Abstract
Recent large--scale association studies, both of genome-wide and candidate gene design, have revealed several single-nucleotide polymorphisms (SNPs) which are significantly associated with risk of developing breast cancer. As both breast and endometrial cancers are considered to be hormonally driven and share multiple risk factors, we investigated whether breast cancer risk alleles are also associated with endometrial cancer risk. We genotyped nine breast cancer risk SNPs in up to 4188 endometrial cases and 11 928 controls, from between three and seven Caucasian populations. None of the tested SNPs showed significant evidence of association with risk of endometrial cancer.
Introduction
Endometrial cancer is the fourth most common cancer in women in the UK, representing 5% of all female cancers, with most cases developing postmenopausally. The most common subtype, ∼80% of cases, is Type 1 estrogen-dependent endometrioid adenocarcinoma, which arises from the lining of the uterus and is associated with excessive estrogen exposure, especially that unopposed by progesterone (1). The known risk factors for this disease are hormone related, e.g. early menarche or late menopause, nulliparity, irregular menstrual periods and obesity. Exogenous hormones, such as oral contraceptives and estrogen hormone replacement therapy, can also increase risk of developing endometrial cancer, although the addition of progesterone counteracts this risk (2). Interestingly, breast cancer risk is increased with the use of progesterone. Furthermore, use of the breast cancer drug, Tamoxifen, is associated with an increased risk of endometrial cancer (3). In normal breast cells and most breast tumors, Tamoxifen acts as an estrogen receptor antagonist and has an antiproliferative effect, but in endometrial cells, it acts in the opposite direction—as an estrogen receptor agonist that increases proliferation and risk of endometrial cancer (4). As there are known similarities and differences in the risk factors for breast and endometrial cancer, it is probably that some but not all genetic risk factors will be shared between these two diseases. A recent genome-wide association study (GWAS) of endometrial cancer (5) revealed a single-nucleotide polymorphism (SNP) at 17q12 (HNF1B), which was associated with risk of endometrial cancer at a genome-wide level of significance [odds ratio (OR) = 0.84, 95% confidence interval (CI) = 0.79–0.89, P = 7 × 10−10]; however, this SNP does not appear to be associated with breast cancer risk (6).
In this study, we assessed risk of endometrial cancer associated with genetic variants that have previously been identified to be associated with breast cancer risk. SNPs were selected for this study, if they fulfilled one or both of the following criteria: (i) At the time this study was initiated, they were SNPs identified to be significantly associated with breast cancer susceptibility in GWAS or candidate studies carried out by the Breast Cancer Association Consortium (BCAC): rs889312 (MAP3KI), rs3817198 (LSP1) and rs13281615 (8q24) (7–9); rs13387042 (2q35) (8,10); rs1045485 (CASP8) (11); and rs3020314 (ESR1) (12). (ii) They showed evidence for significant (P < 0.05) association with endometrial cancer in the Studies of Epidemiology and Risk factors in Cancer Heredity (SEARCH) study during an analysis of 81 SNPs that had originally been selected as candidates for breast cancer susceptibility: rs3803662 (TOX3), rs1045485 (CASP8) and rs3857481 (6q14). We note that rs3857481 was not subsequently confirmed as a breast cancer susceptibility variant (7).
The nine SNPs that fitted these criteria were genotyped in three to six collaborating endometrial studies, comprising 2449–4188 cases and 2711–11928 controls.
Materials and methods
Study populations
Seven studies from Australia, USA and Europe contributed pre-existing data from SNP genotyping at each site to these analyses—hence not all SNPs have been analyzed in all studies. Controls recorded as having had a hysterectomy were excluded from all analyses. The majority of endometrial cancer cases were of endometrioid histology (85.5%). Additional details about each study are provided below.
Australian National Endometrial Cancer Study and Australian Ovarian Cancer Study controls.
