Abstract
Human immunodeficiency virus type 1 (HIV-1), the pathogen of acquired immunodeficiency syndrome (AIDS), causes ~2 millions death every year and still defies an effective vaccine. HIV-1 infects host cells through envelope protein – mediated virus-cell fusion. The transmembrane subunit of envelope protein, gp41, is the molecular machinery which facilitates fusion. Its ectodomain contains several distinguishing functional domains, fusion peptide (FP), N-terminal heptad repeat (NHR), C-terminal heptad repeat (CHR) and membrane proximal extracellular region (MPER). During the fusion process, FP inserts into the host cell membrane, and an extended gp41 prehairpin conformation bridges the viral and cell membranes through MPER and FP respectively. Subsequent conformational change of the unstable prehairpin results in a coiled-coil 6-helix bundle (6HB) structure formed between NHR and CHR. The energetics of 6HB formation drives membrane apposition and fusion. Drugs targeting gp41 functional domains to prevent 6HB formation inhibit HIV-1 infection. T20 (enfuvirtide, Fuzeon) was approved by the US FDA in 2003 as the first fusion inhibitor. It is a 36-residue peptide from the gp41 CHR, and it inhibits 6HB formation by targeting NHR and lipids. Development of new fusion inhibitors, especially small molecule drugs, is encouraged to overcome the shortcomings of T20 as a peptide drug. Hydrophobic characteristics and membrane association are critical for gp41 function and mechanism of action. Research in gp41-membrane interactions, using peptides corresponding to specific functional domains, or constructs including several interactive domains, are reviewed here to get a better understanding of gp41 mediated virus-cell fusion that can inform or guide the design of new HIV-1 fusion inhibitors.
Introduction
Human immunodeficiency virus type 1 (HIV-1) is the pathogen of acquired immunodeficiency syndrome (AIDS), which has caused ~60 million infections and ~25 million deaths worldwide since the disease was first identified in the early 1980s [1–3]. Currently, ~33 million people live with HIV-1/AIDS with another 2 million new infections added yearly, resulting in more than 2 million deaths every year. In the absence of an efficient vaccine in the foreseeable future, antiretroviral therapy (ART), which uses synthetic drugs to prevent the development of AIDS, is the only effective way to treat persons infected with HIV-1 [4]. Currently ~30 antiretroviral drugs are in use for treatment of HIV/AIDS patients, including nucleotide reverse transcriptase inhibitors (NRTI) [5, 6], non-nucleotide reverse transcriptase inhibitors (NNRTI) [7], protease inhibitors (PI) [8, 9], entry/fusion inhibitors [10, 11], and integrase inhibitors [12].
The main challenge to ART is the superior plasticity of the HIV-1 genome and amino acid sequences and the resulting drug resistance which is usually observed at the clinical trial stage and can develop rapidly in patients treated with single drug. The introduction of highly active antiretroviral therapy (HAART), also name cocktail therapy in 1990s [13–15], fundamentally changed the nature of HIV-1/AIDS treatment. Under the HAART regimen, drugs from at least two different classes were recommended. Essentially, it is very hard for viruses to develop resistance against drugs used simultaneously against different targets. As a result, HAART could effectively prevent the development of full-blown AIDS in HIV-1 infected patients under proper treatment and with good patient adherence. Now, persons infected with HIV-1 in the developed world have an expected lifespan close to that of healthy individuals [4]. Because HIV-1 infection is a permanent infection, drug resistance to most available drugs is always possible [16]. Consequently, there is a constant requirement for new drugs, especially new classes of drugs targeting previously unexploited targets. The side effects of the currently used drugs and the high cost of currently available HAART regimens (~10,000 USD/year/patient) also encourage drug developers to provide less toxic and cheaper antiretroviral drugs [17, 18].
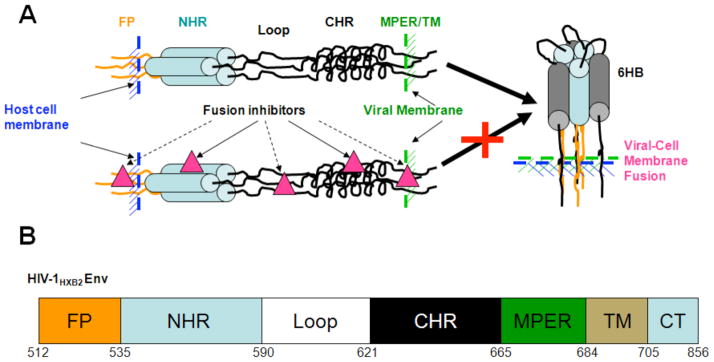
HIV-1 uses a class I fusion protein to enter and infect host cells [19–21]. The fusion machinery HIV-1 envelope protein (ENV) is a complex of non-covalently associated 120 and 41 kilo-Dalton (kD) glycoproteins (gp120 and gp41, respectively). The complex forms a trimer on the HIV surface, with the trimerized metastable gp41 transmembrane subunit sequestered by three gp120 surface subunits. The fusion process is initiated by the binding of HIV-1 gp120 to the primary receptor, CD4, and a co-receptor, CCR5 or CXC4 on the target cell. Receptor and co-receptor binding guides the HIV-1 virion close to the target cell. Receptor binding also causes large conformational changes of gp120, resulting in dissociation of the gp120/gp41 complex and subsequent release of metastable gp41, initiating virus-cell fusion by gp41 ectodomain. Gp41 ectodomain contains several distinguishing functional domains, including fusion peptide (FP), N-terminal heptad repeat (NHR), C-terminal heptad repeat (CHR), and membrane proximal extracellular region (MPER) (Figure 1). In the fusion process, FP first inserts into the target cell membrane to form an extended prehairpin conformation with the C-terminal MPER rooted in the viral membrane; thus gp41 bridges the viral and target cell membranes (Figure 1). The pre-fusion structure undergoes further conformational change with the CHR folding back along the NHR to form a coiled-coil six-helix bundle (6HB) structure. The energetics of 6HB formation drives the apposition of viral and host cell membranes and results in fusion. Fusion inhibitors, targeting gp41 functional domains to prevent the 6HB formation and terminate the HIV-1-cell fusion process, can be used as drugs to prevent HIV-1 infection.
Figure 1.
HIV-1 gp41 mediated virus-cell fusion and fusion inhibitors. Upper panel: The putative HIV-1 gp41 pre-fusion structure is an extended conformation with its N-terminal fusion peptide inserting into target cell, followed sequentially by N-terminal heptad repeat (NHR), Loop region (Loop), C-terminal heptad repeat (CHR), transmembrane domain (TM) and cytoplasmic tail (CT), thus bridging the HIV-1 and target cell membrane. NHR and CHR will automatically fold back to form a low energy stable six-helical bundle (6HB) with NHR trimer as the inner core and anti-parallel binding of three CHRs. The energy from the 6HB formation will drive the juxtaposition of the viral and cell membrane and result in viral-cell fusion. Drugs binding to specific functional domains of gp41 may interfere with the fusogenic 6HB formation thus inhibiting HIV-1 infection. Solid lines indicate established fusion inhibitor targets, dashed lines indicate potential fusion inhibitor targets. Lower panel: cartoon representation of the primary structure of HIV-1 gp41 and the different functional domains. The sequence number was based on HIV-1HXB2 ENV protein sequence.
Peptides derived from gp41 NHR or CHR sequences inhibit HIV-1 infection by interaction with their counterparts in gp41 [22, 23]. The first reported peptide fusion inhibitor was DP107 (16, gp41553–590), an NHR peptide [24], followed by more potent CHR peptides SJ2176 (26, gp41630–659) [25, 26], and DP178 (29, gp41638–673) [27, 28]. In 1997, the X-ray structure of HIV-1 gp41 fusion core was resolved showing a 6HB consisting of three NHRs that form the inner core and three CHRs that are associated with the NHRs in anti-parallel fashion [29–31]. Based on the 6HB core structure, the mechanism of action for this new class of anti-HIV drugs was proposed: by forming unproductive 6HB, the viral-cell fusion process is inhibited, thus preventing HIV-1 infection. The crystallographic structure also showed that the NHR inner core has a deep pocket which interacts critically with CHR, and can therefore be a target for small molecule fusion inhibitors [32].
DP178 (29), later named T20 (29) (brand name: Fuzeon, generic name: enfuvirtide), was approved by the U.S. FDA in 2003 as the first fusion inhibitor [10, 11, 33]. Fusion inhibitors interfere with virus-cell membrane fusion and prevent the virus from infecting the target cells, an earlier HIV-1 life cycle process than NRTI/NNRTI and PI. Thus they are considered more effective in preventing and stopping HIV-1 infections. Also, since new targets are exploited, this class of ART is more effective against HIV-1 strains in NRTI/NNRTI - and PI- experienced patients [23, 34].
T20 (29) is the only currently approved drug targeting gp41; the high cost and inconvenience of twice daily injection of the peptide drug prevents it from being considered as a regular ART drug [34]. Development of new drugs targeting gp41 that overcome the limitation of T20 (29) is of significant importance. NHR and CHR are still the most intensively investigated targets in gp41 [3, 35–58]. Other less exploited functional domains, such as FP [59], MPER [60], and a loop region between NHR and CHR [61], are also potential targets for fusion inhibitors.
As a membrane interacting protein, hydrophobicity and hydrophobic interactions of gp41 are critical for its function. FP and MPER have a direct interaction with virus or cell membrane; the membrane properties also have profound effects on the fusion processes involving the HIV-1 gp41 ectodomain and beyond. Understand these interactions may provide new insight towards fully understanding the mechanism of virus-cell fusion. Also the gp41 membrane interaction may provide new targets for HIV fusion inhibitor design. For this purpose, we review the recent developments in gp41-membrane interactions.
1. Methods used for characterization of gp41-membrane interactions
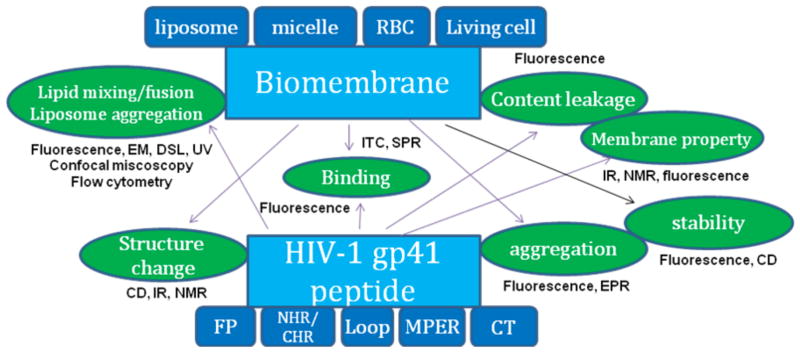
HIV-1 gp41-membrane interactions have been extensively studies by using model peptides from HIV-1 gp41 as well as biomembranes or their mimetics of various types, and their interactions were characterized by different biochemical and biophysical methods. An overview of methods used for gp41 – membrane interaction is depicted in Figure 2, and a detailed description of the different methods is presented below. Abbreviations for techniques, lipids, and peptide domains are delineated in full in the Abbreviations section at the end of this review.
Figure 2.
Methods used to study gp41-membrane interactions. HIV-1 gp41 peptides from different functional domains, including FP, NHR, CHR, Loop, MPER and CT, as shown in Figure 1, were studied with biomembranes and their mimetics, including liposomes, micelles, red blood cells (RBC) and living cells. Property changes incurred by biomembrane - gp41 peptide interactions are shown as icons connected by arrows to the source of the perturbation. These properties changes were studied by various biochemical and biophysical methods as indicated.
1.1 Peptides and membrane mimetics used for gp41-membrane interaction
The peptides used for gp41-membrane interaction studies include model peptides corresponding to a specific HIV-1 gp41 functional domain, well characterized peptide fusion inhibitors, lipopeptides [55, 62–64], and gp41 constructs containing several interactive functional domains up to full length gp41 ectodomain (Tables 1 – 5). Liposomes of various lipid compositions have been prepared to mimic the viral or cellular membrane for studying lipid interaction with gp41 peptides. Liposomes can be constructed bearing different net charge by adding charged lipids. Membrane fluidity can be modulated by altering the gel to liquid crystalline phase transition temperature using different percentages of cholesterol or lipids of different chain lengths.
