Summary
Huntington’s disease (HD) is caused by polyglutamine expansion (exp) in huntingtin (Htt). The type 1 inositol (1,4,5)-triphosphate receptor (InsP3R1) is an intracellular calcium (Ca2+) release channel that plays an important role in neuronal function. In a yeast two-hybrid screen with the InsP3R1 carboxy terminus, we isolated Htt-associated protein-1A (HAP1A). We show that an InsP3R1-HAP1A-Htt ternary complex is formed in vitro and in vivo. In planar lipid bilayer reconstitution experiments, InsP3R1 activation by InsP3 is sensitized by Httexp, but not by normal Htt. Transfection of full-length Httexp or caspase-resistant Httexp, but not normal Htt, into medium spiny striatal neurons faciliates Ca2+ release in response to threshold concentrations of the selective mGluR1/5 agonist 3,5-DHPG. Our findings identify a novel molecular link between Htt and InsP3R1-mediated neuronal Ca2+ signaling and provide an explanation for the derangement of cytosolic Ca2+ signaling in HD patients and mouse models.
Introduction
Huntington’s disease (HD) has onset usually between 35 and 50 years with chorea and psychiatric disturbances and gradual but inexorable intellectual decline to death after 15–20 years (Vonsattel and DiFiglia, 1998). Neuropathological analysis reveals selective and progressive neuronal loss in the striatum (Vonsattel et al., 1985), particularly affecting the GABAergic medium spiny striatal neurons (MSNs). At the molecular level, the cause of HD is a polyglutamine (polyQ) expansion (exp) in the amino terminus of huntingtin (Htt), a 350 kDa ubiquitously expressed cytoplasmic protein (HDCRG, 1993; Nasir et al., 1996). A number of transgenic HD mouse models have been generated, which reproduce many HD-like features (Menalled and Chesselet, 2002; Rubinsztein, 2002). Despite significant progress, cellular mechanisms that link the mutation with the disease remain controversial (Tobin and Signer, 2000).
A number of Htt binding partners have been identified in yeast two-hybrid (Y2H) screens with an Htt amino-terminal fragment (Gusella and MacDonald, 1998; Kalchman et al., 1997; Singaraja et al., 2002). Htt-associated protein-1 (HAP1) was the first identified Htt binding partner (Li et al., 1995; Gutekunst et al, 1998; Page at al, 1998). Importantly, the HD-causing polyQ expansion of Htt (Httexp) promotes Htt-HAP1 association (Li et al., 1995, 1998b). In rodents, two HAP1 protein isoforms differing in their carboxy termini are expressed via alternative splicing —HAP1A and HAP1B—both of which bind Htt (Li et al., 1995; Nasir et al., 1998). Only one HAP1 isoform has been identified in humans, and this is most similar to rodent HAP1A (Li et al., 1998b). Association of HAP1 with hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) (Li et al., 2002), the p150Glued subunit of dynactin (Engelender et al., 1997; Li et al., 1998a), and the Rac1 guanine nucleotide exchange factor Kalirin-7/Duo (Colomer et al., 1997) has been discovered in Y2H screens. Targeted disruption of the HAP1 gene in mice results in postnatal death and depressed feeding behavior, suggesting an important role of HAP1 in hypothalamic function (Chan et al., 2002). Despite all these data, the role of HAP1 in neuronal signaling and the pathogenesis of HD remain unclear (Bertaux et al., 1998).
The inositol (1,4,5)-triphosphate receptor (InsP3R) is an intracellular calcium (Ca2+) release channel that plays an important role in neuronal Ca2+ signaling (Berridge, 1998). Three isoforms of InsP3R have been identified (Furuichi et al., 1994). The type 1 receptor (InsP3R1) is the predominant neuronal isoform. Mice lacking InsP3R1 display severe ataxic behavior (Matsumoto et al., 1996), and mice with a spontaneous mutation in the InsP3R1 gene experience convulsions and ataxia (Street et al., 1997), suggesting a major role of InsP3R1 in neuronal function. To identify novel InsP3R1 neuronal binding partners, we performed a Y2H screen of a rat brain cDNA library and isolated neuronal cytoskeleton 4.1N protein (Maximov et al., 2003) and HAP1A. In the present manuscript, we describe biochemical and functional interactions of InsP3R1 with HAP1A and Htt. The discovered association of InsP3R1 with HAP1A and Htt provides additional insight into HAP1 function in the brain and to our knowledge for the first time links neuronal InsP3R1 function with Htt and HD.
Results
InsP3R1 Binds to HAP1A in the Yeast Two-Hybrid System
We aimed to identify novel proteins that bind to the carboxy-terminal cytosolic region of the InsP3R1. We used the carboxy-terminal region of rat InsP3R1 in the pLexN vector (IC bait, amino acids D2590–A2749) for Y2H screening of a rat brain cDNA library and isolated 16 positive clones. The clones were rescued and tested for their strength of association with the IC bait in a liquid Y2H assay. Three of the 16 isolated clones displayed especially strong interaction with the IC bait. Two of the strongly interacting clones (clones 7 and 8) corresponded to neuronal cytoskeleton protein 4.1N (Maximov et al., 2003). The third strongly interacting clone (clone 11) corresponded to a partial fragment of HAP1A.
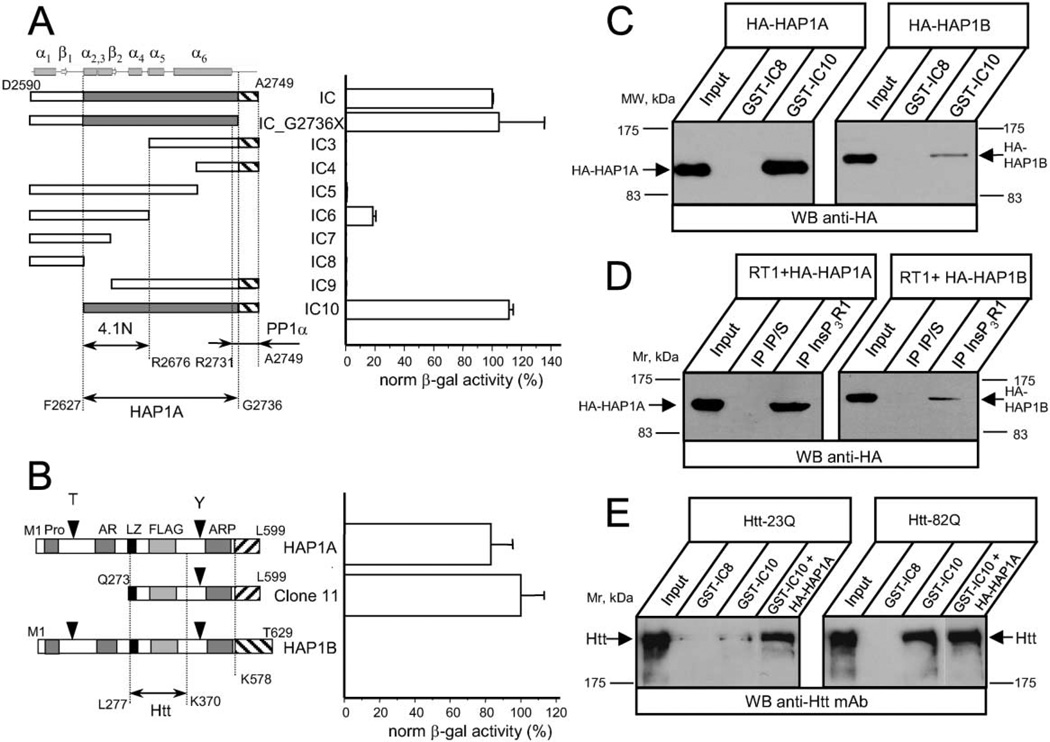
To systematically map the HAP1A-interacting domain, a number of InsP3R1 carboxy-terminal fragments were cloned into pLexN vector, and the strength of interaction with clone 11 (partial HAP1A) was measured in a liquid Y2H assay (Figure 1A). From the obtained results we concluded that the minimal InsP3R1 region required for interaction with HAP1A corresponds to amino acids F2627–G2736 (Figure 1A, shaded). Thus, the HAP1A-interacting domain in the InsP3R1 carboxy-terminal region partially overlaps with the minimal 4.1N-interacting region (F2627–R2676; Figure 1A) and largely complements the minimal protein phosphatase 1α (PP1α) binding region (R2731–A2749; Figure 1A) mapped in our previous studies (Maximov et al., 2003; Tang et al., 2003).
Figure 1. The InsP3R1 Binds HAP1A and Htt in the Yeast Two-Hybrid System and In Vitro.
(A) The strength of interaction between each InsP3R1 IC bait and clone 11 (Q273–L599 of rat HAP1A) was determined by a liquid Y2H assay. The β-galactosiadase activity is plotted as a percentage relative to the IC/clone 11 interaction (mean ± SE, n = 3). On the top, predicted secondary structure elements within the IC sequence are shown. The predicted minimal fragment of the rat InsP3R1 carboxy terminus (F2627–G2736) required for association with HAP1A is shaded. The minimal 4.1N binding domain (F2627–R2676) (Maximov et al., 2003) and minimal PP1α binding region (R2731–A2749) (Tang et al., 2003) are also shown.
(B) The strength of interaction between full-length rat HAP1A, rat HAP1B, and clone 11 with IC10 InsP3R1 bait was determined by liquid Y2H assay. Data are presented in percentages relative to the observed interaction between clone 11 and IC10 (mean ± SE, n = 3). The carboxy-terminal regions unique to HAP1A and HAP1B isoforms are depicted by striped boxes. Also shown are the Htt binding region of HAP1 (L277–K370) (Li et al., 1995, 1998b) and motifs identified in HAP1 by computer analysis.
(C) GST-IC8/IC10 pull-down experiments of HA-HAP1A or HA-HAP1B from COS7 cells extracts.
(D) Lysates from RT1-infected Sf9 cells and COS7 cells overexpressing either HA-HAP1A or HA-HAP1B were mixed for 2 hr and analyzed by immunoprecipitation with anti-InsP3R1 polyclonal antibodies or corresponding preimmune sera (IP/S).
(E) GST-IC8/IC10 pull-down experiments of Htt-23Q or Htt-82Q from HEK293 cells extracts. COS7 lysate containing overexpressed HA-HAP1A was added to GST-IC10 pull-down reactions as indicated. The input lanes on (C)–(E) contain 1/50 of the COS7 and HEK293 cell lysates used in GST pull-down or immunoprecipitation experiments. The precipitated fractions were analyzed by Western blotting with anti-HA monoclonal antibodies (C and D), or anti-Htt monoclonal antibodies (E).