The Australian National Endometrial Cancer Study (ANECS) is an Australian population-based case–control family study of cancer of the uterine corpus (13). Women aged 18–79 years, registered on the Electoral Roll and newly diagnosed with primary cancer of the endometrium between July 2005 and December 2007 were identified through major hospitals nationally and also from state-based cancer registries. Female controls with no personal history of endometrial cancer or hysterectomy were recruited using two sources. A population-based control group comprising women randomly selected using the Australian Electoral Roll (voting is compulsory in Australia) and matched to the age and geographic distribution of the cases. A second control group of female blood donors were recruited with the aid of the Australian Red Cross Blood Service in Queensland. All participants completed a detailed questionnaire providing clinical and epidemiological information including body mass index and ethnicity. In addition, we also accessed available genotype data from controls only (with no prior history of hysterectomy) from the Australian Ovarian Cancer Study (AOCS). This is a national population-based case–control study of ovarian cancer with recruitment strategies almost identical to ANECS, the details of which have been previously published (14).
The Leuven Endometrial Study.
The Leuven Endometrial Study (LES) is a hospital based case–control study (15). Eligible cases, identified by active surveillance of electronic patient files at the Leuven University Hospital, were white women aged 27–80 years diagnosed with endometrial cancer. Clinical and information of endometrial cancer patients was recorded during interview at the time of diagnosis and from pathology reports. All medical records were reviewed by trained abstractors and pathology reports compatible with primary, invasive, epithelial endometrial adenocarcinoma of all stages (I–IV) and all grades were consulted. A control group of healthy female blood donors was recruited with the aid of the Red Cross Blood Service in the University Hospital. Participants completed a detailed questionnaire providing epidemiological information, including age, weight, height and self-reported Belgian (Flemish) ethnicity for three generations.
National Study of Endometrial Cancer Genetics.
NSECG, National Study of Endometrial Cancer Genetics. Cases were identified from collaborating clinicians throughout the UK from 2008 to present, taking care not to recruit from centers involved in SEARCH. Inclusion criteria were adenocarcinomas of the uterus presenting at ≤70 years of age. Almost all cases were incident and sampled within 6 months of diagnosis. Peripheral blood was collected from each participant and DNA extracted using standard methods. Tumor histology was confirmed from routine hospital reports and further details of histopathology and other tumor pathology characteristic was abstracted from these clinical pathology reports. Controls (45% males and 55% females) were drawn from the ColoRectal tumour Gene Identification study (16) and were unaffected by cancer. All cases and controls were of white UK ethnic origin.
The Polish Endometrial Cancer Study.
The Polish Endometrial Cancer Study (PECS) is a population-based case–control study carried out in two cities in Poland (Warsaw and Łódz) during 2001–2003 (17). The cases were women aged 20–74 years old, newly diagnosed with pathologically confirmed endometrial cancer and identified through participating hospitals and local cancer registries. Controls without a history of breast or endometrial cancer and with an intact uterus at time of enrollment were randomly selected from a database of all residents and frequency matched to cases (1:2 case:control ratio) by study site (Warsaw and Łódz) and age in 5 years categories. In-person interviews, including information on demographic characteristics and known or suspected endometrial cancer risk factors, were obtained from 551 cases (79% of the 695 eligible cases) and 1925 controls (68% of the 2843 eligible controls). Trained interviewers collected venous blood from 88% of participating cases and 94% of participating controls. To approximate a 1:1 case–control ratio for genotyping, cases and randomly selected 5 years age categories and study site frequency-matched controls who had donated blood were selected. The study protocol was reviewed and approved by local Polish and US National Cancer Institute institutional review boards. All participants provided written informed consent.
Singapore and Sweden Breast/Endometrial Cancer Study.
Details of the population selection process for the Singapore and Sweden Breast/Endometrial Cancer Study (SASBAC) have been published previously (18). Briefly, this population-based case–control study was conducted among Swedish women aged 50–74 years, who were residing in Sweden between 1 January 1994 and 31 December 1995. Endometrial cancer cases were identified through the nationwide cancer registries in Sweden. Controls, frequency matched for age, were randomly selected from the Swedish Registry of Total Population. The study was restricted to postmenopausal women with an intact uterus and no previous diagnosis of endometrial cancer. All participants provided detailed questionnaire information. For endometrial cancer, histological specimens were reviewed and reclassified by the study pathologist. Genomic analyses were conducted by the Singapore node of the study.
The Studies of Epidemiology and Risk factors in Cancer Heredity.