Table 1.
FP peptide sequences
| Name | Sequence | Literature Name/Ref |
|---|---|---|
| FP12 (1) | AVGIGALFLGFL | [81] |
| FP16 (2) | AVGIGALFLGFLGAAG | [73, 81, 83, 115] |
| FP23 (3) | AVGIGALFLGFLGAAGSTMGARS | [73, 81, 82, 111, 116, 124, 125], gp41 FP, HIVn [68], FP23H [72, 73], HIVArg, [77] FP-1, [87], |
| FP23K3 (3a) | AVGIGALFLGFLGAAGSTMGARSKKK | FPK3 [83] |
| FP23A (3b) | AVGIGALFLGFLGAAGSTMGAS | gp411–23 [76] |
| FP23W8 (3c) | AVGIGALWLGFLGAAGSTMGAS | gp411–23W8 [67] |
| P-FP23A (3d) | PAVGIGALFLGFLGAAGSTMGAAS | P-gp411–23 [76] |
| FP23mut (3e) | AIGLGAMFLGFLGAAGSTMGAS | HIVAla [77] |
| FP23G3K5 (3f) | AVGIGALFLGFLGAAGSTMGARSGGGKKKKK | [117] |
| FP23-H (3h) | NH2–AVGIGALFLGFLGAAGSTMGARS | [73] |
| FP30 (4) | AVGIGALFLGFLGAAGSTMGARSMTLTVQA | [76] |
| FP33 (5) | AVGIGALFLGFLGAAGSTMGARSMTLTVQARQL | [70, 84, 85, 94, 114, 115, 123] |
| FP33V2E (5a) | AEGIGALFLGFLGAAGSTMGARSMTLTVQARQL | [114] |
| FP33-IFFA (5b) | AVGIGALFLGFLGAAGSTMGARSMTLTVQARQL | [94] |
| dFP33 (5c) | AVGIGALFLGFLGAAGSTMGARSMTLTVQARQL | [70] |
| FP33M24C (5d) | AVGIGALFLGFLGAAGSTMGARSCTLTVQARQL | [84] |
| FP34 (6) | AVGIGALFLGFLGAAGSTMGARSMTLTVQARQLL | [75, 126] |
Table 5.
Multidomain constructs and others
| Name | Sequence | Literature Name/Ref |
|---|---|---|
| Gp411–52 (7) | AVGIGALFLGFLGAAGSTMGARSCTLTVQARQLLSGIVQQQNNLLRAIEAQQ | [84] |
| N70 (8) | AVGIGALFLGFLGAAGSTMGARSCTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [78, 84, 85, 92] |
| N70a (8a) | AVGIGALFLGFLGAAGSTMGARSMTLTVQARQLLCGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [75, 126] |
| N70F11G (8b) | AVGIGALFLGGLGAAGSTMGARSCTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [92] |
| N70I62D (8c) | AVGIGALFLGFLGAAGSTMGARSCTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWG KQLQARIL | [84, 92] |
| N70GG (8d) | AVGIGALFLGFLGAAGSTMGARSCGGTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [92] |
| Loop-1 (17) | RILAVERYLKDQQLLGIWGCSGK | Gp41 579~601[152] |
| Loop-2 (18) | RILAVERYLKDQQLLGIWGCSGKLICTTAVPWNAS | Gp41579–613 [80] |
| C56 (41) | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFNITNWLWYIK | [85] |
| N47L6C39 (44) | N47–SGGRGG–C39 | hairpin [78, 126] |
| N70L6C39 (45) | N70–SGGRGG–C39 | FP–hairpin [78, 126] |
| Ectodomain (46) | HIV–1 gp41512–684 | [79] |
| core (47) | HIV–1 gp41538–665 [71, 79] | E. coli recgp41 [71] |
| recgp41 (48) | HIV–1 gp41530–582 | [71] |
| e-core (49) | HIV–1 gp41546–584 | [79] |
| SIV e-gp41 (50) | SIV–1 gp4127–149 | (SIV gp41) [91] |
| FP16-AISb (51) | FP16(2)–Ahx–AISb (31), Ahx was undefined in literature | Hyb [83] |
| Intracell. domain A (52) | RVIEVVQGASRAIRHIPRRIR | [86] |
| CP (42) | GLRILLLKV | Rat TCR [115] |
| TCRα TMD (43) | 251VMGLRILLLKVAGFNLLMTL270 | Rat TCR [115] |
1.2 Peptide-liposome binding assays
Peptide-liposome interactions can be quantified by their binding affinity in the form of association or dissociation constants, and the partition coefficient of peptide between solution and liposome. The binding affinity can be determined directly by using isothermal titration calorimetry (ITC) or surface plasma resonance (SPR) without using a probe [65, 66], although the measurements require expertise in sample preparation and data analysis. Fluorescence measurement provides a fast way to quantify peptide-liposome binding. The intrinsic tryptophan fluorescence of most proteins and peptides (excitation at 280 nm, emission at 330–355nm) is sensitive to liposome membrane binding through the change in the hydrophobic environment of the tryptophan indole ring. This effect results in fluorescence intensity enhancement, fluorescence life time change, and a shift of the emission peak from ~350 nm when fully exposed in solution to shorter wavelengths (blue shift). Through monitoring the fluorescence property changes of a fixed concentration of peptide with different concentrations of liposomes, a binding isotherm can be obtained, from which the association or dissociation constants between peptides and liposomes, and the partition coefficient of peptide between solution and the liposome, can be calculated [67–69].
Most fusion peptides lack a tryptophan residue which can be engineered into the peptide sequence used for binding studies [67]. Another choice is to use fluorescently labeled peptide such as 4-fluoro-7- nitrobenz-2-oxa-1,3-diazole (NBD) labeled peptide. In buffer, NBD-labeled peptides exhibited emission spectra similar to the NBD moiety dissolved in water with low intensity and an emission peak at ~549 nm; in membrane environment, the fluorescence intensity increased by ~10-fold concomitant with a blue shift with an emission peak at ~520 nm [70, 71].
Other methods, such as using fluorescein-phosphatidylethanolamine (FPE) as probe [72–74], or Förster resonance energy-transfer (FRET) between tryptophan of the proteins or peptides and a N-(5-dimethylamino naphthalene-1-sulfonyl)-sn-glycero-3- phosphoethanolamine (DNS-PE) probe included in the liposome, were also reported to study peptide liposome binding [71].
1.3 Lipid mixing and fusion assays
Lipid mixing is the first step of the liposome fusion process. The potency of peptide - induced lipid mixing is assumed to be correlated to its fusogenic activity. FRET between NBD and rhodamine (Rho) fluorescence dye has been widely used for a lipid mixing assay. Liposomes were pre-labeled with NBD-dioleoylphosphatidylethanolamine (PE) or Rho-PE, and the two differently labeled liposomes were co-incubated with peptides. When lipid mixing occurred, FRET was detected between Rho-PE and NBD-PE in the merged liposome. For both lipid mixing and leakage assays, liposomes in 2% Triton X-100 were usually used as a control for complete membrane fusion and leakage [68–70, 72–80].
A self quenching lipid soluble fluorescence probe, octadecyl rhodamine chloride B (R18, excitation at 560 nm, emission at 590 nm), was also used for a lipid mixing assay [81]. Labeled liposomes were prepared by incorporating R18 in dry lipid film at a concentration 5% of the total lipid weight. Labeled and unlabeled liposomes were mixed at a weight ratio 1:4, respectively. Fluorescence intensity enhancement resulting from the dilution of R18 in liposomes indicated lipid mixing.
1.4 Lipid leakage and content mixing assays
Lipid content leakage can be monitored using de-quenching of fluorescence probe enclosed in the liposome. Self quenching of high concentrations of carboxyfluorescein (CF) occurred with liposome inclusion, and was reversed upon leakage of CF from the liposomes, resulting in an increased fluorescence intensity indicating the extent of lipid leakage [74, 77]. FRET quenched fluorescence dyes pairs were also used, such as 8-Aminonaphthalene-1,3,6-trisulfonic acid sodium salt (ANTS)/p-xylyl enebis[pyridinium] bromide (DPX) [68, 80, 82], and 8-hydroxypyrene- 1,3,6-trisulfonic acid (HPTS)/DPX [81, 83].
1.5 Lipid aggregation assays
Lipid mixing and fusion also cause aggregation of liposomes or change the morphology of the liposome membrane. Lipid aggregation can result in cloudiness of the solution, changing the light scattering property of the solution, which can be characterized by dynamic light scattering (DLS). A straightforward method based on absorbance changes at 405 nm was also used for characterizing the lipid aggregations [74, 77, 84]. Electron microscopy (EM) has been used to characterize the morphologic changes of the liposome induced by peptide interactions [69, 75, 79, 84].
1.6 Lipid membrane property changes
Peptide-liposome interaction not only changes the size or morphology, but also changes other properties of liposome membranes. Characterization of these property changes can shed light on the mechanism of peptide-liposome interactions to elucidate their role in gp41 mediated virus-cell fusion. Infrared (IR) and fluorescence methods used to characterize the liposome membrane properties are listed:
Lipid Hydration
The IR carbonyl band of biomembrane mimetics has two main components: carbonyl groups that do not form hydrogen bonds with water and carbonyls hydrogen bonded to water molecules (hydrated carbonyls) [72, 73]. IR can be used to monitor and characterize the lipid hydration properties in the membrane upon peptide binding. IR 1739–1744 cm−1 absorbance is assigned to the stretching vibration of the dehydrated phospholipid carbonyls, 1720–1730 cm−1 is assignable to phospholipid carbonyls which are hydrogen bonded to water molecules, and ~1710 cm−1 corresponds to bihydrated carbonyl groups.
Lipid order
The effect of the peptides on the acyl chain order of membrane lipids can be estimated by comparing the CH2-stretching dichroic ratio of pure multilayers alone with that obtained for identical multilayers with membrane-bound peptide by monitoring the symmetric (vsym ~2850–53 cm−1) and antisymmetric (vantisym ~2920–23 cm−1) vibrations of the lipid methylene C–H bond in attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR) [68, 85].
Other methods have included using 1,6-diphenyl-1,3,5-hexatrienepropionic acid (PA-DPH) and 1-(4-trimethyl ammoniumphenyl)-6-phenyl-1,3,5-hexatriene (TMA-DPH) fluorescence anisotropy to monitor membrane organization dynamics [74], or 1-(3-sulfonatopropyl)-4-[α-[2-(di-n-octylamino)-6-naphthyl]vinyl]pyridinium betaine (di-8-ANEPPS) to probe membrane dipole potential changes [72], and measurements of peptide - induced membrane surface tension changes [77]. Solid state 2D nuclear magnetic resonance (NMR) has been used to obtain site specific order parameters for individual amino acid residues and lipid methylene groups enabling analysis of dynamics at different depths in the membrane [86].
1.7 Peptide-liposome interaction using living biomembrane
Peptide - living membrane interaction can provide direct evidence of the potency of peptides involved in the viral-cell fusion process. The simplest peptide-membrane assay is a hemolysis assay using human red blood cells (hRBC) to assess the potency of peptides to destabilize the cell membrane [63, 87]. Fresh hRBCs were rinsed with phosphate-buffered saline (PBS), followed by centrifugation and resuspension in PBS. Peptides of various concentrations dissolved in PBS were added to the stock hRBC in PBS. The resulting suspension was incubated and the samples were then centrifuged. The release of hemoglobin was monitored by measuring the absorbance of the supernatant at 540 nm. Controls for zero hemolysis (blank) and 100% hemolysis consisted of hRBCs suspended in PBS and PBS with 1% Triton, respectively [63, 87]. A common peptide induced content mixing assay with hRBC uses a self-quenching R18 fluorescence dye, monitoring the fluorescence change upon addition of peptides using excitation and emission wavelengths at 556 and 590 nm, respectively [88].
