Abstract
(−)-Epigallocatechin-3-gallate (EGCG) has been reported to affect many cellular regulatory pathways. This study aims to determine whether EGCG could target microRNA (miRNA), one of the mechanisms for cells to achieve subtle change in multiple targets. We found that, in both human and mouse lung cancer cells in culture, EGCG specifically upregulated the expression of miR-210, a major miRNA regulated by HIF-1α. Furthermore, we found that overexpression of miR-210 led to reduced cell proliferation rate and anchorage-independent growth as well as reduced sensitivity to EGCG. On the mechanisms of miR-210 regulation by EGCG, we demonstrated that the regulation was mediated through the hypoxia-response element in miR-210 promoter. Consistently, the upregulation of miR-210 was found to be correlated with the stabilized HIF-1α in lung cancer cell lines after EGCG treatment. This EGCG-induced stabilization of HIF-1α was further shown by the stabilization of HA-tagged HIF-1α but not the P402A/P564A-mutated HIF-1α by EGCG, suggesting that EGCG targets the oxygen-dependent degradation (ODD) domain. Direct evidence was obtained by affinity binding assay showing that EGCG specifically binds HIF-1α with a Kd = 3.47 μM. This result suggests that EGCG binding interferes with the hydroxylation of key Pro residues in the ODD domain, preventing HIF-1α from the Pro hydroxylation-dependent ubiquitination and subsequent proteosome-mediated degradation. In summary, our results demonstrated, for the first time, the elevation of miR-210 by EGCG in lung cancer cell lines and this is mediated by the stabilization of HIF-1α. This event contributes to the anticancer activity of EGCG.
Introduction
The consumption of green tea (Camellia Sinensis) has been suggested to have beneficial health effect, for example, in cancer prevention. Extensive studies have established that the most abundant and active anticancer constituent in green tea is (−)-epigallocatechin-3-gallate (EGCG). The anticancer activity of EGCG has been demonstrated in many organ sites in different animal models and cell line systems (1,2). Treatment with EGCG leads to the inhibition of tumor cell growth and induction of tumor cell apoptosis (1,2). Such effects have been reported to involve not only the regulation of the expression of specific genes but also the modulations of multiple cellular signaling pathways. The anticancer activity of EGCG may be resulted from a combinatory effects on multiple targets, although the relative importance of the different pathways may depend upon the cell context.
EGCG has been reported to affect the activity of hypoxia-induced factor (HIF), but both suppression of HIF-1α expression (3–6) and accumulation of HIF-1α protein (7–9) have been reported. HIFs are a family of structurally related basic-helix-loop-helix transcription factors that play the master role in controlling the response to hypoxia (10–12). The prototype member is HIF-1, a heterodimer complex consisting of a regulatory HIF-1α subunit and a ubiquitously expressed HIF-1β subunit (also known as aryl hydrocarbon receptor nuclear translocator, ARNT). There are two other HIF-1α isoforms, HIF-2α and HIF-3α. HIF-1α and HIF-2α are rapidly induced in response to hypoxia, whereas the regulation of HIF-3α is not well understood. Both HIF-1α and HIF-2α contain an oxygen-dependent degradation (ODD) domain that is required and sufficient for the proteasome-mediated degradation. Under normoxia conditions (21% oxygen level), HIFα protein is consistently expressed and rapidly degraded and the degradation is controlled by the hydroxylation of HIF-1α Pro residues 402 and 564 (P402 and P564) or HIF-2α Pro residues 405 and 531 (P405 and P531) in the ODD domain. These two key Pro residues are hydroxylated by a family of the egg-laying-defective nine prolyl-hydroxylases (also known as PHDs or HPH hydroxylases), which is activated by the availability of oxygen. Hydroxylated Pro residues create a binding site recognized by the von Hippel–Lindau tumour suppressor protein, the recognition component of an E3 ubiquitin ligase, which consequently results in HIFα protein ubiquitination and proteasome-mediated degradation (13). The activity of PHD is also under the influence of other chemicals, such as iron, 2-oxoglutarate and ascorbate, which cause the accumulation of HIFα protein in normoxia (14–16). Also, the HIFα protein level can be regulated at the transcription level by other signaling (10–12). Thus, the regulation of HIFα is not limited to the availability of oxygen.
An effective mechanism to regulate multiple classes of genes is through small regulatory RNAs, microRNAs (miRNAs) (17,18). MiRNAs are small (∼22 bases), single stranded, endogenous and non-coding RNAs that negatively regulate the translation and/or stability of messenger RNA (mRNA) (17,19). Because miRNA recognizes the short fragment usually in the 3′ untranslated region of mRNA with imperfect complementarity, one miRNA is able to target mRNAs of hundreds of distinct genes and one mRNA can be regulated by different miRNAs (17,20). In addition to the aforementioned gene regulation, miRNA exerts very fine tune regulation on the expression of most protein-coding genes to optimize their expressions (17,18). Upon binding to its target mRNA, miRNA can activates Argonaute-catalyzed targeted cleavage of mRNA (21). Thus, mammalian miRNAs predominantly act to reduce target mRNA species (22). Cellular miRNA pool forms a hierarchical regulatory network that is associated with cell identity and function (23). The cell identity changes resulted from development, differentiation, environmental challenge and tumorgenesis reset the miRNA expression (23).
In lung development, miRNA has been demonstrated to play critical roles. Different groups of miRNAs are expressed in developing and mature lung tissues, suggesting the distinct roles of these miRNAs in regulating lung tissue cell growth and differentiation as well as maintaining normal lung functions (24). Among the miRNAs expressed in lung, the miR-17-92 cluster is required for promoting the proliferation of lung progenitors (24), whereas miR-34c, miR145 and miR-142-5p are required for suppressing lung cell growth (25). There are also evidences supporting the concept that miRNA plays roles in lung tumorgenesis. Firstly, miR-34c, miR145 and miR-142-5p, which suppress lung cell growth, are found to be repressed in both human and mouse lung cancer (25). Secondly, the overall miRNA levels in the oncogenic K-ras-induced mouse lung cancer are found to be reduced (26). Thirdly, lung-specific knockout of Dicer results in the abnormality of lung development and function (27). Since the 22 base-active miRNA is initially expressed in a larger precursor fragment, pre-miRNA or within other mRNA and is generated through the maturation process mediated by Dicer (19), there is no mature functional miRNA without Dicer. This result is consistent with the idea that Dicer is a haploinsufficient tumor suppressor gene and the optimized level of miRNA is necessary to maintain the normal cell functions and phenotype (28).