The SEARCH is an ongoing UK population-based study with cases ascertained through the Eastern cancer Registration and Information Centre (http://www.ecric.org.uk). All women diagnosed with endometrial cancer between the ages of 18–69 years (average age diagnosis 58 years) from 31 July 2001 to 1 September 2007 were eligible for inclusion. Approximately 54% of eligible patients have enrolled in the study. Women taking part in the study were asked to provide a 20 ml blood sample for DNA analysis and to complete a comprehensive epidemiological questionnaire. Controls were matched to cases in geographical profile and age and were drawn from two sources: (i) SEARCH participants (http://www.srl.cam.ac.uk/search/Homepage.htm), who were female with no prior history of cancer at the time of recruitment. Approximately 35% of eligible SEARCH controls enrolled in the study and (ii) European Prospective Investigation of Cancer-Norfolk, a population-based study of diet and cancer (19). There were 1127 endometrial cases and 6824 controls available for genotyping analysis at the time of this study.
The Women’s Insight and Shared Experiences Study.
The Women’s Insight and Shared Experiences Study (WISE) is a population-based case–control study conducted from among residents from a contiguous nine county region around Philadelphia and has been described previously (20). Eligible cases, identified by active surveillance at 61 hospitals, were African-American or Caucasian women aged 50–79 years, who were newly diagnosed with endometrial cancer between 1 July 1999 and 30 June 2002. Pathological reports and medical records were reviewed by trained abstractors and case status was validated by a pathology report that was compatible with primary, invasive, epithelial endometrial adenocarcinoma of all stages (I–IV) and all grades. Frequency-matched control subjects were selected by random digit dialing and restricted to women with no history of endometrial cancer or hysterectomy. Telephone interviews to complete detailed questionnaires were conducted for all participants.
Genotyping
For three studies (ANECS/AOCS, LES, SASBAC), genotyping was performed using matrix-assisted laser desorption/ionization time of flight mass spectrometry for the determination of allele-specific primer extension products using Sequenom’s MassARRAY system and iPLEX technology (Sequenom San Diego, CA). Oligonucleotides were designed using MassARRAY Assay Design software (version 3.1; Sequenom) and results analyzed using TYPER software (version 3.4; Sequenom).
SEARCH genotyping was carried out by nuclease assay (Taqman; Applied Biosystems, Foster City, CA). Taqman genotyping reagents were design by Applied Biosystems (http://www.appliedbiosystems.com/) as Assays-by-Design. Genotyping was performed using the ABI Prism 7900HT Sequence Detection Systems according the manufacturer’s instructions.
PECS genotyping was completed at the Core Genotyping Facility of the National Cancer Institute’s Division of Cancer Epidemiology and Genetics using the Illumina iSelect Custom BeadChip for all the SNPs presented except for two SNPs: rs3857481 and rs4777792 were genotyped using Taqman as described above.
NSECG cases were genotyped using Sequenom iplex methodology, as described above. NSECG control genotype data were extracted from existing Illumina 550K genome-wide scan data (16) or genotyped using the Kaspar method if the SNP was not present on the Illumina 550K platform.
WISE samples were genotyped using the SNaPShot method (Applied Biosystems) and analyzed on an ABI 3130 capillary sequencer according to manufacturer’s instructions.
All studies complied with quality control standards by including ≥2 negative (no DNA) polymerase chain reaction controls per 384-well assay plate and at least 2% of samples in duplicate. A genotyping call rate >95% and at least 98% concordance between duplicated samples for each SNP assay was required. Additionally, the genotype distributions in the control samples of each study were required to conform to Hardy–Weinberg equilibrium (HWE) at P >0.005. Three subsets were excluded from the meta-analyses on this basis: SASBAC: SNPs rs13281615, P = 1 × 10−13 and rs1045485, P = 0.0002; LES: SNP rs3020314, P = 0.002.