Lipid mixing and content mixing of living effector and target cells can be measured in a real-time fashion using fluorescence microsopy, confocal microscopy, or flow cytometry. For lipid mixing, 1,10-dioctadecyl-3,3,30,30-tetramethylindo carbocyanine perchlorate (DiI) labeled target cells (red) were cocultured with 3,30-dioctadecyl oxacarbocyanine perchlorate (DiO) labeled effector cells (green); lipid dye mixing was monitored in real-time using fluorescence microscopy. For content mixing, calcein AM (green) labeled effector were cocultured with ((4-chloromethyl)benzoyl)amino)) tetramethylrhodamin (CMTMR, red) labeled target cells, and dye redistribution was monitored in real time with a fluorescence microscope coupled to a CCD camera [89]. In a confocal microscopy and flow cytometry lipid mixing assay, effector cells labeled with DiO and target cells labeled with DiI were cocultured for 2 h at 37°C, fixed with 4% paraformaldehyde and analyzed on a Bio-Rad MRC1024 microscope for confocal microscopy or on an Epics XL flow cytometer for flow cytometry analysis [90].
1.8 Peptide interaction and aggregation assays
The oligomeric state of peptides can be characterized using intrinsic tryptophan fluorescence which is sensitive to the polarity of the environment [91]. Self quenching of the Rho fluorescence was used to detect oligomerization of Rho-N36 (15) molecules, by the concomitant close association of Rho groups [71]. Rho fluorescence is affected only weakly by the dielectric constant of its environment. Membrane-induced conformational changes that move the N-termini of gp41 trimers away from each other can therefore be studied by fluorescence spectroscopy [91]. Other methods that have been used to characterize peptide aggregation include SDS PAGE, [92] or the probe thioflavin which associates rapidly with aggregated peptides, giving rise to a new excitation maximum at 450 nm and a enhanced emission at 482 nm [74].
The conformational change and solvent accessibility of peptide or protein in the liposome has been monitored by electron paramagnetic resonance (EPR) relaxation rates using an external paramagnetic probe to assess the orientation and depth of immersion of selectively spin labeled residues. [93] Protease (Proteinase-K or trypsin) digest of Rho or NBD labeled peptide or protein has also been applied, in which increasing fluorescence intensity occurs with hydrolysis of the protein or peptide, reflecting conformational changes or the position of insertion in the liposome membrane [70, 75, 88].
The association of the peptides in their membrane-bound state was monitored by measuring NBD-Peptide/Rho-Peptide FRET. Random distribution was calculated assuming a Forster radius R0 of 51Å for the NBD/Rho donor-acceptor pair. A significantly higher level of energy transfer between peptide pairs than those expected for randomly distributed donors and acceptors indicated peptide binding [70, 71, 94].
1.9 Structural characterization of gp41-membrane interactions
Atomic resolution structure of biomacromolecules provides critical information for understanding the biologic processes. Although multiple crystallographic structures of gp41 6HB have been solved, as well as several MPER crystallographic structures in complex with antibodies, which may induce structural changes (see Gach et al in this issue) [29–31, 95], a complete picture of gp41 in its multiple conformational states in still lacking. In HIV-1-cell fusion, gp41 structural changes, especially secondary structure changes, are highly correlated to the function of different gp41 domains, providing a key to understand the role played by gp41. Peptide and protein structure in solution and bound with membrane can be characterized by NMR, circular dichroism (CD), or IR.
NMR
NMR can be used to determine structure of biomacromolecules, and is particularly useful for investigating dynamics and intermolecular interactions. Short range NOE’s and J-coupling, long range dipolar coupling and paramagnetic effects, chemical shift mapping and heteronuclear NOE’s and relaxation rates are utilized in conjunction with computational tools to arrive at molecular structure and dynamics. Both NMR and EPR can play a unique role in the study of protein – lipid interactions and peptide orientation in a bilayer [93, 96, 97]. Up to now, most NMR studies of gp41 have involved short peptides interacting with membrane mimetic solvents or liposomes [98–101], or peptides and small molecules interacting with the hydrophobic pocket on the surface of the gp41 coiled coil [102–105], although NMR has also provided the structure of the longest intact segment of extracellular gp41, which included the NHR and CHR helices and the loop region of SIV gp41 [106]. [93, 96, 97]
CD
CD measurement provides a simple way to monitor macromolecular secondary structure changes and has been widely used to characterize HIV-1 gp41 interactions. Secondary structure elements, mainly α-helix, β-sheet, β-turn, 310-helix, can be deduced from wavelength dependent CD spectra. The percentage of secondary structure can be calculated semi-empirically using a computer program based on established data from well-characterized proteins and peptides of known concentration. α-helix, which plays a critical role in HIV-1 gp41 mediated virus-cell fusion, is characterized by negative peaks at ~208 and 222 nm. The molar ellipticity [θ]222 = −33000 deg cm2 dmol−1 is typically considered to correspond to 100% helicity, from which the percentage of α-helical structure can be obtained from a normalized CD spectrum[78]; Other [θ]222 values for 100% helix, such as −36000 deg.cm2.dmol−1 [22, 107], –30,700 deg.cm2.dmol−1 [108], −40000 × [1-(2.5/n)] where n is the number of peptide bonds, were also used by different groups. For tryptophan rich sequences, the 208/222 nm double minima may switch to 211/217 nm, accompanied by a minor positive band around 230 nm attributed to tryptophan residues whose indole chromophore is known to contribute significant ellipticity in the far-UV CD spectrum of helical peptides or proteins [109].
IR
IR, especially Fourier Transform IR (FTIR) or ATR-FTIR, provides a useful way to monitor protein and peptide secondary structure changes upon interaction with liposomes. The bands between 1600 and 1700 cm−1 are due to the peptide amide group vibration and are sensitive to secondary structure changes. Disordered and α-helical structures were characterized by a band between 1650 and 1660 cm−1 [77] [70]; 310-helix at ~1676 cm−1 [85]; β-aggregates at ~1625 cm−1 [91]; antiparallel β-sheets with a sharp band ~1630 cm−1 [70] and a less intense band near 1690 cm−1 [77]; extended β-strands with strong intermolecular interactions with an intense band of 1621 cm−1 and a less intense band of 1687 cm−1 [77]. IR spectra can be deconvoluted using software such as PEAKFIT (Jandel Scientific, San Rafael, CA), providing sub-peak intensities for calculating the secondary structure components by comparison with accepted literature values.[94] The peaks corresponding to random coil and α-helix partly overlap, but random coils undergo H/D exchange at higher rates than α-helices. Thus, hydration with D2O vapor can improve distinction between the two components [94]. Generally, the secondary structure components determined from CD and IR results are comparable if measured under the same conditions, though discrepancies have been observed in some cases [110].
IR is less compromised by interference from light scattering of aggregated peptides and liposomes than CD [85], so is preferentially used in studying peptide-liposome interactions, especially of the FP and MPER peptides [110]. The IR time scale is faster than NMR, so that different conformational states rather than a time-average over these states can be determined from the IR spectra [73, 85, 111].
2. Effects of membrane composition and physicochemical properties on HIV-1-cell fusion
Membrane composition and chemical and physical properties play an important role in HIV-1-cell fusion. Studying the effects of chemical and physical modification of virus or cell membranes on virus-cell fusion may provide useful clues for understanding the mechanism of HIV-1-cell fusion.
Cholesterol lipids rafts and HIV infection
Lipid rafts formed by cholesterol are important for HIV-1 virions released from the target cells and for ENV protein insertion and binding. The effect of cholesterol lipid rafts on HIV-1 infectivity was studied using cholesterol sequestering reagent 2-hydoxypropyl-β-cyclodextrin (2OH-β-CD). Removal of cholesterol from the membrane of HIV-infected cells by 2OH-β-CD dramatically reduced virus release; virions released from cholesterol-depleted cells were minimally infectious. Exposure of infectious HIV particles to 2OH-β-CD resulted in a dose-dependent inactivation of the virus. In both cases, the effect was attributable to loss of cholesterol and could be reversed by replenishing cholesterol. 2OH-β-CD-treated, non-infectious HIV retained its ability to bind cells. Western blot, p24 ELISA, and reverse transcription assays indicated that virions remained intact after treatment with 2OH-β-CD at concentrations that abolished infectivity. Electron microscopy revealed that the treated HIV had morphology very similar to that of untreated virus. R18 fluorescence dequenching studies showed that the treated HIV did not fuse to the membrane of susceptible cells; dequenching was restored by replenishing virion-associated cholesterol. The results indicated that cholesterol in HIV particles was strictly required for fusion and infectivity. These observations suggested 2OH-β-CD could be an candidate for use as a chemical barrier for AIDS prophylaxis [112].
Membrane fluidity and HIV-1 infection
Correlation between HIV-1 infectivity and membrane fluidity was observed by treatment of fluidity modulators such as increasing temperature or adding xylocaine to intact cells and viruses. The fluidity was measured by a 5-DXSA probe. Higher fluidity correlated to a higher rate of infection. Fluidity of both the plasma membrane and viral envelope was required to form the fusion-pore and to complete the entry of HIV-1. The lipid-bilayer envelope of HIV-1 is highly ordered and rigid, compared with the plasma membrane from which the respective virus was derived, due to high concentrations of cholesterol. These observations suggest that HIV-1 buds exclusively from the cholesterol-rich part or so-called “raft” of the plasma membrane [113].
Dipole potential and hydration/fluidity of membrane on fusogenicity of FP
The dipole potential and hydration/fluidity of membrane effect on the fusogenic and secondary structure of FP was studied by FP23 (3) interaction with PC/PE (1:1) membranes containing different content of cholesterol (Ch) or 6-ketocholestanol (KC) [72]. Both Ch and KC increased the dipole potential of PC/PE membrane. Ch increased the hydration of membrane and reduced the membrane fluidity, while KC reduced the membrane hydration and increased the membrane fluidity. Increasing membrane dipole potential enhances the binding potency of FP with liposome. The Kd changed from ~10 μM in PC/PE LUV to ~5μM when the PC/PE LUV containing 30% KC or 40% Ch [72]. 50 mol % Ch and 15 mol % KC caused a similar increase in the magnitude of the dipole potential of PC/PE membranes. Ch up to 50% does not significantly affect the conversion of FP23 (3) or FP23-H (3h) into β-sheet; in contrast, the conformational transformation was clearly enhanced by the presence of 15 mol % KC. The results suggested that the formation of more fusogenic β-structures was due to an increase in membrane fluidity/hydration, rather than to the variation of the dipole potential [72].
The above observations highlight that cholesterol, which plays an important role in virus-cell fusion processes through modulating the physiochemical properties of the biomembrane, is key to understanding the gp41-membrane interaction and can be a target for fusion inhibitor design. Further details of the role that cholesterol plays in gp41 – membrane interactions are presented in Section 3.
3. HIV-1 gp41-membrane interactions
Gp41-membrane interactions have been extensively studied using model membranes and peptides by the established methods. Below we summarize studies of various gp41 segments and interacting functional domains in the context of a membrane or lipid environment. The results provide insight into domain function in the virus-cell fusion process. We also present a general discussion on the implication for fusion inhibitors design. A detailed description of small molecule fusion inhibitors is presented in another chapter in this issue by Gochin and Zhou, and is not included here.
3.1 Fusion peptides
FP is located at the N-terminus of HIV-1 gp41. Fusion peptide sequences that have been studied are listed in Table 1. The first 12 amino acid residues FP12 (1, gp41512–523) form the minimal sequence to maintain fusion activity [81]. FP16 (2, gp41512–527) contains exclusively flexible and hydrophobic amino acid residues and is considered the core sequence that facilitates insertion into the cell membrane during HIV-1-cell fusion. An 18-residue sequence immediately following FP16 (2) containing polar amino acid residues also plays a significant role in FP interaction with the membrane, and is considered part of the FP sequence. Thus studies of the FP domain have included FP23 (3, gp41512–534), FP33 (5, gp41512–544) and FP34 (6, gp41512–545). The NHR sequence begins at gp41546.