Since we and others have demonstrated that EGCG can prevent lung carcinogenesis and inhibit lung cancer growth (2), we have been wondering whether the cellular change in lung cancer cells treated with EGCG is also associated with the alternation in miRNA expression. Here, we studied the change in miRNA profiles in mouse and human lung cancer cells that were treated with EGCG and found that miR-210 was upregulated. By studying the mechanism of the regulation of miR-210 by EGCG, we further found that the treatment of EGCG led to the accumulation of HIF-1α, an activator of the miR-210 expression. Our results demonstrated the involvement of miR-210 and HIF-1α in the anticancer activity of EGCG.
Material and methods
Cell culture and treatment
Mouse lung adenocarcinoma cell line, CL13 (a gift from Dr Stephen Belinsky, Lovelace Respiratory Research Institute) and human non-small cell lung cancer cell lines, H1299, H460 and A549 (ATCC, Manassas, VA), were maintained in RPMI medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml of penicillin and 100 μg/ml of streptomycin at 37°C in 5% CO2. HEK293T cells (ATCC) were maintained in DMEM medium. Cells were treated with different concentrations of EGCG (a gift from Dr Y.Hara; Mitsui Morin Co., Shizuoku, Japan) in the presence of superoxide dismutase (SOD; 5 U/ml; Sigma, St. Louis, MO) and catalase (30 U/ml; Sigma) for different periods of time after an overnight serum starvation. Unless stated otherwise, SOD and catalase were added to the incubation mixture to prevent the formation of reactive oxygen species due to the autooxidation of EGCG (29,30).
To determine the effect of EGCG using a luciferase reporter, the cells transiently transfected with the luciferase reporter, plus the Rellina luciferase reporter (pRL-CMV; Promega, Madison, WI) as the internal control, were treated with different concentrations of EGCG and analyzed for luciferase activity. In brief, the cells at ∼90% confluency in a 10 cm plate was transfected with a mixture of DNA plasmids (a total amount of 20 μg) using the Effectene transfection reagent (Qiagen, Germantown, MD). Transfected cells were split into 24-well plate for further culture at 12 h after the transfection. After another 12 h culture, the cells were subjected to a 12 h serum starvation. Then, the cells were treated with EGCG in the presence of SOD/catalase for 12 h before being collected for determining luciferase activity using the luciferase assay kit (Promega).
To establish the cells stably expressing miR-210 or HIF-1α, H1299 and H460 cells were infected with the retrovirus expressing miR-210 or HIF-1α and a puromycin selection marker and stable cells were selected using puromycin (5 μg/ml). Retrovirus was produced by HEK293T cells transfected with pBABE-puromycin retroviral expression vector plus the packaging vectors using Polyfect transfection reagent (Qiagen) (31). The cells infected with the virus produced using the empty pBABE-puromycin vector were used as the control.
The changes in cell proliferation in response to EGCG treatment or the miR-210 expression were determined in cells cultured in a medium containing 5% fetal bovine serum and the viable cells were measured by the 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) assay as described previously (29).
The anchorage-independent growth of cells expressing miR-210 was analyzed by the soft-agar assay. In brief, 1000 cells were seeded in 2 ml of RPMI medium containing 10% fetal bovine serum and 0.3% low melting temperature agarose, which was overlaid on a base of 3 ml of medium containing 0.5% low melting temperature agarose in one well of six-well plate. After cultures of H460 cells for 10 days and H1299 cells for 14 days were created, the colonies were stained with 0.5 ml/well of 0.005% Crystal Violet for 1 h and the number and size of colonies were recorded.
Total miRNA purification, microarray and real-time polymerase chain reaction
Total miRNA samples were purified from cells using miRNeasy kit (Qiagen). To obtain miRNA profiles, miRNA samples were analyzed by the miRNA microarray conducted by Ocean Ridge Biosciences (Palm Beach Gardens, FL) using Multispecies MicroRNA Array Chip covering Sanger miRBase v14.0. Data collection and analysis were also conducted by Ocean Ridge Biosciences using the standard method for microarray analysis. Briefly, for the data normalization, the normalization factor (N) for each microarray was obtained by the 20% trim mean of the species (human/mouse) probes intensities above threshold in all samples. The log2-transformed spot intensities for all miRNAs on the array were normalized by subtracting N from each spot intensity and scaled by adding the grand mean of N across all microarrays. P-value and false discovery rate were calculated for the triplicates. The changes were ranked by the smallest P-value and false discovery rate and then by fold of change.
The relative expression levels of miR-210 in samples were determined by real-time reverse transcription–polymerase chain reaction (PCR). In brief, the complementary DNA of the total miRNA was first synthesized by reverse transcription using the Universal cDNA Synthesis kits (Exiquon, Woburn, MA) and miR-210 and the reference gene U6 snRNA were then amplified by real-time PCR using miR-210 and U6 snRNA PCR primer sets (Exiquon), respectively. Real-time PCR was performed using the SYBR Green master mix (Exiquon) in 384-well plates and each sample was repeated three times on the ABI 7900HT real-time PCR system (Applied Biosystems, Carlsbad, CA) at the Bionomics Research and Technology Center at Rutgers University. The relative levels of miR-210 were determined by the ratios of miR-210:U6 snRNA as calculated by the ΔΔCt.
