cross-linking followed by immunoprecipitation have addressed this deficiency to varying degrees(Ule et al., 2003; König et al., 2010; Hafner et al., 2010). One of these techniques, photoactivatable ribonucleoside cross-linking and immunoprecipitation (PAR-CLIP), has multiple advantages attributable to utilizing long wave UV (365 nm) to cross-link photoactive thiouridine incorporated into nascent RNA(Hafner et al., 2010). This cross-linking reaction is more specific and efficient, and does not produce significant photo-damage to nucleic acid and protein compared to short wave UV (254 nm). Advantageously, the occurrence of T to C conversions in cDNAs derived from protected cross-linked RNAs are diagnostic of the RNA-protein interactions providing the ability to discriminate signal versus noise and to delineate the precise binding site.
HuR (ELAVL1) is an essential and ubiquitous protein member of the ELAV/Hu family of RBPs, necessary for proper embryonic development and immune response in mice (Katsanou et al., 2009; Papadaki et al., 2009). ELAV proteins contain three RNA Recognition Motifs (RRMs) that are central components of their RNA-binding domains (Szabo et al., 1991). Previous studies of RNA-targeting suggest that their binding sites consist of short single-stranded stretches of uridines separated by adenosines or, less commonly, other bases (Levine et al., 1993; Meisner et al., 2004). These proteins are part of a broader group of RBPs that are known to interact with RREs known as AU-rich elements (AREs), which occur in the 3′ UTR of many mRNAs encoding important growth and regulatory proteins(Caput et al., 1986; Shaw and Kamen, 1986).
All mammalian members of the ELAV/Hu family, including HuR, positively regulate the stability and translation of their target mRNAs (Jain et al., 1997; Fan and Steitz, 1998; Peng et al., 1998). This positive regulatory effect is a distinctive feature of the Hu family compared to many other RBP-ARE interactions, as well as other modes of post-transcriptional regulation. HuR has also been reported to affect poly-adenylation or splicing for a few mRNA targets (Izquierdo, 2008; Zhu et al., 2007).
RNA targets of HuR have been identified in numerous cell types and cellular conditions, but there has not been a definitive global study identifying the precise RNA-binding sites in vivo (López de Silanes et al., 2004; Mazan-Mamczarz et al., 2008; Mukherjee et al., 2009; Tenenbaum et al., 2000). In this study, we integrated three independent high-throughput approaches for examining HuR-dependent regulation of target gene expression: PAR-CLIP in combination with our own analytical method to identify high-resolution HuR binding sites; RIP-chip analysis as a complementary biochemical method to quantify HuR-mRNA association; and depletion of HuR using RNA interference combined with whole-transcript expression profiling to assess the impact of HuR-binding on mRNA abundance and mRNA isoforms. We observed that the cumulative number of HuR binding sites per transcript was predictive of both the overall association of HuR with the transcript and the HuR-dependent stabilization. Furthermore, we demonstrated that HuR regulates both pre-mRNA processing as well as mRNA stability and may couple these two spatially distinct processes. Finally, we report that HuR can antagonize miRNA-mediated repression of for miRNAs proximal to HuR binding sites.
RESULTS
HuR-RNA interactions and RNA Recognition Element
We applied the recently developed PAR-CLIP technique (Hafner et al., 2010), to precisely identify RNA elements interacting with an epitope-tagged HuR in vivo (Figure S1A). All experiments in this study were performed in the same cell line as the initial PAR-CLIP study (FlpIn T-Rex HEK 293), thereby allowing our data to be integrated with data derived in that study. HuR bound RNA fragments were successfully identified (Figure S1B, ~45 kD band), converted into cDNA after ligation of adapters, and subsequently sequenced using the Illumina platform.
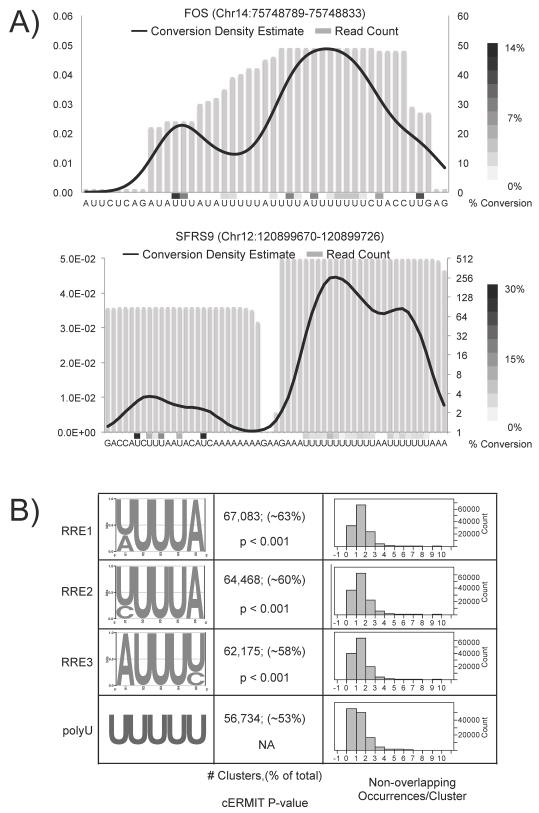
We utilized PARalyzer, an algorithm designed to detect binding sites at a high resolution from PAR-CLIP data (manuscript under review). Briefly, PARalyzer employs kernel density estimation (KDE) of diagnostic T-C conversions deduced from sequence reads mapping uniquely to the genome to delineate HuR protein-RNA cross-linking sites. Read data from Illumina libraries were mapped to the genome, grouped by overlaps, and precise binding sites were derived based on KDE of conversions (Table S1A). We identified 137,305 clusters, including many mapping to known HuR binding sites, such as the ARE in the 3′ UTR of FOS mRNA (Figure 1A, top). The majority of clusters identified mapped to transcripts derived from protein-encoding genes and specifically to 3′ UTRs and intronic regions. Using our approach, the average cluster length for HuR was 25 nucleotides (Figure S1C), a reduction from the 30-nucleotide average cluster length for HuR using the analysis pipeline in (Hafner et al., 2010). We delineated adjacent HuR clusters from the same initial group of reads as seen in the 3′ UTR of SFRS9 mRNA (Figure 1A, bottom) and identified a statistically significant subset of HuR clusters (2,857 pairs) spaced less than 40 nucleotides from adjacent HuR clusters (Figure S1D). The juxtaposed clusters could be attributed to HuR binding on different copies of the same species of transcript or even RNA looping.
