SUMMARY
Protein phosphorylation provides a mechanism for the rapid, reversible control of protein function. Phosphorylation adds negative charge to amino acid side chains, and negatively charged amino acids (Asp/Glu) can sometimes mimic the phosphorylated state of a protein. Using a comparative genomics approach we show that nature also employs this trick in reverse, evolving serine, threonine, and tyrosine phosphorylation sites from Asp/Glu residues. Structures of three proteins where phosphosites evolved from acidic residues (DNA topoisomerase II, enolase, and C-Raf) show that the relevant acidic residues are present in salt bridges with conserved basic residues, and that phosphorylation has the potential to conditionally restore the salt bridges. The evolution of phosphorylation sites from glutamate and aspartate provides a rationale for why phosphorylation sometimes activates proteins, and helps explain the origins of this important and complex process.
INTRODUCTION
Protein phosphorylation is a ubiquitous mechanism for the temporal and spatial regulation of proteins involved in almost every cellular process. Protein phosphorylation is important in prokaryotes, where the best-characterized kinases catalyze the phosphorylation of histidine residues (Laub and Goulian, 2007). Phosphorylation appears to be even more important and more widespread in eukaryotic cells, but while histidine phosphorylation does occur in at least some eukaryotes, most eukaryotic protein phosphorylation occurs at serine, threonine, and tyrosine residues (Manning et al., 2002a). The human genome includes about 500 genes for protein kinases that phosphorylate serine, threonine, and/or tyrosine residues (Manning et al., 2002b) and about 200 genes for phosphoprotein phosphatases that dephosphorylate them (Alonso et al., 2004). Thus, approximately 3.5% of human genes are devoted to proteins that directly regulate protein phosphorylation. The substrates of these kinases and phosphatases are numerous; one commonly cited estimate is that approximately 30% of proteins are phosphorylated (Cohen, 2000; Holt et al., 2009). Since many phosphoproteins are phosphorylated at multiple sites, there are probably tens of thousands of different phosphorylation sites in the human proteome.
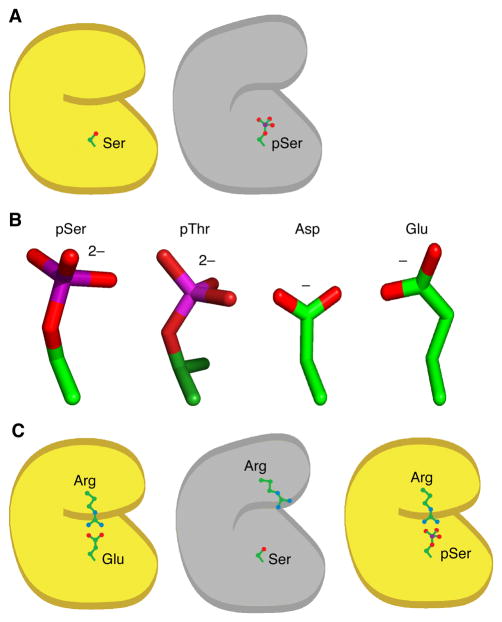
This raises the question of how these myriad phosphorylation sites have evolved. One possibility might be that the protein evolved first, with some structurally or functionally important Ser, Thr, or Tyr residue present on the surface of the protein, and that the phosphorylation of this residue evolved later. A strong prediction of this model is that most phosphorylations should negatively regulate protein function, since a fully functional protein is assumed to be present before the phosphorylation has evolved (Figure 1A).
Figure 1. A Possible Mechanism for the Evolution of Activating Phosphorylation Sites from Acidic Residues.
(A) Phosphorylation of a surface-exposed serine residue in an active protein (yellow) would be expected to decrease the protein’s activity (gray).
(B) Structures of phosphoserine, phosphothreonine, aspartic acid, and glutamic acid. Carbon atoms are represented in green, phosphorus atoms in magenta, and oxygen atoms in red. (C) Phosphorylation of a serine or threonine residue could conditionally restore an important electrostatic interaction originally mediated by an acidic amino acid, thereby activating the protein.
Phosphorylation does sometimes inactivate proteins. For example, phosphorylation of the prokaryotic citric acid cycle enzyme isocitrate dehydrogenase blocks the enzyme’s active site (Hurley et al., 1990; Hurley et al., 1989), providing a rapid and reversible off-switch for the protein. Another example is the eukaryotic cyclin dependent kinase CDK1, which is inactivated by the Wee1 and Myt1 protein kinases through the phosphorylation of two residues, Thr 14 and Tyr 15, in its catalytic cleft (Morgan, 1995).
However, many phosphorylations are activating rather than inactivating. For example, CDK1 activity is positively regulated by the phosphorylation of Thr 161, a residue just outside the catalytic cleft (Brown et al., 1999; Morgan, 1995). It is less intuitive how activating phosphorylations might evolve, since presumably the nonphosphorylated ancestor of the phosphoprotein would be inactive.
One possible mechanism is suggested by mutational studies of isocitrate dehydrogenase. Thorsness and Koshland showed that substituting an acidic aspartate residue for the serine phosphorylation site mimicked the phosphorylated state of the protein (Thorsness and Koshland, 1987). The rationale behind this observation is that acidic residues are negatively charged, like pSer, pThr and pTyr (although Glu and Asp are singly charged whereas pSer, pThr, and pTyr are, nominally, doubly charged at physiological pH (Cooper et al., 1983)), and Asp and Glu are approximately isosteric with pSer and pThr (Figure 1B). This ‘trick’ of substituting Asp or Glu for pSer or pThr turns out to be widely applicable. Although there are examples where two acidic residues are needed to mimic one phosphorylation event (Strickfaden et al., 2007), as well as examples where the activity of the Asp or Glu mutant is more like that of the non-phosphorylated form than the phosphorylated form (Posada and Cooper, 1992), often the Asp or Glu mutants do faithfully mimic the phosphorylated state.
Perhaps nature has been using this same trick in reverse. Imagine that a protein initially evolves with a functionally important glutamate on its surface; for example, in a salt bridge that stabilizes a fold that is necessary for proper function (Figure 1C). A mutation then occurs (perhaps in a non-essential, duplicated gene) and the negatively charged glutamate is replaced by a neutral serine. This destabilizes the salt bridge, rendering the protein inactive. If some kinase is able to phosphorylate the serine, the negatively charged phosphate could reestablish the salt bridge, stabilize the fold, and reactivate the protein. A switchable tyrosine phosphorylation site seems less likely to evolve by this mechanism, since phosphotyrosine is less structurally similar to Glu and Asp than pSer and pThr are, but in cases where net charge is more important than the details of the side chain structure, evolution of a phosphorylated tyrosine from acidic amino acids might be possible.
Here we have examined the sequence record for evidence of the evolution of ancestral glutamate or aspartate residues into phosphorylation sites. Our findings support the idea that some phosphorylation sites—perhaps ~5% of the sites we have examined—have evolved from acidic residues, and that such sites appeared both at the divergence of eukaryotes from prokaryotes and at subsequent times in evolution.