DNA constructs
MiR-210 promoter-luciferase reporters were constructed by inserting the −600 bp of human miR-210 and the −2000 bp of mouse miR-210 promoter fragments into pGL3-basic vector (Promega). Fragments of mouse miR-210 promoters, −480bp, −420bp, HRE (hypoxia response element; −449 to −407 bp), mutHRE (HRE sequence GACGTG within −449 to −407 bp was change to CTGCAC) (as illustrated in Figure 3C) were also cloned into pGL3-basic vector. The well characterized HRE of phosphoglycerate kinase gene and its mutant (mutHRE)-driven luciferase reporters, pGL2-HRE-luciferase and pGL2-mutHRE-luciferase, were described previously (32).
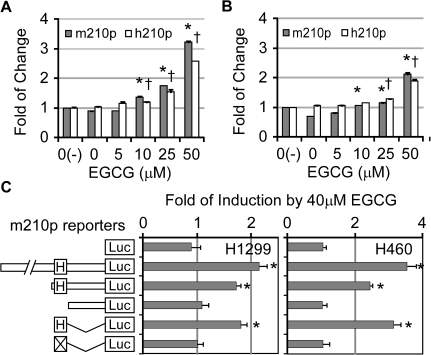
Fig. 3.
HRE in miR-210 promoter was responsive to the EGCG stimulation. H1299 (A) and H460 (B) cells transfected with human and mouse miR-210 gene promoter-luciferase reporter (h210p and m210p) were assayed for reporter activities at 12 h after EGCG treatment in the presence of SOD/catalase (n = 3). * and † indicate statistical difference between levels at different concentrations of EGCG and the control (EGCG = 0) (by one-way analysis of variance; P < 0.05); * is for m210p and † is for h210p.The responses of mouse miR-210 gene promoter fragment to EGCG treatment were analyzed by the luciferase activities in H1299 and H460 cells transfected with different reporter constructs and treated with EGCG for 12 h (n = 3) (C). 0(−) represents for the treatment without SOD/catalase. H and X stand for HRE and mutHRE, respectively. *indicates statistical difference from the control empty vector (by one-way analysis of variance; P < 0.05).
The HA-tagged HIF-1α, Pro402/P564 mutated HIF-1α (mutHIF1α), HIF-2α, Pro405/P531 residue-mutated HIF-2α (mutHIF2α) and ODD-luciferase were expressed using pCDNA and retroviral expression vector pBABE-puromycin as described previously (33,34). The ODD in pCDNA-ODD-luciferase was replaced by Pro402/P564 mutated HIF-1α ODD (mutODD) to construct the mutODD-luciferase reporter. Flag-HIF-1α-ODD and -Pro402/P564 mutant ODD (mutODD) were expressed in HEK293T cells using expression vectors, pCDNA3.1-Flag-ODD and -mutODD, constructed by inserting the wild-type and Pro405/P531-mutated ODD domains (aa303-603) of HIF-1α into pCDNA3.1-Flag vector, respectively.
Human miR-210 was ectoptically expressed in human cell lines, H1299 and H460, using the retroviral pBABE-miR-210 expression vector, which expresses human miR-210 precursor cloned by PCR according to reported sequence information (35).
Western blot and immunoprecipitation
HA- and Flag-tagged proteins were recognized by western blot analysis using anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Flag antibodies (Sigma), respectively. The cellular levels of HIF-1α were analyzed by western blot using anti-HIF-1α antibody (Abcam, Cambridge, MA). To monitor the sample loading, the blot was probed for β-actin using anti-β-actin antibody (Sigma).
To study the potential binding of EGCG to HIF proteins, we used HA-HIF-1α and -HIF-2α proteins as well as Flag-ODD and -mutODD proteins expressed in HEK293T cells. In brief, HA-HIF-1α/HIF-2α and Flag-ODD/mutODD were expressed in HEK293T cells by the transient transfection with the expression vectors using Polyfect transfection reagent (Qiagen). At 36 h after the transfection, cells were cultured in the incubator and saturated with nitrogen for 6 h to reach the hypoxia condition. Then, the cells were collected for the immunopurification using the anti-HA (Santa Cruz) or Flag (Sigma) antibody conjugated on agarose beads as described previously (36,37).
Affinity binding assay
To determine the EGCG binding, purified HA-HIF-1α, HA-HIF-2α, Flag-ODD and Flag-mutODD were applied for the affinity binding using [3H]EGCG (13 Ci/mmol in ethanol containing 8 mg/ml ascorbic acid; a gift from Dr Yukihiko Hara; Food Research Laboratory, Fujieda, Shizuoka, Japan) (38) with modification that the binding reaction was carried out at room temperature for 2 h in the presence of SOD (5 U/ml) and catalase (30 U/ml). The anti-HA or -Flag antibody conjugated on agarose beads alone was applied as the negative control. The Kd value was determined by non-linear regression analysis using the Prizm 4.0 software (Graphpad, La Jolla, CA).
Statistical analysis
One-way analysis of variance followed by Dunnett’s multiple comparison test was used for the comparison of the differences between treatment groups and the control. Statistical significance was indicated by P <0.05 in the two-tailed comparison. Student’s t-test was used for determine the difference between cell line expressing miR-210 and control. Statistical significance was indicated by P <0.05. Linear regression was applied for determining the dose-dependent induction; model fitting was indicated by R2 and the dose-dependent response is determined by that the slope is significantly non-zero (P < 0.05).
Results
MiR-210 expression in lung cancer cells is increased after EGCG treatment
To determine the miRNA expression profile changes in response to EGCG treatment, we first extracted miRNA samples from mouse adenocarcinoma CL13 cells that had been treated with 40 μM EGCG for 0, 1, 3, 6 and 9 h. These samples were subjected to miRNA microarray profile analysis using the Ocean Ridge Multispecies MicroRNA Array Chip covering Sanger miRBase v14.0 (consisting of 1576 probes for 892 Human miRNA and 697 mouse miRNA). The microarray results from three individual experiments showed positive hybridization signals of 484 probes for mouse miRNAs. After analysis, miRNA with significant changes (up- or downregulated by at least 0.7-fold) were ranked by the smallest P-value and false discovery rate of triplicates. MiRNAs with changes (>0.7-fold) on at least two time points were miR-210 and miR-122. MiR-210 was found to be expressed in all samples and upregulated upon EGCG treatment (Figure 1A; P < 0.0001). MiR-122 was found upregulated after EGCG treatment, but the hybridization signals of miR-122 were too weak for positive identities for some time points. No miRNA was found to be significantly downregulated (>0.7-fold). The elevated level of miR-210 was then confirmed by its levels relative to U6 snRNA as determined by real-time PCR (Figure 1B). We therefore concluded that miR-210 is the major EGCG-regulated miRNA in mouse lung adenocarcinoma CL13 cells.