3.1.1 Fusion peptide constructs
FP12(1)
In silico analysis predicted that FP12 (1) should be the minimal FP for membrane destabilization [81]. Based on lipid-mixing and leakage assays using large unilamellar vesicles (LUVs) mimicking the composition of T-cell membrane (1,2-dioleoyl -sn-glycero-3-phosphocholine (DOPC)/cholesterol (Ch)/1,2-dioleoyl -sn-glycerol-3-phosphoethanolamine (DOPE)/1,2-dioleoylphosphatidylglycerol (DOPG)/sphingomyelin (SM) at 34:33:16:10:7 mol ratio), Charloteaux showed that FP12 (1) had the same fusion activity as FP23 (3). 9- and 10-residue peptides did not induce significant fusion; 11 residue long peptide was approximately half as potent as the longer peptides [81].
FP23(3)
FP23(3) contains the 16 residue hydrophobic and flexible FP core with an additional 7 polar residues, and is the most widely used model for FP, perhaps due to its easier handling compared to the exclusively hydrophobic FP16 (2) core. A large number of lipid mixing, leakage, lipid binding and structural assays have been conducted with variants of FP23 (3). In general it was found that FP23 (3) could accommodate mutations such as terminal modification and introduction of a tryptophan residue for fluorescence studies, in some cases with minor effects on kinetics of association with lipids. Lipid structure and composition had a much greater effect on observed binding and structure. These results are discussed in more detail below.
FP33(5)
FP33 (5) contains the full length FP sequence. It includes both FP23 and the fusion peptide proximal region (FPPR). FP33 (5) has similar fusogenic activity to FP23 (3) and FP16 (2), while including FPPR made it more soluble and easier to handle. Studies show that FP33 preferentially partitions into more ordered regions of the membrane, forming β-sheet structures in bilayers and large vesicles.
3.1.2 FP – liposome interactions
FP23 (3) and its Arg to Ala mutant HIVAla (3e) have an effect on negatively charged (1-palmitoyl-2-oleoylphosphatidyl glycerol (POPG) and 1-palmitoyl-2-oleoyl phosphatidylserine (POPS)) LUV and SUV but not on neutral (POPC) liposomes, as tested by lipid mixing assay, leakage assay, light scattering at 400 nm, and a surface tension measurement. FP23 (3) was somewhat more potent than HIVAla (3e), possibly because of an electrostatic interaction between the positively charged Arg side chain and the negatively charged vesicles [77]. A lipid mixing assay with DOPC:DOPE:Ch 1:1:1 LUV indicated that a Pro residue at the N-terminus had no adverse effect on the fusion activity of FP23 [76]. Terminal modification had an effect on kinetics of the FP-membrane interaction. FP23-H (3h) with a free N-terminus displayed relatively slow kinetics of interaction with liposome, compared with FP23 (3) and FP16 (2), which are acetylated at the N-terminus. This was demonstrated by peptide-membrane binding, peptide induced lipid mixing and fusion, lipid content leakage of phosphatidylcholine (PC)/PE liposome, and peptide secondary structure changes upon interaction with liposome, measured by FTIR. All three peptides displayed similar binding affinity (Kd ~ 10 μM) to liposomes, and the final extent of lipid mixing and leakage caused by the three peptides were the same [73].
Tryptophan fluorescence showed that FP-W8 (3c) inserted into sodium dodecyl sulfate (SDS) micelles and 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC)/1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine (DPPS) vesicles. The larger blue shift (30nm) indicated a deeper insertion than the fusion peptide of influenza virus hemagglutinin (~20nm). The binding result was consistent with a decreased molecular mobility τr of 12- and 5- doxyl-stearic acid (DXSA)-labeled FP (3) in micelles, calculated from EPR experiments [67].
FP33 (5) showed self-association in PC/phosphatidyl serine (PS) vesicles. FP33 (5) and an all D-amino acid sequence FP33-D (5c) had similar potency and kinetics of inducing lipid mixing of POPG vesicles and were located at equivalent hydrophobic environments in the POPG vesicle membrane measured by fluorescence using NBD-labeled peptides. FP33 (5) was accessible to complete proteolysis in its membrane-bound state. The lipid samples were well ordered and both FP33 (5) and FP33-D (5c) induced similar small changes in the ordering of the lipid acyl chains, suggesting similar peptide orientation and penetration into the membrane [70]. Similar results were obtained with FP33 (5) and another partially D-amino acid substituted peptide IFFA (5b) in PC:Ch:Rho-PE (10:1:0.001) giant unilamellar vesicles (GUVs) [94].
Fluorescence microscopy showed NBD-FP33 (5) and IFFA (5b) partitioned preferentially into ordered regions of SM:PC:Ch:Rho-PE (1:1:1:0.001) GUVs, suggesting an intrinsic affinity of the peptides for SM/Ch microdomains. The free energy of peptide association with SUV was measured by FRET between Rho-labeled free peptide and NBD-labeled liposome bound peptide, with −9.3 kcal/mol for FP33 (5) and −8.3 kcal/mol for IFFA (5b) [94]. In PC:Ch:Rho-PE (10:1:0.001) GUVs, where no microdomains were present, a uniform binding pattern for IFFA (5b) was observed. However, the time to reach maximal intensity was about four times longer than that observed for SM:PC:Ch GUVs. Confocal fluorescence microscopy showed that both FP33 (5) and IFFA (5b) demonstrated high affinity toward T cells using NBD-labeled peptide as probe. The peptides were located at the plasma membrane and formed patches of high intensity, suggesting both FP33 (5) and FP33 IFFA (5b) preferentially bind to certain domains on the surface of T cells. Z axis scanning indicated no membrane penetration by the peptides, with no peptide found in any other membrane compartment in the cells [94]. Microscopy using Rho-labeled peptide showed that FP33 (5) and FP33V2E (5a) co-localized with T-cell receptor (TCR) and CD4 molecules in the T cell membrane and inhibited T cell activation in vitro and in vivo, suggesting a role for FP in the down regulation of HIV-specific immunity. [114] The same method was used to show that FP binding with TCR involved the α-helical transmembrane domain (TMD) of the TCR α chain (CP (136) or TCRαTMD(137)) [115].
3.1.3 FP structural studies
FP’s appear to display considerable structural plasticity from monomeric α-helical to oligomeric β-sheet conformations [73, 111], depending on sample conditions such as the selection and charge of membrane-mimetic environment, the peptide sequence and the ratio of peptide: lipid, as well as sample preparation methods [116]. FP16 (2) and FP23 (3) formed both α-helical and β-sheet conformations [117–120]; longer constructs (70-residue peptide) displayed only a highly fusogenic β-sheet structure [111]. FTIR showed a mixture of random coil, α-helix and aggregated β-structure for FP23 (3) and FP23-H (3h) dissolved in buffer. The presence of PC/PE model membranes favored transformation into β-sheet aggregates [73]. In other studies, higher loading of FP23 (3) favored the oligomeric β-sheet; lower loading favored monomeric α-helix [121, 122]. IR studies of the longer segment FP33 (5) concluded that the first 16 residues were in a predominantly β-strand conformation in 1:1:1 DOPE:DOPC:Ch [123]. Similarly, FP33 (5) and IFFA (5b) displayed 78 and 70% β-sheet secondary structure respectively in PC:Ch multilayers, adopting either parallel or antiparallel β-sheet structures [94].
1D solid state CP/MAS spectra were conducted in large unilamellar POPC/POPE,/DMPS/Ch vesicles on FP23 (3) labeled at different carbonyl positions [116]. 13C chemical shifts were indicative of non-helical structure, and sharp resonances observed up until Ala-14 indicated well-ordered structure for the N-terminal segment. 2D exchange spectroscopy on doubly carbonyl labeled peptides confirmed β-strand structure, possibly through formation of hairpins or oligomers [124]. This structure may be difficult for a small micelle to accommodate, suggesting that observed FP secondary structure differences may be due to the choice of membrane mimetic. Spectra were sensitive to lipid composition and peptide-lipid ratio. A detailed follow-up study in lipid and Ch mixtures resembling the membrane of HIV susceptible host cells were consistent with antiparallel β-strand structure possibly arising from an aggregate of two trimers [125]. The structural plasticity observed under different conditions may be a reflection of the role of gp41 in rearranging lipid structure during fusion. [126] There is an emerging general consensus that membrane curvature and FP-lipid ratios affect conformation and oligomeric state of FP.[120] β-sheet structure appears to predominate at higher FP concentrations and lower membrane curvatures, explaining the observation of α-helical structure in small micelles and β-sheet structure in large unilamellar vesicles and bilayers [127].
NMR studies of the structure and dynamics of FP23-G3K5 (3a) in dodecylphosphocholine (DPC) micelles confirmed the α-helical content in this system [117]. FP23-G3K5 contained a charged C-terminus in order to maintain solubility and dispersion of the peptides in solution for binding to the micelles. Strong helical propensity was found between Ile4 – Leu12, and a disordered structure from Ala15–Ser23. 60% helical content was observed by CD, in agreement with the NMR results. CD studies in POPC and POPG (4:1) indicated 70% helical content. The authors reconciled their results with reports suggesting that FP forms β-structures in lipid bilayers by performing IR studies of their construct bound to planar supported bilayers, and found predominantly α-helix and random-coil structures with gradually increasing β content as the protein concentration was increased. The authors further concluded that the α-helical form could promote fusion in their system, while the β-sheet form could not.
3.1.4 Inhibition of FP
FP – induced lipid mixing and fusion, as well liposome content leakage, could be inhibited by D-amino acid hexapeptides, which are promising for inhibition of HIV-1 gp41 mediated virus-cell fusion (See Huarte et al in this issue). Ac-GQIDEV-NH2 and Ac-GQIDQV-NH2 inhibited fusion in a dose-dependent manner showing apparent IC50 values of ~20 μM and 150 μM, respectively [82], indicating that FP is a target for fusion inhibitors and that an FP – model membrane interaction assay has the potential to be useful for detecting fusion inhibitors.
FP23 (3) can also interact with human serum albumin (HSA) and the interaction induced α-helix secondary structure as evidence by CD difference spectra [87]. HSA/FP23 (3) interaction blocked FP23 (3)-induced hemolysis of human red blood cells (hRBC) in a dose-dependent manner with EC50 = 1μM (concentration that reduced FP23-induced hemolysis by 50%). Thus FP23 (3) caused only minimal hemolysis of hRBC at HSA concentrations greater than 10 μM, and the inhibitory activity of HSA was maintained for ~24 hours, suggesting strong, irreversible binding to FP [87].
3.1.5 Summary of FP biophysical and biochemical studies
The studies reviewed indicate that the fusion peptide contains a hydrophobic 12-residue N-terminus which is the minimum required sequence for conferring fusogenicity. The 33-residue full-length FP has been more commonly studied, due to the inclusion of a hydrophilic domain that promotes easier handling. Some studies suggested that the FP had a higher affinity for cholesterol-rich regions of membranes. Controversy has arisen over the likely structure of FP in its membrane bound state, although most recent studies suggest it forms β-sheet structure. The fusogenic activity of FP can be inhibited by short D-peptides and HSA, indicating FP can be a target for fusion inhibitors.
3.2 NHR peptides
NHR/CHR is the functional core of the gp41 machinery; interaction between NHR and CHR to fold into antiparallel 6HB provides energy to juxtapose viral and cellular membranes and finally results in virus-cell fusion. 6HB formation is an important target for HIV-1 fusion inhibitor design. The behavior of model NHR peptides, CHR peptides or 6HB in membranes can provide useful information for understanding the HIV-1 gp41 mediated virus-cell fusion process and be useful for fusion inhibitor design. NHR is usually considered the target in fusion inhibitor design, since it forms a distinct subdomain in the interior of the 6HB and is the receptor for highly inhibitory C-peptides. Some engineered NHR domains have shown high activity against HIV-1 ENV mediated virus-cell fusion; none have entered into clinical trials to date. The sequences of gp41 NHR peptides are listed in Table 2.
Table 2.