RESULTS
Glutamate and Aspartate Substitution Rates at Phosphoserines vs. Control Serines
If some phosphosites evolved from acidic amino acids, it might be possible to detect those ancestral origins in homologous proteins that have continued to exist in the unphosphorylatable, acidic form. To test this idea, we asked whether aspartate and glutamate are found at higher rates at the positions of known phosphorylation sites than at sites not known to be phosphorylated. If so, it could mean that phosphosites evolved from Asp/Glu, acidic residues evolved from phosphosites, or both.
We created a database of experimentally verified phosphoproteins collected from the literature (Ballif et al., 2004; Beausoleil et al., 2004; Blom et al., 1999; Ficarro et al., 2002). Because most of the phosphosites were serines (7854 pS, 1688 pT, 566 pY in our analysis) and we would expect different amino acid substitution rates for serines, threonines, and tyrosines, we initially chose to focus on the subset of phosphoproteins containing verified phosphoserines. For each phosphoprotein we obtained its most closely related homologs using BLAST (Altschul et al., 1990). We then generated multiple sequence alignments for each phosphoprotein and its homologs using PROBCONS (Do et al., 2005), a choice made for its high degree of accuracy rather than speed of alignment (Essoussi et al., 2008). For some large alignments (more than 100 sequences), memory limitations inherent to PROBCONS made it necessary to use another alignment tool, MUSCLE (Edgar, 2004). MUSCLE is capable of handling larger alignments and also produces high-quality alignments.
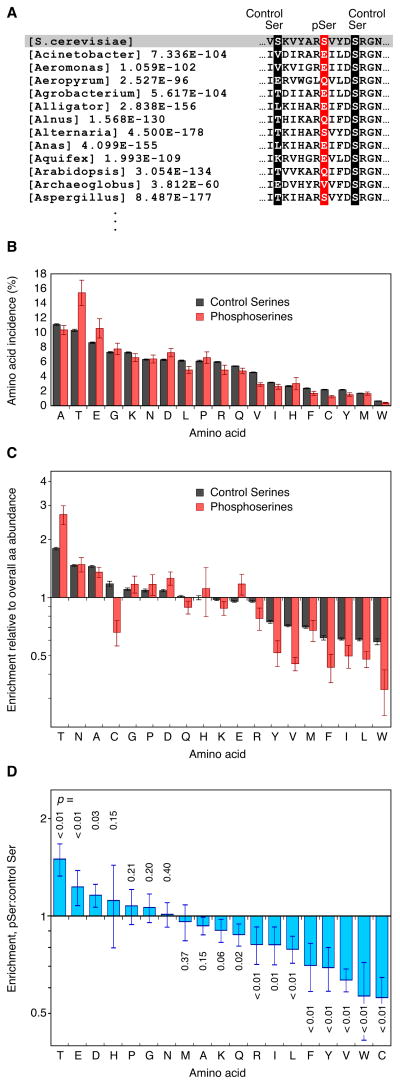
A sample of this procedure is shown in Figure 2A. The phosphoprotein in this case is the S. cerevisiae enolase Eno1, and the phosphorylation site is Ser 10. The alignment included 233 enolase homologs, some with serines at the position of Ser 10. We then determined the frequencies at which the nineteen other amino acids were found at the position of the phosphoserine (Figure 2A, red column). We compared these frequencies to the frequencies at which amino acids were found at the positions of control serines—serines not known to be phosphorylated—in S. cerevisiae enolase (Figure 2A, black columns).
Figure 2. Enrichment of Asp and Glu at the Positions of Phosphoserines.
(A) Construction of a multiple sequence alignment for a phosphorylated protein showing the phosphosite (red) and control serines (serines not known to be phosphorylated, black)
(B) Overall incidences at which various amino acids were found at the position of phosphoserines (N=7584) and control serines (N=257,481) in the multiple sequence alignments. Error bars are standard deviations, obtained by bootstrap resampling.
(C) Replacement percentages normalized to the overall abundance of each amino acid in the multiple sequence alignments. Error bars are standard deviations as calculated by the formula , where A is the replacement percentage at phosphoserines, B is the replacement percentage at control serines, and a and b are the bootstrapped standard deviations of A and B.
(D) Enrichment of various amino acids at phosphoserines relative to control serines. Error bars are standard deviations, calculated by bootstrapping as in (C). P-values were calculated by bootstrapping under the null hypothesis.
The control serine results are shown by the black bars in Figure 2B. Alanine, a common amino acid that is structurally similar to serine (Dayhoff, 1978), was the most common replacement, whereas tryptophan, a rare amino acid that is structurally dissimilar to serine (Dayhoff, 1978), was the least common (Figure 2B).
To see which amino acids were enriched at serine residues relative to their overall abundance in this set of proteins, we divided the replacement percentages plotted in Figure 2B by the abundances of the respective amino acids at all positions in the proteins in our multiple sequence alignments (Figure 2C, black bars). By this measure, Thr, Asn, and Ala were present at the highest relative rates, and isoleucine, leucine, and tryptophan at the lowest (Figure 2C).
The red bars in Figure 2B–C show the incidence of each amino acid at the positions of the phosphoserines in our dataset. Thr, Glu, and Asp were found more frequently at phosphoserines than at control serines. Bootstrapping yielded low p-values for these three enrichments (Figure 2D). The large hydrophobic amino acids, which were underrepresented at the positions of control serines (Figure 2C), were even more underrepresented at phosphoserines (Figure 2D). Another group has also recently shown that Glu and Asp are overrepresented at the positions of phosphoserines relative to control serines (Kurmangaliyev et al., 2011).
Glu and Asp are found preferentially on the surfaces rather than the interiors of proteins (Miller et al., 1987; Tjong et al., 2007). Since phosphoserines might be expected to favor protein surfaces more than control serines do, the enrichment of Asp/Glu residues at phosphoserines vs. control serines could simply reflect this shared preference for the surface. If so, then the other amino acids with a strong preference for surface exposure (Lys, Arg, Gln, and Asn) (Miller et al., 1987; Tjong et al., 2007) should be enriched at phosphoserines. However, Lys, Arg, and Gln were less likely to be found at the position of phosphoserines than at control serines, and Asn was not significantly more likely to be found at phosphoserines than at control serines (Figure 2D). Thus the enrichment of acidic residues at phosphosites appears not to be due to a general enrichment of polar or charged residues.
The conservation of phosphoserines was higher than that of control serines (43±1% vs. 24±0.2%), and the frequency at which gaps were found at the positions of phosphoserines was lower than that of control serines (13±1% vs. 30±0.3%). This suggests that phosphoserines are more likely to be critical to the function of a protein than control serines.