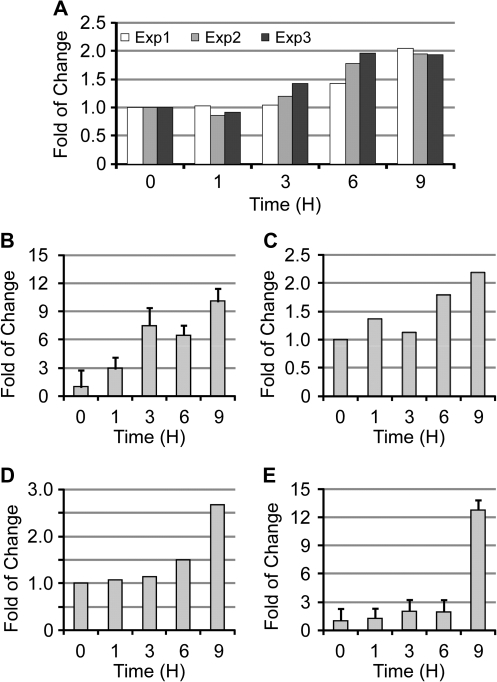
Fig. 1.
MiR-210 was upregulated in lung cancer cells upon EGCG treatement. The expression levels of miR-210 in lung cancer cells after EGCG (40 μM) treatment as compared with the control (0 h). (A). Results of three individual experiments using CL13 cells as determined by miRNA microarray (Exp. 1–3). (B). In CL13 cells as determined by real-time PCR (n = 3). (C) In CL13 cells treated with EGCG in the absence of SOD/catalase as determined by miRNA microarray. (D) In H1299 cells as determined by miRNA microarray. (E) In H1299 cells as determined by real-time PCR (n = 3).
In our experiments, SOD and catalase were routinely added to the cell culture medium to prevent the generation of reactive oxygen species due to the auto-oxidation of EGCG (29,30). The upregulation of miR-210 by EGCG was also observed by microarray using samples from cells treated with EGCG in the absence of SOD and catalase (Figure1C), and we did not find any significant difference between samples treated with or without SOD and catalase, suggesting that the induction of miR-210 is not due to the use of SOD and catalase.
Next, we extended our study to miRNA samples extracted from human lung cancer cell line H1299 cells and found similar results in that miR-210 was upregulated by EGCG (Figure 1D). The microarray detected 544 miRNA-positive hybridization signals of 902 probes covering 892 human miRNA. Again, the only other upregulated miRNA was miR-122 but the signals were too weak to draw any conclusions. Similarly, no miRNA was found significantly downregulated. This result was then validated by the levels of miR-210 relative to U6 snRNA as determined by real-time PCR (Figure 1E). Together with the above results, we found that miR-210 was the major miRNA that was elevated in response to EGCG treatment in lung cancer cells.
Ectopic expression of miR-210 leads to reduced proliferation and anchorage-independent growth
Next, we determined whether miR-210 is involved in the anticancer activity of EGCG by ectopically expressing human miR-210 in H1299 and H460 lung cancer cells and analyzed cell proliferation using the MTT assay. We found that cells expressing miR-210 (H1299-210 and H460-210) grew slower than the control cell lines (infected with the empty virus; H1299-e and H460-e) (Figure 2A and B), suggesting that miR-210 displays inhibitory effect on cell growth. We also found that H1299-210 and H460-210 cells were less sensitive to the inhibition of EGCG (Figure 2C and D), demonstrating that ectopic expression of miR-210 reduces the inhibitory effect of EGCG. These results support that induction of miR-210 by EGCG leads to the inhibition on cell growth. Furthermore, we determined the anchorage-independent growth of miR-210 expressing cells on semi-solid medium. After being cultured for 2 weeks, H1299-e cells displayed anchorage-independent growth and formed colonies in a semi-solid medium; in contrast, H1299-210 cells lost the anchorage-independent growth ability (Figure 2E). H460-210 and its control H460-e cells formed the same number of colonies, but H460-210 cell colonies were smaller (Figure 2F). The smaller colonies formed by H460-210 cells might be resulted from its slower growth rate caused by miR-210, whereas the loss of anchorage-independent growth of H1299-210 cells indicated the inhibitory activity of miR-210 against tumorgenesis. These results were consistent with the reported inhibitory activity of miR-210 on the growth of pancreatic cancer cells (35) and esophageal squamous cell carcinoma (39). Together, we found that miR-210, a major miRNA induced by EGCG treatment, negatively regulates lung cancer cell growth, indicating that the increased miR-210 contributes to the anticancer activity of EGCG.
Fig. 2.
Ectopic expression of miR-210 in lung cancer cells reduced proliferation, sensitivity to EGCG and anchorage-independent growth. H1299 and H460 cells infected with retrovirus expressing human miR-210 were compared with the cells infected with the control virus for their proliferation in 5% fetal bovine serum medium (A and B; n = 8), responses to different concentrations of EGCG at 48 h (C and D; n = 8) and growth abilities on the semi-solid medium (E and F; n = 3). *indicates statistical difference from the controls at each time point (A and B) or concentration of EGCG (C and D) (Student’s t-test; P < 0.05).