Figure 1.
Identification of HuR regulatory elements. A) For binding sites detected in FOS 3′ UTR (top) and SFRS9 3′UTR (bottom), the KDE of conversion events (black line) and percent conversion (heatmap) were superimposed on read depth information (grey bars) for each nucleotide in the initial group. B) RREs identified applying cERMIT to clusters ranked by CLIs (left). Comparison of quantile plots, number of occurrences, and p-values for each RRE shown (right).
The initial PAR-CLIP study demonstrated that ranking clusters by the number of T to C conversion reads within a cluster resulted in enrichment of RRE at the top of the rank-ordered list (Hafner et al., 2010). Similarly, we calculated a cross-linking index (CLI) using the number of T to C conversions detected in all reads underlying a cluster and ranked clusters by the CLI (see methods for details). Comparing the distribution of CLIs for clusters mapping to different regions of the transcriptome revealed a hierarchy, in which clusters with higher CLIs were proportionally more often derived from regions representing mature mRNA (3′ UTR, CDS, and 5′ UTR), followed by introns, and finally intergenic regions (Figures S2 and ST2). Disparities in the abundance of RNA from each of these regions likely contributed to the observed CLI hierarchy. For the remainder of the study, we focused on clusters mapping to RNA transcripts from protein-encoding genes (n=106,840).
We applied cERMIT, an evidence-ranked motif discovery tool designed to investigate quantitative genome-wide binding data, to CLI-ranked clusters to define the in vivo RRE for HuR (Georgiev et al., 2010). The three highest scoring motifs contained a stretch of 3–4 U’s separated by an A or C and occurred in over 58–63% of clusters (Figure 1B). In comparison, clusters containing poly-U stretches of 5 nucleotides or longer occurred in 53% of clusters. Collectively, these RREs were present in ~84% (90,231/106,840) of all clusters. Interestingly, multiple core RREs were often found arranged adjacently forming higher-order arrays of RREs as seen in the FOS ARE (Fig 1A top). The high scoring RREs had more non-overlapping occurrences than 5-mers of poly-U (Figure 1B). These higher-order arrays may reflect different modes of binding through contributions from multiple RRMs or cooperative assembly of HuR molecules on the RNA ligand (Wang and Hall, 2001; Fialcowitz-White et al., 2007).
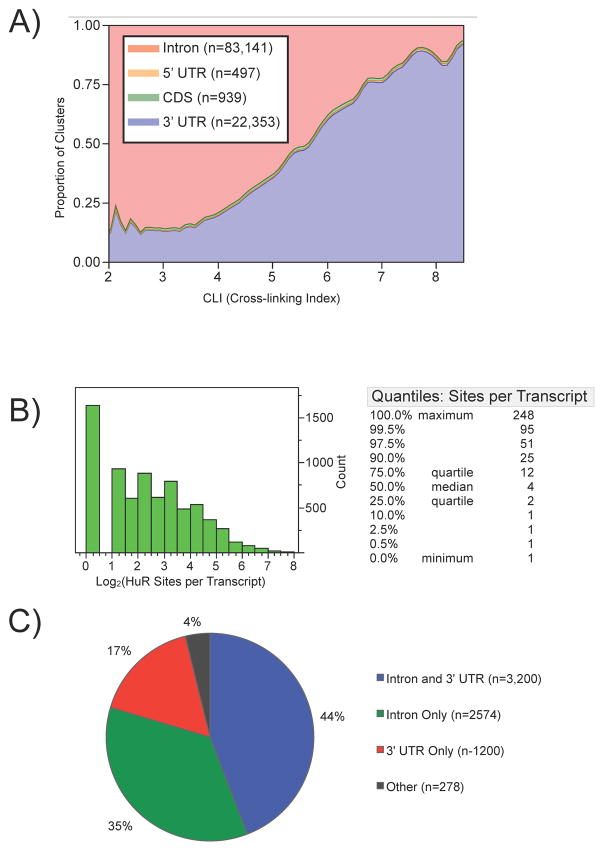
The distribution of binding sites across individual transcripts provided insights into HuR targeting. Mapping clusters to specific transcript regions revealed that introns (n=83,141) and 3′ UTRs (n=22,353) were the regions predominantly targeted by HuR (Figure 2A). Furthermore, clusters derived from transcript regions mapping to both pre-mRNA and mature mRNA (5′ UTR, CDS, and 3′ UTR) exhibited higher CLIs than those clusters derived from transcript regions mapping only to pre-mRNA (intron). The lower abundance of pre-mRNAs compared to their mature mRNA counterparts may partially explain this observation.
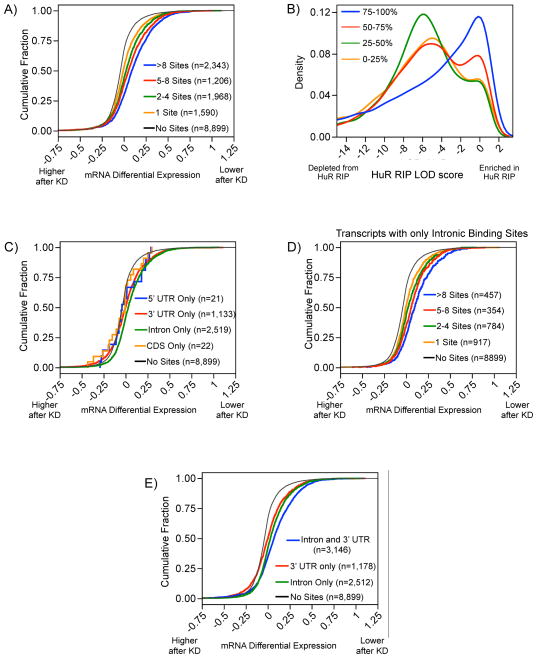
Figure 2.
Landscape of HuR binding sites in transcript regions. A) The proportion of densities for HuR binding clusters derived from each region of an mRNA as a function of cross-linking index (CLI). B) Histogram (log2) and quantiles of the distribution for the number of HuR binding sites per transcript. C) Pie-chart depicting transcripts with specific regions of HuR binding sites.