A Subset of Phosphoserines Has Very High Percentages of Asp/Glu Replacement
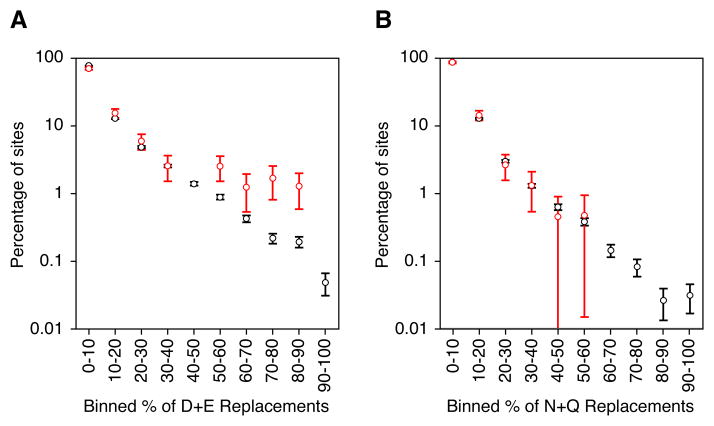
We next asked whether the overrepresentation of Asp/Glu residues was evenly distributed over all of the phosphoproteins, or arose from a subset of phosphosites with very high replacement frequencies. To address this question we binned our alignments according to the combined Asp+Glu perecentages at the position of phosphoserines or control serines.
We carried out this binning on all of the sequence alignments with at least 60 homologs; these alignments contained 234 phosphoserines and 16198 control serines. The fraction of the alignments with a given Asp/Glu percentage declined approximately exponentially for the control serines. For the phosphoserines (Figure 3A, red) there was a similar decline up until the 50% bin, above which the distribution leveled off. Approximately 6.8% of phosphoserine sites had at least 50% Asp or Glu at their position in the multiple sequence alignments, as opposed to just 1.8% for control serines (p = 4 × 10−6). Thus, there is a group of proteins with a very high frequency of replacement of Glu or Asp for phosphoserines, suggesting that phosphosites evolved from acidic residues (or vice versa) in these protein families. Similar trends were seen when cutoffs of more or less than 60 homologs were used for inclusion in the binning. Thus, approximately 5% more of the bona fide phosphorylation sites had Asp+Glu replacement percentages of >50% than did the control serines.
Figure 3. A Subset of the Phosphosites Have Very High Asp/Glu Replacement Percentages.
(A) Percentage of phosphosites (red) and control serine sites (black) which had various Asp/Glu replacement percentages in the alignments.
(B) Percentage of phosphosites (red) and control serine sites (black) which had various Asn/Gln replacement percentages in the alignments.
Replacement percentages were calculated from the 234 multiple sequence alignments that contained at least sixty homologs. Standard deviations and p values were calculated by boostrapping. See also Table S1.
As a further control, we looked for an indication that the high Asp/Glu replacement percentages might be due to the structural similarity of Asp and Glu to pSer rather than the charge similarity. If so, we might expect more phosphosites than control serine sites to have high percentages of Asn+Gln replacement, since Asn and Gln are structurally similar to Asp and Glu. As seen in Figure 3B, we do not find an overabundance of high Asn+Gln replacements percentages at the position of phosphoserines. In fact, only 0.46% of phosphoserines have >50% replacement by Asn or Gln, compared to 0.65% for control serines (p = 0.69). This suggests that it is the acidic nature of Asp and Glu that promoted their evolutionary overrepresentation at the position of phosphoserines.
Phylogenetic Analysis
The high frequency at which Asp/Glu residues were found at the positions of phosphoserines suggests that acidic amino acids may have often evolved into phosphosites, or vice versa, during evolution; however, pairwise alignments are insufficient to identify evolutionary events and therefore cannot distinguish these hypotheses from alternatives. Phylogenetic analysis and reconstruction of ancestral sequence states allow the timing and direction of historical sequence changes to be inferred, providing a direct test of our hypothesis.
We began by examining the 16 serine-phosphorylated proteins with the highest Asp/Glu replacement percentages; later we extended the analysis to phosphothreonine and phosphotyrosine sites with high Asp/Glu replacement percentages as well. Phylogenetic trees were inferred from the multiple sequence alignments using a maximum likelihood method implemented in the software package PhyML (Guindon and Gascuel, 2003). The leaves of the trees were colored to indicate which amino acid was present in each homolog at the position of the phosphosite. Green denoted the phosphorylatable amino acids Ser, Thr, and Tyr; red denoted the acidic residues Asp and Glu, and gray represented any other amino acid. We used the maximum likelihood trees inferred by PhyML as input to a second maximum likelihood implementation, FASTML (Pupko et al., 2002), to infer the sequences of the hypothetical ancestral proteins represented by the internal nodes, and colored these nodes according to the amino acid hypothesized to be present at the position of the phosphosite.
Most of the 16 trees displayed one of three basic patterns of Ser/Thr/Tyr versus Asp/Glu residues. Examples of each are shown in Figures 4–6 and described below.
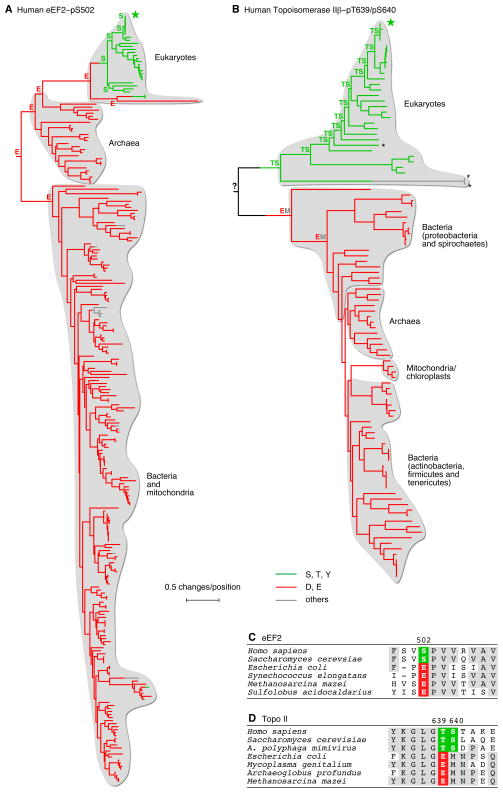
Figure 4. Evolution of Phosphorylation Sites from Acidic Residues at the Divergence of Eukaryotes from Prokaryotes.
(A) Homologs of human eEF2, which is phosphorylated at S502. The tree is colored to depict the amino acids present in homologs of human eEF2 at the positions corresponding to pS502. Human eEF2 is denoted by the star. eEF2 homologs were identified using BLAST. Homologs with E-values < 10−16 were aligned. Trees were inferred by PhyML using maximum likelihood. The tree was rooted using E. coli EF4 as an outgroup. Leaves were colored according to the amino acid at the relevant position. Internal nodes were colored based on the maximum likelihood amino acid inferred for those ancestral sequences using FASTML. The amino acids inferred for the hypothetical ancestors of human eEF2 are shown.
(B) Homologs of human topoisomerase II (Topo II) β were identified and aligned, and trees were inferred and colored as described for panel A. Human Topo II is phosphorylated at T639 and S640. The former residue is replaced by a Glu in all of the prokaryotic gyrase B proteins. A larger version of this figure, which includes species names and bootstrap values, is available as Figure 1B. The asterisks denote three viral Topo II homologs.