The regulation of miR-210 by EGCG is mediated through the HRE in the promoter of miR-210
MiR-210 is the major miRNA that is induced in response to hypoxia (35). HRE is a predominant regulatory element in miR-210 gene promoter and conserved among animal species (35). To explore whether HRE plays a role in the response to EGCG, we examined the responses of the −2 kb of mouse and −600 bp of human mi-210 gene promoter-driven luciferase reporters, both including HRE, to EGCG treatment. Because it is difficult to transfect CL13 cells, we transfected human lung cancer cell lines, H1299 and H460, which are known to be responsive to EGCG (29,40), with these reporters and found that EGCG treatment increased the activity of both mouse and human miR-210 gene promoters in H1299 and H460 (Figure 3A and B) cells in a dosage-dependent manner. This result is consistent with the above microarray results and suggests that EGCG activates both mouse and human miR-210 by a common mechanism.
To further define the involvement of HRE in the EGCG response, we analyzed the responses of truncated/mutated mouse miR-210 promoters, including a shorter fragment containing mouse miR-210 HRE, a shorter fragment without HRE, a 40 bp fragment containing HRE and a same 40 bp fragment with mutated HRE (mutHRE) (as illustrated in Figure 3C). By comparing their responses to EGCG in both H1299 and H460 cells, we found that the fragment without HRE lost response to EGCG, whereas a 40 bp fragment with intact HRE remained to be responsive (Figure 3C). In contrast, when the HRE was mutated, the 40 bp fragment did not response to EGCG. These results supported the concept that EGCG activates the major hypoxia miRNA, miR-210, through the HRE in its gene promoter.
Next, we determined whether EGCG activates putative HRE by examining the response of a luciferase reporter driven by a well-characterized HRE in phosphoglycerate kinase gene in H1299 cells. We found that EGCG treatment increased HRE activity but not its mutated form (mutHRE) (Figure 4A). The upregulation of HRE activity by EGCG displayed the dose-dependent response by linear regression analysis (R2 = 0.996; P = 0.0001). To assess the involvement of HIF proteins, we performed experiments with the addition of HIF-1β plus HIF-1α or HIF-2α. The activities of HRE-luciferase in responses to EGCG were increased from 2000–4500 U to 8000–40 000 U with the addition of HIF-1α expression vector or 12 000–33 000 U with the addition of HIF-2α expression vector, which suggests that the HRE-luciferase response in the contransfection experiment is mainly resulted from exogenous HIFs. The result showed that EGCG enhanced the activities of both HIF-1α and HIF-2α in dose-dependent manner by linear regression analysis (R2 = 0.989, P < 0.0001 for HIF-1α and R2 = 0.938, P = 0.0066 for HIF-2α) (Figure 4B). These data demonstrated that the upregulation of HRE reporter by EGCG treatment involves the EGCG-enhanced HIF activity.
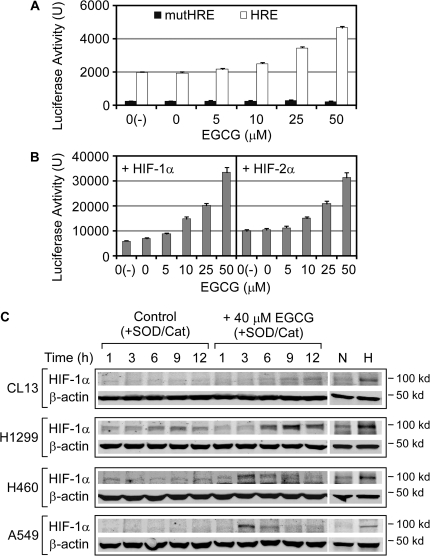
Fig. 4.
EGCG enhanced the activities of HIF-1α and HIF-2α. The upregulation of the HRE-luciferase reporter by EGCG was determined in H1299 cells: (A) transfected with phosphoglycerate kinase HRE or mut-HRE-luciferase reporter plus Rellina-luciferase as internal control (n = 3) and (B) transfected with the HIF-1α or HIF-2α expression vector. (C) HIF-1α levels in lung cancer cell lines after treatment with EGCG (40 μM) were determined by western blots. The cells without treatment were also collected at the match time points as the controls. Sample loading was monitored by β-actin level. As negative and positive controls for determining the hypoxia-induced accumulation of HIF-1α, protein samples collected from cells grown under normoxia (N) and hypoxia (H) were used.
Based on above result, we then investigated HIFα protein levels by western blots in lung cancer cell lines, CL13, H1299 and H460, which displayed the upregulated miR-210 and/or HRE activity in the above experiments. We found that EGCG treatment increased HIF-1α level, whereas HIF-1β level remained unchanged (Figure 4C). Even in cells such as A549 that is less responsive to EGCG, a shorter transient induction of HIF-1α was found. As for HIF-2α, we could not detect any signal in all these samples by western blots, which might be due to the sensitivity of the antibody. Overall, these results supported the hypothesis that EGCG increases HIF-1α protein, which subsequent activates miR-210.
