Abstract
Background
CD5 is a pan-T cell surface marker that is also present on a subset of B cells, B-1a cells.Functional and developmental subsets of T cells express characteristic CD5 levels that vary over roughly a 30-fold range. Previous investigators have cloned a 1.7 Kb fragment containing the CD5 promoter and showed that it can confer similar lymphocyte-specific expression pattern as observed for endogenous CD5 expression.
Results
We further characterize the CD5 promoter and identify minimal and regulatory regions on the CD5 promoter. Using a luciferase reporter system, we show that a 43 bp region on the CD5 promoter regulates CD5 expression in resting mouse thymoma EL4 T cells and that an Ets binding site within the 43 bp region mediates the CD5 expression. In addition, we show that Ets-1, a member of the Ets family of transcription factors, recognizes the Ets binding site in the electrophoretic mobility shift assay (EMSA). This Ets binding site is directly responsible for the increase in reporter activity when co-transfected with increasing amounts of Ets-1 expression plasmid.
We also identify two additional evolutionarily-conserved regions in the CD5 promoter (CD5X and CD5Y) and demonstrate the respective roles of the each region in the regulation of CD5 transcription.
Conclusion
Our studies define a minimal and regulatory promoter for CD5 and show that the CD5 expression level in T cells is at least partially dependent on the level of Ets-1 protein. Based on the findings in this report, we propose a model of CD5 transcriptional regulation in T cells.
Introduction
The murine CD5 protein (Ly-1), a 67 kD membrane-associated glycoprotein found on all T cells and at low levels on B-1a cells, a CD5-expressing subset of B lymphocytes [1,2,3,4]. CD5 levels on the developmental and functional subsets range are ordered in a characteristic fashion: Thymic CD4+ T > Splenic CD4+ T > Thymic CD8+ T > Splenic CD8+ T > Thymic CD4+CD8+ T >> B-1a cells [5, 6]. The CD5 surface and mRNA expression levels in these subsets are highly correlated, suggesting that CD5 expression is primarily regulated at the transcriptional level.
Structurally, CD5 belongs to the scavenger receptor family of proteins based on the homology of its three extracellular scavenger receptor cysteine-rich (SRCR) domains to other family members [7]. Several potential CD5 ligands have been reported so far: the pan-B cell marker, CD72 [8]; an as yet unidentified lymphocyte specific inducible glycoprotein [9]; and, VH framework determinants on immunoglobulins [10]. While none of these potential ligands have been unequivocally shown to physiologically interact with CD5, these potential CD5 receptor/ligand pairs suggest that CD5 may play a role in T-B or B-1a-B cell-cell communication.
CD5 has been shown to be physically associated with the T cell receptor (TCR)/CD3 complex in T cells and with B cell receptor (BCR) in B-1a cells [11,12,13]. Although several laboratories reported that crosslinking CD5 on the surface augments T cells signaling by inducing calcium flux and enhancing mitogenic response [14,15,16], recent studies using CD5-deficient mice indicate that CD5 may be more important as a negative modulator of TCR and BCR signal transduction [17, 18]. Consistent with this idea, the positive and negative selection in the thymus of CD5-deficient mice is altered in that their thymocytes are hyperresponsive and hyperproliferative to activation induced by anti-CD3 antibodies [17]. Similarly, CD5-deficient B-1a cells show higher proliferative responses to surface IgM ligation than heterozygote littermates [18].
CD5 most likely exerts its negative modulation of receptor signal transduction by associating with SH-2-containing signaling molecules. This association could be directed by an imperfect immunoreceptor tyrosine-based activating motif (ITAM) [19] or by a motif similar to immunoreceptor tyrosine-based inhibitory motif (ITIM) located in the CD5 cytoplasmic domain [20]. Consistent with SH-2 interaction, CD5 cytoplasmic tyrosine residues are phosphorylated by p56lck kinases upon TCR/CD3 ligation and serve as targets for association with proteins containing SH2 domains in both T and B-1a cells [12, 21, 22].
In human Jurkat cells and phytohemagglutinin-expanded T lymphoblasts, the CD5 ITIM-like motif is crucial for association with SHP-1, a cytosolic tyrosine phosphotase implicated in the negative regulation of antigen receptor-mediated signaling [23]. However, in murine B lymphoma cells, the CD5 pseudo-ITAM motif mediates its inhibitory action via a SHP-1 independent mechanism [24].
Several studies have examined the CD5 promoter. Comparison of the sequences of the CD5 promoters cloned from man and mouse [25, 26] revealed conserved transcription factor binding sites. In addition, deletion analysis of the murine CD5 promoter indicated the presence of lymphoid-specific regulatory elements [25].
Here, we present a detailed analysis of the CD5 promoter. Using unstimulated EL4 thymoma cells as a model system, we demonstrate that a CD5 expression is regulated by a 43 bp region (-172 to -215 bp) upstream of the methionine start codon for CD5. The presence of the Ets binding site in this regulatory region is particularly interesting because levels of Ets-1, a lymphoid-specific member of the Ets family [27, 28], have been shown to closely resemble the mRNA and protein levels observed for CD5 in different T cell subsets [29] [5,6]. In addition, CD5 surface protein expression is greatly reduced in ets-1-/- T cells [30, 31].
We show here that Ets-1 protein recognizes and binds the Ets site in the 43 bp regulatory region and that CD5 expression is positively correlated with Ets-1 levels in co-transfection studies. Thus, we closely link Ets-1 to CD5 regulation. In addition, we show that two other evolutionarily conserved regions located close to the Ets site (provisionally named CD5X and CD5Y) play a role in CD5 expression. Based on these data, we propose a model for CD5 transcriptional control in resting T cells that includes interaction between CD5X, CD5Y and Ets-1.
Results
Characterization of regions on the CD5 promoter necessary for CD5 transcription
We constructed luciferase reporter constructs containing progressively deleted fragments of a 3 kb genomic DNA clone which contains the CD5 promoter [1, 32] (Figure 1a). Each deletion (luciferase) construct and a control LacZ reporter construct (pON405) were co-transfected into the EL4 T cell line and lysates prepared after cells were grown for 36-48 hours were assayed for luciferase activity.