Two carboxy-terminal splice variants of HAP1 protein are expressed in the rodent brain (Li et al., 1995). Clone 11 corresponds to a carboxy-terminal portion of the rat HAP1A isoform. To address whether both HAP1 isoforms bind InsP3R1 with similar affinity, we cloned full-length rat HAP1A and HAP1B cDNAs into the Y2H vectors and tested their ability to associate with IC10 bait. We observed strong interaction between IC10 and either full-length HAP1A or clone 11 (Figure 1B). In contrast, the HAP1B isoform did not display any detectable inter-action with IC10. Thus, unique carboxy-terminal sequences present in HAP1A protein (K578–L599) appear to robustly modulate association with the IC10 bait.
InsP3R1 Binds to HAP1A and Htt In Vitro
To confirm the InsP3R1-HAP1 association, we performed a series of in vitro binding experiments. In the first experiments, we expressed GST-IC8 and GST-IC10 InsP3R1 carboxy-terminal proteins in E. coli and performed GST pull-downs with extracts from COS7 cells transiently expressing hemagglutinin-tagged HAP1 proteins (HA-HAP1A and HA-HAP1B). In agreement with Y2H data, GST-IC10, but not GST-IC8, specifically associated with HA-HAP1A protein (Figure 1C). In further agreement with Y2H results, GST-IC10 association with HA-HAP1B was much weaker than that with HA-HAP1A (Figure 1C).
Do HAP1 proteins bind to full-length InsP3R1? To address this, we expressed full-length InsP3R1 (RT1) in Sf9 cells by baculovirus infection and performed in vitro binding experiments using lysates from COS7 cells transiently transfected with HA-HAP1A- or HA-HAP1B-expressing plasmids. The Sf9 and COS7 cell lysates were incubated together, precipitated with anti-InsP3R1 polyclonal anti-bodies, and blotted with anti-HA monoclonal antibodies. In agreement with Y2H and GST pull-down experiments, full-length InsP3R1 associated more strongly with HA-HAP1A than with HA-HAP1B (Figure 1D).
Since HAP1 interacts with Htt (Li et al., 1995), we investigated whether an InsP3R1-HAP1A-Htt ternary complex might be formed. To explore this, we expressed full-length Htt-23Q or Htt-82Q proteins (Cooper et al., 1998) in HEK293 cells and performed GST pull-down experiments. We discovered that GST-IC10 protein, but not GST-IC8, efficiently precipitated Htt-82Q proteins from HEK293 cell lysates (Figure 1E). Compared to Htt-82Q, much smaller amounts of Htt-23Q were precipitated by GST-IC10 (Figure 1E). The addition of COS7 cell lysates containing overexpressed HA-HAP1A into the GST-IC10 pull-down reaction significantly increased the amount of precipitated Htt-23Q protein (Figure 1E), but had only a minor effect on the amount of precipitated Htt-82Q protein (Figure 1E). In control experiments we detected the expression of endogenous HAP1A protein in HEK293 cells (data not shown). One explanation of our findings is that Htt-82Q bound endogenous HAP1A with much higher affinity than Htt-23Q. As a result, endogenous levels of HAP1A in HEK293 cells were sufficient to support robust Htt-82Q, but not Htt-23Q, precipitation by GST-IC10. Alternatively, Htt-82Q, but not Htt-23Q, may be exhibiting strong and direct binding to the InsP3R1 carboxy terminus, even in the absence of HAP1A (see Discussion).
InsP3R1-HAP1-Htt Association In Vivo
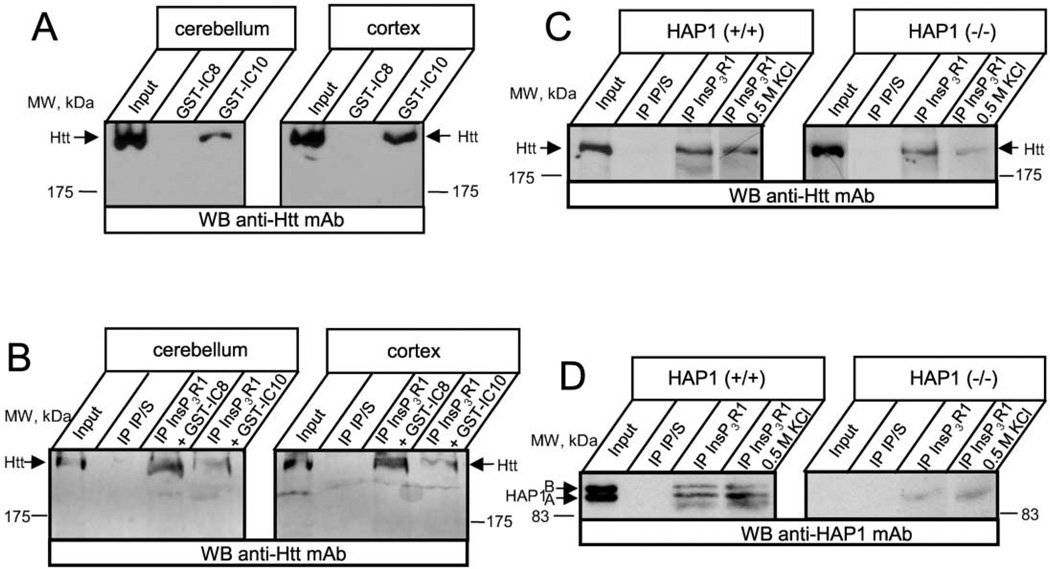
We performed additional GST pull-down experiments with rat cerebellar and cortical lysates. In agreement with data using cultured cells, GST-IC10, but not GST-IC8, precipitated endogenous Htt from rat brain cerebellar and cortical lysates (Figure 2A). To address whether InsP3R1 and Htt interacted in vivo, we performed coimmunoprecipitation experiments with rat brain cerebellar and cortical lysates using anti-InsP3R1 polyclonal antibodies. In agreement with the pull-down data, endogenous Htt protein is coimmunoprecipitated with the InsP3R1, even in the presence of GST-IC8 protein, which was added in the reaction (Figure 2B). In contrast to GST-IC8, inclusion of GST-IC10 protein into anti-InsP3R1 immunoprecipitation reactions significantly reduced the amounts of precipitated Htt (Figure 2B). These results are consistent with the formation of an InsP3R1-HAP1-Htt ternary complex in neurons within rat cerebellum and cortex regions. Similar results were obtained in experiments with rat striatal lysates (data not shown).
Figure 2. InsP3R1 Is Associated with Htt In Vivo.
(A) Rat cerebellar (left) or cortical (right) lysates were used in GST-IC8/IC10 pull-down experiments.
(B) Rat cerebellar (left) or cortical (right) lysates were used in coimmunoprecipitation experiments with anti-InsP3R1 polyclonal antibodies or corresponding preimmune sera (IP/S). GST-IC8 and GST-IC10 proteins (200 µg/ml final concentration) were included in the immunoprecipitation reactions as indicated. The precipitated fractions on (A) and (B) were analyzed by Western blotting with anti-Htt monoclonal antibodies.
(C and D) Cortical lysates from wild-type (left panel) or HAP1−/− (right panel) mice (at postnatal day 3) were used in immunoprecipitation experiments with the anti-InsP3R1 polyclonal antibodies or with the corresponding preimmune sera (IP/S). The precipitated proteins were analyzed by Western blotting with the anti-Htt monoclonal antibodies (C) or with the anti-HAP1 monoclonal antibodies (D). An additional 0.5 M KCl wash step was included in the immunoprecipitation protocol as indicated. The input lanes on (A)–(D) contain 1/50 of lysates used in immunoprecipitation and GST pull-down experiments.
To determine the requirement of HAP1 for the InsP3R1-Htt association, we studied brain extracts from mice with targeted disruption of HAP1 (Chan et al., 2002). In these experiments, brain extracts from HAP1−/− mouse pups and wild-type control littermates were used in immunoprecipitaion experiments with anti-InsP3R1 polyclonal antibodies. The precipitated fractions were blotted with anti-Htt monoclonal antibodies. In agreement with the rat brain coimmunoprecipitation results, we found that anti-InsP3R1 polyclonal antibodies specifically precipitated Htt from wild-type mouse cortical lysates (Figure 2C, left panel, third lane). Slightly reduced amounts of Htt protein were precipitated by anti-InsP3R1 polyclonal antibodies from HAP1−/− cortical lysates (Figure 2C, right panel, third lane). To investigate preferentially strong protein-protein interactions, an additional 0.5 M KCl washing step was employed. The 0.5 M KCl wash did not appreciably decrease the amount of Htt precipitated by anti-InsP3R1 antibodies from wild-type mouse brain extract (Figure 2C, left panel, fourth lane). In contrast, the high-salt wash drastically reduced the amount of Htt precipitated from HAP1−/− brain extracts (Figure 2C, right panel, fourth lane). From these results we concluded that the presence of HAP1 proteins is required for the formation of a high-affinity salt-resistant InsP3R1-Htt association. Western blotting of anti-InsP3R1 immunoprecipitates with HAP1 monoclonal antibodies confirmed the preferential binding of InsP3R1 to the HAP1A isoform (Figure 2D, left panel) and the absence of both HAP1 isoforms in HAP1−/− samples (Figure 2D, right panel). The binding of InsP3R1 to HAP1 was not significantly affected by the 0.5 M KCl wash (Figure 2D, left panel).