EGCG is able to target the ODD domain of HIFα protein
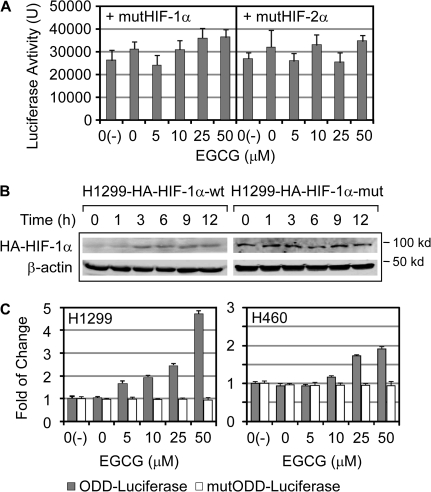
Previous studies suggested that EGCG can either activate (7–9) or inhibit (3–6) the activity of HIF-1α. We performed a series of experiments to determine the effect of EGCG treatment on HIF-1α and to investigate whether the ODD domain is involved. Firstly, we performed the HRE-luciferase experiment by using the P402A/P564A mutant form of HIF-1α (mutHIF1α) and P405A/P531A mutant form of HIF-2α (mutHIF2α). Because the hydroxylation of these Pro residues in ODD domain is required for the proteasome-mediated degradation of HIFα, the protein levels of mutHIF1α and mutHIF2α are not expected to be affected this modification. We found that mutHIFαs lost their responses to EGCG (Figure 5A), suggesting that the increased activity of HIFα by EGCG requires the participation of these key Pro residues. To obtain additional evidence, we established cells stably expressing HA-tagged wild-type HIF-1α and mutHIF1α so that the transcription level of HIF protein is not driven by its native/endogenous promoter. We found that EGCG treatment increased the HA-HIF-1α but not the HA-mutHIF1α level (Figure 5B). Therefore, we concluded that the protein stabilization controlled by Pro hydroxylation is affected by EGCG. Furthermore, we determined whether EGCG can stabilize ODD-luciferase fusion reporter, of which the fusion protein level indicated by the luciferase activity is controlled by ODD domain. After ODD-luciferase was introduced into H1299 and H460 cells, we determined the luciferase activity and found that the activity of ODD-luciferase was upregulated by increasing doses of EGCG (Figure 5C) and this upregulation is dose dependent by linear regression analysis (R2 = 0.969, P = 0.002 in H1299 cells and R2 = 0.895, P < 0.015 in H460 cells). In contrast, EGCG treatment showed no effect on the activity of Pro402A/Pro564A mutant (mutODD)-luciferase fusion protein (Figure 5C). Together, our results supported the conclusion that protein degradation regulated by Pro hydroxylation in ODD domain is targeted by EGCG.
Fig. 5.
The ODD domain was responsive to EGCG. (A) H1299 cells transfected with the mutHIF1α or mutHIF2α expression vector with HRE-luciferase reporter were incubated with EGCG for 12 h and luciferase activity were measured. (B) H1299 cells stably expressing HA-tagged HIF-1α or mutHIF1α (H1299-HA-HIF-1α-wt and -mut) were treated with EGCG (40 μM) and analyzed for HA-HIF-1α and -mutHIF1α levels by western blots using anti-HA antibody. Sample loading was monitored by β-actin level. (C) The ODD-luciferase fusion protein or mutODD-luciferase expression vector was transfected into H1299 and H460 cells with Renilla-luciferase as the internal control. Luciferase activity of ODD- or mutODD-luciferase was normalized by Renilla-luciferase activity (n = 3).
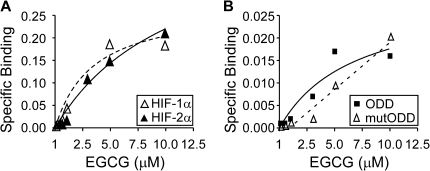
Pro residues were shown involved in binding polyphenols, including EGCG, in studies on the interaction between salivary Pro-rich proteins and polyphenols (41–43). Previously, we also suggested that Pro-rich domain is the target for EGCG to interfere Raf1-MEK1 and Erk1/2-Elk1 interactions (44). Therefore, we hypothesized that EGCG can directly bind to these key Pro residues in HIF-1α. In affinity binding assay, we found that EGCG bound to HIF-1α and 2α with Kd of 3.47 and 11.1 μM, respectively (Figure 6A). Importantly, we found that EGCG only display specific binding to HIFα proteins prepared using mammalian cells but not GST-HIFα proteins prepared in bacteria, possibly due to the incorrect folding. To further demonstrate that EGCG targets the ODD domain, we performed affinity binding assays using the purified Flag-HIF-1α-ODD and the Pro402A/Pro564A mutant (mutODD) and found that EGCG bound to Flag-ODD with Kd of 6.77 μM, but not to Flag-mutODD (Figure 6B). These results demonstrated the physical binding of EGCG to HIFα, which involves the key Pro residues in the ODD domain of HIF-1α that regulate the stabilization of HIFα proteins.
Fig. 6.
EGCG directly bound HIFα proteins. (A) HA-tagged HIF-1α and 2α proteins bound to the anti-HA agarose beads were used to study the binding of EGCG. The beads without HIF protein were used to determine the non-specific binding. Specific binding after correction for non-specific binding was plotted in the figure. The empty and solid triangles represent the specific binding of EGCG on HIF-1α and 2α, respectively. The dashed and solid line lines represent the saturation curves by non-linear regression analysis. (B) The specific binding of EGCG to Flag-ODD or Flag-mutODD bound to the anti-Flag agarose beads was measured. The solid square and solid line represent the specific binding of EGCG on ODD. The empty triangle and dashed line represent the binding of EGCG on mutODD.
Discussion
In this study, we demonstrated, for the first time, that EGCG increases the level of miR-210 in human and mouse lung cancer cells, and the enhanced level of miR-210 resulted in a significant reduction in proliferation and anchorage-independent growth. The increased level of miR-210 was found to be due to the stabilization of HIF-1α protein through EGCG binding to its ODD domain, presumably interfering with Pro hydroxylation that is required for the degradation of HIF-1α. The result revealed a novel route for EGCG to regulate cellular activity by targeting HIFα protein and miR-210.
Ectopical expression of miR-210 in lung cancer cell line H1299 shows a major role of miR-210 in inhibiting cell proliferation and anchorage-independent growth. This result is consistent with the tumor suppressor activity of miR-210 found in the xenograft models with pancreatic tumor cells (35) and esophageal squamous cell carcinoma (39). Some miRNAs display tumor suppression or oncogenic activity by suppressing the expression of oncogenes (e.g. K-ras) or tumor suppressor (e.g. p53 and PTEN) (45). MiR-210 is a new player and it directly targets 51 genes, including FGFRL1, HOXA1 and HOXA9 (35), which are known to play roles in regulating cell growth, differentiation and proliferation. In particular, FGFRL1 is recently reported to be targeted by miR-210 for inhibiting cancer by inducing cell cycle arrest and cell death (39). At present, it is not clear which miR-210 target plays predominant role in mediating the action of EGCG. In addition to the studies of miR-210, we also performed a microarray experiment aiming to identify EGCG-targeted genes. In lung cancer H460 cells treated with EGCG, there are only 30 genes being upregulated by at least 1-fold at 24 h and it includes several known HIF-1α upregulated genes such as vascular endothelial growth factor, very low-density lipoprotein receptor, MIX1, and BNIP3 (unpublished data, H. Wang and C.S. Yang). Thus, the stabilization of HIF-1α by EGCG is part of mechanism of EGCG regulation on the expression of genes including miRNA. More, miR-210 targets could co-operate with other HIF-1α targets to inhibit cell proliferation. Further study toward this direction is needed. Thus, the activation of HIF-1α and miR-210 could at least in part contribute to the anticancer activity of EGCG.