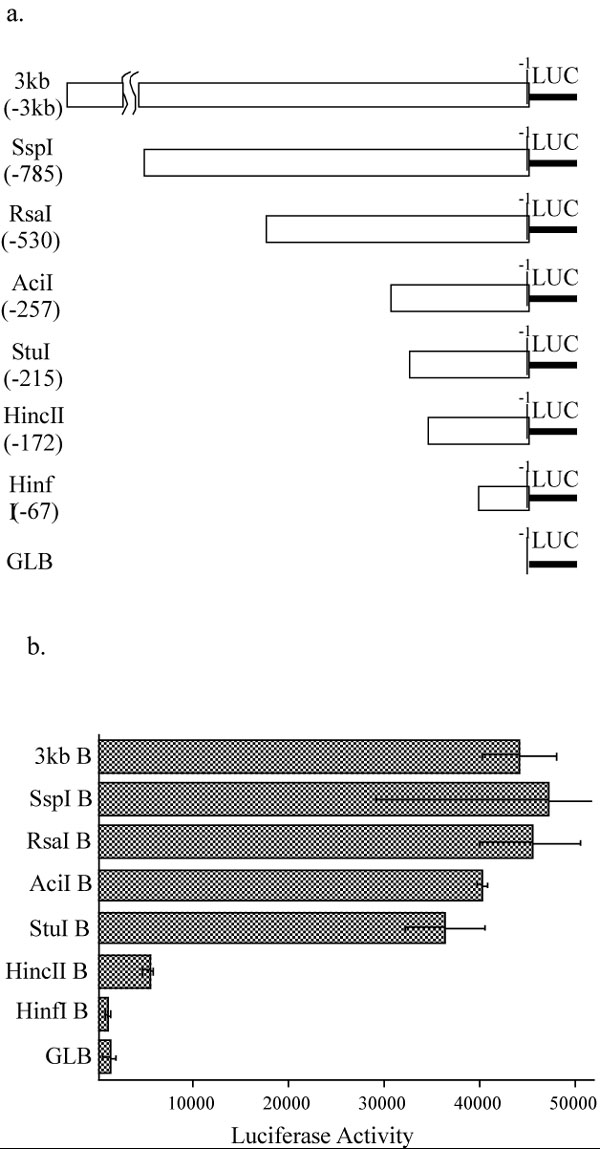
Figure 1.
a. CD5 promoter deletion constructs. The 3 kb CD5 5-flanking region fragment was excised from p12-1 and subject to 5' to 3' deletion with SspI (-785), RsaI (-530), AciI (-257), StuI (-215), HincII (-172), HinfI (-67). The deleted CD5 5'-flanking regions were cloned into a promoterless luciferase construct, pGLB (Promega). The DNA base pair on the CD5 promoter, before the translation start, is labeled as -1.
b. A 43 bp region in the CD5 promoter is responsible for normal expression in EL4 (T) cells. EL4 cells were transiently co-transfected with each deletion construct and pON405, a LacZ reporter construct, as described in Materials and Methods. Luciferase activity in cell extracts was measured between 36 and 48 hours after transfection. The measurement for luciferase activity was normalized based on the β-galactosidase activity in each cell extract. Each bar value represents the average of three independent transfections in one experiment, with standard deviation as error bars. Each experiment was repeated several times.
Results show that the 3 kb fragment induces a large increase in luciferase activity, which we define as the full level of CD5 expression in resting T cells (Figure 1b). Transfections with constructs containing successive deletions of the 3 kb fragment show that removal of DNA regions upstream of a StuI site (at -215, relative to the CD5 protein initiation methionine codon) does not significantly decrease luciferase expression from the level obtained with the full length fragment. However, the deletion of an additional 43 bp (from StuI to HincII site at -172) reduces the reporter activity by more than five-fold to basal level, which is somewhat higher than the background level obtained with a promoterless luciferase construct (GLB = pGLBasic). Additional deletion from the HincII to the HinfI site at -67 further reduced the basal reporter activity down to the background level. These deletion constructs define two regions as important for CD5 expression: a minimal/core promoter region (HincII to HinfI site) that is required for basal activity and a regulatory region necessary for the full CD5 expression (StuI to HincII) site in resting T cells.
The Ets binding site is important for full CD5 expression
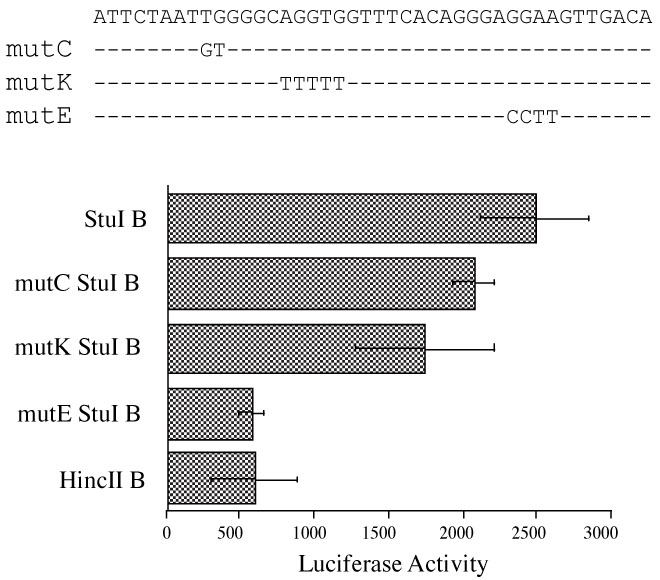
Three potential transcription factor binding sites - CCAAT, κE2, and Ets - were previously identified by database sequence comparison in the 43 bp region described above (StuI to HincII) [25]. However, only the κE2 and Ets site are conserved between mouse and human [26]. To determine which of these potential transcription factor binding sites is responsible for regulating normal CD5 expression, we performed site-directed mutagenesis on the CCAAT (mutC), κE2 (mutK) and Ets (mutE) sites from the StuI B reporter construct. Wild-type control and the mutant constructs were tested for their promoter activity in EL4 cells. As figure 2 shows, mutations in the CCAAT or κE2 binding site resulted in minor reductions in reporter activity. In contrast, the mutation in the Ets binding site completely abolished the full induction of transcription and reduced the luciferase activity to the basal level.
Figure 2.
Site-directed mutagenesis of the 43 region (StuI to HincII) identified an Ets binding site on the CD5 promoter as necessary for maximal CD5 promoter. Each of the three potential transcription factor binding sites- CCAAT, κE2, and Ets, was mutated on the StuI B reporter construct as described in Materials and Methods. EL4 cells were transiently transfected with StuI B, mutC StuI B, mutK StuI B, mutE StuI B, or HincII B constructs, and with pON405 LacZ reporter plasmid as internal control. Each bar value represents the average of three independent transfections in one experiment, with standard deviation as error bars.
The Ets binding site is recognized by Ets-1 protein
The above experiments demonstrate that a potential Ets binding site on the CD5 promoter is necessary for the full induction of transcription. Since Ets-1 is lymphoid-specific, and its expression levels in T cell subsets correlate with the CD5 expression levels, we focused our study on Ets-1 interaction with this Ets binding site.
For the following experiments we used an Ets-1 expression plasmid (pEVRF-ets-1) which encodes Ets-1 protein [33, 34]. We then performed electrophoretic mobility shift assays (EMSA) using radiolabelled Ets oligonucleotide with nuclear extracts prepared from pEVRF-ets-1 transfected M12 cells (figure 3a, lane 3-9). This olignucleotide corresponds to the Ets binding site in the 43 bp regulatory region of the CD5 promoter. It has the core Ets binding concensus with and additional 6 bases on each side.