Imputation
Rs3857481 was the only SNP that had not been genotyped in the ANECS study. We were able to obtain imputed genotype dosages for this SNP for 599 ANECS cases with endometriod histology genotyped as part of a GWAS of endometrial cancer using an Illumina Infinium 610k array (5) and for 5190 UK control subjects who had been genotyped using an Illumina Infinium 1.2M array as part of the Wellcome Trust Case Control Consortium (21). Non-genotyped SNPs were imputed using the HapMap Phase 3 data as a reference panel (22). Imputed genotype dosages were compared between cases and controls, adjusting for the first three principal components of the genomic kinship matrix to take into account any differences in population structure between cases and controls. This part of the analysis was performed using GenABEL (23), ProbABEL (24) and MACH (25).
Statistical methods
For each SNP, deviation of genotype frequencies in controls from Hardy–Weinberg equilibrium was assessed by a χ2 test with one degree of freedom. Genotype risks were estimated as ORs with 95% CIs, and significance levels were calculated using logistic regression. Meta-analysis was carried out and Forest plots generated using the Metan command within Stata™(http://www.stata.com).
Results and discussion
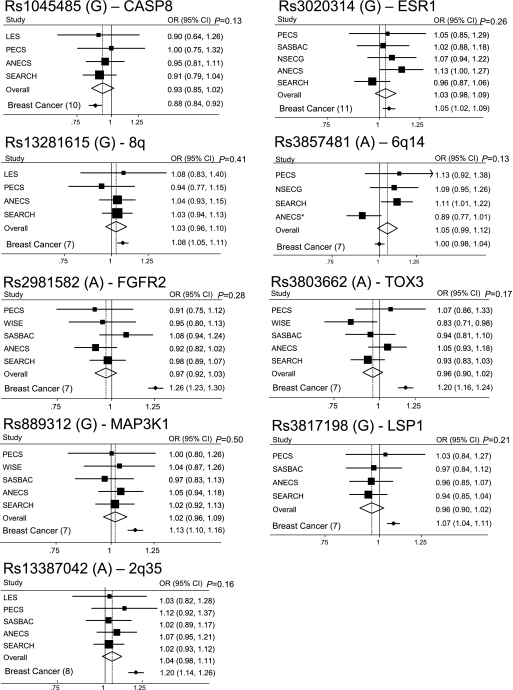
A total of nine breast cancer-associated SNPs were genotyped in up to six different endometrial cancer case/control studies of European ancestry from Sweden, UK, Belgium, Poland, Australia and the USA. Genotype counts and ORs (95% CIs) for individual studies and overall minor allele frequencies and risk estimates are shown in Supplementary Table 1, available at Carcinogenesis Online. Per allele ORs (95% CIs) for each SNP are shown as Forest plots in Figure 1 with the original reported breast cancer per allele risk estimates also shown for comparison. There were no significant heterogeneities between studies for any SNP (P > 0.05) apart from rs3857481 (P = 0.041).
Fig. 1.
Forest plots showing per allele OR and 95% CIs for each SNP (risk allele) by study. The size of the black box is proportional to the study size. The diamond represents the OR and 95% CIs for the meta-analysis of each SNP. Per allele ORs and 95% CIs for breast cancer (reference in brackets) are shown for comparison at the bottom of each Forest plot. *Results for ANECS rs3857481 were imputed using a subset of the ANECS cases (n = 599) (5) matched to an alternative control group (1958 Birth Cohort controls n = 2694 and National Blood Donor controls n = 2496) (21).
The SNPs in CASP8 (rs1045485), ESR1 (rs3020314) and in the GWAS-identified 8q24 region (rs13281615) were not significant, but their point estimates lie within the 95% CIs for the corresponding breast cancer risk estimates (Figure 1). Further collaboration may clarify the effects of these SNPs on endometrial cancer risk.
The other six GWAS-identified breast cancer SNPs: rs3857481 (6q14), rs2981582 (FGFR2), rs3803662 (TOX3), rs889312 (MAP3KI), rs3817198 (LSP1) and rs13387042 (2q35) had no apparent association with risk of endometrial cancer in these studies (Supplementary Table 1 is available at Carcinogenesis Online, Figure 1). The CIs for the overall endometrial cancer risk do not overlap with the point estimates for breast cancer risk, indicating that these SNP alleles cannot have similar effects on breast and endometrial cancer risk. We have >90% power to detect associations of the same magnitude as those observed for breast cancer for FGFR2, TOX3, 2q35 and MAP3K1, but the equivalent power is considerably lower for SNPs with more modest effects on breast cancer risk, e.g. 45% for LSP1 and 28% for ESR1.