Since the activation of HIF-1α and miR-210 is a direct action of EGCG, they are likely to be immediate cellular responses to EGCG treatment. The effective concentrations of EGCG used in this study, 25–50 μM, are higher than those can be observed in blood and tissues of animals after administration of EGCG or green tea preparations (2). As we discussed previously, the effective concentrations of EGCG, against the same cell lines, observed in cell culture system were much higher than those observed in a xenograft model, possibly due to the short expose time and other conditions of the cell culture system which do not mimic the in vivo situation (29). Therefore, the concept that the upregulation of miR-210 contributes to the anticancer activity of EGCG should be further evaluated in animal model.
Our result, along with other reports (35,39), demonstrates the tumor suppressor activity of miR-210. This finding also suggests an anticancer role of HIFα protein. Published results, however, suggest that HIFα protein plays dual roles in tumorigenesis. On one hand, the overexpression or accumulation of HIFα protein is associated with oncogenic events by upregulating vascular endothelial growth factor expression and promoting the tumor-associated angiogenesis, which is a mechanism for cancer cells to survive through the hypoxia in the tumor microenvironment (10–13). On the other hand, there is an evidence for the protective roles of HIFα in against tumorgenesis. For example, a study using HIF-1α null cells has found that HIF-1α is required for the induction of p27 and cell cycle arrest (46) and a study using HIF-2α knockout animal has shown that loss of HIF-2α dramatically increases tumor progression in K-ras-driven mouse lung cancer model (47). In addition, HIF-1α can co-operate with either p53-related apoptotic machinery to mediate tumor cell death in response to stress (48) or with Myc to induce p21 and cell cycle arrest (49). It appears that, depending on the cellular context such as the presence and the absence of p53 or Myc, HIF-1α can promote or inhibit tumorigenesis (50). In our experimental system, the stabilization of HIF-1α and upregulation of miR-210 by EGCG represents one type of the cellular responses and could contribute to the anticancer activity of EGCG.
Concerning molecular mechanism of EGCG regulation of the HIF-1α level, our result shows that EGCG binds to the ODD domain, resulting in the stabilization of HIF-1α presumably by inhibiting the hydroxylation of key Pro residues. This is consistent with previous reporters supporting that EGCG induces HIF-1α by inhibiting of the prolyl-hydroxylation of HIF-1α and prevents the interaction between HIF-1α and von Hippel–Lindau, resulting in the stabilization of HIF-1α (7–9). EGCG at concentrations >20 μM inhibits PHD activity in prostate cancer cell line PC3 presumably by chelating iron required for PHD activity (7). In neuroblastoma cell line SH-SY5Y, 0.1–1 μM EGCG has been reported to increase the levels of HIF-1α by decreasing the expression of β subunit of prolyl-4-hydroxylase, an enzyme that controls the hydroxylation of HIFα protein (8). An analog of EGCG, epicatechin-3-gallate, has been reported to inhibit prolyl-4-hydroxylase by chelating iron in breast cancer T47D cells at 100 μM (9). On the other hand, it has been reported that 50 μM EGCG inhibits the induction of HIF-1α by hypoxia due to blocking the activation of PI3K/Akt and extracellular signal-regulated kinase 1/2 by hypoxia in Hela and HepG2 cells (5) as well as in colon cancer SW837 cells (4). Therefore, the effect of EGCG on HIF-1α is dependent on the cell line and experimental conditions used.
In summary, we demonstrated, for the first time, that EGCG induces the expression of miR-210 in lung cancer cells and this is due to stabilization of HIF-1α protein. We have further provided the evidence that overexpressed miR-210 inhibits the proliferation and anchorage-independent growth of lung cancer cells. The upregulation of miR-210 reflects an earlier response when cancer cells are exposed to EGCG and could contribute to the anticancer activity of EGCG. Whether the upregulation of miR-210 is a key cancer preventive or anticancer mechanism of EGCG in animal models remains to be investigated.
Funding
This work was supported by the National Institutes of Health (CA120915, CA122474, CA128613 and CA133021) and by core facilities supported by National Cancer Institute Cancer Center Support Grant (P30CA72720) and the National Institute of Environmental Health Sciences center grant (ES05022).