(C, D) Amino acid sequences close to the phosphosites for a few selected eEF2 and Topo II homologs.
For larger versions of the trees shown in panels A and B, and control trees, see Table S2.
Figure 6. Evolution of Phosphorylation Sites from Acidic Residues in Particular Eukaryotic Paralogs.
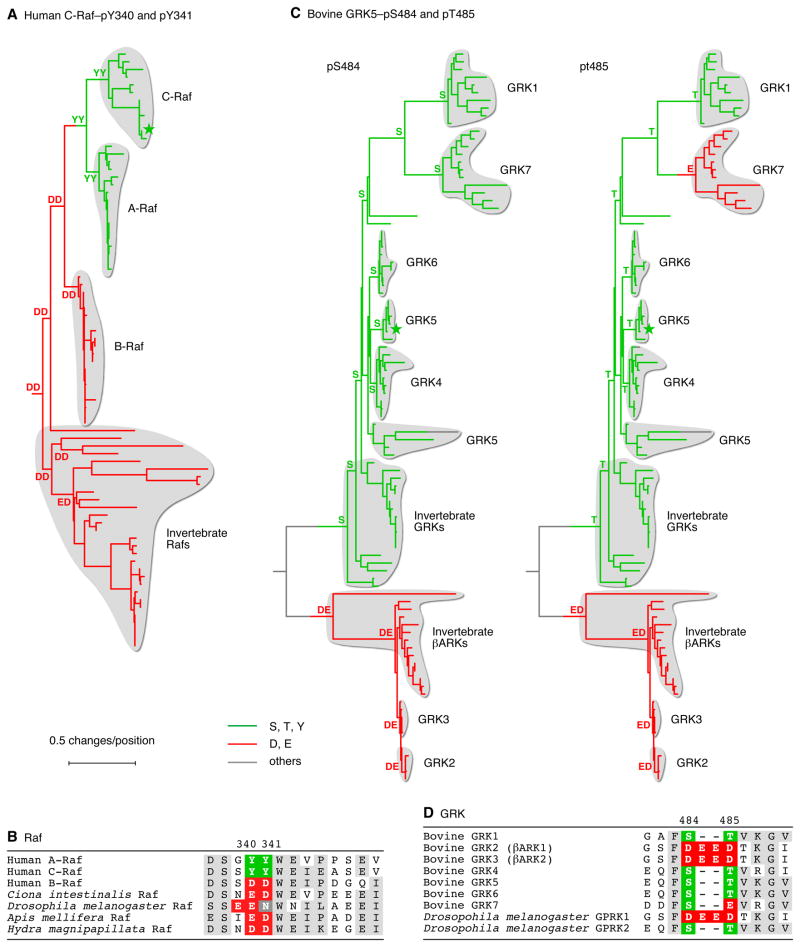
(A) Homologs of human C-Raf were identified and aligned, trees were inferred as described for Figure 4. The human Ksr2 protein was used as an outgroup to define the root of the tree.
(B) Local alignments of various Raf proteins in the vicinity of phosphosites Y340 and Y341.
(C) Homologs of bovine GRK5 were identified and aligned, and trees were inferred as described for Figure 4. The human Akt2 protein was used as an outgroup to define the root of the tree.
(D) Local alignments of the seven bovine GRK proteins and two Drosophila GRK proteins in the vicinity of phosphosites S484 and T485.
See also Table S4.
Emergence of Phosphorylation Sites at the Prokaryotic/Eukaryotic Split
Elongation Factor eEF2
The protein eEF2 is a translation factor required for the translocation step in protein synthesis (Jorgensen et al., 2006). Human eEF2 is reported to be phosphorylated at Ser 502 (Beausoleil et al., 2004). The functional significance of this phosphorylation is uncertain.
The EF2 tree included eukaryotic eEF2 proteins, archaeal aEF2 proteins, and bacterial EF-G proteins (Figure 4A). The amino acids in the vicinity of the Ser 502 were well conserved throughout the tree (Figure 4C). The tree was rooted using the related E. coli protein EF4 as an outgroup, which placed the root of the tree between the archaea and the bacteria (Figure 4A), consistent with current views of deep phylogenetics (Ciccarelli et al., 2006; Pace, 2009).
Almost all (27/28) of the eukaryotic eEF2 homologs had a phosphorylatable residue at the position corresponding to Ser 502. Most (211/224) of the bacterial, mitochondrial, and archaeal homologs, had either Asp or Glu at that position. All of the eukaryotic eEF2 proteins with serine residues at the position of Ser 502 appear to have been derived from a single Glu –> Ser change at the divergence of eukaryotes from archaea, which has been maintained ever since (Figure 4A). The two eEF2 proteins with Thr residues at the position of Ser 502 may have arisen from an independent Glu –> Thr amino acid change (Table S2A).
DNA Topoisomerase II
DNA topoisomerase II (Topo II) is a conserved enzyme that catalyzes the passage of one DNA duplex through another (Schoeffler and Berger, 2008). Eukaryotic Topo II is a homodimer. Its prokaryotic homolog, gyrase, is an A2B2 tetramer, with gyrase B corresponding to the N-terminal half of eukaryotic Topo II and gyrase A corresponding to the C-terminal half. It has been reported that human topoisomerase IIβ is phosphorylated at Thr 639 and Ser 640 (Figure 4B) (Wang et al., 2010), with the functional consequences of these phosphorylations unknown.
The BLAST search using human Topo IIβ as query yielded sequences including vertebrate topoisomerase IIα and IIβ proteins, other eukaryotic Topo II proteins, and prokaryotic gyrase proteins, with the position of the phosphosite present in the gyrase B chains. The amino acids surrounding the phosphosite were well conserved throughout the tree (Figure 4D). We were unable to identify a satisfactory paralog to use as an outgroup; the rooting shown in Figure 4B is arbitrary.
The Topo II tree consisted of two main clades (Figure 4B). The first included the eukaryotic Topo II proteins plus three viral Topo II proteins whose genes were likely to have been transduced from their eukaryotic hosts. The second included all of the bacterial, archaeal, and chloroplastic/mitochondrial Topo II proteins. All of the animal, fungal, plant, and protist Topo II homologs had phosphorylatable Thr and Ser residues at the sites corresponding to Thr 639 and Ser 640, respectively. All of the bacterial, archaeal, and chloroplastic/mitochondrial Topo II homologs had a glutamate residue at the position of Thr 639 and a Met or Ile residue at the position of Ser 640 (Figure 4B). If one assumes that bacteria and archaea diverged before eukaryotes and archaea did (Ciccarelli et al., 2006; Pace, 2009), then the Glu 639/Met 640-containing Topo II proteins probably came first, with the Thr 639/Ser 640 arising later.