Httexp, but Not Normal Htt, Activates InsP3R1 In Vitro
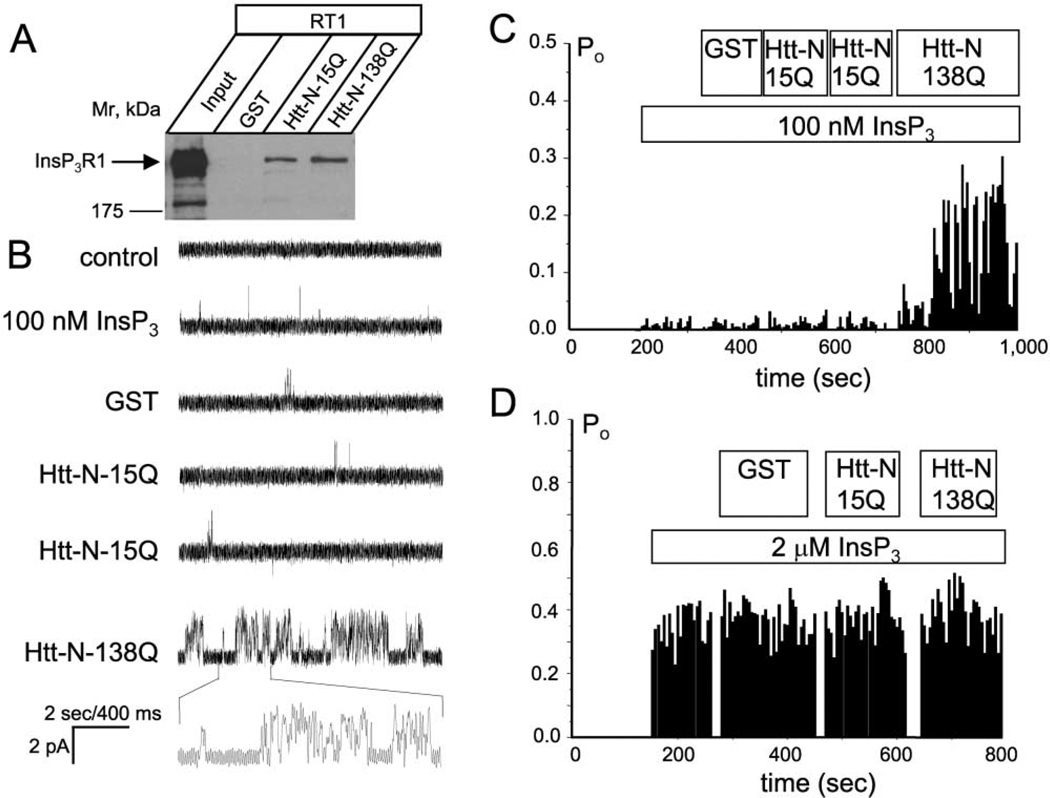
The HAP1 binding site and the site of polyQ expansion are localized to the most amino-terminal region of Htt protein (Li et al., 1995). We expressed amino-terminal fragments (Htt-N, amino acids 1–158) of Htt-15Q and Htt-138Q as GST-fusion proteins and confirmed association of Htt-N-15Q and Htt-N-138Q with HA-HAP1A (data not shown). We further demonstrated that bacterially expressed Htt-N-15Q and Htt-N-138Q proteins specifically precipitated recombinant full-length InsP3R1 from Sf9 cell lysates (Figure 3A). Since Sf9 lysates do not contain any HAP1 (data not shown), this Htt-N interaction with full-length InsP3R1 appears to be direct. Our GST-IC10 pull-down experiments (Figure 1E) and immunoprecipitation experiments from HAP1−/− cortical lysates (Figure 2C) also support the existence of a direct Htt-InsP3R1 interaction. InsP3R1 associated with Htt-N-138Q more strongly than with Htt-N-15Q (Figure 3A), although this difference was less dramatic than the difference between Htt-23Q and Htt-82Q in GST-IC10 pull-down experiments (Figure 1E).
Figure 3. The Httexp Amino Terminus Sensitizes the InsP3R1 to InsP3.
(A) Lysates from full-length InsP3R1 (RT1)-infected Sf9 cells were used in pull-down experiments with GST, Htt-N-15Q, and Htt-N-138Q GST-fusion proteins as indicated. The precipitated fractions were analyzed by Western blotting with anti-InsP3R1 polyclonal antibodies
(B) Effects of GST, Htt-N-15Q, and Htt-N-138Q on activity of recombinant InsP3R1 in planar lipid bilayers at 100 nM InsP3. Each current trace corresponds to 10 s (2 s for expanded traces) of current recording from the same experiment.
(C) The average InsP3R1 open probability (Po) in the presence of 100 nM InsP3 is calculated for a 5 s window of time and plotted for the duration of an experiment. Data from the same experiment are shown on (B) and (C). Similar results were obtained in four independent experiments.
(D) The average InsP3R1 Po plot for the experiment performed in the presence of 2 µM InsP3. Similar results were obtained in two independent experiments. The times of InsP3, GST, Htt-N-15Q, and Htt-N-138Q additions on (C) and (D) are shown above the Po plot.
What are the functional consequences of InsP3R1 association with HAP1 and Htt? To address this, we expressed InsP3R1 in Sf9 cells and reconstituted recombinant InsP3R1 into planar lipid bilayers as previously described (Tang et al., 2003; Tu et al., 2002). Addition of 100 nM InsP3 to the cis (cytosolic) chamber induced low levels of InsP3R1 activity (Figure 3B, second trace, and Figure 3C). Further addition of GST protein or repetitive additions of Htt-N-15Q protein directly to the bilayer had no effect on InsP3R1 activity (Figure 3B, traces 3–5, and Figure 3C). In contrast, addition of Htt-N-138Q protein to the same bilayer resulted in facilitation of InsP3R1 activity (Figure 3B, trace 6, and Figure 3C). On average, InsP3R1 open probability was equal to 0.020 ± 0.008 (n = 16) in the presence of 100 nM InsP3, 0.02 ± 0.01 (n = 6) after addition of Htt-N-15Q, and 0.19 ± 0.05 (n = 4) after addition of Htt-N-138Q. The facilitation of InsP3R1 activity by Htt-N-138Q was most pronounced at low InsP3 concentrations. When InsP3R1 was activated by 2 µM InsP3, additions of GST, Htt-N-15Q, and Htt-N-138Q proteins did not have a significant effect on InsP3R1 open probability (Figure 3D). From these results, we concluded that Htt-N-138Q, but not Htt-N-15Q, sensitizes InsP3R1 to activation by submaximal doses of InsP3.
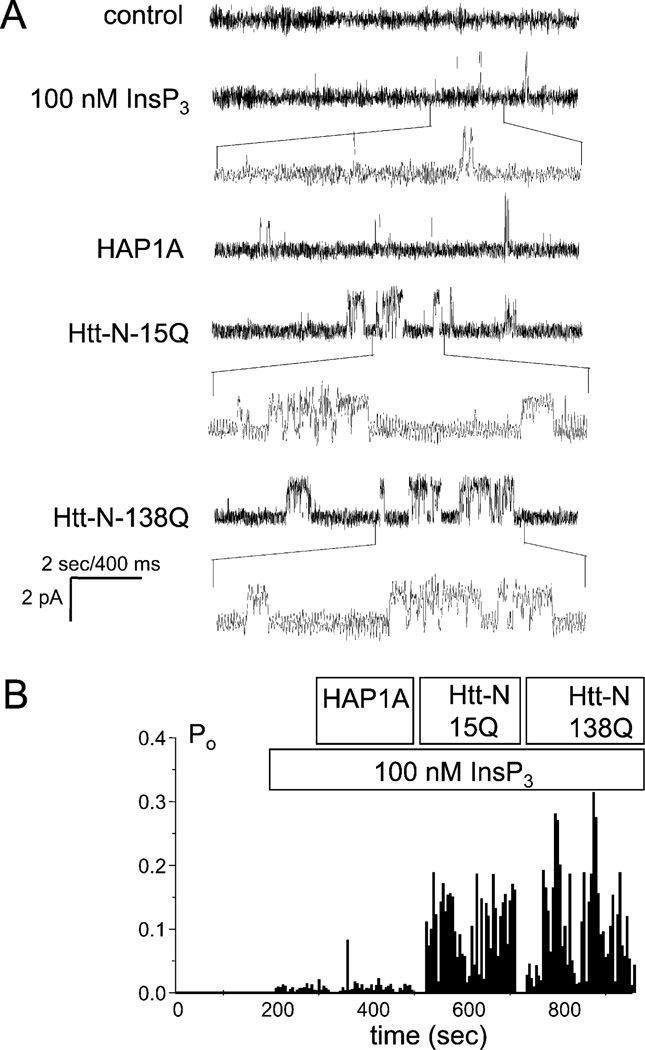
To evaluate the role of HAP1 in InsP3R1 activation by Htt, we expressed and purified full-length HAP1A as a GST-fusion protein. Addition of HAP1A to the bilayer had no effect on the activity of InsP3R1 in the presence of 100 nM InsP3 (Figure 4A, third trace, and Figure 4B). In contrast to previous experiments (Figures 3B and 3C), addition of Htt-N-15Q to the bilayer pre-exposed to HAP1A facilitated InsP3R1 activity (Figure 4A, fourth trace, and Figure 4B). The following addition of Htt-N-138Q to the same membrane had an additional potentiating effect (Figure 4A, trace 5, and Figure 4B). Similar results were obtained in the experiments when HAP1A was premixed with Htt-N-15Q or Htt-N-138Q prior to addition to the bilayer (data not shown). On average, InsP3R1 open probability was equal to 0.020 ± 0.008 (n = 16) in the presence of 100 nM InsP3, 0.14 ± 0.04 (n = 5) after addition of HAP1A + Htt-N-15Q, and 0.26 ± 0.11 (n = 5) after addition of HAP1A + Htt-N-138Q. From these in vitro functional experiments and our biochemical data, we concluded that HAP1A facilitates activation of InsP3R1 by Htt, most likely by promoting Htt association with InsP3R1 carboxy terminus (Figure 1E).
Figure 4. The Htt Amino Terminus Facilitates InsP3R1 Activity in the Presence of HAP1A.
(A) Effects of HAP1A, Htt-N-15Q, and Htt-N-138Q on activity of recombinant InsP3R1 in planar lipid bilayers in the presence of 100 nM InsP3. Each current trace corresponds to 10 s (2 s for expanded traces) of current recording from the same experiment.
(B) The average InsP3R1 open probability (Po) in the presence of 100 nM InsP3 is calculated for a 5 s window of time and plotted for the duration of an experiment. The times of InsP3, HAP1A, Htt-N-15Q, and Htt-N-138Q additions to the bilayer are shown above the Po plot. Data from the same experiment are shown on (A) and (B). Similar results were obtained in two independent experiments or in experiments when HAP1A and Htt-N-15Q (n = 4) or HAP1A and Htt-N-138Q (n = 4) were premixed.