NHR peptide sequences
| Name | Sequence | Literature Name/Ref |
|---|---|---|
| N54 (9) | STMGARSMTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | FPPR+HR1, 17–70 [69] |
| C10-N54 (9a) | C10–STMGARSMTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [69] |
| C16-N54 (9b) | C16–STMGARSMTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [69] |
| N47 (10a) | CTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [126], Gp4124–70 [84] |
| pFP23 (11) | QARLLLSGIVQQQNNLLRAIEAQ | [74] |
| N51 (12) | QARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQ | [71] |
| N62 (13) | QARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQQLLGIWGCSGK | [71] |
| pFP15 (14) | LLSGIVQQQNNLLRA | [74] |
| N36 (15) | SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [71, 75] |
| C10-N36 (15a) | C10–SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [69] |
| C16-N36 (15b) | C16–SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL | [69] |
| N36I62D (15c) | SGIVQQQNNLLRAIEAQQHLLQLTVWG KQLQARIL | [71, 75] |
| SN36 (15d) | AGIVQQQQQLLDVVKRQQELLRLTVWGTKNLQTRVT | SIV N36 [110] |
| DP107 (16) | NNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQ | T21 [130] |
3.2.1 NHR constructs
The most extensively studied NHR peptide is N36 (15, gp41546–581); it forms a 6HB with CHR peptide C34 (23, gp41628–661) in the crystallographic structure. It has been studied along with fatty acid conjugated N36 (15) in association with lipids. A coiled-coil lipopeptide of SIV N36 (SN36 (15d)) was also constructed and used as a model for a potential fusion inhibitor assay. Other NHR peptides include DP107, the first peptide fusion inhibitor, and N54 (9, gp41528–581), pFP23 (11, gp41540–562) and pFP15 (14, gp41544–558). Studies of the interaction between NHR peptides and liposomes are presented below.
3.2.2 NHR – liposome interactions
N36 (15)
N36 (15) is the most extensively characterized gp41 NHR model peptide. It forms a stable and well characterized 6HB with a CHR partner C34 (23, gp41628–661) as evidenced by crystallographic structure and other methods [29, 128, 129]. It bound zwitterionic PC:Ch (9:1) mimicking the outer leaflet and negatively charged PC:PS:Ch (4.5:4.5:1) phospholipids mimicking the inner leaflet of target cells with similar affinity. The mutant peptide N36I62D (15c), although non-fusogenic, shared a similar partition coefficient in zwitterionic membranes as the wild-type ((1.1±0.1)×104), and it bound slightly less tightly with negatively charged lipids ((0.7±0.1)×104). Transmission electron microscopy showed that N36 (15) caused notable PC:PS:Ch vesicle enlargement compared to the untreated control image (p<0.0007), whereas N36I62D (15c) did not affect the mean vesicle diameter significantly. The N36(15)/C34(23) core, which is protease-resistant in solution, was destabilized in electronegative liposomes but not in zwitterionic membranes. It dissociated rapidly upon binding to membrane as measured by rhodamine fluorescence. The secondary structure of N36 (15) was shifted from a predominantly α-helical conformation in zwitterionic membranes (29% β-sheet) to 84% β-sheet in negatively charged membranes mimicking the inner-leaflet. N36I62D (15c) demonstrated a 32% to 70% β-sheet secondary shift under the same conditions [75].
Another study showed that N36 (15) bound tightly with both PC and PC/PS (1:1) LUV with surface partition coefficients 1×105 M−1 and 3.3×104 M−1 respectively [71]; C34 (23) displayed ~10 fold reduced binding compared to N36 (15), with surface partition coefficients of 5×103 M−1 and 3×103 M−1 to PC and PC/PS respectively. No interaction was observed between NBD-N36 (15) and Rho-C34 (23) after NBD-N36 (15) was already bound to the membrane. In 1:1 PC/PS SUV, the experimental CD spectrum of N36 (15)/C34 (23) mixture was only marginally different from the theoretical non-interacting spectrum, suggesting that most N36(15)/C34(23) complexes had dissociated. Similar results were observed using N51 (12, gp41540–590) and C34 (23). Rho fluorescence indicated that the N36 (15) oligomer, as well as N36 (15)/C34 (23) complex, dissociated upon binding to SUV membranes [71].
Fatty acid conjugated N36 (15) or N54 (9)
To have a better understanding of the role of hydrophobic FP in gp41 – mediated virus-cell fusion, N36 (15) and N54 (9), a peptide including NHR and a polar region between FP core and NHR, were modified at the N-terminus with long chain fatty acids as a substitute for the FP, and the effects on membrane interactions were tested [69]. N36 (15) was unable to induce PC LUV lipid mixing, C10-N36 (15a) and C16-N36 (15b) were active at 0.07 peptide:lipid ratio; and C16-N54 (9b) had significantly increased fusogenic activity compared with N54 (9). The importance of the amino acid sequence of the gp41 peptides was confirmed using GCN4 and C16-GCN4 control peptides, which showed no activity. Atomic Force Microscopy (AFM) showed N54 (9) did not induce distinctive pores but had a global effect on PC LUV membrane morphology. In contrast, C10–N36 (15a) created nanoholes in the phospholipid surface in a slow kinetic phase. Both unmodified and fatty-acid modified N36 peptides had similar surface partition coefficients in the range (1.1–1.7)×104 M−1, as measured using tryptophan fluorescence binding. The secondary structure of the lipopeptides was similar to N36 (15) in PC micelles with ~70% α-helix content as determined by ATR-FTIR, or by CD using [θ]222 of −37,000 deg. cm2 dmol−1 for 100% α-helix [69]. These results suggested that N36 binding with the biomembrane was not enough to elicit its fusogenic activity, which required inclusion of hydrophobic FP or fatty acid, although the specific amino acid sequence of NHR was necessary to facilitate this activity. The role of the NHR in interacting with biomembrane in the presence of FP will be discussed in section 3.6.
DP107 (16)
DP107 (16) was the first identified peptide HIV-1 fusion inhibitor. It contains a C-terminal segment of the NHR, including part of the loop region [24]. EPR measurements indicated that the peptide bound parallel to POPC/POPG (9:1) LUV membrane surface, at the water-membrane interface. DP107 (16) in its liposome bound state was a monomer, in contrast to a tetramer in solution as measured by analytical ultracentrifuge and EPR. When Asp, Pro, or Ser was substituted for Ile at the core “a” position of the heptad repeat in the middle of the peptide, the coiled coil was destabilized and showed reduced membrane-binding affinity. These results suggested two functions for the heptad repeat of gp41 after CD4 binding: to form an extended coiled coil, and to provide a hydrophobic face that binds to the host-cell membrane, bringing the viral and cellular membranes closer and facilitating fusion [130].
pFP23 (11) and pFP15 (14)
pFP23 (11), a peptide overlapping the sequence of FP and NHR; and pFP15 (14), containing the N-terminal part of NHR, were synthesized and studied with model membranes [74]. Both peptides showed aggregation in solution. Addition of liposomes reduced the aggregation, as evidenced by CD and FTIR, as well as a thioflavin T assay; in SDS micelles, aggregation of pFP15 (14) was reduced by ~ 75%, and aggregation of pFP23 (11) completely disappeared. Both peptides bound tightly to negatively charged phospholipids egg L-R-phosphatidylcholine (EPC): bovine brain L-R-phosphatidylserine (BPS):Ch (5:4:1) and a complex membrane mimicking the lymphocyte plasma membrane EPC: egg transesterified L-R-phosphatidylethanolamine (TPE):SM: bovine brain L-R-phosphatidylinositol (BPI):BPS: egg L-R-phosphatidic acid (EPA):Ch (46.4:21.2:8.8:4.4:9.3:0.81:9.1). The peptides had very little or no effect on EPC/Ch and EPC/SM/Ch membranes, as measured using an FPE membrane binding assay. The peptide induced liposome aggregation was correlated to binding activity, with more liposome aggregation observed in negatively charged membranes. pFP23 (11) was capable of binding to membranes to a greater extent than pFP15 (14), inducing a higher extent of liposome aggregation. pFP15 (14) induced significant lipid mixing and content leakage in negatively charged phospholipids; the activity was completely abolished in zwitterionic lipids. pFP23 (11) was unable to induce lipid mixing in either negatively charged or zwitterionic membrane; it induced content leakage only on negatively charged lipids. Both peptides reduced the mobility of the phospholipid acyl chains above but not below the Tm as measured by fluorescence anisotropy assay using PA-DPH and DPH probe. The anisotropy was larger with negatively charged membrane than with zwitterionic membrane. The effect on membrane fluidity suggested that both pFP23 (11) and pFP15 (14) should be located at the lipid-water interface [74]. The authors concluded that the N-terminal part of the NHR domain could be working synergistically with other membrane-active regions of the gp41 glycoprotein to boost the fusion process. This region of the NHR domain could be stabilized in an extended conformation by its interaction with negatively charged phospholipid headgroups, prior to 6HB formation.
3.2.3 Coiled-Coil Lipopeptides as a target for a fusion inhibitor assay
A coiled-coil lipopeptide of SN36 (15d) was constructed to mimic the gp41 prehairpin and to test its potential application in a fusion inhibitor assay [110]. The assay was based on covalently attached lipid anchor 1,2-dioleoyl-sn-glycero-3- phosphoethanol amine-N-[4-(p-maleimidomethyl) cyclohexanecarboxamide] (MCC-POPE), which was substituted for FP, to drive SN36 (15d) into the desired coiled-coil conformation on a solid-supported model POPC membrane. Formation of the DOPC/MCC-DOPE-SN36 (9:1) conjugate was monitored by CD, FTIR, ellipsometry and AFM. SN36 (15d) in solution adopted a predominantly random-coil conformation. CD and FTIR spectroscopy strongly supported the formation of coiled-coil structures when SN36 (15d) was anchored to the bilayer. In situ AFM showed a ribbon-like structure of SN36 (15d) assemblies within a POPC bilayer upon coupling on mica [110].
Binding of antagonists to this prehairpin mimetic is thought to model the natural formation of a trimer-of-hairpin conformation. Addition of T20 (29) changed the ribbon-like MCC-DOPE-SN36 (15d) assemblies, forming a homogeneous coverage of peptide aggregates over the bilayer surface as evidenced by AFM. After heating the sample to 65°C for 20 min and rinsing with buffer, the characteristic ribbon structure of the SN36 (15d) assemblies re-emerged and the additional peptide domains, which resulted from addition of T20 (29), disappeared. A binding constant of 0.058 μM−1 for T20 (29)/SN36 (15d) was observed, corresponding to a half-maximal binding at 17 μM. Control experiments using pure DOPC bilayers without SN36 (15d) did not show any inhibitor adsorption on solid-supported membranes, suggesting T20 (29) bound specifically and partially reversibly to the prehairpin mimetic. It was possible to repeat these steps several times without losing the binding ability of the SN36 (15d) coiled-coil receptors, suggesting potential application of the lipopeptide prehairpin construct for a fusion inhibitor assay [110].
3.2.4 Summary of NHR biophysical and biochemical studies
The literature reviewed here indicates that NHR peptides form highly α-helical structures, which interact strongly with their CHR-peptide counterparts. There is evidence that the NHR/CHR association is weakened in a membrane-mimetic environment, although this may depend on the presence and composition of the loop region (see Sections 3.5 and 3.6). The typical construct representing the NHR is N36 (15), which binds to liposomes, losing some of its α-helical character, but it does not induce fusion. Addition of part of the FP or a lipophilic group at the N-terminus rescued the fusogenicity, indicating the gp41 environment has a profound effect on NHR function; a coiled coil lipopeptide including NHR and lipid membrane mimetic may be a more suitable model for assessing the potency of fusion inhibitors targeting gp41 NHR.
3.3 CHR peptides and peptide fusion inhibitors
Most of the highly potent HIV-1 fusion inhibitors are C-peptides. These include T20, an approved fusion inhibitor drug for clinical use, and T1249 and sifuvirtide, potent peptide fusion inhibitors in clinical trials. The sequences of CHR peptides are listed in Table 3.
Table 3.