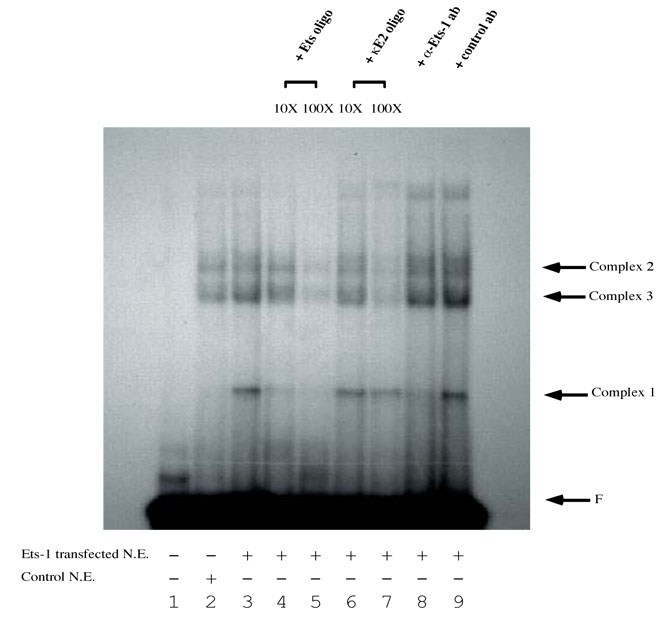
Figure 3.
EMSA analysis of the ets binding site on the CD5 promoter. The 32P-labeled oligonucleotide probe containing the Ets binding site (5' CGC GTA CAG GGA GGA AGT TGA CCT CGA 3') on the CD5 promoter was incubated with nuclear extracts prepared from M12 cells. The Ets-1 protein:DNA complex is indicated as complex 1. Lane 1 is the probe alone; lane 2, probe plus M12 nuclear extract from transfection with pEVRF0; and lane 3-9: probe plus M12 nuclear extract from transfection pEVRF-ets-1. Competitive EMSA was performed using molar excess unlabeled olionucleotides as indicated: in lane 4, ten-fold molar excess of unlabeled homologous Ets oligonucleotides; Lane 5, 100-fold molar excess of unlabeled Ets oligonucleotides; Lane 6, ten-fold molar excess of unlabeled κE2; and lane7, 100-fold molar excess of unlabeled κE2 oligo. Inhibition EMSA was also performed using antibodies as indicated: lane 8, anti-Ets-1 antibody; and lane 9, pre-immune antibody. F indicates the migration of the free probe.
The pEVRF-ets-1 transfected M12 nuclear extracts form an additional protein:DNA complex (complex 1, figure 3, lane 3) compared with the nuclear extracts from M12 cells transfected with the control plasmid, pEVRF0 (figure 3, lane 2). This complex is easily competed away by increasing amount of unlabeled Ets oligos (figure 3, lanes 4 and 5) but not by increasing amounts of irrelevant κE2 oligos (figure 3, lanes 6 and 7). Furthermore, its formation is prevented by pre-incubating the nuclear extract with anti-Ets-1 polyclonal antibodies, which recognize the Ets-1 DNA binding domain (figure 3, lane 8), but not by pre-incubation with pre-immune polyclonal antibodies (figure 3, lane 9). Thus, this complex represents true Ets-1 binding.
We observed two additional endogenous complexes (complex 2 and 3) that were competed away with increasing amount of unlabeled Ets oligos. However, these two complexes were also competed away with increasing amounts of irrelevant κE2 oligos. Furthermore, complex 1 and 2 are not inhibited by pre-incubation with anti-Ets-1 antibodies. Therefore, we conclude that complex 1 and 2 are not Ets-1 specific.
These competition and inhibition experiments demonstrate that Ets-1 specifically binds to the oligonucleotide used for these studies. As indicated above, this olignucleotide corresponds precisely to the Ets binding site in the 43 bp regulatory region of the CD5 promoter with the core Ets binding concensus and only an additional 6 bases on each side. Therefore, we conclude that Ets-1 binds to the Ets site in the 43 bp regulatory region of the CD5 promoter or very closely to that site.
Ets-1 affects CD5 expression in T cells
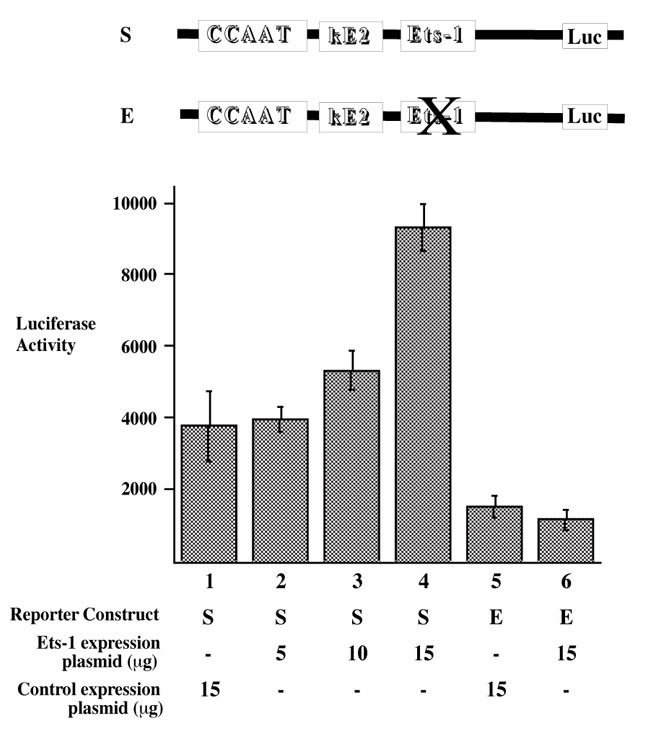
To understand the effect of Ets-1 on CD5 expression, we co-transfected EL4 cells with increasing amounts of Ets-1 expression plasmid (pEVRF-ets-1) along with a StuI B reporter construct containing either the wild type (S) or mutated Ets binding site (E). As figure 4 shows, the luciferase reporter activity from the wild type StuI B reporter construct increased in a dose-dependent fashion with increasing amounts of Ets-1 expression plasmid (lanes 1-4). Importantly, the increase in reporter activity from Ets-1 expression was mediated by the Ets binding site on the CD5 promoter, as the increase in reporter expression did not occur when the Ets binding site on the CD5 promoter was mutated (lane 4 and 5).
Figure 4.
Ets-1 affects CD5 expression levels. EL4 cells were transiently transfected with StuI B reporter construct (S) plus different amounts of Ets-1 expression plasmid (pEVRF-ets-1): 5 μg (lane 2), 10 μg (lane 3), or 15 μg (lane 4); or with StuI B construct and 15 μg control plasmid pEVRF0 (lane 1). EL4 cells were also co-transfected with mutE StuI B reporter construct (E) and 15 μg of pEVRF-ets-1 or pEVRF0 (lanes 5 and 6). Each bar value represents the average of three independent transfections in one experiment, with standard deviation as error bars.
Based on these transfections results and the EMSA observations described above, we conclude that Ets-1 regulates CD5 expression by binding to the Ets site located with in the 43 bp regulatory region.
The CD5Y region is essential for basal transcription in T cells
Using the Intelligenetics sequence analysis program to compare the human and murine CD5 minimal promoter regions, we found two conserved regions, provisionally designated CD5X (-172 to -154) and CD5Y (-133 to -119) (figure 5a), that abut the Ets binding site. These sequences did not show any homology to known transcription factor binding sites. Transfection studies show that deletion upstream of CD5X abolishes the regulatory promoter activity but does not hamper the basal promoter activity. In contrast, deletion of the region containing CD5X and CD5Y eliminates both the regulatory and basal promoter activity (figure 1b).