More than half of the targeted transcripts had 4 or more binding sites, with 10% of transcripts having over 25 sites (Figure 2B and ST3). The many 3′ UTR binding sites identified was consistent with the long-held hypothesis that HuR targets the 3′ UTR to execute its primary function as a regulator of mRNA stability and/or translation. However, the amount of intronic binding indicated an additional unexpected role for HuR in pre-mRNA processing. Nearly all targeted transcripts (96%) had a HuR binding site in either the 3′ UTR or an intron and almost half (44%) had binding sites in both introns and 3′ UTRs (Figure 2C), suggesting HuR may couple pre-mRNA processing and mRNA stability for a subset of mRNAs as discussed below.
We utilized RIP-chip as a complementary method to further explore principles of HuR-mRNA targeting in vivo. After confirming enrichment of positive control HuR targets, RNA from these samples was interrogated using Affymetrix Human Exon 1.0 ST Arrays to obtain whole transcript targeting data complementary to PAR-CLIP data (Figure S3A,B). Similar to previous HuR RIP-chip results (Mukherjee et al., 2009), we detected two populations of mRNAs: an enriched population representing HuR-associated mRNAs and a non-enriched population representing background mRNAs (Figure S3C). We estimated a Gaussian mixture to calculate log odds ratios (LOD) representing the probability of an mRNA being associated with HuR (Figure S3D). Transcripts with HuR LOD scores greater than zero were considered a discrete population of HuR-bound mRNAs.
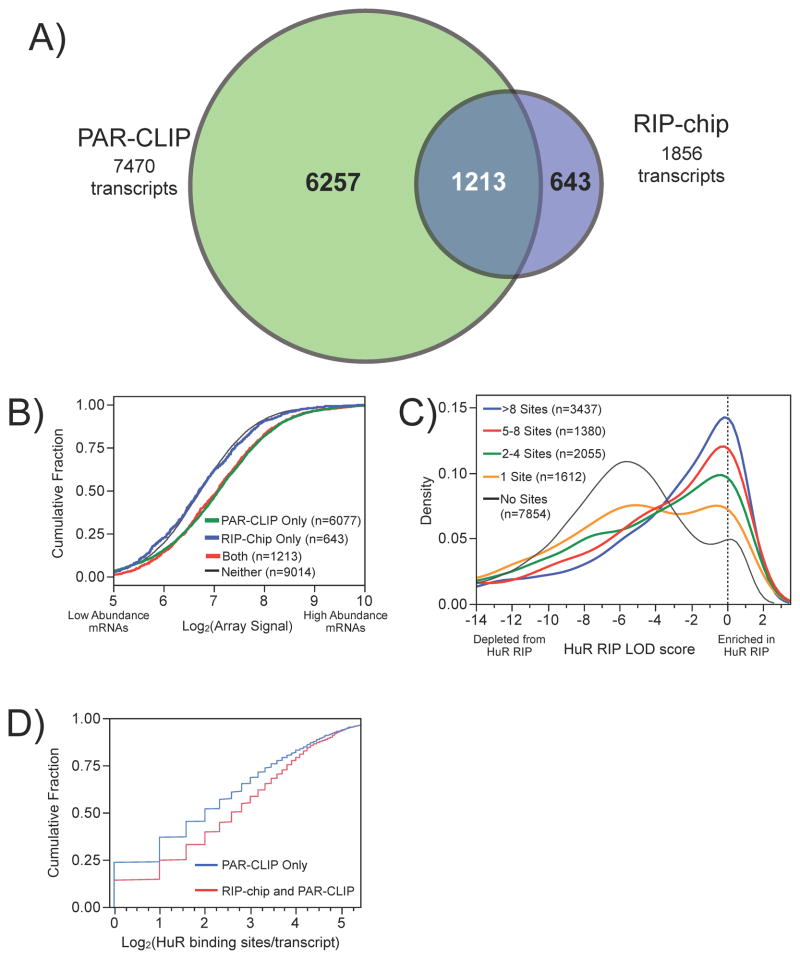
Transcript level analysis of both datasets revealed that 7,470 and 1,856 mRNA targets of HuR were identified using PAR-CLIP and RIP-chip, respectively (Figure 3A and ST3). Over 65% of the HuR targets identified by RIP-chip were also identified by PAR-CLIP (1,213/1,856). Conversely, only ~16% of the HuR targets identified by PAR-CLIP were also identified by RIP-chip (1,213/7,470). Targets identified by both methods were significantly more enriched than PAR-CLIP only targets based on the number of HuR binding sites derived from the PAR-CLIP data (Figure 3D and S4A), but were not significantly more enriched in the RIP-chip than RIP-only targets based on LOD scores derived from the RIP-chip data (Figure S4B).
Figure 3.
Comparison of PAR-CLIP and RIP-chip HuR interaction data. A) Venn diagram of the overlap between transcripts defined as HuR targets using PAR-CLIP and RIP-chip, which were also functionally responsive (see figure S5C). B) Comparison of the cumulative distribution of transcript abundance for targets detected by RIP-chip only (blue), PAR-CLIP only (green), and RIP-chip and PAR-CLIP (red) using microarray data to quantify transcript abundance. C) Comparison of the HuR LOD score distributions of PAR-CLIP data categorized by number of binding sites per transcript. D) Comparison distribution of PAR-CLIP binding sites/transcript for targets identified only by PAR-CLIP or by both PAR-CLIP and RIP-chip.
Non-overlapping HuR targets identified by only one of the methods (PAR-CLIP only or RIP-chip only) could be explained by differences in the IP protocols, detection strategies, and/or the stability of the RNA-protein interactions. Because we did not utilize cross-linking in our RIP protocol, the 643 HuR targets identified only by the RIP-chip were inspected to determine if they were true-positives. Many of these targets contained the U-rich RREs identified above and were functionally responsive to HuR knockdown (Figure S5C). The 643 transcripts could potentially represent transcripts from multi-copy genes or other transcripts filtered out during PAR-CLIP analysis. However, we found that the HuR targets identified only by RIP-chip were significantly lower in abundance than remaining targets as quantified by microarray (Figure 3B) or even more noticeably by RNA-seq (Figure S3E), implying we may not have reached sufficient sequencing depth in the PAR-CLIP analysis to identify all HuR targets.