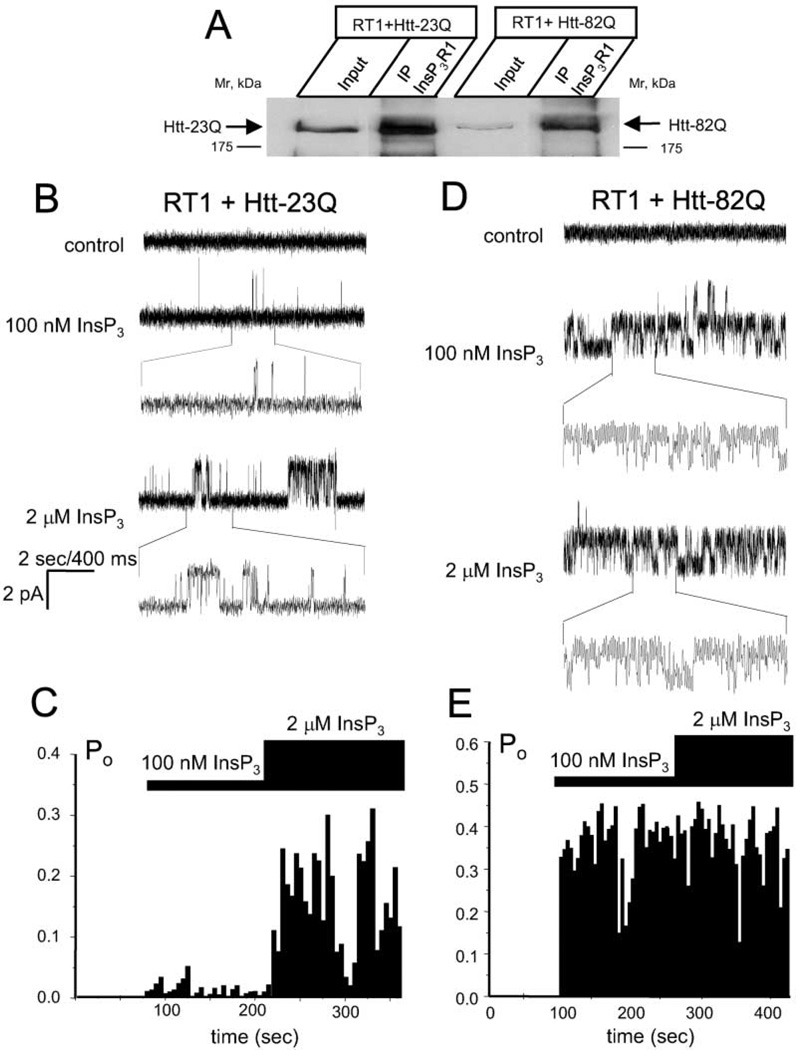
Since amino-terminal Htt fragments were used in bilayer experiments shown above, we determined the effect of full-length Htt on InsP3R1 activity. We generated baculoviruses expressing full-length Htt-23Q and Htt-82Q and used them in coinfection experiments with RT1 baculovirus encoding InsP3R1. In immunoprecipitation experiments we found that InsP3R1 and Htt-23Q/82Q formed a complex when coexpressed in Sf9 cells (Figure 5A), which do not contain any detectable HAP1. As such, interaction of the InsP3R1 with Htt23Q/82Q in Sf9 cells appears direct, consistent with our earlier findings. Similar to pull-down data gathered with the GST-Htt-N protein (Figure 3A), binding of Htt to full-length InsP3R1 was only modestly modulated by polyQ expansion.
Figure 5. Full-Length Httexp Sensitizes InsP3R1 to Activation by InsP3.
(A) Microsomes from Sf9 cells coinfected with RT1 and Htt-23Q or Htt-82Q baculoviruses were solubilzed in 1% CHAPS, precipitated with anti-InsP3R1 polyclonal antibodies, and blotted with the anti-Htt monoclonal antibodies. The input lanes contain 1/10 of lysates used in the immunoprecipitation experiments.
(B) Activity of InsP3R1 coexpressed in Sf9 cells with Htt-23Q and reconstituted into planar lipid bilayers. Responses to application of 100 nM InsP3 and 2 µM InsP3 are shown. Each current trace corresponds to 10 s (2 s for expanded traces) of current recording from the same experiment.
(C) The average InsP3R1 open probability (Po) was calculated for a 5 s window of time and plotted for the duration of an experiment. The times of 100 nM and 2 µM InsP3 additions to the bilayer are shown above the Po plot. Data from the same experiment are shown on (B) and (C). Similar results were obtained in four independent experiments.
(D and E) Same as (B) and (C) for InsP3R1 coexpressed in Sf9 cells with Htt-82Q. Similar results were obtained in six independent experiments.
Microsomes prepared from Sf9 cells coinfected with RT1 and Htt-23Q or Htt-82Q baculoviruses were fused to planar lipid bilayers. No channel activity was observed in control experimental conditions (Figures 5B and 5D, first trace). Addition of 100 nM InsP3 induced only low levels of channel activity in InsP3R1 coexpressed with Htt-23Q (Figure 5B, second trace, and Figure 5C), but resulted in dramatic activation of InsP3R1 coexpressed with Htt-82Q (Figure 5D, second trace, and Figure 5E). Elevation of InsP3 concentration to 2 µM increased activity of InsP3R1 coexpressed with Htt-23Q (Figure 5B, third trace, and Figure 5C) and had no additional effect on activity of InsP3R1 coexpressed with Htt-82Q (Figure 5D, third trace, and Figure 5E). From these experiments we concluded that sensitivity of InsP3R1 to activation by InsP3 is increased by formation of a complex with Htt-82Q full-length protein, but not with Htt-23Q full-length protein.
Httexp Activates InsP3R1 in Cultured MSNs
Striatal MSN neurons are the most selectively and severely affected in HD (Vonsattel et al., 1985). To test the in vivo functional effects of Httexp on InsP3R1, we established primary MSN cultures from E18 embryonic rats (Mao and Wang, 2001). Over 90% of striatal neurons are projection GABAergic MSNs (Gerfen, 1992). A large fraction of cells in our cultures (>90%) were strongly positive for GAD65 marker in immunostaining experiments (data not shown), confirming their identity as MSNs (Chesselet et al., 1993; Mao and Wang, 2001). We transfected MSNs at 20 days in vitro (DIV) with full-length Htt-23Q, Htt-82Q, or Htt-138Q expression plasmids. To identify transfected cells, the Htt plasmids were cotransfected with enhanced green fluorescent protein (EGFP)-expressing plasmid. During transfections, the Htt:EGFP plasmid ratio was kept at 3:1 to ensure that every GFP-positive cell was transfected with Htt-expressing plasmid. In control experiments, MSNs were transfected with the EGFP plasmid alone. Only GFP-positive cells were compared in our analysis of different Htt constructs.
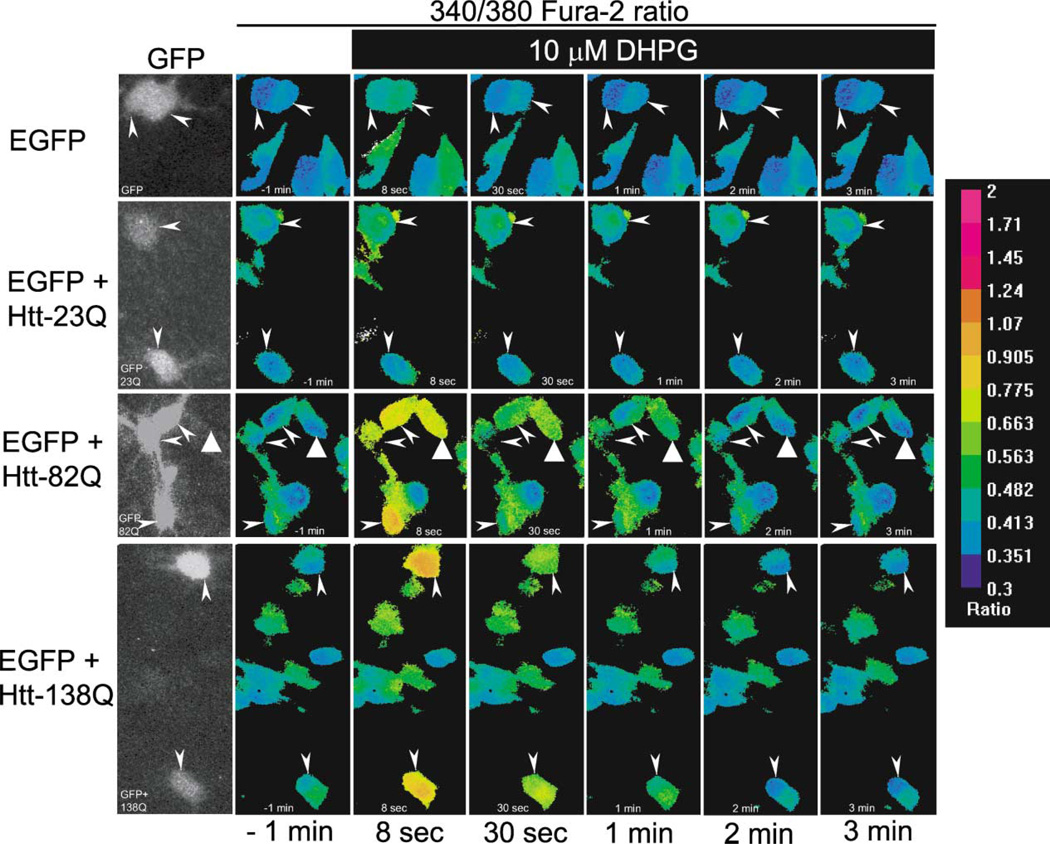
In contrast to striatal interneurons, MSNs abundantly express phospholipase C (PLC)-linked mGluR1/5 receptors (Mao and Wang, 2001, 2002; Tallaksen-Greene et al., 1998). To stimulate InsP3R1-mediated Ca2+ release, we challenged Fura-2-loaded MSN neurons with 10 µM 3,5-dihydroxyphenylglycine (DHPG), a specific mGluR1/5 receptor agonist (Mao and Wang, 2002; Schoepp et al., 1999). To exclude the contribution of N-methyl-D-aspartate receptors (NMDAR) and L-type Ca2+ channels to the observed Ca2+ signals and to simplify the analysis, the imaging experiments were performed in Ca2+-free media containing 100 µM EGTA (see Experimental Procedures for details). The local Ca2+ concentration in these experiments is estimated from the ratio of Fura-2 signals at 340 nm and 380 nm excitation wavelengths as shown by pseudocolor images. Representative data with EGFP, EGFP + Htt-23Q, EGFP + Htt-82Q, and EGFP + Htt-138Q transfected MSN neurons are shown on Figure 6. The transfected cells were identified by GFP imaging (Figure 6, first column, arrows) prior to collecting quantitative Fura-2 340/380 ratio data. We noticed that prior to application of DHPG, the basal Ca2+ levels were slightly elevated in Htt-transfected cells when compared to control cells (Figure 6, second column). On average, the basal 340/380 ratio was equal to 0.43 ± 0.02 (n = 14) for EGFP-transfected cells (Figures 7A and 7I), 0.51 ± 0.02 (n = 18) for EGFP + Htt-23Q, 0.55 ± 0.02 (n = 29) for EGFP + Htt-82Q, and 0.54 ± 0.015 (n = 21) for EGFP + Htt-138Q (Figures 7C, 7E, 7G, and 7I). The basal Ca2+ levels were significantly (p < 0.01, unpaired t test) higher in Htt-transfected MSNs as compared to control MSNs. These results are in agreement with elevated basal Ca2+ levels observed in hippocampal neurons from YAC46 transgenic HD mice (Hodgson et al., 1999).