Emergence of Phosphorylation Sites Later in Evolution
Enolase
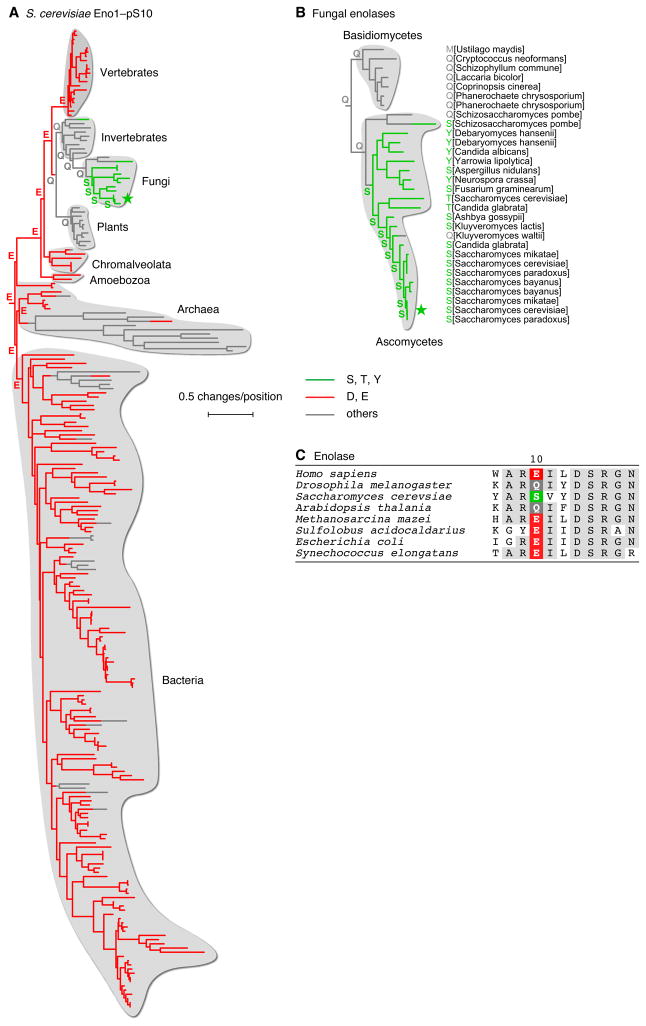
Some phosphosites apparently evolved after the divergence of eukaryotes from bacteria and archaea. One example of this is provided by enolase, a key enzyme in glycolysis and gluconeogenesis. The S. cerevisiae enolases Eno1 and Eno2 have been reported to be phosphorylated at Ser 10 (Ficarro et al., 2002), and the region surrounding this residue is well conserved among prokaryotes and eukaryotes (Figure 5C).
Figure 5. Evolution of Phosphorylation Sites from Acidic Residues Later in Evolution.
(A, B) S. cerevisiae enolase Eno1. Homologs of the S. cerevisiae enolase Eno1 were identified and aligned, and trees were inferred as described for Figure 4. The root of the tree was placed between the archaea and eukaryotes. An expanded view of thirty fungal enolases is shown in panel (B). The fungus Cryptococcus neoformans was used as an outgroup to define the root of the tree.
(C) Amino acid sequences close to the phosphosite for a few selected enolase homologs. See also Table S3.
Enolase homologs with phosphorylatable residues at this position were largely confined to the fungal clade of the phylogenetic tree (Figure 5A). Of the non-fungal sequences, most contained either Asp or Glu at this position.
Figure 5B shows a tree inferred for 30 fungal enolases from 23 dikaryal fungal species. All seven basidiomycetes enolases had non-acidic, non-phosphorylatable amino acids at the Ser 10 position, with Gln being the most common. Almost all of the ascomycetes enolases had phosphorylatable residues at this site, and the remaining two (one of the two S. pombe enolase paralogs and the K. waltii enolase) had Gln residues.
Reconstruction of the sequences of the ancestral species suggests that the primordial enolase protein possessed a Glu residue at the position of pSer 10, with this Glu residue persisting in most present day prokaryotes, protists, and vertebrates (Figure 5A, B). This residue apparently then became a Gln in some eukaryotes, and persists as a Gln in invertebrates, plants, and basidiomycetes fungi. Finally, the Gln residue became a phosphorylatable amino acid shortly after the divergence of the ascomycetes fungi from the basidiomycetes fungi. This dates the evolution of the phosphosite to approximately 400 mya (Taylor and Berbee, 2006), the time when the ascomycetes and basidiomycetes are thought to have diverged, and raises the possibility that amide amino acids like Gln can act as intermediaries in the evolution of phosphosites.
Emergence of Phosphosites in Particular Paralogs of a Multi-Paralog Protein Family
Raf
Some of the BLAST searches yielded families of paralogous proteins where a phosphosite was present in some modern paralogs and a Asp/Glu residue was present in others. One example is provided by the Raf family of protein kinases. Most vertebrates possess three Raf homologs: A-Raf, B-Raf, and C-Raf. The C-Raf proteins have a pair of adjacent tyrosine phosphorylation sites (Tyr 340 and Tyr 341 in human C-Raf) just N-terminal to its kinase domain, and these phosphorylations are critical for C-Raf activation (Fabian et al., 1993; Marais et al., 1995). These Tyr phosphorylation sites are present in A-Raf as well, but the corresponding sites in B-Raf are Asp residues, and the Asp residues are critical for B-Raf function (Fabian et al., 1993; Marais et al., 1995).
To investigate the evolutionary relationships among these Raf proteins, we aligned the sequences of all of the Raf homologs present in the KEGG database and inferred a maximum likelihood tree. As shown in Figure 6A, B and Table S4A, almost all of the C-Raf proteins have YY residues at the positions of Tyr 340 and 341. The one exception is from the horse Equus caballus, which has a duplicated C-Raf gene; one of its C-Raf proteins has the typical YY residues, and the other has CY in their place. All of the A-Raf sequences contain YY residues. All of the B-Raf proteins have DD residues, and all of non-vertebrate Raf proteins have some variation on DD (DD, ED, DE, EG, or EN). Interestingly, the EN variation is confined to a sub-clade of the insects, and all of those Raf proteins have not only lost the second acidic residue, but have also gained a new acidic residue just N-terminal to the EN site (Figure 6A, B). These findings indicate that C-Raf’s activating Tyr phosphorylations evolved from a primordial Raf protein with a pair of acidic residues in their place, with phosphorylation conditionally restoring a function formerly provided by the acidic residues.
The G-Protein-Coupled Receptor Kinases (GRKs)
The GRK family of protein kinases provide another example of paralogs where some possess phosphorylation sites and others possess acidic residues. Bovine GRK5 is reported to be autophosphorylated at Ser 484 and Thr 485, two sites C-terminal to the GRK5 kinase domain, with these phosphorylations increasing the activity of GRK5 towards the β2-adrenergic receptor by 15–20-fold (Kunapuli et al., 1994). The corresponding residues are also known to be phosphorylated in GRK1 and GRK6 (Olsen et al., 2006; Palczewski et al., 1992), and the region around these residues is well aligned in all of the GRK-family proteins (Figure 6E).