Transcripts with more HuR binding sites were significantly more enriched by the RIP-chip than those with fewer binding sites. (Figure 3C). Targets identified by both RIP-chip and PAR-CLIP (median number of sites/transcript=6) contained ~2 more binding sites than targets identified by PAR-CLIP only (Figure 3D; median number of sites/transcript=4). Assuming that more binding sites per transcript result in a more stable interaction, these results support the hypothesis that RIP-chip enriches for stable interactions, while PAR-CLIP identifies both stable and transient interactions, possibly attributable to the high cross-linking efficiency. Transcripts containing intronic binding sites only were significantly less enriched in the RIP-chip than mRNAs with sites in the intron and 3′ UTR or 3′ UTR only (Figure S4C); thus, the utility of this specific RIP-chip protocol may be limited for investigating RBPs interacting exclusively with introns. Differences in transcript coverage of the microarrays compared to deep sequencing may have contributed to the difference; more likely explanations include the difference in pre-mRNA and mature mRNA abundance or the pre-IP RNase digestion step used in PAR-CLIP.
HuR regulated gene expression regulators
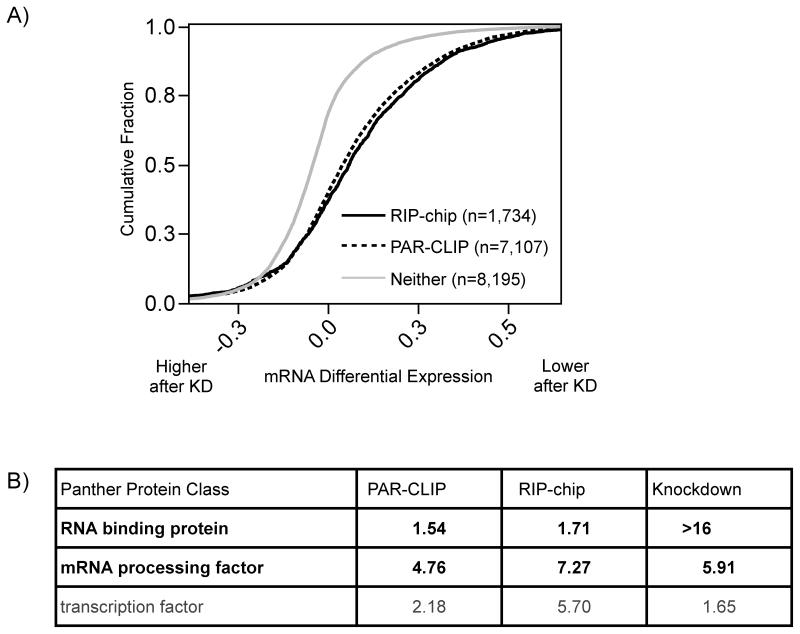
As a third independent genomic approach to identify HuR target mRNAs, we conducted transcriptomic analysis of cells treated with HuR siRNA or negative control scrambled siRNA. This resulted in ~20% remaining HuR protein (Figure S5A), as well as depletion of a few known mRNA targets resulting in 2–4 fold less mRNA, specifically VEGF (~28%), TNF-alpha (~32%), and CCNA2 (46%) (Figure S5B). RNA from these samples was analyzed with exon microarrays. Since HuR is a know regulator of mRNA stability, we assumed depletion of targeted transcripts was a result of decreased mRNA stability. Over 77% of the 154 transcripts exhibiting at least a 1.5-fold decrease were identified as HuR targets by PAR-CLIP or RIP-chip. All targets identified by either method were significantly depleted following HuR knockdown relative to non-targets (Figure 4A and ST3). These results supported our use of epitope-tagged HuR in the PAR-CLIP and RIP-chip. Moreover, RIP-chip targets were slightly more responsive than PAR-CLIP targets, consistent with the hypothesis that RIP-chip enriches for more stable interactions and thus targets that are more functionally responsive with regards to mRNA stability.
Figure 4.
HuR-dependent mRNA stabilization. A) Comparison of cumulative distribution of mRNA differential expression after HuR knockdown for targets identified by RIP-chip (black), PAR-CLIP (dashed), and neither method (grey). The depletion of RIP-chip and PAR-CLIP targets were statistically significantly compared non-targets (p < 0.001). B) Protein classes enriched in HuR PAR-CLIP, RIP-chip, and knockdown targets. After each dataset was percentile rank transformed, the enrichment of significant Panther defined gene sets was reported using the −log10(enrichment p-value).
To look for common functional relationships among mRNA targets enriched by the different methods used in this study, the number of HuR binding sites (PAR-CLIP), the HuR LOD scores (RIP-chip), and the HuR-dependent fold change (HuR knockdown) were individually examined for classes of proteins significantly enriched at the top of each list (Figure 4B). Protein classes representing RBPs, mRNA processing factors, and transcription factors exhibited the most statistically significant enrichment across all datasets. The enrichment of gene expression regulators is a common feature noted previously in RIP-chip studies (Mansfield and Keene, 2009); the “regulator of regulators” concept has been demonstrated in more detail for HuR (Pullmann et al., 2007).
HuR regulated pre-mRNA and mature mRNA processing
There was a positive correlation between the cumulative number of binding sites per transcript and the degree of HuR-dependent stabilization (Figure 5A). The degree of HuR-dependent stabilization was also correlated with higher HuR RIP-chip LOD scores (Figure 5B). Therefore, both PAR-CLIP and RIP-chip provide quantitative binding data predictive of the degree of regulation by HuR. We examined the extent to which intronic binding events contributed to the stability of the mRNA targets. To not conflate effects potentially mediated by different regions of the transcript it was necessary to restrict this analysis to transcripts containing binding sites exclusive to a certain transcript region. Transcripts containing only intronic binding sites were as responsive to HuR knockdown as transcripts containing only 3′ UTR binding sites (Figure 5C) and the degree of stabilization was proportional to the number of binding sites (Figure 5D). Intriguingly, mRNAs containing both intronic and 3′ UTR binding sites were significantly more stabilized than mRNAs with binding sites only in the 3′ UTR or intron (Figure 5E) and exhibit the same degree of stabilization as observed for transcripts with over 8 binding sites. (Fig 5A, blue curve). Thus, the positive regulation of stability may be accomplished through pre-mRNA sites not limited to the mature mRNAs, and specifically the 3′ UTR. Futhermore, these results indicate a mechanism by which HuR couples pre-mRNA processing and mRNA stability.