Figure 6. Effects of Httexp on DHPG-Induced Ca2+ Release in Transfected Medium Spiny Neurons.
Representative images showing Fura-2 340/380 ratios in transfected rat medium spiny neurons (MSNs). The pseudocolor calibration scale for 340/380 ratios is shown on the right. Ratio recordings are shown for 10 µM DHPG-induced Ca2+ transients in MSN neurons transfected with EGFP (first row), EGFP + Htt-23Q (second row), EGFP + Htt-82Q (third row), and EGFP + Htt-138Q (fourth row). The recordings were performed in Ca2+-free ACSF containing 100 µM EGTA. GFP images (1st column) were captured before Ca2+ imaging to identify transfected cells (arrowheads). 340/380 ratio images are shown for MSN neurons 1 min before (2nd column), and 8 s, 30 s, 1 min, 2 min, and 3 min after application of 10 µM DHPG as indicated. The GFP-negative cell that responds to 10 µM DHPG in the third row (triangle arrow) is interpreted to correspond to a neuron transfected with Htt-82Q plasmid alone. We used Htt:EGFP plasmids at a 3:1 ratio during transfections, which probably resulted in the expression of Htt in some GFP-negative MSNs. Only GFP-positive MSNs were considered for quantitative analyses shown on Fig 7.
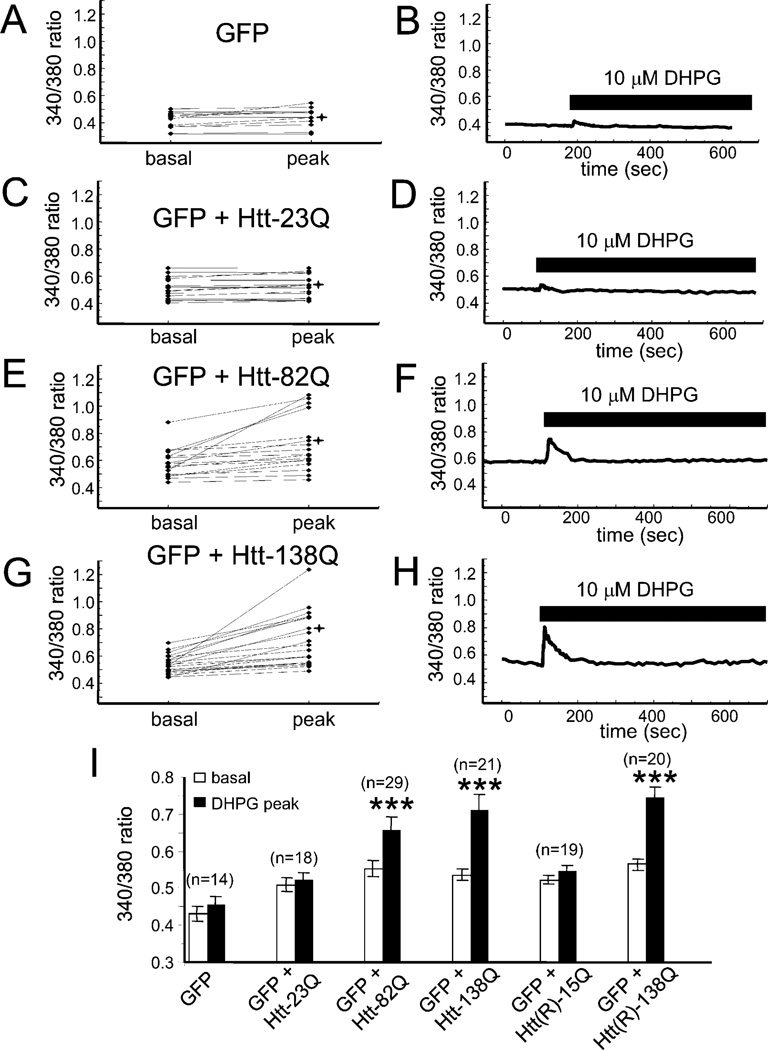
Figure 7. Httexp Facilitates DHPG-Induced Ca2+ Release in Medium Spiny Neurons.
Basal and peak 340/380 ratios are shown for individual MSN neurons transfected with (A), EGFP, (C), EGFP+Htt-23Q, (E), EGFP+Htt-82Q, (G), EGFP+Htt-138Q. The experiments were performed as described in Figure 6. Only GFP-positive MSNs were considered for quantitative analysis for each group of cells. The basal ratios were determined 1 min prior to DHPG application (−1 min). The peak ratios were measured from maximal signals observed within 30 s after DHPG application. 340/380 ratio traces for representative cells (marked *) are shown in (B), (D), (F), and (H). Time of DHPG application is shown. Similar results were obtained in four independent transfections. (I) Summary of MSN Ca2+ imaging experiments with Htt and Htt(R) constructs. Average basal and DHPG-evoked peak 340/380 ratios from four independent transfections are shown as mean + SEM (n = number of cells). These averages represent the data shown in this figure in addition to data from other experiments. The peak ratios in MSNs transfected with EGFP + Htt-82Q, EGFP + Htt-138Q, or EGFP + Htt(R)-138Q are significantly (***p < 0.001, paired t test) higher than the basal ratios in the same cells.
Ten micromolar DHPG corresponds to a threshold concentration for mGluR1/5 receptor activation in MSN neurons, and only a small response to DHPG application at this concentration was observed in control MSNs transfected with EGFP plasmid alone (Figure 6, first row, and Figures 7A and 7B) and in MSN neurons transfected with Htt-23Q plasmid (Figure 6, second row, and Figures 7C and 7D). In contrast, significant response to 10 µM DHPG was observed in MSNs transfected with Htt-82Q plasmid (Figure 6, third row, and Figures 7E and 7F). Even stronger response to this agonist was observed in MSNs transfected with Htt-138Q plasmid (Figure 6, fourth row, and Figures 7G and 7H). Paired t test analysis revealed that peak 340/380 ratios were significantly (p < 0.001) higher than basal 340/380 ratios in Htt-82Q and Htt-138Q transfected neurons, but not in Htt-23Q and EGFP-only transfected neurons (Figure 7I). Thus, we concluded that overexpression of full-length Httexp sensitizes InsP3R1 to InsP3 in MSN neurons. The effects of Httexp on InsP3R1 function were most pronounced at threshold levels of stimulation, as control, Htt-, and Httexp-transfected MSN neurons responded in a similar manner to 500 µM DHPG (data not shown).
Htt is a substrate for cleavage by caspases, and Htt proteolysis may be an early step in the pathogenesis of HD (Wellington et al., 2000, 2002). Removal of caspase 3 and 6 cleavage sites in the Htt quintuple caspase-resistant mutants [Htt(R)] reduces Httexp proteolysis and toxicity in apoptotically stressed neurons (Wellington et al., 2000). Does sensitization of InsP3R1 by Httexp depend on prior cleavage by caspases? To answer this question, we cotransfected MSN neurons with EGFP + Htt(R)-15Q or EGFP + Htt(R)-138Q and analyzed responses of transfected cells to 10 µM DHPG. We discovered that Htt(R)-138Q, but not Htt(R)-15Q, significantly potentiated DHPG-induced Ca2+ release in MSNs (Figure 7I). Importantly, the potentiating effects of caspase-resistant Htt(R)-138Q and the cleavable form of Htt-138Q were similar (Figure 7I). From these results, we concluded that Httexp does not require proteolytic cleavage by caspases 3 and 6 to have potentiating effects on InsP3R1-mediated Ca2+ release in MSN neurons.
Discussion
Huntington’s Disease and Abnormal Ca2+ Signaling
Programmed neuronal death (apoptosis) underlies the symptoms of many neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Huntington’s disease (Mattson, 2000). Ca2+ plays an important role in neuronal signaling (Berridge, 1998), and perturbed Ca2+ homeostasis is one of the key steps during initiation of the apoptotic program in affected neurons (Mattson and Chan, 2001). Abnormalities in ER-mediated Ca2+ signaling due to a mutation of presenilin 1 have been linked to the development of Alzheimer’s disease (Mattson and Chan, 2001). Several lines of experimental evidence point to a connection between HD and aberrant neuronal Ca2+ signaling. Abnormally high cytosolic Ca2+ levels were detected in CA1 pyramidal neurons from the YAC46 HD mouse model (Hodgson et al., 1999). The lymphoblast mitochondria of HD human patients and brain mitochondria from the YAC72 HD mouse model display abnormal Ca2+ homeostasis (Panov et al., 2002). The derangement in Ca2+ signaling occurs early (Panov et al., 2002) and can therefore be invoked as an initiating event in the pathogenesis of HD, followed by activation of caspases in response to high Ca2+ levels (Juin et al., 1998; Zeron et al., 2002). In addition to promoting apoptosis, increased caspase activity would be expected to catalyze Httexp proteolysis (Wellington et al., 2002), resulting in the generation of toxic amino-terminal Httexp fragments (Hackam et al., 1998). Abnormally high Ca2+ levels would also be expected to promote Httexp proteolysis via calpain-dependent mechanisms (Goffredo et al., 2002; Kim et al., 2001).
What are the molecular mechanisms linking the Httexp mutation with increased Ca2+ levels in affected neurons? Httexp alters mitochondrial Ca2+ homeostasis by directly interacting with the mitochondrial membrane (Panov et al., 2002). While this interaction likely contributes to mitochondrial dysfunction, it remains unclear how intracellular Ca2+ is affected by Httexp at the mitochondria. An alternative molecular mechanism has been suggested by the recent description of a Htt-PSD95-NMDAR complex (Sun et al., 2001) and the ability of Httexp to potentiate NMDAR function (Chen et al., 1999; Sun et al., 2001; Zeron et al., 2002). In addition to influx via NMDAR and voltage-gated Ca2+ channels, neuronal Ca2+ is also largely influenced by release from intracellular stores in the endoplasmic reticulum (ER) upon activation of class 1 metabotropic glutamate receptors (mGluR1/5) (Pin and Duvoisin, 1995). The alterations in ER enzymes that have been observed in HD brains (Cross et al., 1985) might be caused by abnormal Ca2+ release from the ER (Korkotian et al., 1999). Here, we describe a novel mechanism linking Httexp to InsP3R1-mediated Ca2+ release from the ER in striatal MSNs.