The GRK tree divides into two main clades. One contains the vertebrate GRK1 and 4–7 proteins together with their closest invertebrate relatives, and the other contains GRK2 and 3 and their closest invertebrate relatives (Figure 6C, D). Most of the GRK1, 4, 5 and 6 proteins have a Ser residue at the position of Ser 484 and a Thr residue at the position of Thr 485 (Figure 6C, D, and Figures S3B and S3C). Most of the GRK2 and 3 proteins have four acidic residues (DEED) in place of Ser 484 and Thr 485. The GRK7 proteins possess a conserved serine in the position of Ser 484 and a single acidic amino acid in the position of Thr 485. Thus, as was the case with Raf, phosphosites and acidic residues are confined to particular paralogs, with both the phosphosites and acidic residues generally being maintained throughout the subsequent evolution of the paralogs.
Figure 6E shows the residues around the positions of Ser 484/Thr 485 in the several GRKs. Note that different GRK paralogs employ different combinations of fixed charges (acidic residues) and conditional charges (phosphorylation sites) in this region.
Evolution of a Phosphosite into an Acidic Residue
The GRK trees (Figure 6C, D, and Figures S3B and S3C) were rooted using human Akt2, the closest human non-GRK homolog of the GRKs, as an outgroup. With the root defined this way, it was difficult to determine whether the DEED or ST pattern is likely to have come first, or whether both evolved independently from non-acidic, non-phosphorylatable residues.
However, the tree illustrates another interesting phenomenon. It seems likely that one subgroup of the GRKs replaced Thr 485 with a glutamate residue, which is now present in the GRK7 proteins (Figure 6C, D). Thus, the GRKs provide an example of evolution of a phosphosite into an acidic amino acid, changing a conditional negative charge to a fixed negative charge.
Another such example is provided by the GMGC family of protein kinases. The primordial GMGC kinase is predicted by maximum likelihood reconstruction to have had a T-X-Y dual phosphorylation motif, as the present-day MAP kinase proteins do, whereas a small clade of CDK1-like kinases appear to have replaced the Tyr residue with a Glu (Table S4H).
Trees for Control Serine Sites
We next examined several “control trees”— phylogenetic trees where the leaves, branches and internal nodes were colored based on the amino acids present at serine sites that are not known to be phosphosites (Tables S2B–C, S3C–E, and S4D–G). In the case of eEF2 we identified two control serines, Ser 530 and Ser 774, that had Asp/Glu replacement percentages that were roughly equal to those of the phosphosite, pSer 502. The Ser 530 tree (Table S2B) was quite similar to the phosphosite tree (Figure 4A); all of the eukaryotic eEF2 homologs had phosphorylatable residues at this site, whereas almost all of the prokaryotic homologs possessed acidic residues. According to our hypothesis Ser 530 could be a good candidate for an undiscovered phosphosite. Several other control serine sites with high Asp/Glu replacement percentages are shown in Table S1. It will be of interest to determine whether high Asp/Glu replacement percentages can be used to successfully predict phosphorylation sites.
The Ser 774 tree (Table S2C) was qualitatively different, with both acidic and phosphorylatable residues scattered throughout the eukaryotes, bacteria, archaea, and mitochondria. Most of the control trees constructed for the enolase and GRK families also showed scattered distributions of Asp/Glu vs. Ser/Thr/Tyr residues (Tables S3C–E, S4D–G). These findings suggest that patterns of acidic residues vs. phosphorylatable residues seen at phosphosites (Figures 4–6) were not adventitious; instead it appears that both types of residues have been actively maintained.
Topo II, Enolase, and Raf Protein Structures
The original rationale for this work was that some phosphorylation sites have arisen from structurally important Asp/Glu residues (Figure 1). We were therefore interested in investigating the structural contexts of the phosphosites and corresponding acidic residues for the protein families shown in Figures 4–6. Were the acidic residues present in salt bridges? If so, was the hydrogen bond donor a conserved basic residue? Once the phosphosite had evolved, was the salt bridge lost in the dephospho-form, and might it be regained through phosphorylation?
In the case of Topo II, crystal structures are available for the S. cerevisiae Topo II homodimer, which possesses the phosphosite conserved among eukaryotes (Thr 607), and for the E. coli gyrase A2B2 tetramer, which has an acidic residue (Glu 744) in its place. Glu 744 resides in the gyrase B chains and participates in an interchain salt bridge with Lys 65 in gyrase A (Figure 7A). The distance between the Lys side chain nitrogen and the closest Glu oxygen is 3.1 Å, compatible with a salt bridge (Vogt et al., 1997). Lys 65 is well conserved throughout the Topo II/gyrase family.
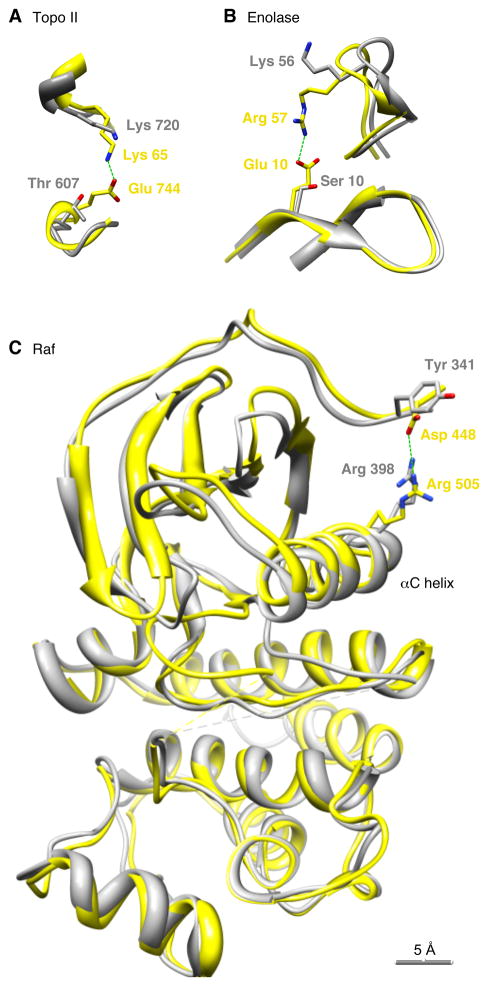
Figure 7. Salt Bridges in Phosphoprotein Homologs Possessing Glu Residues at the Positions of the Phosphosites.
(A) Topoisomerase II. Yellow represents the E. coli gyrase A2B2 tetramer (PDB ID 3NUH). Gray represents the S. cerevisiae Topo II homodimer (2RGR).
(B) Enolase. Yellow represents E. coli enolase (1E9I). Gray represents S. cerevisiae Eno1 protein (1ONE).
(C) Raf. Yellow represents human B-Raf (1UWJ). Gray represents human C-Raf (3OMV). In all three panels, the structures were superimposed so as to minimize the overall RMS deviation of the positions of the aligned residues, using UCSF Chimera (Pettersen et al., 2004).
We superimposed the S. cerevisiae Topo II homodimer structure on the E. coli gyrase tetramer structure (Figure 7A). The conserved basic residue (Lys 720) resides close to Lys 65, and the phosphosite (Thr 607) sits close to where the Glu residue in gyrase resides, but the salt bridge is lost; the distance from the side chain oxygen in Thr 607 to the closest nitrogen in Lys 720 is 7.3 Å. It is structurally plausible that the phosphorylation of Thr 607 could restore the salt bridge present in bacterial gyrase.