Figure 5.
Degree of HuR binding and functional outcome. A) Transcripts with more HuR binding sites exhibit a greater decrease in response to HuR knockdown. B) Transcripts were divided into 4 categories based on their percentile of decrease after knockdown, 75–100th representing mRNAs that decreased the most. Transcripts with higher HuR LOD scores correlated with a greater decrease in mRNA levels following HuR knockdown. C) Transcripts with binding sites only in introns (green) or only in 3′ UTRs (red, behind green line) were significantly destabilized by HuR knockdown. D) More intronic HuR binding sites exhibit greater destabilization after HuR knockdown. E) Transcripts with HuR binding sites in both 3′ UTRs and introns are significantly more destabilized by HuR knockdown than transcripts with HuR binding sites only in 3′ UTRs or introns.
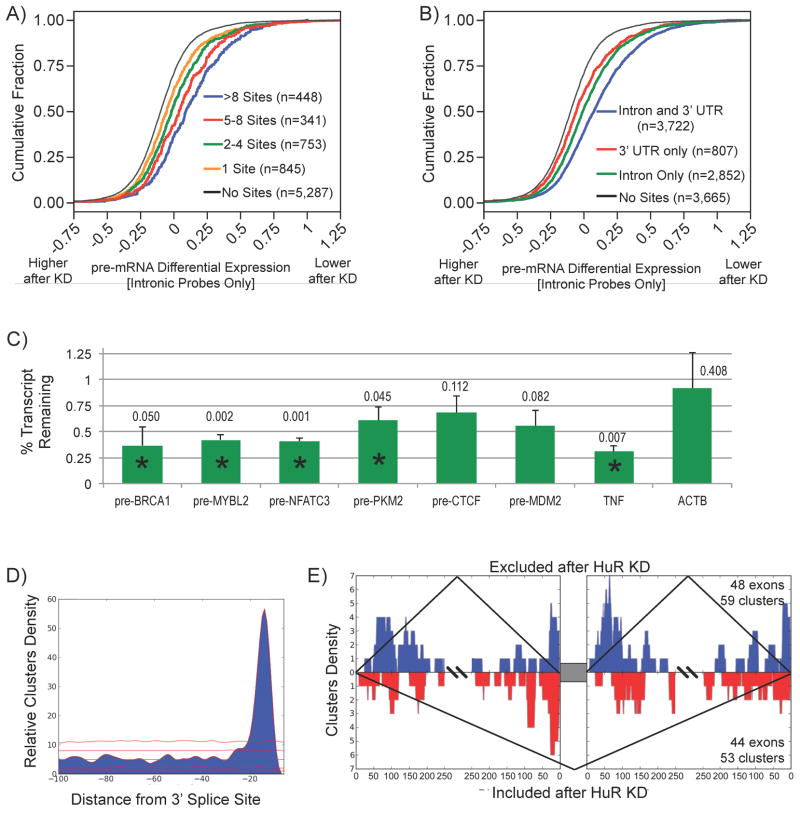
In order to examine the extent of HuR-dependent stabilization at the pre-mRNA level, we recalculated whole transcript expression estimates solely from probes designed to hybridize to intronic regions (for coverage and expression controls see Figure S6A,B). For transcripts with only intronic binding sites, we again observed a correlation between the number of binding sites and the degree of pre-mRNA stabilization (Figure 6A), and transcripts with intronic and 3′ UTR binding sites still exhibited augmented pre-mRNA stabilization (Figure 6B). HuR-dependent stabilization for individual candidate pre-mRNAs was validated using qRT-PCR with pre-mRNA-specific primers (Figure 6C). Collectively, these results indicate that many of the 3′ UTR binding sites are deposited on the pre-mRNA and together with intronic sites can stabilize targeted pre-mRNAs.
Figure 6.
HuR regulated pre-mRNA processing. A) Transcripts (restricted to those with only intronic sites) with more intronic binding sites exhibited more pre-mRNA destabilization upon HuR depletion. B) Transcripts with both intronic and 3′ UTR sites were more destabilized upon HuR depletion than transcripts with either intronic or 3′ UTR sites. C) Effect of HuR depletion on individual pre-mRNAs were validated using pre-mRNA specific primers (* indicates paired t-test p < 0.05, actual p-values listed above bar). D) Overrepresentation of binding sites in the Py-tract exons (red lines represent 1st, 10th, 50th, 90th, and 99th percentiles of the null distribution). E) HuR RNA splicing map exhibiting consistent binding pattern for both included and excluded exons with adjacent HuR binding sites.
Upon examining the spatial position of intronic binding sites, a significant overrepresentation of intronic HuR binding sites within the first 50 nucleotides upstream of the 3′ splice sites implicated a role for HuR in splicing (Figure 6D and S6C). Both the position of these intronic binding sites and the underlying sequence are consistent with the poly U/C motif, suggesting that HuR interacts with the Py-tract located between the 3′ splice site and the branchpoint to affect splicing. To examine HuR-dependent splicing events associated with the observed intronic binding sites, we created HuR RNA splicing map by integrating HuR PAR-CLIP clusters with HuR knockdown exon array splicing data, specifically cassette-exons. AltAnalyze was used to identify splicing events in response to HuR knockdown and detected both exon-inclusion and -exclusion events (Salomonis et al., 2009). Many of the alternative processing events detected upon HuR depletion did not have adjacent intronic HuR binding sites. Since functionally responsive HuR targets were enriched for pre-mRNA processing factors (Fig 4B), the observed changes in exon usage may represent downstream regulatory events. Both of these exonic events exhibited similar spatial patterns of HuR binding in adjacent intronic regions (Figure 6E), with most binding occurring in flanking Py-tracts. Altogether the observed spatial pattern was associated with promoting both exon-inclusion events (48 exons) and exon-exclusion events (44 exons) (Figure 6E and S6D,E).