Figure 5.
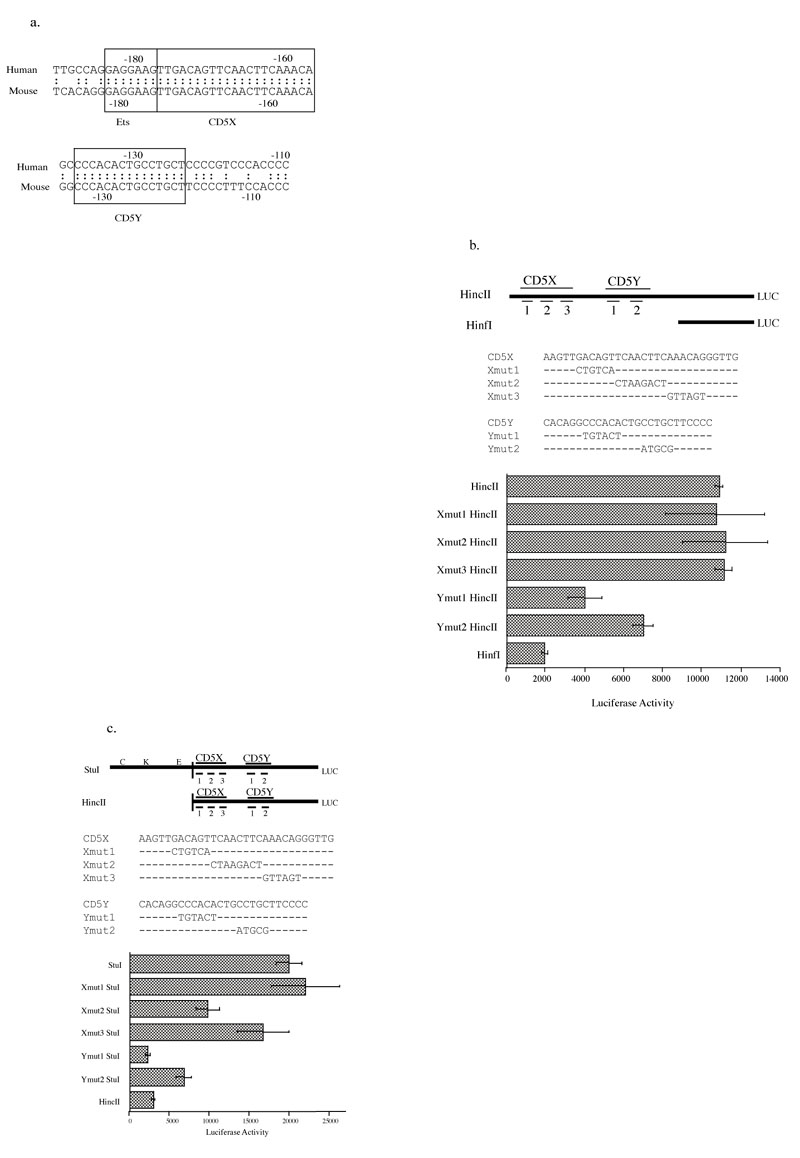
a: Sequence comparison of the Ets site, the CD5X region, and the CD5Y region between human and mouse. Partial sequence alignment of the homologous regions on the mouse and human CD5 promoter, as determined by sequence analysis program from Intelligenetics Corporation. The consensus sequences for the Ets, CD5X, and CD5Y regions are shown in boxes.
b: Site-directed mutagenesis analysis showed that CD5Y region is important for basal CD5 transcription. Three areas in CD5X region (Xmut1, Xmut2, and Xmut3) and two in CD5Y region (Ymut1 and Ymut2) were mutated by site-directed mutagenesis on HincII B construct as described in Materials and Methods. Each mutant construct was cotransfected with pON405 LacZ construct in EL4 cells. Wild-type HincII B and HinfI B constructs were also transfected as controls. Each bar value represents the average of three independent transfections in one experiment, with standard deviation as error bars.
c: Site-directed mutagenesis analysis revealed that both the CD5X and CD5Y regions affect overall CD5 expression. Three sites in the CD5X region (Xmut1, Xmut2, and Xmut3) and two in the CD5Y region (Xmut1 and Xmut2) on the StuI B construct were mutated. Each X and Y mutant StuI B construct was co-transfected into EL4 cells along with pON405 LacZ construct. Wild-type StuI B and HincII constructs were also transfected as controls.
To determine whether the CD5X or CD5Y site is responsible for basal level expression, we generated three mutations encompassing the CD5X region (Xmut1, Xmut2, and Xmut3) and two mutations on the CD5Y region of the HincII B reporter plasmid (Ymut1 and Ymut2). These mutant reporter constructs were transiently transfected into EL4 cells and subsequently analyzed for luciferase reporter activity.
None of the three mutated sites on the CD5X region had any effect on the basal level of transcription (figure 5b). In contrast, both mutations in the CD5Y region resulted in reduction of the reporter activity. In fact, the luciferase activity was reduced almost to background levels with the Ymut1 mutation. Thus, we conclude that the CD5Y, particularly the Ymut1 site, is important for basal transcription.
Both CD5X and CD5Y regions affect full CD5 expression
To determine whether either the CD5X or the CD5Y region can affect the full level of transcription controlled by the 43 bp CD5 regulatory promoter, we generated the mutations described above in the CD5X and CD5Y regions in the StuI B reporter construct. The Xmut2 mutation had a measurable effect (2-fold decrease) on the full level of transcription and the Ymut2 mutation reduced the reporter activity by more than three-fold (figure 5c). However, the Ymut1 mutation decreased the luciferase activity more than five fold, and brought the transcription activity to slightly below basal levels of CD5 transcription (figure 5c). These results demonstrate that both CD5X and CD5Y regions contribute to overall CD5 transcription by modifying the transcriptional activation from the 43 bp regulatory region and that the most important regulation comes from the CD5Y region.