Acknowledgments
We thank Dr S.Belinsky and Dr Y.Hara for providing research materials. We are also grateful to Dr Andrew Brooks at Bionomics Research and Technology Center, EOSHI at Rutgers, the State University of New Jersey for technical support.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- EGCG
(−)-epigallocatechin-3-gallate
- HIF
hypoxia-induced factor
- HRE
hypoxia-response element
- mRNA
messenger RNA
- miRNA
microRNA
- MTT
3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide
- ODD
oxygen-dependent degradation
- PCR
polymerase chain reaction
- PHD
prolyl-hydroxylase
- SOD
superoxide dismutase
References
- 1.Khan N, et al. Targeting multiple signaling pathways by green tea polyphenol (-)-epigallocatechin-3-gallate. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 2.Yang CS, et al. Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat. Rev. Cancer. 2009;9:429–439. doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang G, et al. Green tea catechins augment the antitumor activity of doxorubicin in an in vivo mouse model for chemoresistant liver cancer. Int. J. Oncol. 2010;37:111–123. [PubMed] [Google Scholar]
- 4.Shimizu M, et al. (-)-Epigallocatechin gallate inhibits growth and activation of the VEGF/VEGFR axis in human colorectal cancer cells. Chem. Biol. Interact. 2010;185:247–252. doi: 10.1016/j.cbi.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q, et al. Green tea extract and (-)-epigallocatechin-3-gallate inhibit hypoxia- and serum-induced HIF-1alpha protein accumulation and VEGF expression in human cervical carcinoma and hepatoma cells. Mol. Cancer Ther. 2006;5:1227–1238. doi: 10.1158/1535-7163.MCT-05-0490. [DOI] [PubMed] [Google Scholar]
- 6.Domingo DS, et al. Anti-angiogenic effects of epigallocatechin-3-gallate in human skin. Int. J. Clin. Exp. Pathol. 2010;3:705–709. [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas R, et al. Epigallocatechin gallate inhibits HIF-1alpha degradation in prostate cancer cells. Biochem. Biophys. Res. Commun. 2005;334:543–548. doi: 10.1016/j.bbrc.2005.06.114. [DOI] [PubMed] [Google Scholar]
- 8.Weinreb O, et al. A novel approach of proteomics and transcriptomics to study the mechanism of action of the antioxidant-iron chelator green tea polyphenol (-)-epigallocatechin-3-gallate. Free Radic. Biol. Med. 2007;43:546–556. doi: 10.1016/j.freeradbiomed.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Zhou YD, et al. Hypoxia-inducible factor-1 activation by (-)-epicatechin gallate: potential adverse effects of cancer chemoprevention with high-dose green tea extracts. J. Nat. Prod. 2004;67:2063–2069. doi: 10.1021/np040140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaelin WG, Jr., et al. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Poon E, et al. Targeting the hypoxia-inducible factor (HIF) pathway in cancer. Expert Rev. Mol. Med. 2009;11:e26. doi: 10.1017/S1462399409001173. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 13.Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 14.Bruick RK. Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev. 2003;17:2614–2623. doi: 10.1101/gad.1145503. [DOI] [PubMed] [Google Scholar]
- 15.Bruick RK, et al. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 16.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du T, et al. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs. genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 22.Guo H, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosik KS. MicroRNAs and cellular phenotypy. Cell. 2010;143:21–26. doi: 10.1016/j.cell.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, et al. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, et al. Uncovering growth-suppressive microRNAs in lung cancer. Clin. Cancer Res. 2009;15:1177–1183. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 27.Harris KS, et al. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl Acad. Sci. USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li GX, et al. Pro-oxidative activities and dose-response relationship of (-)-epigallocatechin-3-gallate in the inhibition of lung cancer cell growth: a comparative study in vivo and in vitro. Carcinogenesis. 2010;31:902–910. doi: 10.1093/carcin/bgq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou Z, et al. Mechanism of action of (-)-epigallocatechin-3-gallate: auto-oxidation-dependent inactivation of epidermal growth factor receptor and direct effects on growth inhibition in human esophageal cancer KYSE 150 cells. Cancer Res. 2005;65:8049–8056. doi: 10.1158/0008-5472.CAN-05-0480. [DOI] [PubMed] [Google Scholar]
- 31.Pear WS, et al. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernardi R, et al. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 33.Kondo K, et al. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 34.Yan Q, et al. The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol. Cell. Biol. 2007;27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, et al. Positive and negative modulation of the transcriptional activity of the ETS factor ESE-1 through interaction with p300, CREB-binding protein, and Ku 70/86. J. Biol. Chem. 2004;279:25241–25250. doi: 10.1074/jbc.M401356200. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, et al. RhoH plays distinct roles in T-cell migrations induced by different doses of SDF1 alpha. Cell Signal. 2010;22:1022–1032. doi: 10.1016/j.cellsig.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Shim JH, et al. (-)-Epigallocatechin gallate regulates CD3-mediated T cell receptor signaling in leukemia through the inhibition of ZAP-70 kinase. J. Biol. Chem. 2008;283:28370–28379. doi: 10.1074/jbc.M802200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchiya S, et al. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J. Biol. Chem. 2010;2010:2. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu G, et al. Synergistic inhibition of lung tumorigenesis by a combination of green tea polyphenols and atorvastatin. Clin. Cancer Res. 2008;14:4981–4988. doi: 10.1158/1078-0432.CCR-07-1860. [DOI] [PubMed] [Google Scholar]
- 41.Murray NJ, et al. Study of the interaction between salivary proline-rich proteins and a polyphenol by 1H-NMR spectroscopy. Eur. J. Biochem. 1994;219:923–935. doi: 10.1111/j.1432-1033.1994.tb18574.x. [DOI] [PubMed] [Google Scholar]
- 42.Baxter NJ, et al. Multiple interactions between polyphenols and a salivary proline-rich protein repeat result in complexation and precipitation. Biochemistry. 1997;36:5566–5577. doi: 10.1021/bi9700328. [DOI] [PubMed] [Google Scholar]
- 43.Canon F, et al. Ability of a salivary intrinsically unstructured protein to bind different tannin targets revealed by mass spectrometry. Anal. Bioanal. Chem. 2010;398:815–822. doi: 10.1007/s00216-010-3997-9. [DOI] [PubMed] [Google Scholar]
- 44.Chung JY, et al. Mechanisms of inhibition of the Ras-MAP kinase signaling pathway in 30.7b Ras 12 cells by tea polyphenols (-)-epigallocatechin-3-gallate and theaflavin-3,3'-digallate. FASEB J. 2001;15:2022–2024. doi: 10.1096/fj.01-0031fje. [DOI] [PubMed] [Google Scholar]
- 45.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goda N, et al. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol. Cell. Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazumdar J, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc. Natl Acad. Sci. USA. 2010;107:14182–14187. doi: 10.1073/pnas.1001296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sumiyoshi Y, et al. Overexpression of hypoxia-inducible factor 1alpha and p53 is a marker for an unfavorable prognosis in gastric cancer. Clin. Cancer Res. 2006;12:5112–5117. doi: 10.1158/1078-0432.CCR-05-2382. [DOI] [PubMed] [Google Scholar]
- 49.Koshiji M, et al. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang CV, et al. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]