Combinatorial regulation by HuR and miRNAs
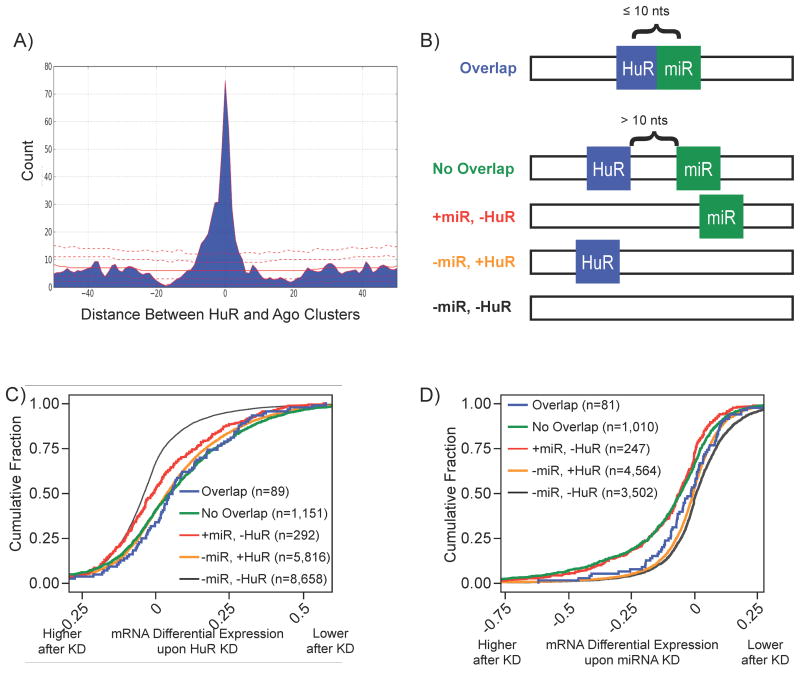
Previous studies have demonstrated combinatorial regulation by HuR and miRNAs (Bhattacharyya et al., 2006; Kim et al., 2009). We inspected the positional relationship between HuR binding sites and binding sites from members of the Argonaute family (Ago) (Table S1B). Over 75% (3,031/3,942) of mRNAs with Ago binding sites in 3′ UTR also contained HuR binding sites in the 3′ UTR, as indicated by previous work (Mukherjee et al., 2009). We found a significant enrichment for overlapping/adjacent HuR and Ago binding sites in 3′ UTRs of targeted mRNAs (Figure 7A), with 443 pairs of HuR and Ago sites (in the 3′ UTR of 331 mRNAs) that were within 10 nts of one another, indicating the potential for competition for effective binding and regulation.
Figure 7.
Combinatorial regulation by HuR and miRNAs. A) Distribution of distances between HuR and Ago sites restricted to the 3′ UTR identified by PAR-CLIP. Red lines indicate distribution of null model (1st, 10th, 50th, 90th, and 99th percentile). B) Based on the distribution of distances, transcripts were classified by HuR and miRNA binding profiles depicted. Only transcripts exclusively belonging to one of the classes depicted and targeted by one of the depleted miRNAs were considered. C) Transcript classes with HuR binding sites were destabilized upon HuR knockdown regardless of miRNA sites. D) De-repression of miRNA and HuR overlap transcripts compared to no-overlap transcripts upon miRNA depletion (see ST5 for p-values).
To look for functional antagonism between HuR and miRNAs, transcripts were classified based on the presence of at least one instance of the following five interaction profiles (detailed in figure 7B): overlap(s), no-overlap(s), miRNA site(s) and no HuR site(s), no miRNA site(s) and HuR site(s), and no miRNA site(s) and no HuR site(s). We compared the fate of each class of transcripts in response to HuR depletion or depletion of 25 highly expressed miRs. The destabilization profiles for these transcript classes in the HuR KD was explained by the presence of HuR sites regardless of joint or positional effects from miRs (Figure 7C). However, transcripts with overlapping miRNA (restricted to depleted miRs) and HuR sites were significantly less upregulated than transcripts with only miRNA sites or with miRNA and HuR sites that do not overlap (Figure 7D). In fact, the overlap class responded like transcripts without miRNA sites, while the no-overlap class responded like transcripts containing only miRNA sites. There are two models of combinatorial regulation by HuR and miRs. In one model HuR antagonizes miRNA mediated repression (Bhattacharyya et al., 2006); and in the other model HuR promotes miRNA-mediated repression (Kim et al., 2009). Our data indicate that the model of relieving miRNA-mediated repression would most likely be due to competition for physical access to the respective binding sites that are overlapping or adjacent.
DISCUSSION
We presented a systematic analysis of RNA targeting and consequent RNA processing by the RBP HuR. Integration of quantitative features derived from high-throughput biochemical HuR-RNA interaction data was informative of specific mechanisms underlying HuR-mRNA targeting and functional outcomes. All targets were not quantitatively equivalent with respect to their cumulative HuR-interaction stability (more binding sites corresponded with more enrichment in the RIP-chip) and subsequently the degree of HuR-dependent regulatory outcome. Although we employed state-of-the-art technologies, they all provided data for populations of cells and mRNAs and none yet have the resolution to provide information regarding individual copies of mRNA. Therefore, we cannot definitively know that HuR concurrently occupied multiple distinct interaction sites in a given copy of mRNA. Nevertheless, our data supported the notion that RIP-chip enriches for stable interactions, while PAR-CLIP captures both stable and transient interactions (Hafner et al., 2010).
RIP-chip targets were slightly more functionally responsive than PAR-CLIP targets, demonstrating that the potential for post-lysis reassociation was negligible. The only study demonstrating post-lysis reassocation with HuR utilized very different RIP conditions, including sonication of lysates (Mili and Steitz, 2004). Based on these data and published results from many different laboratories [reviewed in (Morris et al., 2010; Keene et al., 2006), the evidence demonstrates that cross-linking is not a de facto necessity to legitimize RNA-protein interactions.