A smaller study of 692 endometrial cancer cases and 1723 controls by McGrath et al. (26) also assessed endometrial cancer risk associated with the six GWAS-identified breast cancer SNPs examined in this study. These authors similarly report no statistically significant associations with endometrial cancer risk that are of similar magnitude and/or direction to that reported for breast cancer, although two of the risk estimates for endometrial cancer (rs2981582, rs13281615) might be interpreted to be consistent with the direction of effect we observed for endometrial cancer in this study.
Given their known associations with breast cancer risk, the SNPs examined here might be considered to have a higher prior probability of association with endometrial cancer than randomly chosen SNPs. The point estimates of the effect sizes of three of these alleles in endometrial cancer were comparable with those in breast cancer but did not reach statistical significance, whereas for breast cancer, they are highly significant due to the large numbers (currently ∼80 000) contributing to the Breast Cancer Association Consortium (BCAC). This emphasizes the need for further collaboration between studies in order to provide the sufficiently large sample sizes needed for confirmation of common low-penetrant associations at appropriate levels of significance.
Supplementary material
Supplementary Table 1 can be found at http://carcin.oxfordjournals.org/
Funding
The genotyping and analysis of this study, and the conduct of the SEARCH study, was funded by Cancer Research UK grants (C490/A11021, C8197/A10123, C1287/A7497, C1287/A10118), BCC grant (2007NovPR17) and EU FP7 COGS (HEALTH–F2-2009–223175). A.M.D. was supported by Cancer Research UK grant (C8197/A10865) and The Joseph Mitchell Trust.
ANECS was supported by a project grants from the National Health and Medical Research Council (NHMRC) of Australia (ID #339435), The Cancer Council Queensland (ID #4196615) and Cancer Council Tasmania (ID #403031 and ID #457636). A.B.S. is supported by an NHMRC Senior Research Fellowship. T.M. is supported by an Australian Postgraduate Award, an Institute of Health and Biomedical Innovation PhD Top-Up and a Smart State PhD Award.
LES was supported by the Verelst Foundation for Endometrial Cancer.
NSECG was supported by funding from Cancer Research UK and the Oxford Comprehensive Biomedical Research Centre.
PECS was funded by the intramural research program at the US National Cancer Institute, Division of Cancer Epidemiology and Genetics in the Hormonal and Reproductive Epidemiology Branch.
SASBAC is supported by the Agency for Science, Technology and Research of Singapore (A*STAR).
WISE was supported by a Public Health Service Grant (P01-CA77596).
Supplementary Material
Acknowledgments
The authors would like to thank the many individuals who participated in this study and the numerous institutions and their staff who have supported recruitment.
In Particular, ANECS would like to thank Felicity Lose, Jyotsna Batra, Xiaoqing Chen and Jonathan Beesley from The Molecular Cancer Epidemiology and Cancer Genetic laboratories at QIMR for technical assistance. We also thank the Australian Red Cross Blood Services (ARCBS) donors who participated as healthy controls in this study. We are grateful to the staff at ARCBS for their assistance with the collection of risk factor information and blood samples, and Mary-Anne Kedda and members of the Molecular Cancer Epidemiology Laboratory for their assistance with collection and processing of blood samples.
ANECS would also like to gratefully acknowledge the co-operation of the following institutions: NSW: John Hunter Hospital, Liverpool Hospital, Mater Misericordiae Hospital (Sydney), Mater Misericordiae Hospital (Newcastle), Newcastle Private Hospital, North Shore Private Hospital, Royal Hospital for Women, Royal Prince Alfred Hospital, Royal North Shore Hospital, Royal Prince Alfred Hospital, St George Hospital; Westmead Hospital, Westmead Private Hospital; Queensland: Brisbane Private Hospital, Greenslopes Hospital, Mater Misericordiae Hospitals, Royal Brisbane and Women's Hospital, Wesley Hospital, Queensland Cancer Registry; South Autralia: Adelaide Pathology Partners, Burnside Hospital, Calvary Hospital, Flinders Medical Centre, Queen Elizabeth Hospital, Royal Adelaide Hospital, South Australian Cancer Registry; Tasmania: Launceston Hospital, North West Regional Hospitals, Royal Hobart Hospital; Victoria: Freemasons Hospital, Melbourne Pathology Services, Mercy Hospital for Women, Royal Women's Hospital, Victorian Cancer Registry; WA: King Edward Memorial Hospital, St John of God Hospitals Subiaco and Murdoch, Western Australian Cancer Registry.