Crystal structures are also available for S. cerevisiae Eno1 in its non-phosphorylated form, and for E. coli enolase, which has a Glu residue at the position of Ser 10. As shown in Figure 7B, the E. coli enolase possesses a 2.7 Å salt bridge between the Glu residue and a conserved basic residue (Arg 57). In non-phosphorylated yeast Eno1, a basic residue (Lys 56) is present at the position of Arg 57, but the distance between this residue’s amino nitrogen and the Ser 10 oxygen is 12.8 Å, (Figure 7B). Phosphorylation of Ser 10 could plausibly restore the lost salt bridge.
A third example is provided by the Raf family proteins. In human B-Raf, one of the two Asp residues (Asp 448) forms a 3.5 Å salt bridge with a conserved Arg residue, Arg 505. This residue is part of the αC helix, a structural element critical for protein kinase activation (Figure 7C). The interaction between Asp 448 and Arg 505 has been hypothesized to stabilize the active conformation of B-Raf (Wan et al., 2004). The salt bridge is lost in C-Raf; the distance from the hydroxyl oxygen of the corresponding tyrosine (Tyr 341) to the nearest nitrogen in the corresponding arginine (Arg 398) is 7.7 Å. It seems plausible that phosphorylation of Tyr 341 could again restore the salt bridge.
Thus, crystal structures provide support for the underlying scenario (Figure 1) that motivated our hypothesis that some phosphosites have evolved from acidic residues.
DISCUSSION
Here we have used comparative genomic analysis to demonstrate that some phosphorylation sites have evolved from acidic amino acids. The acidic amino acids Asp and Glu were found to be present at the positions of phosphoserines more often than they were at control serines (Figure 2). Additionally, other charged and polar amino acids did not show increased incidence, indicating the importance of the negative charge in this enrichment. We did not see a similar enrichment of Asp and Glu at Thr or Tyr phosphorylation sites. This may have been simply due to the smaller number of Thr and Tyr phosphorylation sites in our database, as other evidence, discussed below, supports the hypothesis that all three types of phosphorylation sites (Ser, Thr, and Tyr) can evolve and have evolved from acidic residues.
For our high-confidence set of multiple sequence alignments—those with at least 60 homologs—6.8% of the 234 phosphosites had very high (>50%) incidences of Asp or Glu at the position of the phosphosite, while only 1.8% of the 16198 control serine sites did (Figure 3). This suggests that ~5% of the phosphorylation sites we examined may have evolved from acidic residues.
Phylogenetic analysis demonstrates that the switch between acidic and phosphorylatable residues occurred for some proteins at the evolutionary division between eukaryotes and prokaryotes (Figure 4). In the two examples examined in detail here (eEF2 and Topo II), most or all of the bacterial and archaeal species possessed a Glu residue, and most or all of the eukaryotic species possessed a Ser or Thr residue at the position of the phosphosite. If one assumes that the split between bacteria and archaea occurred prior to the split between prokaryotes and eukaryotes, the phosphorylation site probably evolved from the acidic residue rather than vice versa. This interpretation is supported by the maximum likelihood (FASTML) reconstruction of the ancestral sequences for the eEF2 tree. Reconstruction of the ancestors for the Topo II tree is also consistent with this direction of evolution (Glu –> Thr), although we have less confidence in the rooting of this tree. The phosphosite appears to have evolved once (Topo II) or possibly twice (eEF2), and to have then been maintained over long evolutionary times.
At other times the switch between an acidic amino acid and a phosphosite occurred later in evolution. For example, it occurred at the divergence of ascomycetes from basidiomycetes fungi (~400 mya) in the case of enolase (Figure 5A–C). In the GRK and Raf families, the switch occurred early in animal evolution, with modern species retaining Asp/Glu-residues in some paralogs (GRK2/3, B-Raf) and acquiring phosphorylatable residues in others (GRK1, 4–7, A-Raf, and C-Raf). It appears that Raf gene duplication has allowed a primordial DD-containing Raf to evolve new conditional regulation. For GRKs it is not clear whether the acidic residues or phosphosites came first.
With the GRKs, it appears that two phosphorylation sites have replaced four acidic residues, consistent with the observation that sometimes a pair of acidic amino acids better mimics a phosphorylation than a single amino acid does (Strickfaden et al., 2007). In most of the examples examined here, however, phosphorylation sites appear to have replaced single amino acids, in line with Thorsness and Koshland’s classic study (Thorsness and Koshland, 1987).
Our original rationale for hypothesizing that phosphorylation sites evolved from acidic residues provided an explanation for the fact that phosphorylation sometimes activates proteins. In two of the seven examples examined in detail here (Raf, GRK), the phosphorylation is in fact known to be activating. In the other cases the functional significance of the phosphorylations remains untested. We predict that many of these will prove to be activating phosphorylations.
Finally, we found examples where a phosphorylation site appears to have become fixed as a glutamate residue. In the GRK7 protein kinases, a glutamate residue appears to have evolved from a threonine phosphosite that remains present in the GRK1, 4, 5, and 6 proteins (Figure 6C). A second example is provided by the CMGC family of protein kinases, where one clade of the kinases (the CDK1-like kinases) appears to have evolved a glutamate residue in the place of an activating tyrosine phosphorylation site (Table S3H).
Phosphorylation sites are sometimes found in conserved, structured regions of proteins, but more frequently they are found in poorly conserved, putatively unstructured regions (Holt et al., 2009; Moses et al., 2007). The prevailing hypothesis is that the former type of site is more likely to be involved in complicated allosteric regulation, and the latter type of site is more likely to be involved in simpler types of regulation such as bulk electrostatic effects or the generation of phosphoepitopes (Holt et al., 2009; Moses et al., 2007; Pawson et al., 2001; Serber and Ferrell, 2007). Our original rationale (Figure 1) seems more applicable to well-conserved phosphorylation sites in structured regions. Indeed, despite the fact that ~60–90% of phosphorylation sites are believed to be the poorly conserved type (Holt et al., 2009; Kurmangaliyev et al., 2011; Moses et al., 2007), all five of the phosphosites examined in Figures 4–6 are evolutionarily well-conserved and present in structured regions of the proteins. This raises the interesting possibility that phosphosites that evolved from Asp/Glu residues will be more likely than average to be involved in allosteric regulation.
The crystal structures of Topo II, enolase, and Raf show that in the Asp/Glu-containing versions of these protein, the acidic residue is involved in a salt bridge with a conserved basic residue, and in the Ser/Thr/Tyr-containing versions, the salt bridge is lost, with phosphorylation having the potential to restore it (Figure 7). These findings provide strong support for the rationale that motivated these studies (Figure 1).
Taken together, this work provides insight into the evolution of phosphorylation, and in particular provides a rationale for how well-conserved, activating phosphorylations have evolved.