EMSA analysis revealed specific proteins binding to the CD5X and CD5Y regions
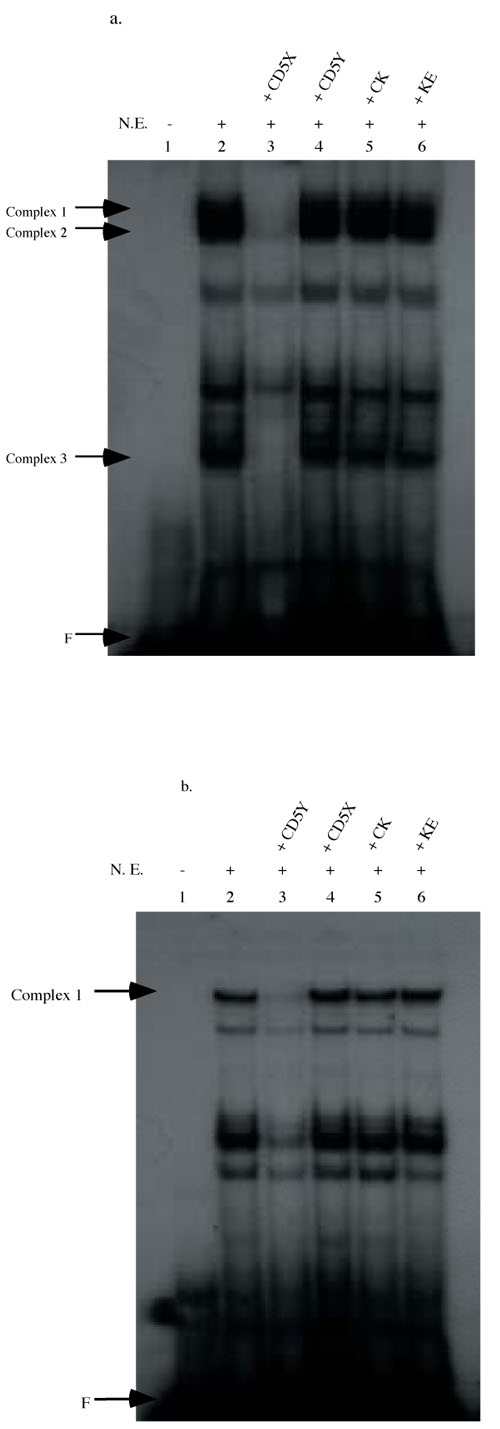
We performed EMSA using oligonucleotide probes containing either the CD5X or the CD5Y region to characterize protein binding to the respective regions. As figure 6a shows, several protein complexes can be seen binding to the 32P-labeled CD5X oligo (lane 2). Competition with 100-fold excess unlabeled CD5X oligo eliminated complexes 1-3 (figure 6a, lane 3). These complexes are specific to the CD5X probes, as they can not be competed away with the addition of excess irrelevant oligos, CD5Y, CK, and KE (figure 6a, lanes 4-6).
Figure 6.
EMSA analysis with oligonucleotides corresponding to the CD5X region. The 32P-labeled CD5X oligonucleotide (5' AGG CCT AAG TTG ACA GTT CAA CTT CAA ACA CTC GAG 3') was incubated with EL4 nuclear extract (lane 2-6). Competitive EMSA was performed with 100-fold excess specific oligonucleotide, X oligos (lane 3), or irrelevant oligos: Y oligos (lane 4); CK oligos (lane 5); and KE (lane 6). The protein:DNA complexes specific to the CD5X oligonucleotides are indicated with arrows. F indicates the migration of the free probe.
EMSA analysis with oligonucleotides corresponding to the CD5Y region. The 32P-labeled CD5Y oligonucleotide (5' AGG CCT CAC AGG CCC ACA CTG CCT GCT TCC CTG GAG 3') was incubated with EL4 nuclear extract and subsequently resolved in EMSA. The protein:DNA complexes specific to the CD5Y oligonucleotides were indicated with arrows. In lane 1 is 32P-labeled CD5Y oligos alone; lanes 2-6, 32P labeled CD5Y oligos plus EL4 nuclear extract. Competitive EMSA are shown on lanes 3-6 with addition of 100-fold molar excess oligonucleotides for competition: CD5Y (lane 3), CD5X (lane 4), CK (lane 5), and KE (lane 6). The protein:DNA complex specific to the CD5Y oligonucleotides are indicated with arrows. F indicates the migration of the free probe.
Similarly, several protein:DNA complexes can be seen in EMSA with 32P-labeled CD5Y oligo (figure 6b, lane 2). Competition analysis demonstrated that complex 1 can specifically be competed away by excess unlabeled CD5Y oligo (figure 6b, lane 3) but not by excess, irrelevant CD5X, CK, and KE oligos (figure 6b, lanes 4-6).
Thus we conclude that there are distinct intracellular proteins that specifically bind to CD5X and CD5Y and likely mediate the transcriptional activity of these newly recognized sites.
Discussion
Studies here define the regions within the CD5 promoter that control transcriptional regulation in resting (unstimulated) T cells growing in culture. By constructing a deletion series of CD5 promoter constructs using luciferase as the reporter gene, we demonstrate that a 43 bp region between the StuI and HincII sites at -215 to -172 is necessary for normal, resting CD5 expression level in T cells. Further, we show that this region contains an Ets binding site that regulates CD5 transcription, two previously unrecognized regions that also control CD5 expression, one of which is responsible for controlling basal expression of the promoter.
Weichert and Schwartz [25] have also reported a deletion analysis of the CD5 promoter. Several of their deletions overlap the deletions we report. For those constructs which were similar in both studies, our results agree. However, these workers observed negative regulation with a construct that that we did not have. None of our constructs showed negative regulation. We investigated three potential transcription factor binding sites (CCAAT, κE2, and Ets) within the 43 bp regulatory promoter region and showed that only the Ets binding site influences CD5 expression in EL4 T cells. Further, we showed that the Ets-1 protein binds to the Ets site and that overexpression of Ets-1 leads to increased promoter activity. Thus we conclude that Ets-1 positively regulates CD5 expression levels in T cells.
Theses findings are consistent with previous evidence showing that Ets-1 levels in T cell subsets correlate with the expression of CD5 on these subsets (see Introduction to this paper). In addition, they are consistent with evidence demonstrating that CD5 expression is drastically reduced in gene targeted mice in which the Ets-1 locus is disrupted. While the possibility that the decreased CD5 surface expression that was demonstrated in the gene targeted mice is due to post-translational rather than transcriptional effects, our co-transfection studies with the Ets-1 expression plasmid and the CD5 promoter constructs demonstrate argue that Ets-1 is regulating transcription in all cases. Thus, the weight of our findings, in combination with findings with presented by others, leads us to conclude that the distinctive CD5 expression levels observed in T cell subsets is due to the different amounts of Ets-1 protein in the cells in these subsets. This conclusion does not eliminate the possibility that Ets family members other than Ets-1 also participate in the regulation of CD5 expression. PU.1, for example, could be important in the regulation of CD5 expression on B-1 cells, since it is predominantly expressed in B cells and macrophages [35, 36]. In fact, preliminary evidence from our laboratory suggests that this may indeed be the case. Ets-2, which is ubiquitously expressed, could also contribute to CD5 regulation [29, 37, 38]. However, the data from Ets-1 gene targeted mice argues against this possibility since disruption of the Ets-1 gene is sufficient to prevent CD5 expression on T cells. Therefore, Ets-1 is the likely regulatory molecule in T cells.
We also characterized two regions (CD5X and CD5Y) that contribute to CD5 expression. These two regions, which contain previously unrecognized transcription binding factor sequence, are conserved between mouse and human. Our results showed that the CD5Y region is necessary for basal level of CD5 transcription. Computer analysis of the CD5Y region did not reveal any potential transcription factor binding site or a canonical TATA box for establishing the typical RNA polymerase II transcription complex. However, the Ymut1 site contains a very weak homology to the TdT/ML intiator consensus sequence, 5'-PyPyCAPyPyPyPyPy-3' [39]. The Ymut2 site, which is the most important site for controlling basal expression, overlaps a potential initiator site fitting the consensus (5' CTT CCC TTT TCC 3') for the ribosomal protein Inr family with only one mismatched base [40]. Interestingly, both Ymut1 and Ymut2 are positioned near the most upstream transcriptional start site [25].
The initiator (Inr) element has been shown to be critical in positioning RNA polymerase II complex for transcription initiation in TATA-less promoters [39]. Since the CD5 promoter does not contain a TATA box, these initiator binding sites may play a more important role in establishing transcription initiation.