The ANECS Group comprises A.B.S., P.Webb, J.Young (Queensland Institute of Medical Research); Consumer representative: L.McQuire; clinical collaborators: New South Wales: S.Baron-Hay, D.Bell, A.Bonaventura, A.Brand, S.Braye, J.Carter, F.Chan, C.Dalrymple, A.Ferrier (deceased), G.Gard, N.Hacker, R.Hogg, R.Houghton, D.Marsden, K.McIlroy, G.Otton, S.Pather, A.Proietto, G.Robertson, J.Scurry, R.Sharma, G.Wain, F.Wong; Queensland: J.Armes, A.Crandon, M.Cummings, R.Land, J.Nicklin, L.Perrin, A.Obermair, B.Ward; South Australia: M.Davy, T.Dodd, J.Miller, M.Oehler, S.Paramasivum, J.Pierides, F.Whitehead; Tasmania: P.Blomfield, D.Challis; Victoria: D.Neesham, J.Pyman, M.Quinn, R.Rome, M.Weitzer; Washington: B.Brennan, I.Hammond, Y.Leung, A.McCartney, C.Stewart, J.Thompson; project managers: S.O'Brien, S.Moore; laboratory manager: K.Ferguson; pathology support: M.Walsh; admin support: R.Cicero, L.Green, J.Griffith, L.Jackman, B.Ranieri; laboratory assistants: M.O'Brien, P.Schultz; research nurses: B.Alexander, C.Baxter, H.Croy, A.Fitzgerald, E.Herron, C.Hill, M.Jones, J.Maidens, A.Marshall, K.Martin, J.Mayhew, E.Minehan, D.Roffe, H.Shirley, H.Steane, A.Stenlake, A.Ward, S.Webb, J.White.
Full membership of the Australian Ovarian Cancer Study Group is listed at http://www.aocstudy.org/.
LES gratefully acknowledges the contribution of Drs Evelyn Despierre, Thomas Van Brussel and Gilian Peuteman.
The NSECG Group comprises Ian Tomlinson (Oxford University); M.Adams, A.Al-Samarraie, S.Anwar, R.Athavale, S.Awad, A.Bali, A.Barnes, G.Cawdell, S.Chan, K.Chin, P.Cornes, M.Crawford, J.Cullimore, S.Ghaem-Maghami, R.Gornall, J.Green, M.Hall, M.Harvey, J.Hawe, A.Head, J.Herod, M.Hingorani, M.Hocking, C.Holland, T.Hollingsworth, J.Hollingworth, T.Ind, R.Irvine, C.Irwin, M.Katesmark, S.Kehoe, G.Kheng-Chew, K.Lankester, A.Linder, D.Luesley, C.BLynch, V.McFarlane, R.Naik, N.Nicholas, D.Nugent, S.Oates, A.Oladipo, A.Papadopoulos, S.Pearson, D.Radstone, S.Raju, A.Rathmell, C.Redman, M.Rymer, P.Sarhanis, G.Sparrow, N.Stuart, S.Sundar, A.Thompson, S.Tinkler, S.Trent, A.Tristram, N.Walji, R.Woolas.
PECS would like to thank Neonila Szeszenia-Dabrowska of the Nofer Institute of Occupational Medicine (Lodz, Poland) and Witold Zatonski of the Maria Sklodowska-Curie Institute of Oncology and Cancer Center (Warsaw, Poland) for their contribution to the PECS; Pei Chao and Michael Stagner (IMS, Silver Spring, MD) for their invaluable management of the PECS and the physicians, nurses, interviewers and study participants for their dedicated efforts.
SEARCH would like to thank Caroline Baynes, Craig Luccarini, Don Conroy, Patricia Harrington, Rebecca Mayes and Hannah Munday.