HuR-dependent RNA processing
Our data suggest HuR is involved in coupling pre-mRNA processing and mature mRNA stability. HuR-dependent stabilization was evident at the pre-mRNA level suggesting that HuR-binding is a very early step in the life of an mRNA. We propose that the stabilization of pre-mRNA by HuR ensures proper mRNA processing, akin to the effect of Pol II transcription (Hicks et al., 2006). Over half of the targeted transcripts contained both intronic and 3′ UTR binding sites and these are significantly more stabilized than transcripts with only 3′ UTR or only intronic binding sites. It has been known for decades that the presence of an intron can greatly increase the expression of certain gene constructs (Buchman and Berg, 1988). Later studies revealed that the intron-dependent effects on mRNA expression have a greater effect on enhancing 3′ end formation and translation, rather than nuclear export (Lu and Cullen, 2003). Exon junction complexes (EJC) have been implicated in coupling pre-mRNA processing events in the nucleus with export and translation of mature mRNA in the cytoplasm and non-sense mediated decay (reviewed in (Moore and Proudfoot, 2009)). We speculate that HuR binding sites in pre-mRNA stabilize the transcript and promote appropriate and efficient splicing and/or 3′ end formation in the nucleus, and that retained mature mRNA binding sites contribute to stabilization in the cytoplasm. The dependence of our observations on EJC deposition and transcription represent areas of future investigation.
Speculation that Hu proteins may play a role in pre-mRNA processing date to their initial discovery (Szabo et al., 1991; Robinow et al., 1988). Prior to our study, across all four mammalian ELAV/Hu proteins there were a few known splicing and poly-adenylation substrates (Hinman and Lou, 2008). Perhaps, the discovery that these proteins could bind 3′ UTRs of growth factor and cytokine mRNAs shifted the emphasis of investigation in the field to cytoplasmic mRNA stability and translation (Levine et al., 1993). Since early studies found roles in nuclear export and mRNA stability (Brennan and Steitz, 2001), the role of HuR in nuclear pre-mRNA events has by and large not been investigated. In this study, we revealed a significant function for HuR in both pre-mRNA stability and splicing.
The inclusion of exon 6 of Fas was the only known example demonstrating a direct role for HuR in mRNA splicing (Izquierdo, 2008). The proposed model describes inhibition of exon definition and subsequent U2AF binding to the upstream 3′ exon splice site (SS) and Py-tract through multimeric HuR interactions with an exonic splicing silencer. We observed relatively few CDS binding sites and could not detect binding sites in the Fas transcript, although its expression was not not detected in our RNA-seq data. While the proposed model for Fas pre-mRNA splicing was related to exonic HuR regulatory elements promoting exon exclusion, our observations focused on intronic HuR regulatory elements, for which those upstream of the 3′ exon SS were associated with both exon inclusion and exclusion.
RNA splicing maps have indicated positional relationships between intronic binding sites and exon usage decisions for several RBPs [reviewed in (Witten and Ule, 2011)]. This type of spatial analysis for HuR did not indicate an obvious relationship between different spatial binding patterns correlating with different exon usage decisions. Yet the cases in which exon inclusion was promoted, there was slightly more cluster density at the downstream 5′ splice site (Figure 6E, blue). Interestingly, differentially expressed exons without any adjacent HuR binding sites exhibited a slight preference for exon exclusion (Figure S6D, dashed line). This could be due to secondary effects from HuR depletion and consequent down-regulation of splicing factors like FOX2, NOVA, PTB, and HNRNPC; all of which were targets of HuR with decreased mRNA expression after the HuR knockdown. The majority of differentially expressed exons did not have adjacent HuR binding sites. However, there are a subset of exons for which we detected both alternative usage and adjacent binding sites suggesting direct splicing regulation by HuR. Similar to its function of promoting expression of negative regulators of mRNA stability, HuR may promote the expression of other regulatory RBPs with antagonistic functions in pre-mRNA splicing to achieve appropriate exon usage decisions.
RNA-RBP dynamics and interaction stability
Previous RIP-chip studies have demonstrated dynamic changes in the association of mRNAs with ELAV/Hu proteins (Mazan-Mamczarz et al., 2008; Mukherjee et al., 2009; Tenenbaum et al., 2000). In these examples, mRNAs exhibited quantitative differences in their RBP association, demonstrating remodeling of the RNPs in response to stimuli. The mechanisms underlying these dynamics are not understood; however, the data in this study are consistent with the idea that remodeling of stable interactions enriched by the RIP-chip may derive from the larger pool of transient interactions only observed in the PAR-CLIP (Figure 3A). Recent studies have indicated RNA accessibility partially determines if RBP-mRNA interactions occur (Kazan et al., 2010). Computational methods accounting for sequence specificity as well as structural accessibility combined with the PAR-CLIP and RIP-chip data may help determine such RNA binding preferences.
Combinatorial regulation by HuR and miRNAs
Our results generalize and describe a physical mechanism for HuR-mediated antagonism of miRNA-mediated repression at proximal sites. Specifically, we only observed a differential functional outcome between transcripts containing overlapping binding sites compared to transcripts containing non-overlapping binding sites in response to miRNA depletion, but not in response to HuR depletion (Figure 7). Thus, the presence of proximal HuR binding sites explains why a subset of miRNA targets did not respond in the expected manner to miRNA depletion. Given that HuR is capable of relieving miR-122 mediated translational repression of the CAT-1 mRNA after environmental stresses (Bhattacharyya et al., 2006), in all likelihood HuR molecules outcompete overlapping miRNA binding sites upon environmental stresses. Our results predict this to be a more widespread phenomenon affecting mRNA stability as well. Thus, it will be necessary to investigate RBP-RNA interaction dynamics accounting for multiple factors in a quantitative manner to understand apparently unexpected regulatory outcomes. Integration of global technologies similar those described here may lead to computational simulations of mRNA regulatory dynamics and the coherent outcomes predicted by posttranscriptional RNA operon/regulon model.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK 293 Flip-In T-REx cells containing a FLAG-HA-tagged HuR were cultured in DMEM supplemented with 10% tet-reduced FBS and appropriate selection antibiotics. For induction, cells were treated overnight with 1 ug/mL doxycycline (Sigma).
PAR-CLIP
Protocol was performed as detailed in (Hafner et al., 2010) for a FLAG-HA-tagged HuR and sequenced using the Illumina platform. Previously derived Ago1–4 PAR-CLIP libraries were analyzed with PARalyzer. Read mapping statistics for both are provided in Table S1A, B. Cluster and gene level data are provided in Table S2 and S3, respectively.