InsP3R1-HAP1A-Htt Functional Complex
We have identified a protein complex that contains InsP3R1, Htt, and HAP1. This complex was discovered through the identification of HAP1A as a binding partner of the InsP3R1 cytosolic carboxy-terminal tail in the Y2H screen (Figures 1A and 1B). In biochemical experiments, we found that Htt directly interacts with the InsP3R1 cytosolic carboxy-terminal tail and that binding to this limited region of InsP3R1 was highly dependent on both the presence of HAP1A and polyQ expansion within Htt (Figure 1E). Htt also binds to full-length InsP3R1, but this interaction is not strongly modulated by polyQ expansion (Figures 3A and 5A) or the presence of HAP1A (Figures 2C, 3A, and 5A).
The effects of Htt and HAP1A on InsP3R1 function are largely consistent with our biochemical analysis. In planar lipid bilayer experiments, the Httexp amino terminus or full-length Httexp, but not wild-type Htt, greatly facilitated InsP3R1 activity at 100 nM InsP3 (Figures 3B, 3C, 5D, and 5E). However, the amino terminus of wild-type Htt facilitated InsP3R1 activity in the presence of HAP1A (Figures 4A and 4B). Thus, InsP3R1 is sensitized to InsP3 in conditions when Htt binds to the InsP3R1 carboxy terminus (Httexp or Htt with HAP1A), but not in conditions when Htt does not bind to InsP3R1 carboxy terminus (Htt alone). From these results we concluded that association of Httexp or Htt-HAP1A with InsP3R1 carboxy terminus sensitizes InsP3R1 to activation by InsP3.
Sensitization of InsP3R1 and NMDAR by Httexp and Huntington’s Disease
The molecular and cellular basis for selective vulnerability of MSNs in HD remains elusive. One proposed mechanism is centered on the finding that MSNs primarily express the NR1A/NR2B subtype of NMDAR (Kuppenbender et al., 1999; Landwehrmeyer et al., 1995; Zeron et al., 2002). Importantly, Httexp appears to selectively enhance the activity of NR1A/NR2B channels, relative to other NMDAR subtypes, suggesting that increased NMDAR function might be specifically occurring in MSNs of HD patients (Chen et al., 1999; Sun et al., 2001; Zeron et al., 2001). Our results suggest another possible explanation for selectivity in HD involving the enrichment of mGluR5, a member of the group I mGluRs, in MSNs (Kerner et al., 1997; Mao and Wang, 2001, 2002; Tallaksen-Greene et al., 1998). Stimulation of group I mGluR in MSNs leads to the generation of InsP3 and release of Ca2+ via InsP3R1 (Mao and Wang, 2002). In addition, stimulation of group I mGluR is known to potentiate NMDAR activity in neurons, most likely via a PKC-dependent pathway (Calabresi et al., 1999; Pisani et al., 2001; Skeberdis et al., 2001).
In striatal MSNs, the presence of full-length Httexp sensitizes Ca2+ release in response to subthreshold concentrations of the mGluR1/5 agonist DHPG (Figures 6 and 7). We propose that sensitizing influences of Httexp on InsP3R1 and the NR1A/NR2B subtype of NMDAR (Chen et al., 1999; Sun et al., 2001; Zeron et al., 2001) have a synergistic effect on glutamate-induced Ca2+ signals in MSNs of HD individuals (Figure 8). In MSNs of HD patients, the activation of both mGluR5 and NMDAR by low concentrations of glutamate released by corticostriatal projection neurons leads to supranormal Ca2+ signals (Figure 8B) when compared to normal MSNs (Figure 8A), resulting in neuronal dysfunction and apoptosis. Overall, our findings provide support for the hypothesis that perturbation of neuronal Ca2+ signaling mediated by NMDAR and InsP3R1 may be a primary cause of MSN selectivity in HD. In our model, the toxic “gain of Httexp function” (Tobin and Signer, 2000) corresponds to the polyQ-dependent ability of Httexp to sensitize NMDAR and InsP3R1-mediated Ca2+ signals in MSN neurons (Figure 8B). In addition, our model points to mGluR5 as potential target for pharmacological treatment of HD.
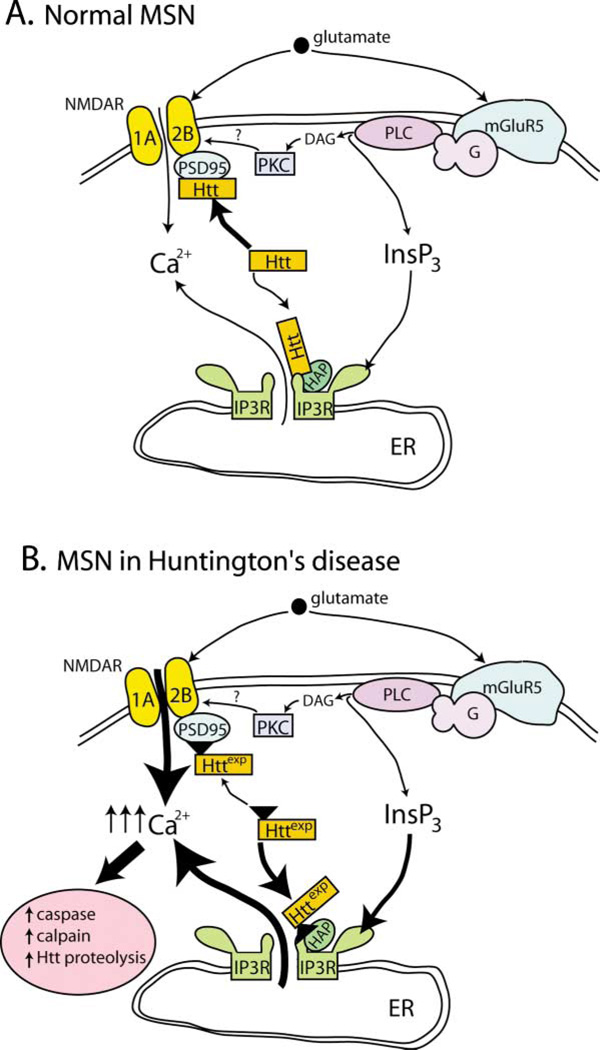
Figure 8. Proposed Mechanism Linking Httexp to Increased InsP3R1- and NMDAR-Mediated Neuronal Ca2+ Signaling in Medium Spiny Neurons of HD Patients.
Glutamate released from corticostriatal projection neurons stimulates NR1A/NR2B NMDAR and mGluR5 receptors on striatal medium spiny neurons (MSNs).
(A) In normal MSNs, NMDAR activation triggers Ca2+ influx from extracellular sources. Activation of mGluR5 stimulates phospholipase C (PLC) via a heterotrimeric G protein (G) pathway to generate diacylglycerol (DAG) and inositol triphosphate (InsP3) second messengers. Activation of mGluR5 cooperates with NMDAR activation largely through the actions of DAG, which activates PKC. The InsP3 generated downstream of mGluR5 does not trigger robust Ca2+ release from the endoplasmic reticulum (ER) in response to low levels of glutamate, as InsP3R1 sensitivity to InsP3 is low. The association of Htt with the cytosolic carboxy-terminal tail of the InsP3R1 is weak and with the PSD95-NR1A/NR2B is strong. In this schematic, only the interaction of Htt with the InsP3R1 carboxy-terminal tail is represented. This interaction appears to be stabilized by the presence of HAP1A (labeled as HAP1) in normal MSNs.
(B) In MSNs of Huntington’s disease (HD) patients, Httexp perturbs Ca2+ signaling via two synergistic mechanisms. Httexp enhances NMDAR function, possibly through decreased interaction with the PSD95-NR1A/NR2B complex. In addition, Httexp strongly binds to InsP3R1 carboxy terminus and sensitizes the InsP3R1 to activation by InsP3. As a result, low levels of glutamate released from corticostriatal projection neurons lead to supranormal Ca2+ influx via NMDAR and Ca2+ release via the InsP3R1. The net result is elevated intracellular Ca2+ levels in MSNs that eventually trigger pathogenic Ca2+-dependent downstream pathways such as increased caspase and calpain activity, increased Htt proteolysis, activation of the apoptotic program, and MSN degeneration.
Experimental Procedures
Yeast Two-Hybrid Methods
The carboxy-terminal region of rat InsP3R1 (Mignery et al., 1990) (amino acids D2590–A2749) was amplified by PCR and cloned into pLexN vector (IC bait). The yeast two-hybrid screen of a rat brain (P8-P9) cDNA library (kind gift of Dr. T. Südhof) with the IC bait and liquid yeast two-hybrid assays were performed as previously described (Maximov et al., 2003).
Plasmids
The following rat InsP3R1 (Mignery et al., 1990) baits in pLexN vectotor were generated by PCR (listed by encoded amino acid and residue numbers): IC = D2590–A2749, IC_G2736X = D2590–G2736, IC3 = R2676–A2749, IC4 = Q2714–A2749, IC5 = D2590–Q2714, IC6 = D2590–R2676, IC7 = D2590–L2646, IC8 = D2590–F2627, IC9 = L2646–A2749, IC10 = F2627–A2749. The InsP3R1 expression constructs in pGEX-KG are GST-IC8 = D2590–F2627 and GST-IC10 = F2627–A2749. The full-length clones of rat HAP1A and HAP1B (Li et al., 1995) were amplified by RT-PCR from rat brain mRNA and cloned into the pVp16-3 yeast two-hybrid prey vector, pCMV-HA mammalian expression vector, and pGEX-KG bacterial expression vector. Full-length Htt plasmids Htt-23Q (HD-FL-23Q) and Htt-82Q (HD-FL-82Q) in pRc/CMV expression vector were kindly provided by Dr. Christopher A Ross (Cooper et al., 1998). The full-length Htt-15Q and Htt-138Q plasmids (Wellington et al., 2000) were cloned into the pCI expression vector (Promega). The caspase 3 and 6 resistant Htt quintuple mutant plasmids Htt(R)-15Q and Htt(R)-138Q in pRc/CMV vector have been previously described (Wellington et al., 2000). The Htt-N expression constructs in pGEX-KG are Htt-N-15Q/138Q = M1–K158 of human Htt.