CHR peptide sequences
| Name | Sequence | Literature Name/Ref |
|---|---|---|
| CHR1 (19) | WNHTTWMEWDREINNYTSLIHSLIEESQNQQEKNEQ | [65] |
| C52 (20) | NHTTWMEWDREINNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFNI | [71] |
| Sifuvirtide (21) | SWETWEREIENYTRQIYRILEESQEQQDRNERDLLE | [146, 147] |
| T1249 (22) | WQEWEQKI–––––––TALLEQAQ QQEKNE ELQKLDKWASLWEWF | [139] |
| C34 (23) | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELL | [71, 75] |
| C36 (24) | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELLEL | [65] |
| C39 (25) | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELLELDKW | [126] |
| SJ2176 (26) | EWDREINNYTSLIHSLIEESQNQQEKNEQE | DP-219 [25, 26] |
| CHR3 (27) | REINNYTSLIHSLIEESQNQQEKNEQELLELDKWAS | [65] |
| DP (28) | YTSLIHSLIEESQNQQEKNEQELLE | [63] |
| DP–C12 (28a) | YTSLIHSLIEESQNQQEKNEQELLE–C12 | [63] |
| DP–C16 (28b) | YTSLIHSLIEESQNQQEKNEQELLE–C16 | [63] |
| T20 (29) | YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF | DP178 [65] |
| SIV–T20 (29a) | LEENITALLEEAQIQQEKNMYELQKLNSWDVFGNWF | [66] |
| C8–SIV–T20 (29b) | C8–LEENITALLEEAQIQQEKNMYELQKLNSWDVFGNWF | [66] |
| SIV–T20–C8 (29c) | LEENITALLEEAQIQQEKNMYELQKLNSWDVFGNWF–C8 | [66] |
| MutSIV–T20 (29d) | LEENITALLEEAQIQQEKNMYELQKLNSWDVFANAA | [66] |
| Mut–C8–SIV–T20 (29e) | C8–LEENITALLEEAQIQQEKNMYELQKLNSWDVFANAA | [66] |
| Mut–SIV–T20–C8 (29f) | LEENITALLEEAQIQQEKNMYELQKLNSWDVFANAA–C8 | [66] |
| CHR5 (30) | HSLIEESQNQQEKNEQELLELDKWASLWNWFNITNW | [65] |
3.3.1 CHR fusion inhibitor – liposome interactions
T20 (29)
T20 (29) is the first FDA approved HIV-1 fusion inhibitor [28, 33, 34, 131]. It contains the C-terminal CHR sequence and a hydrophobic tail extending ~10 residues into the MPER which is considered to play an important role in binding with lipids and contributing to its high activity. [63] To investigate the function of the T20 (29) hydrophobic tail, a truncated T20 (29) construct, DP (28, gp41638–662), and its C-terminal fatty acid conjugates were prepared and tested for antiretroviral activity and membrane binding potency. DP (28) is ~5000 fold less active than T20 (29), while the activity of DP-C16 (28b) was almost restored to that of T20 (29), as assessed by a cell-cell fusion assay. The hydrophobic tail contributed activity which could be replaced by a long chain lipid. A flow cytometry assay using NBD-labeled peptides showed T20 (29) and DP-C16 (28b) have a similar binding profile to cell membrane, and that binding was increased proportionately with the size of the lipid chain. The results also showed that none of the peptides T20 (29), DP-C12 (28a), DP-C16 (28b) were hemolytic to red blood cells at up to 50 μM concentration [63].
Shai et al also studied the interaction of SIV T20 (29a) and various mutants and conjugates of the C-terminal hydrophobic residues with PC/SM/PE/Ch (4.5:4.5:1:1) SUV by SPR and fluorescence spectroscopy. Tryptophan fluorescence showed that T20 (29), octyl-T20 (29), and T20-octyl (29) have similar oligomeric states in PBS. The SPR results showed similar binding affinity to the membrane for all peptides despite their different fusion inhibitory activities, suggesting that biological activity differences were not simply the result of higher peptide concentrations in the proximity of the membrane. The C-terminal fatty acid labeled SIV T20 (29a) peptides had a much higher antiretroviral potency than N-terminal labeled peptides suggested the existence of a pre-hairpin intermediate in which the CHR must insert into the membrane with a certain orientation [66].
To identify the lipid binding sequence of gp41 CHR, Liu et al evaluated a series of 36 amino acid CHR peptides spanning residues 623 to 678, designated CHR1 (19, gp41623–658), C36 (24, gp41628–663), CHR3 (27, gp41633–668), T20 (29), and CHR5 (30, gp41643–678) from the N to the C-terminus. A significant blue shift of the fluorescence spectra was observed for C-terminal peptides T20 (29) (354 to 340 nm) and CHR5 (30) (348 to 339 nm) and the most N-terminal peptide CHR1 (19) (355 to 345 nm) in the presence of POPC LUVs. C36 (24) and CHR3 (27) showed no blue shift under the same conditions. The binding constant to POPC LUVs was 1.66 ×105 M−1 for CHR-5 and 5.41 × 104 M−1 for T20 (29) measured by ITC. The other peptides bound too weakly with POPC LUVs for binding constants to be determined [65]. These observations indicated that both the specific interaction with NHR and the hydrophobic interaction with lipid membrane were critical for T20’s antiretroviral activity.
T1249 (22)
T1249 (22) is a secondary generation peptide HIV-1 fusion inhibitor developed by Trimeris. The 39-residue peptide fusion inhibitor was designed to contain the critical amino acid sequence that interacts with both NHR and the lipid membrane [132–138]. T1249 (22) and a 20-residue gp41 peptide CTP (33, gp41660–679) containing the T1249 (22) hydrophobic C-terminus with 6 residues expanded into the MPER of gp41 were included in the studies for comparison. T1249 (22) is essentially in a random coil conformation in the liposome, although a small α-helix contribution is present. Both peptides partitioned extensively into liquid-crystalline POPC and were located at the interface of the membrane, as evidenced by 5NS and 16NS quenching. When other lipid compositions were used (DPPC, POPG/POPC, and POPC/Ch) partition decreased, the most severe effect being the presence of Ch, which resulted an apparently kp close to 0. After a detailed analysis of the previous literature, the authors suggested that the low partition coefficient of T1249 (22) apparently observed in Ch-rich membrane may be an artifact of the fluorescence measurement. A decrease in fluorescence intensity upon peptide insertion into these liposomes may preclude the use of fluorescence intensity to measure binding. New partition experiments and fluorescence resonance energy transfer analysis showed that T1249 (22) adsorbed to Ch-rich membranes, although a broad range of partition coefficients (kp ~ 0.4 to 4.6 × 103 M−1) was obtained with different methods [139]. The authors concluded that the improved clinical efficacy of T1249 (22) relative to T20 (29) may be related to its larger partition coefficient and ability to adsorb to rigid lipidic areas on the cell surface where most receptors are inserted, thus increasing the local concentration of the inhibitor peptide at the fusion site [139].
Sifuvirtide (21)
Sifuvirtide (21) is another 36-residue new generation peptide HIV-1 fusion inhibitor derived from gp41 CHR sequences [50, 58, 140–145]. It showed high activity against T20 (29) resistant HIV-1 strains and a longer in vivo half-life than T20 (29). Phase IIb clinical trials have been completed. Sifuvirtide (21) was studied in aqueous solution and interacting with LUV using fluorescence spectroscopy techniques (both steady-state and time-resolved) [146, 147]. Sifuvirtide (21) showed no significant aggregation in aqueous solution below 60 μM; a fluorescence assay suggested aggregation at higher concentration, although aggregates were undetectable by DLS. No significant interaction was observed between the peptide and the liposome, either with zwitterionic fluid lipid membranes (liquid-disordered phase), or with Ch-rich membranes (liquid-ordered phase), by using POPC, POPC:Ch (2:1), POPC:PSM:Ch (1:1:1), SM/ceramide and DPPC:Ch (2:1) vesicles [146, 147]. However, significant partitioning was observed with the positively charged lipid models POPC:EPOPC (1:1) in fluid phase. Adsorption of the peptide at the surface of gel-phase DPPC membranes was observed by tryptophan fluorescence measurements and was further confirmed by FRET experiments. These results indicated that the peptide targeted more rigid gel phase domains, and increased fluidity reduced this interaction. The larger affinity and selectivity of sifuvirtide (21) toward more rigid areas of the membranes, where most receptors are found, or to viral membrane, may help explain its improved clinical efficacy - by providing a local increased concentration of the peptide at the fusion site. Fluorescence quenching experiments using 5NS and 16NS probes showed sifuvirtide (21) was preferentially located near the lipid bilayer interface. The superficial interaction of the peptide with gel-phase membranes was further confirmed by using tPnA, a fluorescent tetraene fatty acid which preferentially partitions into gel-phase membranes and which has been commonly used as a probe in phase-transition studies. The results showed that the interaction of the peptide did not lead to a significant perturbation of membrane fluidity [146]. Ionic strength dependence of the partition coefficient of sifuvirtide (21) with PC membranes also suggested superficial binding between the peptide and the liposome [147]. The effects of sifuvirtide (21) on the lipid membranes’ structural properties were further evaluated using dipole-potential membrane probes, zeta-potential, DLS and AFM measurements. The results showed that sifuvirtide (21) did not cause a noticeable effect on lipid bilayer structure, except for membranes composed of cationic phospholipids [147].
3.3.2 Summary of CHR biophysical and biochemical studies
A myriad of CHR peptides have been constructed, and excellently reviewed previously [40, 148]. CHR peptide – biomembrane interactions appear to enhance the inhibitory activity, i.e. CHR peptides more prone to partition into membranes are more active as fusion inhibitors. This is an important feature that should be built into future fusion inhibitor design.
3.4 MPER peptides
The Membrane Proximal Extracellular Region (MPER) is located at the C-terminal end of the HIV-1 gp41 ectodomain, directly followed by the transmembrane domain. In the HIV-1 gp41 prehairpin structure, the MPER and FP are the functional links by which the gp41 prehairpin bridges viral and target cell membranes. Therefore, MPER has also been considered the counterpart of FP, and has been called the internal fusion peptide (IFP).
3.4.1 MPER constructs
MPER studies have involved sequences of just five residues at the C-terminus up to the full-length sequence MP22 (40, gp41662–683), which contains the epitopes for neutralizing antibodies 2F5 and 4E10. The amino acid sequence of MPER peptides is listed in Table 4. The five-residue C-terminus LWYIK is called the cholesterol recognition amino acid consensus (CRAC) sequence, which promotes binding to cholesterol-rich domains in membranes. The tryptophan-rich MP19 sequence is required for induction of fusion. Structural studies have mainly been conducted on MP22 (40) in the absence or presence of liposomes and/or antibodies.
Table 4.