METHODS
Identifying homologs
The list of proteins closely related to each of phosphoprotein in our database was generated using stand-alone protein BLAST (Altschul et al., 1990) (blastp) against the non-redundant “nr” protein database using default options. From these results, only those that were closely related (E-value ≤ 10−16) were used for subsequent alignment and analysis.
Alignments
For each phosphoprotein and its close relatives, a multiple sequence alignment of the protein sequences was generated using PROBCONS 1.09 (Do et al., 2005). Default options were used with the exception that iterative refinement was set to the maximum of 1000 iterations using the argument --iterative-refinement 1000. For very large alignments MUSCLE 3.6 (Edgar, 2004) was used for its high quality alignment generation as well as reduced memory requirements.
Binning and Bootstrapping
The bins for glutamate/aspartate replacement and their error bars were generated by bootstrap sampling, using the phosphosites for which there were at least 60 homologs in the alignment (N=234). Thus 234 phosphosites were chosen with replacement from this set, and each site in the sample was placed in the appropriate bin based on what percentage of replacement amino acids were either glutamate or aspartate. This was repeated 1000 times. The percentage replacement in each bin was calculated as the mean of the percentage of sites in each bin in the bootstrap samples. The error bars are the standard deviation of the bootstrap sample percentages from this mean. The same procedure was repeated for control serine sites (N=16,198).
The p values reported in Figures 2 and 3 were also calculated by bootstrapping. The p-values for the amino acid replacement ratios (Figure 2) were obtained using the following method: All phosphoserine sites and control serine sites were grouped together into a new, combined data set. This set (representing the null hypothesis that amino acids are found at the same frequency at the positions of phosphoserines and control serines) was sampled with replacement, drawing a bootstrap phosphoserine set and control serine set, each the same size as the original sets. This was repeated 10,000 times. The p-value for each amino acid was calculated as the percentage of bootstrap samples which resulted in a replacement ratio at least as great as the ratio for that amino acid calculated using the true phosphoserine and control sets.
For Figure 3, we pooled the phosphoserine and control serine alignments, and randomly sampled with replacement 234 as mock phosphoserines and 16000 as mock control serine sites. For the Asp+Glu distribution, the sampling was repeated 106 times and we calculated the fraction of the bootstrap samples that showed at least 5% (6.8% - 1.8%) difference in Asp+Glu replacement percentages between mock phosphoserine alignments and the mock control serine alignments. For the Asn+Gln distribution, we calculated the fraction of the bootstrap samples that showed at least 0.19% (0.46% – 0.65%) difference in Asn+Gln replacement perentages between mock phosphoserine alignments and the mock control serine alignments.
Trees
Phylogenetic trees depicting the relationship of the aligned sequences were inferred using PhyML 3.0 (Guindon and Gascuel, 2003). Gapped regions that were poorly conserved were first removed from each alignment. Each alignment was then tested using ProtTest 3.0 (Abascal et al., 2005) to determine the most appropriate protein evolution model and associated parameters for phylogenetic inference. The models, parameters and ungapped alignments were then used in PhyML to infer the trees. The leaf nodes in each tree were colored to depict the amino acid present at the position of interest for each taxon in the alignment from which the tree was inferred.
The internal nodes were colored according to the amino acid at the predicted ancestral sequence at that node. These sequences were reconstructed using maximum likelihood by FASTML (Pupko et al., 2002). FASTML uses the inferred maximum likelihood tree, the alignment from which the tree was inferred, and the protein evolution model and parameters to calculate the most likely ancestral sequence at each internal node.
Supplementary Material
Acknowledgments
We thank Kurt Thorn, Peter Chien, Michael Reese, Arend Sidow, Serafim Batzoglou, Rob Tibshirani, Bill Weis, the participants of the 2011 Evolution of Protein Phosphorylation Keystone Meeting, our colleagues in the Stanford Dept. of Chemical and Systems Biology, and members of the Ferrell lab for many helpful comments. This work was supported by grants from the National Institutes of Health (R01 GM046383 and T15 LM007033) and the Arnold Beckman Foundation. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Sefton BM, Hunter T. Detection and quantification of phosphotyrosine in proteins. Methods Enzymol. 1983;99:387–402. doi: 10.1016/0076-6879(83)99075-4. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, editor. Atlas of Protein Sequence and Structure. Silver Springs M.D: National Biomedical Research Foundation; 1978. [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essoussi N, Boujenfa K, Limam M. A comparison of MSA tools. Bioinformation. 2008;2:452–455. doi: 10.6026/97320630002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian JR, Daar IO, Morrison D. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Molecular and Cellular Biology. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Dean AM, Thorsness PE, Koshland DE, Jr, Stroud RM. Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme. J Biol Chem. 1990;265:3599–3602. doi: 10.2210/pdb4icd/pdb. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Thorsness PE, Ramalingam V, Helmers NH, Koshland DE, Jr, Stroud RM. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci U S A. 1989;86:8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Merrill AR, Andersen GR. The life and death of translation elongation factor 2. Biochem Soc Trans. 2006;34:1–6. doi: 10.1042/BST20060001. [DOI] [PubMed] [Google Scholar]
- Kunapuli P, Gurevich VV, Benovic JL. Phospholipid-stimulated autophosphorylation activates the G protein-coupled receptor kinase GRK5. J Biol Chem. 1994;269:10209–10212. [PubMed] [Google Scholar]
- Kurmangaliyev YZ, Goland A, Gelfand MS. Evolutionary patterns of phosphorylated serines. Biol Direct. 2011;6:8. doi: 10.1186/1745-6150-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002a;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002b;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J Mol Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Moses AM, Heriche JK, Durbin R. Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 2007;8:R23. doi: 10.1186/gb-2007-8-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pace NR. Mapping the tree of life: progress and prospects. Microbiol Mol Biol Rev. 2009;73:565–576. doi: 10.1128/MMBR.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Buczylko J, Van Hooser P, Carr SA, Huddleston MJ, Crabb JW. Identification of the autophosphorylation sites in rhodopsin kinase. J Biol Chem. 1992;267:18991–18998. [PubMed] [Google Scholar]
- Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Posada J, Cooper JA. Requirements for phosphorylation of MAP kinase during meiosis in Xenopus oocytes. Science. 1992;255:212–215. doi: 10.1126/science.1313186. [DOI] [PubMed] [Google Scholar]
- Pupko T, Pe’er I, Hasegawa M, Graur D, Friedman N. A branch-and-bound algorithm for the inference of ancestral amino-acid sequences when the replacement rate varies among sites: Application to the evolution of five gene families. Bioinformatics. 2002;18:1116–1123. doi: 10.1093/bioinformatics/18.8.1116. [DOI] [PubMed] [Google Scholar]
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- Serber Z, Ferrell JE., Jr Tuning bulk electrostatics to regulate protein function. Cell. 2007;128:441–444. doi: 10.1016/j.cell.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- Thorsness PE, Koshland DE., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987;262:10422–10425. [PubMed] [Google Scholar]
- Tjong H, Qin S, Zhou HX. PI2PE: protein interface/interior prediction engine. Nucleic Acids Res. 2007;35:W357–362. doi: 10.1093/nar/gkm231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.