Several Inr binding protein have been identified: TFII-I [41], YY1 [42, 43], and a 150 kDa TATA binding protein (TBP) associated factor (TAFII150) [44]. Recognition of the Inr element by these Inr binding proteins (ITF) provides a way for formation of transcription competent complex via their interaction with TBP or with other subunits of RNA polymerase II. Thus we suggest that CD5Y region interacts with these ITFs to regulate transcription.
Like CD5Y, CD5X region does not contain any consensus transcription factor binding sites. This region is not necessary for basal CD5 transcription but it affects the overall level of CD5 transcription obtained with the full length 3 Kb promoter. At least three specific protein:DNA complexes are formed in EMSA using the CD5X oligonucleotides. We speculate that proteins binding to the CD5X region may interact with other upstream transcription factors, e.g., Ets-1, rather than with proteins binding to the CD5Y region. This interaction, we suggest, acts to stabilize a transcriptional competent complex and thereby to increase the CD5 expression to normal levels.
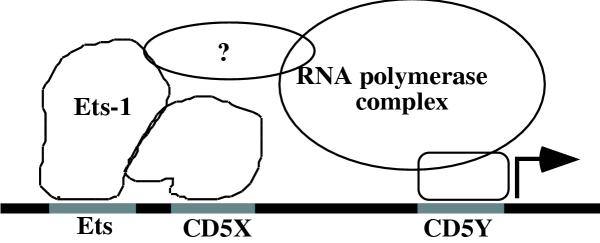
This suggestion underlies the model proposed in figure 6 for regulating CD5 expression in T cells. In this model, the CD5Y region sets up the basal transcription machinery by interacting with components in the RNA polymerase II transcription complex and/or with transcription initiators, since CD5 lacks a TATA box for establishing a typical RNA polymerase II trancription initiation complex. The CD5X region recruits transcription factors to the upstream 43 bp region identified as necessary for overall CD5 expression.
The proteins binding to the CD5X region in this model recruit Ets-1 protein (and perhaps other Ets family members) to the Ets binding site. Alternatively, these proteins help to alleviate the internal Ets-1 inhibition domains and stabilize the binding of Ets-1 to its cognitive site [33, 34, 45]. In any event, they facilitate the interaction of Ets-1 with other factors to form activate transcription and increase it from the basal to the overall level.
Our studies here focus on characterizing the promoter elements responsible for regulating the constitutive CD5 expression in unstimulated T cells. These elements, we have shown, exist in a region proximal to the translation start and quite distant from an enhancer element shown in other studies [46,47,48] to regulate the activation of CD5 expression in BCR-stimulated LPS-induced B cell blasts. This enhancer element, located at approximately 2 kb upstream of the translation start on the CD5 promoter, is dependent on NF-AT activation to induce CD5 expression in anti-IgM stimulated primary splenic cells cultured with LPS for 3 days [48]. It is reported to interact with proteins in the proximal region of the CD5 promoter, which contains the all three regions characterized here.
An Ets binding site also has been implicated in the activity of the enhancer discussed above. In addition, this CD5 response to BCR stimulation has been shown to be mediated in part by vav-2, which activates of NF-AT [49]. Our studies with constitutive expression of CD5 on B cells are still in progress. However, preliminary evidence suggests that unlike the stimulated CD5 expression in these other studies, the low-level constitutive expression of CD5 on the M12 B cell line is enabled by the same 43 bp regulatory region that we have shown here is sufficient to induce full constitutive CD5 expression on T cells.
Materials and Methods
Plasmids
The reporter constructs were created as follows: The 3 kb genomic fragment upstream of the CD5 translation start was excised by restriction digest with HindIII and NcoI from p12-1 [32], a genomic DNA plasmid containing the first two CD5 exons and the 3 kb upstream region. Various deletion constructs were created by first digesting the 3 kb genomic fragment (HindIII-NcoI) with various restriction enzymes, blunted with Klenow DNA polymerase, and then cloned into pGL2Basic luciferase reporter plasmid. The resulting deletion reporter plasmids were further modified by mutating the blunted Nco I sites to Bgl II sites. The structure of all reporter constructs were confirmed by DNA sequencing (Amplitaq sequencing kit, Perkin-Elmer).
The Ets-1 expression plasmid, pEVRF-ets-1, contains the parental expression plasmid pEVRF0 and the insertion of the Ets-1 cDNA. The control expression plasmid, pEVRF0, contains the human CMV enhancer/promoter, the translation initiation from HSV thymidine kinase, splicing and polyadenylation signals from the rabbit β-globin gene, and the SV40 origin of replication in the pSP65 bacterial plasmid.
For internal control in transient transfections, we used PON405, which contains LacZ under the transcriptional control of human CMV promoter. The resulting β-galactosidase activity was used to normalize transient transfection results. Mutagenized constructs based on StuI B or HincII plasmid are described in the site-directed mutagenesis section.
Cell culture
Murine thymoma EL4 cells and murine CD5+ B cells, M12, were maintained in RPMI 1640 medium supplemented with 10% (v/v) 1:1 fetal calf serum and horse serum, 2 mM glutamine, 100 U/ml penicillin, 0.05 mM streptomycin, and 50 nM 2-mercaptoethanol. All cell cultures were maintained at 37°C with 7% CO2.
Transient transfections
EL4 or M12 cells were grown to 1 - 2 × 106 cells per ml, centrifuged and resuspended at a concentration of 107 cells/300 μl in complete RPMI medium. Each aliquot (300 μl) of the cells was electroporated along with 15 μg of reporter constructs and 5 μg of PON405, in a Bio-Rad gene pulser at 250 volts with 960 μD capacitance. The electroporated cells were diluted in 3 ml of complete RPMI 1640 medium and maintained at 37°C for 36-48 hours. At the time of analysis, 1.5 ml of the transfected cells were transferred to eppendorf tubes, centrifuged and lysed in 320 μl of lysis buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 0.1% Triton X-100 at pH7).
Luciferase activity was measured using a luminometer (Analytical Luminescence Laboratory). The β-galactosidase activity in each cell lysate was measured in the 4-Methylumbelliferyl-β-D-Galactoside (MUG) assay and was used to normalize the luciferase activity analyzed in each cell lysate. The reported values represent the average of three independent transfections, with standard deviation as error bars. Each experiment was repeated at least twice.
Nuclear extract isolation
Nuclear extracts were isolated from EL4 and M12 as follows. The cultured cells were grown to 1-2 × 106 cells/ ml, then collected and hypotonically swollen in buffer A (10 mM Hepes pH7.9, 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and 1 mM PMSF) before lysis with NP40. The nuclear proteins were extracted by resuspending the nuclear pellet with buffer C (50 mM Hepes pH7.9, 300 mM NaCl, 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and 1 mM PMSF). Nuclear extracts were quantitated using Bio-Rad protein assay (Bio-Rad Laboratories, CA).