GST Pull-Down Assays
GST-IC8 and GST-IC10 proteins were expressed in the BL21 E. coli strain and purified on glutathione-agarose beads. HA-HAP1A and HA-HAP1B were expressed in COS7 cells by DEAE-dextran transfection (Sambrook et al., 1989). Htt-23/82Q proteins were expressed in HEK293 cells by calcium-phosphate transfection (Sambrook et al., 1989). 48 hr after transfection, COS7 or HEK293 cells were collected with ice-cold PBS and solubilized for 30 min at 4°C in extraction buffer A (1% CHAPS, 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.2], 5 mM EDTA, 5 mM EGTA, and protease inhibitors). Extracts were clarified by centrifugation for 20 min at 100,000 × g and incubated for 1 hr at 4°C with GST-IC8 or GST-IC10 proteins. Beads were washed three times with the extraction buffer A. Attached proteins were analyzed by Western blotting with anti-Htt or anti-HA monoclonal antibodies.
In Vitro Binding Assay
Full-length rat InsP3R1 (RT1)-encoding baculoviruses were previously described (Tu et al., 2002). Full-length Htt plasmids Htt-23Q and Htt-82Q (Cooper et al., 1998) were subcloned into pFastBac1 vector (Invitrogen), and Htt-23Q/82Q baculoviruses were generated using Bac-to-Bac system (Invitrogen). The Spodoptera frugiperda (Sf9) cells (100 ml) were infected with RT1 viruses or coinfected with RT1:Htt-23Q/82Q viral mixtures (3:1) and collected by centrifugation 72 hr postinfection. The RT1-infected cells were solubilized in extraction buffer A, cleared by centrifugation (100,000 × g), and mixed with an equal volume of HA-HAP1A or HA-HAP1B-expressing COS7 lystates prepared as described above for 2 hr at 4°C. The mixture was precipitated with anti-InsP3R1 polyclonal antibody (T443) attached to protein A-Sepharose beads and analyzed by Western blotting with anti-HA monoclonal antibodies. The cells coinfected with RT1 and Htt-23Q/82Q baculoviruses were used to prepare microsomes for bilayer experiments as previously described (Tu et al., 2002). Obtained microsomes were solubilized in extraction buffer A, cleared by centrifugation (100,000 × g in TL-100), used in immunoprecipitation experiments with anti-InsP3R1 polyclonal antibodies, and analyzed by Western blotting with anti-Htt monoclonal antibodies.
Brain Pull-Downs and Immunoprecipitations
Rat and mouse brain tissues were isolated, homogenized, and solubilized for 1.5 hr at 4°C in extraction buffer A. The lysate was clarified by 20 min centrifugation at 100,000 × g and utilized in pull-down experiments with GST-IC8 and GST-IC10 proteins or in the immunoprecipitation experiments with anti-InsP3R1 polyclonal antibodies (T443) performed as described above. GST-IC8 and GST-IC10 proteins (200 µg/ml final concentration) were included in the immunoprecipitation reactions as indicated. An additional 0.5 M KCl wash step was included in the immunoprecipitation experiments with mouse samples as indicated in the text. The precipitated fractions were analyzed by Western blotting with anti-Htt and anti-HAP1 monoclonal antibodies.
Planar Lipid Bilayer Experiments
Single-channel recordings of recombinant InsP3R1 (RT1) expressed in isolation or coexpressed with Htt-23Q/82Q proteins were performed as previously described (Tang et al., 2003; Tu et al., 2002) at 0 mV transmembrane potential using 50 mM Ba2+ (trans) as a charge carrier. The cis (cytosolic) chamber contained 110 mM Tris dissolved in HEPES (pH 7.35), 0.5 mM Na2ATP, pCa 6.7 (0.2 mM EGTA + 0.14 mM CaCl2) (Bezprozvanny et al., 1991). InsP3R1 were activated by addition of 100 nM InsP3 or 2 µM InsP3 (Alexis) to the cis chamber as indicated in the text. GST, Htt-N-15Q, Htt-N-138Q, and HAP1A proteins were expressed in BL21 E. coli, purified on gluthathione beads, eluted with reduced glutathione, dialyzed overnight against cis recording buffer (110 mM Tris/HEPES [pH 7.35]), and added in 1 µl volume (0.3 mg/ml protein with addition of 0.02 mM ruthenium red) directly to the cis side of the bilayer containing InsP3R1 without stirring. Exposure of InsP3R1 to the test proteins was terminated 2–3 min after addition by stirring the cis chamber for 30 s (1:3000 dilution of test protein stocks). The InsP3R1 single-channel currents were amplified (Warner OC-725), filtered at 1 kHz by a low-pass eight-pole Bessel filter, digitized at 5 kHz (Digidata 1200, Axon Instruments), and stored on computer hard drive and recordable optical discs. For off-line computer analysis (pClamp 6, Axon Instruments), currents were filtered digitally at 500 Hz. For presentation of the current traces, data were filtered at 200 Hz.
Ca2+ Imaging Experiments
The rat medium spiny neuronal (MSN) cultures on poly-D-lysine (Sigma) coated 12 mm round glass coverslips were established by following published procedures (Mao and Wang, 2001). The 5 µM of cytosine arabinoside (AraC, Sigma) was added at 2–4 DIV to inhibit glial cell growth. At 20 DIV the MSN cultures were transfected by the calcium-phosphate method (Maximov and Bezprozvanny, 2002) with EGFP-C3 plasmid (Clontech) or a 1:3 mixture of EGFP:Htt plasmids as indicated in the text. 48 hr after transfection, the MSN neurons were loaded with 5 µM Fura2-AM (Molecular Probes) in artificial cerebrospinal fluid (ACSF) (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM HEPES [pH 7.3]) for 45 min at 37°C. For imaging experiments the coverslips were mounted onto a recording/perfusion chamber (RC-26G, Warner Instrument) maintained at 37°C (PH1, Warner Instrument), positioned on the movable stage of an Olympus IX-70 inverted microscope, and perfused with ACSF media by gravity flow. Following GFP imaging, the coverslip was washed extensively with Ca2+-free ACSF (omitted CaCl2 from ACSF and supplemented with 100 µM EGTA). In Ca2+ imaging experiments the MSN cells were intermittently excited by 340 nm and 380 nm UV light (DeltaRAM illuminator, PTI) using a Fura-2 dichroic filter cube (Chroma Technologies) and 60× UV-grade oil-immersed objective (Olympus). The emitted light was collected by an IC-300 camera (PTI), and the images were digitized by ImageMaster Pro software (PTI). Baseline (6 min) measurements were obtained prior to bath application of 10 µM or 500 µM 3,5-DHPG (Tocris) dissolved in Ca2+-free ACSF. The DHPG solutions were prewarmed to 37°C before application to MSNs. Images at 340 and 380 nm excitation wavelengths were captured every 5 s and shown as 340/380 image ratios at time points as indicated. Background fluorescence was determined according to manufacturer’s (PTI) recommendations and subtracted.
Antibodies
The following monoclonal antibodies were used: anti-HA (HA.11) from Covance, anti-Htt (mAB2166) from Chemicon International, Intramonoclonal anti-HAP1 1B6 is a kind gift of Dr. Claire-Anne Gutekunst (Chan et al., 2002), GAD65 antibodies from BD Pharmingen. Polyclonal anti-InsP3R1 T443 antibody was previously described (Kaznacheyeva et al., 1998). Secondary HRP-conjugated anti-rabbit and anti-mouse antibodies were from Jackson Imunoresearch.
Acknowledgments
We thank Thomas C. Südhof for advice with yeast two-hybrid screen and the gift of a rat brain cDNA library and Phyllis Foley for administrative assistance. We thank Thomas C. Südhof for rat InsP3R1 cDNA, Claire-Anne Gutekunst for HAP1 monoclonal antibodies, and Christopher A Ross for HD-FL-23Q and HD-FL-82Q plasmids. I.B. is supported by the Robert A. Welch Foundation, the Huntington’s Disease Society of America, the Hereditary Disease Foundation, and NIH R01 NS38082. M.R.H. is supported by the Canadian Institutes of Health Research, the Hereditary Disease Foundation, and the Huntington’s Disease Society of America and holds a Canada Research Chair in Human Genetics.