MPER peptide sequences
| Name | Sequence | Literature Name/Ref |
|---|---|---|
| AISb (31) | NEQELLELDKWASLWN | [83] |
| MPER–2F5 (32) | ELLELDKWASLWN | [99] |
| CTP (33) | LLELDKWASLWNWFDITNWL | [139] |
| LASWIK (36) | LASWIK | [157] |
| MP5 (37) | LWYIK | [157] mutations [150] |
| MP19 (38) | KWASLWNWFNITNWLWYIK | (Gp41W) [100, 157] |
| MP20 (39) | DKWASLWNWFNITNWLWYIK | [85] preTM, HIVc [68] |
| MP20M (39a) | DKAASLANAFNITNWLWYIK | HIVW(1–3)A [68] |
| MP22 (40) | ELDKWASLWNWFNITNWLWYIK | C22 [93, 100, 109] |
| C56 (41) | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFNITNWLWYIK | [85] |
3.4.2 MPER – lipid interactions
LWYIK (37, gp41679–683)
The 5-residue peptide at the C-terminal end of the gp41 ectodomain LWYIK (37) was considered to play an important role in sequestering gp41 into Ch-rich domains in the membrane. [149, 150]. It contains the CRAC sequence motif L/V-(X)1–5-Y-(X)1–5-K/R, where X can be any amino acid residue, for eliciting cholesterol sequestration. DSC was used to assess the extent to which the peptide caused phase separation of 1-stearoyl-2-oleoyl phosphatidylcholine (SOPC)/Ch membrane. Two criteria were used to identify phase separation of Ch-rich domains: 1) Formation of a Ch-rich domain has to be accompanied by depletion of Ch from another region of the membrane, resulting in increased enthalpy of the chain melting transition of the Ch-depleted region; 2) Beyond its solubility limit in the Ch-rich domain, Ch will associate to form crystallites. LWYIK (37) effectively sequestered Ch in SOPC/Ch membrane, as evidenced by a marked increase in the enthalpy of the chain melting transition of SOPC [150]. A variety of single and double amino acid substitutions of LWYIK were tested in order to identify the critical residues. Ala, Val and Ile substitutions of Leu resulted in peptides with reduced potential to sequester Ch, in the order Leu > Ala, Val > Ile [149]. Double mutants of LWYIK at Trp and Ile indicated that the most flexible peptide LGYGK had the greatest effect on Ch redistribution in membranes [150]. The corresponding mutation in gp41 resulted in a protein retaining 72% of the fusion activity of the wild-type protein. However, the CRAC Leu to Ile mutant retained similar fusogenicity [150]. These results confirmed that the CRAC motif is not the sole critical factor for the cholesterol recognition function of LWYIK. The authors showed that LASWIK (36), the corresponding HIV-2 and SIV peptide, which does not fulfill the CRAC motif criteria, showed potency to sequester Ch in between that of LWYIK and MP19 (38, gp41665–683), the latter containing almost the full length MPER sequence [150].
3.4.3 MPER structure and membrane orientation
MP22(40. Gp41662–683)
MP22 (40) is important as the antigenic determinant of broadly neutralizing antibodies (BNAb’s) 4E10, 2F5, Z13E1 [100, 109]. Structural studies with antibodies have been described in detail in this issue by Gach and colleagues. The MPER was found to be insoluble with poor quality NMR spectra in SDS micelles, but gave excellent spectra in DPC micelles, an indication of a preference for a zwitterionic membrane-like environment [100]. A strongly helical amphipathic structure was observed.
In aqueous solution, the 2F5 epitope which comprises the more soluble N-terminal region of MPER was found to form a 310-helix [99]. NMR solution studies of the full length MPER in DPC micelles revealed an “L”-shaped strongly amphiphilic helix–hinge–helix structure with a membrane binding face containing the 4th or the 5th Trp residues as well as the critical Phe673 residue [93]. The degree to which the peptide was immersed in POPC/POPG liposomes was studied using ESR on peptides spin labeled at selected sites. Differential relaxation rates in the absence or presence of a relaxation reagent revealed the solvent accessibility/depth of immersion of each residue. A periodicity of 4 residues supported the existence of helical peptide lying on the membrane surface with its hydrophobic face embedded into the lipid membrane, and with a distinct break in the periodicity at the hinge region. Significant changes in immersion depths and amide chemical shifts occurred in the presence of 4E10. The system was in fast exchange, and cross saturation experiments were used to show that the C-terminal segment 671–683 interacted with the antibody, in agreement with the crystal structure. Phe673 swung from the lipid interior into the antibody binding pocket. The authors constructed a model in which 4E10 extracts its epitope from the lipid membrane. It appears that the ability of the antibody to induce a structural rearrangement, rather than just its affinity, is a key determinant of neutralization potency. Immersion depths and amide chemical shift changes were also studied for MPER interactions with 2F5 and Z13E1, indicating that 2F5 extracts the residues N-terminal to the hinge, while Z13E1 freezes the MPER in its hinge conformation [151].
3.4.4 Summary of MPER biophysical and biochemical studies
As the counterpart of FP, MPER peptides show similar hydrophobic character and are sequestered into cholesterol rich areas of membranes. MPER peptides adopt helical structure in a biomembrane environment. MPER harbors epitopes of some BNAbs, so it is an important target for vaccine design; the high conserved antibody epitopes in MPER may also be a target for fusion inhibitors.
3.5 Loop Region and Intracellular Tail
A ~30-residue HIV-1 gp41 sequence between NHR and CHR is unstructured and was designated the loop region. It contains two cysteines that can form a disulfide bridge; and is an immunodominant domain of gp41. The loop region functions both as a linker between NHR and CHR of gp41 and as a molecular recognition element during the fusion process, and it can be a target for fusion inhibitors [61]. Another less exploited part of gp41 is its cytoplastic tail. It contains about half of the gp41 sequence, much longer than the cytoplasmic domain of other viral envelope proteins, and plays a role in regulating the function of gp41. Peptide sequences studied that correspond to these domains are included in Table 5.
3.5.1 Loop peptides
Loop-1 (17, gp41579–601)
The highly conserved sequence gp41 Loop-1 (17) is a major immunodominant region, recognized by antibodies from approximately 98% of AIDS patients; it also contributes to the fusion inhibitory activity of DP107 (16) [24, 152]. The interaction of gp41 Loop-1 (17) in its disulfide chain linked dimeric form and in its reduced monomeric form were studied by different fluorescence methods including energy migration, red edge excitation shift (REES), fluorescence lifetime, fluorescence quenching using 5NS or 16NS lipophilic probes, fluorescence intensity and spectral shifts (blue shift). The monomeric form incorporated in the membrane model systems POPC/DMPG (80:20), and pure DMPG LUV and SUV, with the tryptophan residue located in a shallow position near the interface. The dimeric form (17), however, didn’t interact with these vesicles. The results suggested a “two step” model for the gp41 fusion mechanism similar to the one proposed for influenza virus hemagglutinin [152]. After dissociation from gp120 upon CD4 and co-receptor binding, gp41 undergoes several conformational changes, including oligomerization, formation of the coiled-coil domain (6HB), and “projection” of the fusion domain toward the target cell. The insertion of the fusion peptide in the target membrane might lead to a second conformational change, with the dissociation of the gp41 oligomer. The coiled-coil precursor region of each gp41 monomer is inserted in the target cell bilayer interface, bringing the two membranes together and contributing to the formation of the fusion pore [152].
Loop-2 (18, gp41579–613)
A peptide containing part of the gp41 NHR and loop sequence Loop-2 (18) was designed and its interaction with negatively charged liposomes was studied.[80] Loop-2 (18) bound tightly to negatively charged phospholipid-containing model membranes (EPA, EPG, BPS), based on anisotropy values of the tryptophan fluorescence. The peptide had almost no effect on content leakage of zwitterionic EPC liposomes, but significant leakage was observed for LUVs composed of negatively charged phospholipids. Similar results were obtained in lipid mixing assays and aggregation assays based on light scattering. The destabilization and fusion activity of Loop-2 (18) towards negatively charged model membranes suggested an essential role of the loop domain in the membrane fusion process induced by gp41 [80].
W596L/W610F mutations in the loop region
The effect of mutation of tryptophan residues in the loop region on cell-cell fusion was studied using confocal microscopy and flow cytometry technologies. W596L and W610F mutations retained wild type levels of gp120-anchoring ability in a virion context but abolished virus entry functions of gp41, based on the following observations using living cells: 1) Confocal microscopy with DiO-labeled 293T effector cells transfected with Env-expression vectors, and DiI-labeled 293T target cells co-transfected with pcDNA.T4 and pc.FUSIN showed W596L and W610F mutations almost completely abolished the entry activity of HIV-1 Env; 2) Flow cytometry lipid mixing assay using Env-expressing HeLa effector cells and HeLa-P4 target cells showed Env glycoproteins bearing W596L/W610F mutations were unable to mediate lipid mixing [90].
3.5.2 Intracellular domain A (52, gp41828–848)
Highly amphipathic positively charged cytolytic peptide from the carboxy terminal end of gp41 was studied in negatively charged unilamellar lipid vesicles by solid state NMR and in SDS micelles by solution state NMR [86]. In solid state, both peptide and lipid signals were studied. Quadrupole splitting of perdeuterated isoleucines were consistent with penetration of the N-terminus into the hydrophobic core of the bilayer and with the C-terminus lying at the water-lipid interface. Lipid chain order decreased towards the center of the bilayer. Partial helical conformation was obtained for the N-terminal segment in SDS micelles. The changes in lipid order profiles determined by NMR suggested that membrane curvature stress is the driving force for peptide translocation and pore formation.
3.5.3 Summary of Loop and CT peptide biophysical and biochemical studies
The loop area, connecting the N-terminal and C-terminal halves of the gp41 ectodomain, is usually not considered a functional domain in HIV-1 gp41 mediated virus-cell fusion; the CT, containing ~ half the length of the HIV-1 gp41 sequence, is assumed to regulate gp41 function, while its actual role in gp41 mediated virus-cell fusion is not yet clear. The interactions between the loop area or CT and biomembrane indicate that these less exploited parts of gp41 may play a role in gp41 – membrane interaction; detailed study of these interactions may provide new insight into understanding the mechanism of gp41 mediated virus-cell fusion and uncover clues for fusion inhibitor design. In addition, recent small molecule inhibitor development has targeted the loop region as a possible site for inhibition (see review in this issue by Gochin and Zhou).
3.6 Constructs containing multiple interactive gp41 functional domains
Cooperative association between different gp41 domains plays an important role in HIV-1 gp41 mediated virus-cell fusion, and it has been assessed in studies using constructs containing multiple gp41 functional domains. These studies of gp41-membrane interaction using multiple-functional-domain constructs may provide insight into understanding this cooperation. Multi-domain constructs are listed in Table 5.
3.6.1 MPER – FP – membrane interactions
FP23 (3) - MP20 (39) interaction
MP20 (39) is considered a counterpart of FP. Tryptophan fluorescence intensity of MP20 (39) and FP23-8W (3c) in solution decreased with time, suggesting self-aggregation. Both peptides penetrated into DOPC-DOPE-Ch LUV with an intensity enhancement and blue shift of tryptophan fluorescence. The apparent mole fraction partition coefficients (Kp) were 7.3 ×105 for MP20 (39) and 3.0×104 for FP23-8W (3c). MP20 (39) was >10 times more efficient than FP23-W8 (3c) in inducing DOPC/DOPE/CH LUV lipid mixing and fusion, and lipid content leakage, perhaps attributed to its tighter binding with the membrane. After correcting for the difference in bound peptide, membrane-associated FP23-8W (3c) was as effective as MP20 (39) at perturbing liposome membranes. The existence of MP20-FP23 interactions was confirmed by detecting changes of MP20 (39) tryptophan fluorescence in the presence of the non-fluorescent FP23 (2). This interaction lacked a correlation to fusogenic activity with lipid membranes. A mixture of MP20 (39) and FP23-8W (3c) was not more active than MP20 (39) alone [68].
FP16 (2) - AISb
Increasing concentrations of FP16 (2) effectively permeabilized liposomes and induced merging of POPG LUV bilayers in the presence of Ca2+. In comparison, an amphipathic-at-interface sequence (AIS) AISb (31, gp41656–671), a conserved aromatic-rich membrane proximal region which includes the MPER-2F5 (32, gp41659–671) immunodominant sequence, did not induce a significant release of the aqueous contents of liposomes, and it was unable to induce vesicle fusion. The capacity of FP16 (2) to induce permeabilization was diminished and its fusogenic effect was completely abolished in a hybrid peptide FP16-AISb (51). Thus, the FP16 (2) –AISb (31) interactions appear to reduce the capacity of FP (2) to perturb membranes. CD and IR spectroscopy showed the existence of a stable FP-AIS complex with a specific β-turn- and β-sheet-rich secondary structure [83]. The interactions between FP and MPER suggest these fusogenic segments of gp41 may engage in crosstalk during gp41 mediated virus-cell fusion, but the exact role of the interaction still requires clarification.