Sequence analysis and site-directed mutagenesis
The consensus transcription factor binding sites were identified by comparing the CD5 promoter sequences against Transcription Factor Database using Quest program (Intelligenetics Corporation). The mutagenesis primers corresponding to each of the five regions, Ets (mutE), κE2 (mutK), CCAAT (mutC), CD5X (Xmut1, 2, and 3), and CD5Y (Ymut1 and 2) were synthesized at the Protein and Nucleic Acid Facility, Stanford University. The mutC, mutK, mutE, Xmut, and Ymut mutagenesis primers were first PCR amplified with GL2 primer (5' CTT TAT GTT TTT GGC GTC TTC CA 3') using StuI B plasmid as template to generate megaprimers. For the HincII B plasmid, Xmut and Ymut mutagenesis primers were used in conjunction with GL2 for PCR amplification.
The mutagensis primers were as follows: for mutC, 5' GCTCGACCT CCA TTC TAA TGT GGG CAG GTG GTT TCA CAG 3'; mutK, 5' CCA TTC TAA TTG GGG CTT TTT GTT TCA CAG GGA GGA AGT 3'; mutE, 5' TTT CAC AGG GAC CTT GTT GAC AGT TCA ACT TCA AAC 3'; Xmut1, 5' CAC AGG GAG GAA GTT CTG TCA TCA ACT TCA AAC AGG 3'; Xmut2, 5' GAG GAA GTT GAC AGT CTA AGA CTA AAC AGG GTT GGC AGT 3'; Xmut3, 5' TGA CAG TTC AAC TTC GTT AGT GGT TGG CAG TGA CAC 3'; Ymut1, 5' TGG CAG TGA CAC AGG TGT ACT CTG CCT GCT TCC CCT 3'; and Ymut2, 5' ACA CAG GCC CAC ACT GAT GCG TTC CCC TTT CCA CCC CTG 3'. The underlined sequences denote the consensus transcription factor binding site. Boldfaced sequences indicate the mutated base pairs.
Each of the megaprimers was purified and subsequently amplified with GL1 primer (5' TGT ATC TTA TGG TAC TGT AAC TG 3'), using StuI B or HincII B plasmid as a template. The resulting PCR products were gel isolated, digested with BglII and MluI, and cloned into the pGLBasic luciferase reporter construct. E. coli clones carrying the mutagenized plasmids were identified by colony lifts and each mutagenized plasmid was sequenced to confirm the mutations.
EMSA
The double stranded oligonucleotide probes were end-filled with Klenow DNA polymerase and α-32P-dGTP. The double stranded oligos used were as follows: for the Ets binding site on the CD5 promoter, 5' CGC GTA CAG GGA GGA AGT TGA CCT CGA 3'; for the oligonucleotides containing the CD5X region, 5' AGG CCT AAG TTG ACA GTT CAA CTT CAA ACA CTC GAG 3'; and for the CD5Y oligo, 5' AGG CCT CAC AGG CCC ACA CTG CCT GCT TCC CTG GAG 3'. The probes were purified using the Qiaquick nucleotide removal kit (Qiagen, Germany), and 5 × 105 cpm of the probe were incubated with nuclear extracts at room temperature for 30 minutes. Competitor oligonucleotides or inhibition antibodies were added 20 minutes before the addition of the radiolabeled probe DNA.
The competitor oligos are as follows: CD5 κE2, 5' GTA CCT AGG GGC AGG TGG TTT CAC GCG 3'; CD5 CK, 5' CCT CCA TTC TAA TTG GGG CAG GTG GTT TCA CAG G 3'; and CD5 KE, 5' AGG CCT GGG GCA GGT GGT TTC ACA GGG AGG AAG TTC TCG AG 3'. The CK oligo contains CD5 κE2 and CCAAT sites. The KE oligo contains κE2 and Ets sites. The binding reaction consisted of 10 mM Hepes pH7.9, 10 mM Tris-HCl pH 7.5, 100 mM NaCl, 15 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 2 μg dIdC.
The protein:DNA complexes were resolved by electrophoresis through 5% nondenaturing polyacrylamide gels in 0.25 X TBE buffer. Electrophoresis was conducted for 2 1/2 hours at 150 V. The gels were then vacuum dried and exposed to X-ray film overnight with an intensifying screen screen at -80°C.
Figure 7.
Proposed model for CD5 transcription. Proposed model depicting the potential interaction between Ets-1 and proteins binding to the CD5X and CD5Y regions in forming a transcriptional competent complex.
Acknowledgments
Acknowledgments
We thank Dr. B. Graves for the pEVRF-ets-1 and pEVRF0 expression plasmids. We also thank Dr. Yang Yang for criticism and discussion in the framing of this manuscript. We thank Mr. John Mantovani for help with the preparation of the manuscript, and we thank Ms. Ometa Herman and other members of the Herzenberg laboratory for technical and other help as this study progressed.
Contributor Information
James W Tung, Email: james.tung@genemed.com.
Shaun S Kunnavatana, Email: skunn@stanford.edu.
Leonard A Herzenberg, Email: lenherz@darwin.stanford.edu.
Leonore A Herzenberg, Email: leeherz@darwin.stanford.edu.
References
- Huang HJ, Jones NH, Strominger JL, Herzenberg LA. Molecular cloning of Ly-1, a membrane glycoprotein of mouse T lymphocytes and a subset of B cells: molecular homology to its human counterpart Leu-1/T1 (CD5). Proc Natl Acad Sci U S A. 1987;84:204–208. doi: 10.1073/pnas.84.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The "Ly-1 B" cell subpopulation in normal immunodefective, and autoimmune mice. J Exp Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor AB, Herzenberg LA. Origin of murine B cell lineages. Annu Rev Immunol. 1993;11:501–538. doi: 10.1146/annurev.iy.11.040193.002441. [DOI] [PubMed] [Google Scholar]
- Herzenberg LA, Stall AM, Lalor PA, Sidman C, Moore WA, Parks DR, Herzenberg LA. The Ly-1 B cell lineage. Immunol Rev. 1986;93:81–102. doi: 10.1111/j.1600-065x.1986.tb01503.x. [DOI] [PubMed] [Google Scholar]
- Tung JW. Molecular characterization of CD5 expression: study of the murine CD5 promoter Stanford University; Thesis. 1997.
- Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick D, Pearson A, Krieger M. The SRCR superfamily: a family reminiscent of the Ig superfamily. Trends Biochem Sci. 1994;19:5–8. doi: 10.1016/0968-0004(94)90165-1. [DOI] [PubMed] [Google Scholar]
- Van de Velde H, von Hoegen I, Luo W, Parnes JR, Thielemans K. The B-cell surface protein CD72/Lyb-2 is the ligand for CD5. Nature. 1991;351:662–665. doi: 10.1038/351662a0. [DOI] [PubMed] [Google Scholar]
- Biancone L, Bowen MA, Lim A, Aruffo A, Andres G, Stamenkovic I. Identification of a novel inducible cell-surface ligand of CD5 on activated lymphocytes. J Exp Med. 1996;184:811–819. doi: 10.1084/jem.184.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisil R, Fitts MG, Mage RG. CD5 is a potential selecting ligand for B cell surface immunoglobulin framework region sequences. J Exp Med. 1996;184:1279–1284. doi: 10.1084/jem.184.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyers AD, Spruyt LL, Williams AF. Molecular associations between the T-lymphocyte antigen receptor complex and the surface antigens CD2, CD4, or CD8 and CD5. Proc Natl Acad Sci U S A. 1992;89:2945–2949. doi: 10.1073/pnas.89.7.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess KE, Yamamoto M, Prasad KV, Rudd CE. CD5 acts as a tyrosine kinase substrate within a receptor complex comprising T-cell receptor zeta chain/CD3 and protein-tyrosine kinases p56lck and p59fyn. Proc Natl Acad Sci U S A. 1992;89:9311–9315. doi: 10.1073/pnas.89.19.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankester AC, van SG, Cordell JL, van NC, van LR. CD5 is associated with the human B cell antigen receptor complex. Eur J Immunol. 1994;24:812–816. doi: 10.1002/eji.1830240406. [DOI] [PubMed] [Google Scholar]
- June CH, Rabinovitch PS, Ledbetter JA. CD5 antibodies increase intracellular ionized calcium concentration in T cells. J Immunol. 1987;138:2782–2792. [PubMed] [Google Scholar]
- Ceuppens JL, Baroja ML. Monoclonal antibodies to the CD5 antigen can provide the necessary second signal for activation of isolated resting T cells by solid-phase- bound OKT3. J Immunol. 1986;137:1816–1821. [PubMed] [Google Scholar]
- Ledbetter JA, June CH, Grosmaire LS, Rabinovitch PS. Crosslinking of surface antigens causes mobilization of intracellular ionized calcium in T lymphocytes. Proc Natl Acad Sci U S A. 1987;84:1384–1388. doi: 10.1073/pnas.84.5.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N, Rajewsky K. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- Bikah G, Carey J, Ciallella JR, Tarakhovsky A, Bondada S. CD5-mediated negative regulation of antigen receptor-induced growth signals in B-1 B cells. Science. 1996;274:1906–1909. doi: 10.1126/science.274.5294.1906. [DOI] [PubMed] [Google Scholar]
- Reth M. Antigen receptor tail clue [letter]. Nature. 1989;338:383–384. doi: 10.1038/338383b0. [DOI] [PubMed] [Google Scholar]
- Unkeless JC, Jin J. Inhibitory receptors, ITIM sequences and phosphatases. Curr Opin Immunol. 1997;9:338–343. doi: 10.1016/S0952-7915(97)80079-9. [DOI] [PubMed] [Google Scholar]
- Davies AA, Ley SC, Crumpton MJ. CD5 Is Phosphorylated on Tyrosine After Stimulation of the T-Cell Antigen Receptor Complex. Proc Natl Acad Sci USA. 1992;89:6368–6372. doi: 10.1073/pnas.89.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Yamamoto M, Rudd CE. The T-cell antigen CD5 acts as a receptor and substrate for the protein- tyrosine kinase p56lck. Mol Cell Biol. 1994;14:2862–2870. doi: 10.1128/mcb.14.5.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Villar JJ, Whitney GS, Bowen MA, Hewgill DH, Aruffo AA, Kanner SB. CD5 negatively regulates the T-cell antigen receptor signal transduction pathway: involvement of SH2-containing phosphotyrosine phosphatase SHP-1. Mol Cell Biol. 1999;19:2903–2912. doi: 10.1128/mcb.19.4.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary-Gouy H, Bruhns P, Schmitt C, Dalloul A, Daeron M, Bismuth G. The pseudo-immunoreceptor tyrosine-based activation motif of CD5 mediates its inhibitory action on B-cell receptor signaling. J Biol Chem. 2000;275:548–556. doi: 10.1074/jbc.275.1.548. [DOI] [PubMed] [Google Scholar]
- Weichert TR, Schwartz RC. Cloning of the murine CD5 promoter and its tissue-specific regulation. J Immunol. 1995;154:4603–4612. [PubMed] [Google Scholar]
- Calvo J, Sole J, Simarro M, Vives J, Lozano F. Evolutionarily conserved transcription regulatory elements within the 5'-flanking region of the human CD5 gene. Tissue Antigens. 1996;47:257–261. doi: 10.1111/j.1399-0039.1996.tb02551.x. [DOI] [PubMed] [Google Scholar]
- Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Gunther CV, Nye JA, Bryner RS, Graves BJ. Sequence-specific DNA binding of the proto-oncoprotein ets-1 defines a transcriptional activator sequence within the long terminal repeat of the Moloney murine sarcoma virus. Genes Dev. 1990;4:667–679. doi: 10.1101/gad.4.4.667. [DOI] [PubMed] [Google Scholar]
- Bhat NK, Komschlies KL, Fujiwara S, Fisher0 RJ, Mathieson BJ, Gregorio TA, Young HA, Kasik JW, Ozato K, Papas TS. Expression of ets genes in mouse thymocyte subsets and T cells. J Immunol. 1989;142:672–678. [PubMed] [Google Scholar]
- Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- Huang HS. Molecular cloning and characterization of the murine lymphocyte differentiation antigen Ly1-1 (murine CD5)Stanford University; 1987.
- Jonsen MD, Petersen JM, Xu QP, Graves BJ. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J, Grosschedl R. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci U S A. 1992;89:8889–8893. doi: 10.1073/pnas.89.19.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene [see comments]. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- Hromas R, Orazi A, Neiman RS, Maki R, Van Beveran C, Moore J, Klemsz M. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood. 1993;82:2998–3004. [PubMed] [Google Scholar]
- Kola I, Brookes S, Green AR, Garber R, Tymms M, Papas TS, Seth A. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc Natl Acad Sci U S A. 1993;90:7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9:1551–1565. [PubMed] [Google Scholar]
- Weis L, Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. Faseb J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- Hariharan N, Perry RP. Functional dissection of a mouse ribosomal protein promoter: significance of the polypyrimidine initiator and an element in the TATA- box region. Proc Natl Acad Sci U S A. 1990;87:1526–1530. doi: 10.1073/pnas.87.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL, Meisterernst M, Pognonec P, Roeder RG. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- Hariharan N, Kelley DE, Perry RP. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci U S A. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Yokomori K, Chen JL, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- Skalicky JJ, Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;5:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong YZ, Rabin E, Wortis HH. Treatment of murine CD5- B cells with anti-Ig, but not LPS, induces surface CD5: two B-cell activation pathways. Int Immunol. 1991;3:467–476. doi: 10.1093/intimm/3.5.467. [DOI] [PubMed] [Google Scholar]
- Teutsch M, Higer M, Wang D, Wortis HW. Induction of CD5 on B and T cells is suppressed by cyclosporin A, FK- 520 and rapamycin. Int Immunol. 1995;7:381–392. doi: 10.1093/intimm/7.3.381. [DOI] [PubMed] [Google Scholar]
- Berland R, Wortis HH. An NFAT-dependent enhancer is necessary for anti-IgM-mediated induction of murine CD5 expression in primary splenic B cells. J Immunol. 1998;161:277–285. [PubMed] [Google Scholar]
- Doody GM, Billadeau DD, Clayton E, Hutchings A, Berland R, McAdam S, Leibson PJ, Turner M. Vav-2 controls NFAT-dependent transcription in B- but not T-lymphocytes. Embo J. 2000;19:6173–6184. doi: 10.1093/emboj/19.22.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]