WISE wish to acknowledge Rita Schinnar, Anita L.Weber, Greta Bunin, Jesse A.Berlin, Mona Baumgarten, Angela DeMichele, Stephen C.Rubin, Michelle Berlin, Andrea B.Troxel, Elene Turzo and Deisree Burgh.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ANECS
Australian National Endometrial Cancer Study
- AOCS
Australian Ovarian Cancer Study
- CI
confidence interval
- GWAS
genome-wide association study
- LES
Leuven Endometrial Study
- NSECG
National Study of Endometrial Cancer Genetics
- OR
odds ratio; PECS, Polish Endometrial Cancer Study
- SASBAC
Singapore and Sweden Breast/Endometrial Cancer Study
- SEARCH
Studies of Epidemiology and Risk factors in Cancer Heredity
- SNP
single-nucleotide polymorphism
- WISE
Women’s Insight and Shared Experiences Study
Biographies
LES—
PECS—
SASBAC—
SEARCH—
WISE—
References
- 1.Kaaks R, et al. Obesity, endogenous hormones, and endometrial cancer risk: a synthetic review. Cancer Epidemiol. Biomarkers Prev. 2002;11:1531–1543. [PubMed] [Google Scholar]
- 2.Beral V, et al. Endometrial cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2005;365:1543–1551. doi: 10.1016/S0140-6736(05)66455-0. [DOI] [PubMed] [Google Scholar]
- 3.Fisher B, et al. Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J. Natl Cancer Inst. 2005;97:1652–1662. doi: 10.1093/jnci/dji372. [DOI] [PubMed] [Google Scholar]
- 4.Shang Y, et al. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 5.Spurdle AB, et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat. Genet. 2011;43:451–455. doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott KS, et al. Evaluation of association of HNF1B variants with diverse cancers: collaborative analysis of data from 19 genome-wide association studies. PLoS One. 2010;5:e10858. doi: 10.1371/journal.pone.0010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 9.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne RL, et al. Risk of estrogen receptor-positive and -negative breast cancer and single nucleotide polymorphism 2q35-rs13387042. J. Natl Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 12.Dunning AM, et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum. Mol. Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spurdle A, et al. Re: Excess of early onset multiple myeloma in endometrial cancer probands and their relatives suggests common susceptibility. Gynecol. Oncol. 2008;109:153. doi: 10.1016/j.ygyno.2007.12.010. author reply 154. [DOI] [PubMed] [Google Scholar]
- 14.Beesley J, et al. Association between single-nucleotide polymorphisms in hormone metabolism and DNA repair genes and epithelial ovarian cancer: results from two Australian studies and an additional validation set. Cancer Epidemiol. Biomarkers Prev. 2007;16:2557–2565. doi: 10.1158/1055-9965.EPI-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Mara TA, et al. Progesterone receptor gene variants and risk of endometrial cancer. Carcinogenesis. 2011;32:331–335. doi: 10.1093/carcin/bgq263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houlston RS, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudet MM, et al. Genetic variation in CYP17 and endometrial cancer risk. Hum. Genet. 2008;123:155–162. doi: 10.1007/s00439-007-0454-8. [DOI] [PubMed] [Google Scholar]
- 18.Weiderpass E, et al. Risk of endometrial cancer following estrogen replacement with and without progestins. J. Natl Cancer Inst. 1999;91:1131–1137. doi: 10.1093/jnci/91.13.1131. [DOI] [PubMed] [Google Scholar]
- 19.Day N, et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br. J. Cancer. 1999;80:95–103. [PubMed] [Google Scholar]
- 20.Strom BL, et al. Case-control study of postmenopausal hormone replacement therapy and endometrial cancer. Am. J. Epidemiol. 2006;164:775–786. doi: 10.1093/aje/kwj316. [DOI] [PubMed] [Google Scholar]
- 21.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aulchenko YS, et al. GenABEL. an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 24.Aulchenko YS, et al. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGrath M, et al. Novel breast cancer risk alleles and endometrial cancer risk. Int. J. Cancer. 2008;123:2961–2964. doi: 10.1002/ijc.23862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.