Mapping reads
The reads from the HuR and Ago deep sequencing library were stripped of the 3′ adapter sequence using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Reads that were less than 13nt in length or contained an ambiguous nucleotide were discarded. The remaining reads were aligned to the human genome (hg19), with up to 2 mismatches allowed, by the Bowtie algorithm (Langmead et al., 2009). Mapped locations were only reported for those with the minimum number of observed mismatches for each read. All T to C mismatches between the RNA fragment and the genome were subtracted from the mismatch count for each mapped location. After subtraction only reads that mapped to a single genomic location at the minimum number of mismatches were used for further analysis. The location that a read mapped, relative to a known transcript, was determined based on the ENSEMBL v57 database 1(Hubbard et al., 2007). If a read mapped to a location representing multiple categories, it was reported to belong to the category based on the following order of preference: 3′UTR, coding sequence, 5′UTR, intron, non-coding RNA, intergenic.
Defining HuR binding sites by PARalyzer
Overlapping reads were grouped together for further analysis. A group must have contained ≥5 reads with conversions at two or more locations. Separate kernel density estimates were calculated across all positions based on read counts both with and without T to C conversions, as long as there was a minimum read depth of 5 at a position to ensure a robust density estimate. Clusters were defined as regions for which the conversion density was greater than the non-conversion density for at least 5 consecutive nucleotides, and extended until the group read depth fell below 5. Following filtering (see supplemental methods), clusters were scored by a cross-linking index (CLI) = log2(1+T to C conversion events for all reads within a cluster). PARalyzer is available at http://www.genome.duke.edu/labs/ohler/research/PARalyzer.
Motif finding
All groups mapping to mRNA were ranked using CLI scores. cERMIT (Georgiev et al., 2010) was run on a list ranked by CLI scores (see supplemental methods for details).
RIP-chip
Protocol was performed as described in (Keene et al., 2006) with anti-FLAG M2-conjugated beads or mouse IgG-conjugated beads was used for immunoprecipitation reaction. Appropriate amounts of total RNA from biological replicates of HuR and Mock RIPs were submitted to the Microarray Facility at Duke University for analysis on GeneChip Human Exon 1.0 ST Arrays (Affymetrix).
siRNA knockdown
were analyzed by microarray and qRT-PCR (see supplement for details).
Array data processing and analysis
RIP-chip microarray data was normalized with PLIER using Affymetrix Power Tools. Probes with DABG p-values > 0.05 in all three HuR RIP replicates were excluded. For HuR RIP-chip experiments, t-scores were calculated with GSEA and HuR LOD scores were computed using weighted probability distribution functions based on mixture model as described in (Mukherjee et al., 2009; Subramanian et al., 2005). Knockdown data was normalized with RMA and expression changes were compared using t-scores and log-fold change in GSEA. The same transcript annotation used for mapping PAR-CLIP data was used to categorize probes as either intronic or exonic based on their annotated genomic coordinates. Gene expression values for all intronic probes mapping to the same gene were collapsed to the median. Splicing analysis was performed using AltAnalyze (Salomonis et al., 2009), using the FIRMA metric on extended probes to analyze exon usage. Normalized antagomiR microarray data was obtained from GEO (GSE21577). Log fold expression changes were calculated after filtering for detection, and collapsing expression data to the median for all probes mapping to a gene.
GO analysis
Each data set was percentile-rank transformed (background list not necessary) and analyzed as gene expression values in PantherDB (Thomas et al., 2006).
Comparison of spatial patterns
Distance between HuR binding sites, HuR and Ago binding sites, and the RNA splicing map analyses were performed using custom python scripts (see supplemental methods for details). To generate RNA splicing maps, clusters falling within 250 nts (divided in 50-nt bins) of an exon of interest and its flanking exons were mapped into the corresponding introns. The number of clusters that cover each position in the intron were then summed to define the density at for the 50-nt bin. HuR and Ago binding site comparisons were restricted to the 3′ UTR. The RNA splicing map was created using cluster data across introns upstream and downstream of the central exon. Annotation of exons as constitutive was determined using AltAnalyze.
Comparisons of distributions
JMP 9.0 (SAS) was utilized for all comparisons of distribution and densities for various categories and metrics including cumulative distribution fraction plots, comparison of densities, and proportion of densities. Student’s t-test comparing the means of different categories was also calculated using JMP 9.0 to assess statistically significant differences.
Supplementary Material
Acknowledgments
We thank members of the Tuschl, Ohler, and Keene labs for discussions and advice, particularly Matt Friedersdorf. This work was supported by National Science Foundation grants 0842621 (J.D.K.) and 0822033 (U.O.), and National Institutes of Health grant DA030086 (U.O.). Experiments conducted in the Tuschl laboratory were funded by HHMI and a grant from the Starr Foundation. M.H. is supported by a fellowship from the Charles H. Revson, Sr. Foundation. J.D.K. has a financial relationship with Ribonomics, Inc. and MBL, Inc. that hold licenses to technologies used in this study. T.T. is cofounder and scientific advisor to Alnylam Pharmacetuticals and advisor to Regulus Therapeutics.
Footnotes
Accession Numbers.
All datasets are available through the Gene Expression Omnibus, GSE29780.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev S, Boyle AP, Jayasurya K, Ding X, Mukherjee S, Ohler U. Evidence-ranked motif identification. Genome Biol. 2010;11:R19. doi: 10.1186/gb-2010-11-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MJ, Yang C, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–7. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J Biol Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comp Biol. 2010;6:e1000832. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Molecular Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mansfield KD, Keene JD. The ribonome: a dominant force in coordinating gene expression. Biol Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Hagner PR, Dai B, Wood WH, Zhang Y, Becker KG, Liu Z, Gartenhaus RB. Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach. Cancer Research. 2008;68:7730–7735. doi: 10.1158/0008-5472.CAN-08-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner N, Hackermüller J, Uhl V, Aszódi A, Jaritz M, Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chem Bio Chem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip Rev Syst Biol Med. 2010;2:162–180. doi: 10.1002/wsbm.54. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullmann R, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, Campos AR, Yao KM, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, Ta L, Mercola M, Conklin BR. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comp Biol. 2009;5:e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Wang X, Hall TMT. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nature Structural & Molecular Biology. 2001;8:141. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou H, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.