References
- Berridge MJ. Neuronal calcium signaling. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- Bertaux F, Sharp AH, Ross CA, Lehrach H, Bates GP, Wanker E. HAP1-huntingtin interactions do not contribute to the molecular pathology in Huntington’s disease transgenic mice. FEBS Lett. 1998;426:229–232. doi: 10.1016/s0014-5793(98)00352-4. [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature. 1991;351:751–754. doi: 10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- Calabresi P, Centonze D, Pisani A, Bernardi G. Metabotropic glutamate receptors and cell-type-specific vulnerability the striatum: implication for ischemia and Huntington’s disease. Exp. Neurol. 1999;158:97–108. doi: 10.1006/exnr.1999.7092. [DOI] [PubMed] [Google Scholar]
- Chan EY, Nasir J, Gutekunst CA, Coleman S, Maclean A, Maas A, Metzler M, Gertsenstein M, Ross CA, Nagy A, Hayden MR. Targeted disruption of Huntingtin-associated protein-1 (Hap1) results in postnatal death due to depressed feeding behavior. Hum. Mol. Genet. 2002;11:945–959. doi: 10.1093/hmg/11.8.945. [DOI] [PubMed] [Google Scholar]
- Chen N, Luo T, Wellington C, Metzler M, McCutcheon K, Hayden MR, Raymond LA. Subtype-specific enhancement of NMDA receptor currents by mutant huntingtin. J. Neurochem. 1999;72:1890–1898. doi: 10.1046/j.1471-4159.1999.0721890.x. [DOI] [PubMed] [Google Scholar]
- Chesselet MF, Mercugliano M, Soghomonian JJ, Salin P, Qin Y, Gonzales C. Regulation of glutamic acid decarboxylase gene expression in efferent neurons of the basal ganglia. Prog. Brain Res. 1993;99:143–154. doi: 10.1016/s0079-6123(08)61343-6. [DOI] [PubMed] [Google Scholar]
- Colomer V, Engelender S, Sharp AH, Duan K, Cooper JK, Lanahan A, Lyford G, Worley P, Ross CA. Huntingtin-associated protein 1 (HAP1) binds to a Trio-like polypeptide, with a rac1 guanine nucleotide exchange factor domain. Hum. Mol. Genet. 1997;6:1519–1525. doi: 10.1093/hmg/6.9.1519. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum. Mol. Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- Cross AJ, Crow TJ, Johnson JA, Dawson JM, Peters TJ. Loss of endoplasmic reticulum-associated enzymes affected brain regions in Huntington’s disease and Alzheimer-type dementia. J. Neurol. Sci. 1985;71:137–143. doi: 10.1016/0022-510x(85)90055-3. [DOI] [PubMed] [Google Scholar]
- Engelender S, Sharp AH, Colomer V, Tokito MK, Flanagan A, Worley P, Holzbaur EL, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- Furuichi T, Kohda K, Miyawaki A, Mikoshiba K. Intra-cellular channels. Curr. Opin. Neurobiol. 1994;4:294–303. doi: 10.1016/0959-4388(94)90089-2. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 1992;15:133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- Goffredo D, Rigamonti D, Tartari M, De Micheli A, Verderio C, Matteoli M, Zuccato C, Cattaneo E. Calcium-dependent cleavage of endogenous wild-type huntingtin in primary cortical neurons. J. Biol. Chem. 2002;277:39594–39598. doi: 10.1074/jbc.C200353200. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Huntingtin: a single bait hooks many species. Curr. Opin. Neurobiol. 1998;8:425–430. doi: 10.1016/s0959-4388(98)80071-8. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Ferrante RJ, Li XJ, Hersch SM. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. J. Neurosci. 1998;18:7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J. Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HDCRG (The Huntington’s Disease Collaborative Research Group) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, et al. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- Juin P, Pelletier M, Oliver L, Tremblais K, Gregoire M, Meflah K, Vallette FM. Induction of a caspase-3-like activity by calcium in normal cytosolic extracts triggers nuclear apoptosis in a cell-free system. J. Biol. Chem. 1998;273:17559–17564. doi: 10.1074/jbc.273.28.17559. [DOI] [PubMed] [Google Scholar]
- Kalchman MA, Koide HB, McCutcheon K, Graham RK, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn FC, Wellington C, Metzler M, et al. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat. Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- Kaznacheyeva E, Lupu VD, Bezprozvanny I. Single-channel properties of inositol (1,4,5)-trisphosphate receptor heterologously expressed in HEK-293 cells. J. Gen. Physiol. 1998;111:847–856. doi: 10.1085/jgp.111.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner JA, Standaert DG, Penney JB, Jr, Young AB, Landwehrmeyer GB. Expression of group one metabotropic glutamate receptor subunit mRNAs in neurochemically identified neurons in the rat neostriatum, neocortex, and hippocampus. Brain Res. Mol. Brain Res. 1997;48:259–269. doi: 10.1016/s0169-328x(97)00102-2. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N, DiFiglia M. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. USA. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkotian E, Schwarz A, Pelled D, Schwarzmann G, Segal M, Futerman AH. Elevation of intracellular glucosylceramide levels results in an increase in endoplasmic reticulum density and in functional calcium stores in cultured neurons. J. Biol. Chem. 1999;274:21673–21678. doi: 10.1074/jbc.274.31.21673. [DOI] [PubMed] [Google Scholar]
- Kuppenbender KD, Albers DS, Iadarola MJ, Landwehrmeyer GB, Standaert DG. Localization of alternatively spliced NMDAR1 glutamate receptor isoforms in rat striatal neurons. J. Comp. Neurol. 1999;415:204–217. [PubMed] [Google Scholar]
- Landwehrmeyer GB, Standaert DG, Testa CM, Penney JB, Jr, Young AB. NMDA receptor subunit mRNA expression by projection neurons and interneurons in rat striatum. J. Neurosci. 1995;15:5297–5307. doi: 10.1523/JNEUROSCI.15-07-05297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Li SH, Sharp AH, Nucifora FC, Jr, Schilling G, Lanahan A, Worley P, Snyder SH, Ross CA. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- Li SH, Gutekunst CA, Hersch SM, Li XJ. Interaction of huntingtin-associated protein with dynactin P150Glued. J. Neurosci. 1998a;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Hosseini SH, Gutekunst CA, Hersch SM, Ferrante RJ, Li XJ. A human HAP1 homologue. Cloning, expression, and interaction with huntingtin. J. Biol. Chem. 1998b;273:19220–19227. doi: 10.1074/jbc.273.30.19220. [DOI] [PubMed] [Google Scholar]
- Li Y, Chin LS, Levey AI, Li L. Huntingtin-associated protein 1 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate and functions in endosomal trafficking. J. Biol. Chem. 2002;277:28212–28221. doi: 10.1074/jbc.M111612200. [DOI] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Upregulation of preprodynorphin and preproenkephalin mRNA expression by selective activation of group I metabotropic glutamate receptors in characterized primary cultures of rat striatal neurons. Brain Res. Mol. Brain Res. 2001;86:125–137. doi: 10.1016/s0169-328x(00)00276-x. [DOI] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Glutamate cascade to cAMP response element-binding protein phosphorylation in cultured striatal neurons through calcium-coupled group I metabotropic glutamate receptors. Mol. Pharmacol. 2002;62:473–484. doi: 10.1124/mol.62.3.473. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Nakagawa T, Inoue T, Nagata E, Tanaka K, Takano H, Minowa O, Kuno J, Sakakibara S, Yamada M, et al. Ataxia and epileptic seizures in mice lacking type 1 inositol 1,4,5-trisphosphate receptor. Nature. 1996;379:168–171. doi: 10.1038/379168a0. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Chan SL. Dysregulation of cellular calcium homeostasis in Alzheimer’s disease: bad genes and bad habits. J. Mol. Neurosci. 2001;17:205–224. doi: 10.1385/JMN:17:2:205. [DOI] [PubMed] [Google Scholar]
- Maximov A, Bezprozvanny I. Synaptic targeting of N-type calcium channels in hippocampal neurons. J. Neurosci. 2002;22:6939–6952. doi: 10.1523/JNEUROSCI.22-16-06939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A, Tang T-S, Bezprozvanny I. Association of the type 1 inositol (1,4,5)-trisphosphate receptor with 4.1N protein in neurons. Mol. Cell. Neurosci. 2003;22:271–283. doi: 10.1016/s1044-7431(02)00027-1. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Chesselet MF. Mouse models of Huntington’s disease. Trends Pharmacol. Sci. 2002;23:32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- Mignery GA, Newton CL, Archer BT, Sudhof TC. Structure and expression of the rat inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 1990;265:12679–12685. [PubMed] [Google Scholar]
- Nasir J, Goldberg YP, Hayden MR. Huntington disease: new insights into the relationship between CAG expansion and disease. Hum. Mol. Genet. 1996;5:1431–1435. doi: 10.1093/hmg/5.supplement_1.1431. [DOI] [PubMed] [Google Scholar]
- Nasir J, Duan K, Nichol K, Engelender S, Ashworth R, Colomer V, Thomas S, Disteche CM, Hayden MR, Ross CA. Gene structure and map location of the murine homolog of the Huntington-associated protein, Hap1. Mamm. Genome. 1998;9:565–570. doi: 10.1007/s003359900819. [DOI] [PubMed] [Google Scholar]
- Page KJ, Potter L, Aronni S, Everitt BJ, Dunnett SB. The expression of Huntingtin-associated protein (HAP1) mRNA in developing, adult and ageing rat CNS: implications for Huntington’s disease neuropathology. Eur. J. Neurosci. 1998;10:1835–1845. doi: 10.1046/j.1460-9568.1998.00185.x. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- Pisani A, Gubellini P, Bonsi P, Conquet F, Picconi B, Centonze D, Bernardi G, Calabresi P. Metabotropic glutamate receptor 5 mediates the potentiation of N-methyl-D-aspartate responses in medium spiny striatal neurons. Neuroscience. 2001;106:579–587. doi: 10.1016/s0306-4522(01)00297-4. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. Lessons from animal models of Huntington’s disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Edition. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schoepp DD, Jane DE, Monn JA. Pharmacological agents acting at subtypes of metabotropic glutamate receptors. Neuropharmacology. 1999;38:1431–1476. doi: 10.1016/s0028-3908(99)00092-1. [DOI] [PubMed] [Google Scholar]
- Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, Warby S, Yanai A, Gutekunst CA, Leavitt BR, Yi H, et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- Skeberdis VA, Lan J, Opitz T, Zheng X, Bennett MV, Zukin RS. mGluR1-mediated potentiation of NMDA receptors involves a rise in intracellular calcium and activation of protein kinase C. Neuropharmacology. 2001;40:856–865. doi: 10.1016/s0028-3908(01)00005-3. [DOI] [PubMed] [Google Scholar]
- Street VA, Bosma MM, Demas VP, Regan MR, Lin DD, Robinson LC, Agnew WS, Tempel BL. The type 1 inositol 1,4,5-trisphosphate receptor gene is altered in the opisthotonos mouse. J. Neurosci. 1997;17:635–645. doi: 10.1523/JNEUROSCI.17-02-00635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Savanenin A, Reddy PH, Liu YF. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D-aspartate receptors via post-synaptic density 95. J. Biol. Chem. 2001;276:24713–24718. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- Tallaksen-Greene SJ, Kaatz KW, Romano C, Albin RL. Localization of mGluR1a-like immunoreactivity and mGluR5-like immunoreactivity in identified populations of striatal neurons. Brain Res. 1998;780:210–217. doi: 10.1016/s0006-8993(97)01141-4. [DOI] [PubMed] [Google Scholar]
- Tang TS, Tu H, Wang Z, Bezprozvanny I. Modulation of type 1 inositol (1,4,5)-trisphosphate receptor function by protein kinase a and protein phosphatase 1alpha. J. Neurosci. 2003;23:403–415. doi: 10.1523/JNEUROSCI.23-02-00403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin AJ, Signer ER. Huntington’s disease: the challenge for cell biologists. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- Tu H, Miyakawa T, Wang Z, Glouchankova L, Iino M, Bezprozvanny I. Functional characterization of the type 1 inositol 1,4,5-trisphosphate receptor coupling domain SII(+/−) splice variants and the opisthotonos mutant form. Biophys. J. 2002;82:1995–2004. doi: 10.1016/S0006-3495(02)75548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, Cattaneo E, Hackam A, Sharp A, Thornberry N, et al. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J. Biol. Chem. 2000;275:19831–19838. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S, Graham RK, Loubser O, van Raamsdonk J, Singaraja R, Yang YZ, et al. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington’s disease. J. Neurosci. 2002;22:7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron MM, Chen N, Moshaver A, Lee AT, Wellington CL, Hayden MR, Raymond LA. Mutant huntingtin enhances excitotoxic cell death. Mol. Cell. Neurosci. 2001;17:41–53. doi: 10.1006/mcne.2000.0909. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]