3.6.2 N70 (8) and its components
N70 (8, gp41512–581) contains the N-terminal 70 residues of gp41, including two functional domains FP and NHR. The interaction between membrane and N70 (8), as well as various mutations and composite parts, were studied and compared. N70 (8) mutations in the NHR (N70I62D) (8c), FP (N70F11G) (8b), as well as an N70 construct engineered to disrupt secondary structure in a region between the FP and NHR regions (N70GG) (8d) were designed. The three mutations retained 35–60% of wild-type fusogenic activity toward PC LUV. The mutations had the same effect on LUV’s mimicking the target membrane (38% SM, 34% PC, 14% PE, 10% Ch, and 3% negatively charged PS). The I62D mutant was lethal in vivo while the F11G mutant retained low cell-cell fusion activity (10%). The relative α-helical content of each peptide decreased as follows: N70F11G (8b) > N70 (8) > N70GG (8d) > N70I62D (8c) as measured by CD and FTIR spectra. 22~30% β-aggregates were also observed for the mutant peptides. All peptides were predominantly monomeric in SDS PAGE, with a significant population of dimer and higher order oligomers in N70 (8), N70F11G (8b) and N70GG (8d). [92].
The cooperativity between FP and NHR in membrane destabilization was studied by investigating the fusogenic properties of N70 (8) and component FP and NHR peptides [75, 84]. PC LUV is difficult to fuse and represents a highly stringent test of membrane fusogenic function; inclusion of PE or negatively charged lipids creates vesicles more facile to fusion. FP23 (3) was unable to fuse PC vesicles; FP33 (5) and FP33M24C (5d), bearing the mutation for facilitating the synthesis of longer peptide N70 (8) using native chemical ligation, were equally able to mix membranes of PC LUV; N47 (10a, gp41534–580) had similar fusogenicity to FP33 (5). The continuous FP/NHR composite N70 (8) was 10 fold more fusogenic than N47 (10a), either alone or in a mixture with FP23 (3), demonstrating a synergy between the FP and NHR domains in mediating membrane fusion. The direct contribution of NHR to fusogenic activity was also confirmed by N70I62D (8c), which had dramatically reduced fusogenicity. The similar partition coefficient with PC LUV (~104 M−1) of N70 (8) and N70I62D (8d) does not explain their different fusogenic potencies. Negative stain electron microscopy showed N70 (8) caused a substantial increase in vesicle size of PC LUV while FP23 (2) induced no change, confirming that the lipid mixing induced by N70 (8) is a result of membrane fusion [84].
Another study showed that N36 (15) has significant fusogenic activity, similar to FP34 (6) [75]. Its activity was phospholipid-dependent; decreasing the relative amount of PS to 5% reduced the peptide’s activity significantly and fusogenic capabilities were completely lost in a zwitterionic environment. N70 (8a) was ~4 times more fusogenic than its deconstructed components (FP34 (6) and N36 (15)). N36I62D (15c), known to be lethal in vivo and to disrupt the helical structure in NHR fragments, caused a substantial loss of function. CHR-derived fusion inhibitors C34 (23) reduced the fusogenic activity of N-terminal peptides in a dose-dependent manner. Real-time AFM experiments showed that 6 μM FP34 (6) induced a homogeneous distribution of ~5 nm deep holes in electronegative PC:PS:Ch (4.5:4.5:1) LUV with a diameter of ~200 nm, while not affecting the global shape of the bilayer surface; N36 (15) did not induce distinctive pores, but rather had a global effect on the membrane morphology [75].
3.6.3 C56 (41, gp41628–683) and its fragments
C56 (41) contains the sequence of MP20 (39, gp41664–683) and a CHR sequence C34 (23) with an additional two residues. C56 (41) and MP20 (39) preferentially destabilized Ch-rich LUV mimicking the virion surface (PC:SM:Ch at 1:1:1), whereas the N-terminal counterparts N70 (8) and FP33 (5d) preferentially fused vesicles that were either Ch-free (PC LUV) or contained 10% Ch mimicking the outer leaflet of blood cells (SM:PC:PE:Ch:PS in a ratio of 11:10:4:3:1) [85]. NHR appears to play a direct role in the fusogenic properties of the N-terminus, while CHR plays an indirect role, by stabilizing the IFP that is primarily responsible for membrane destabilization induced by the C-terminal subdomain. This correlated with the findings that the IFP effectively partitioned into and self-assembled in membranes, whereas the CHR (C34 (23)) bound poorly to membranes. Acting alone, both C56 (41) and N70 (8) perturbed PC LUV, and in combination they acted additively to induce lipid mixing of PC LUV [85].
The structure of fusogenic peptides has been shown to be important for their ability to promote fusion. C56 (41) was more effective than MP20 (39) in inducing lipid mixing of both PC and PC: SM:Ch (1:1:1) LUV. Lysophosphatidylcholine (LPC) at micellar concentrations (1% w/v) was chosen for peptide secondary structure measurements to avoid light scattering caused by liposomes. CD data demonstrated that the α-helical content of C56 (41) was significantly higher than that of MP20 (39). FTIR showed that MP20 (39) and C56 (41) were predominantly α-helical (~55%) while C34 (23) was largely unstructured in deuterated membranes, indicating that helical structure was induced in the CHR when conjugated to the largely helical MP20, known as IFP. FTIR data revealed that C34 (23), MP20 (39) and C56 (41) disrupted the acyl chain order of 1:1:1 PC:SM:Ch membranes to a similar degree, suggesting the differential lipid-mixing ability of C56 (41) and MP20 (39) was not a function of their ability to disrupt the order of the membranes. These findings suggest that the IFP, stabilized in the context of C56 (41), is primarily responsible for the membrane fusion function of the C-terminal subdomain [85]. Cooperative stabilization of the CHR by MP20 (39) could simultaneously enhance the ability of the CHR to interact with the coiled-coil NHR regions to facilitate hairpin formation. The same is true of the N-terminal subdomain. FP33(5) was ineffective, while N70 (8, gp41512–581), a peptide construct including FP33 (5) and NHR, disrupted PC multilayers.
3.6.4 gp41 ectodomain
N70-CHR constructs
A gp41 construct contains N70 (8) and C39 (25, gp41628–666) linked by a 6-residue flexible amino acid linker N70(L6)C39 (47) was designed and designated FP-hairpin, and was used to study gp41-liposome interactions using PC:PG:Ch (8:2:5) (mimicking HIV-1 and host cell membrane) or PC:PG (4:1) [126]. Lipid mixing activity increased in the order N70(L6)C39 (45) < FP34 (6) < N70 (8a). Both β-sheet and α-helical membrane-associated structure was observed. FP34 (6) had the greatest β-sheet population which increased with protein loading. β-sheet population was positively correlated with membrane Ch content for FP34 (6) and N70 (8a), while for N70(L6)C39 (45), β-sheet and α-helical populations were approximately independent of Ch content. This suggested that β-sheet FP conformation is more fusogenic than the α-helical conformation. NHR coiled-coil and NHR-CHR 6HB regions retained helical structure following membrane association [126].
Another study showed N70 (8) had significant α-helical content in buffer and detergent at 37°C measured by CD. N70 (8) had a Tm ~45°C in buffer and higher stability and α-helical content in detergent. Extremely rapid lipid mixing of POPC:POPG:Ch vesicles was observed for N70 (8) that had been initially dissolved in either buffer or detergent. Dissolving FP34 (6) in detergent accelerated it potency to induce lipid mixing. N47(L6)C39 (44) containing NHR and CHR without the FP sequence was 100% α-helical in buffer and detergent with Tm ~ 100°C; it induced neither aggregation nor lipid mixing between these vesicles. Approximately 20 residues of N70(L6)C39 (45) and N70 (8) were non-helical in buffer; in detergent both constructs had fewer non-helical residues. The correlation of membrane fusion function with prehairpin conformation of the studied constructs suggested that one of the roles of the final hairpin conformation is to sequester membrane-perturbing gp41 regions, reducing the membrane disruption induced earlier by the prehairpin structure [78].
Recgp41 (47 gp41538–665) and (48 gp41540–682)
Recgp41 (47) is a recombinant gp41 construct including NHR, CHR and an intact loop area, and 540–682 (48) is a recombinant gp41 ectodomain lacking the fusion peptide. Both gp41 constructs bound to membranes, consequently undergoing a major conformational change. The interactions were similar with both negatively charged PS/PC/Ch/DNS-PE (8:8:2:1, w/w) and zwitterionic PC/Ch/DNS-PE (16:2:1, w/w) SUV, indicating that non-electrostatic forces were involved in the gp41-membrane interactions. Trypsin digest of recgp41 in aqueous buffer resulted in a protease-resistant core consisting of two tryptic fragments: N62 (13, gp41540–601) and C52 (20, gp41624–675); digestion was complete when recgp41 (47) was fully bound to phospholipid vesicles. Combined with the C34 (23)/N36 (15) interaction, which showed decomposition of the 6HB structure upon membrane binding, these results suggested that, after the prehairpin conformation was formed, membrane binding induced opening of the gp41 core complex [71].
Ectodomain (46, gp41512–584), E-core (49, gp41546–584), Core (47)
E. coli expressed recombinant HIV-1 gp41 ectodomains were constructed, including ectodomain (46), e-core (49) and core (47). All the three proteins folded into a stable coiled-coil core in aqueous solution and retained a stable helical fold with reduced coiled-coil characteristics in a zwitterionic and negatively charged membrane mimetic environment as measured by CD. In contrast to an extended exposed N-terminal domain, the folded gp41 ectodomain (46) did not induce lipid mixing of zwitterionic membranes while strongly disrupting and inducing lipid mixing of negatively charged PC/PS LUV. It was ~100-fold more effective against negatively charged lipids than FP23 (3) and 10-fold more effective than N70 (8). The kinetics of membrane fusion revealed that all three proteins acted immediately after addition to the liposome solution. Real-Time AFM showed nanoscale holes in negatively charged membranes PC:PS (3:2) induced by the gp41 ectodomain (46) and its mutants. The results supported a model in which one of the roles of gp41 folding into the 6HB conformation was to slow down membrane disruption effects induced by early exposed gp41. However, it could further affect membrane morphology once exposed to negatively charged membranes during later stages [79].
SIVmac239 e-gp41 (50, SIV gp4127–149)
To test whether covalently linking the NHR and CHR segments via the immunodominant loop affected the conformational change, a SIVmac239 gp41 ectodomain 27–149 named e-gp41 (50) was designed. SPR, tryptophan and rhodamine fluorescence, ATR-FTIR spectroscopy, and DSC results suggested that the presence of the loop stabilized the trimeric helical hairpin both when e-gp41 (50) was in aqueous solution and when it was bound to the membrane surface of egg-PC or DOPC SUV or LUV. The protein-liposome association constant (KA) was 4.33×105 M−1 and the corresponding ΔG was −10.1 kcal/mol. SIV e-gp41 (50) had a monomer-trimer association constant Ka of 3×1011M−1, so >90% monomer and trimer were presented at 0.2 μM and 5 μM concentration respectively, which were selected for testing. Binding with PC SUV did not result in dissociation of e-gp41 (50) trimers to a significant extent as monitored by tryptophan fluorescence, and by using rhodamine fluorescence change following trypsin digest. Tm of e-gp41 (50) was independent of lipids added as measured by DSC, with a small secondary structure shift from 76±6% helix in buffer to 68±1% helix in lipids as measured by ART-IR. The authors concluded that the immunodominant loop stabilizes gp41 helical hairpin conformation. [91].
3.6.5 Summary of membrane interaction of gp41 constructs containing multi-functional domains
The above observations show that there is cooperativity between different functional domains in gp41 – membrane interactions. Some dual domains, such as FP/NHR and CHR/MPER, were synergistic; others, such as ectodomain constructs containing both N-terminal and C-terminal part of gp41, were antagonistic; there are also interactions, such as between FP and MPER, with unclear function. Fully understanding how different gp41 functional domains work in concert to regulate gp41 mediated virus-cell fusion processes may be helpful for fusion inhibitor design.
Conclusion and perspectives on gp41 fusion inhibitor development
In this review, we have attempted to convey key aspects of the literature covering the effect of lipids on gp41 domain structure and interactions. Most of the studies have involved biological and biophysical assays on isolated domains. We have elected to present findings from multiple different studies, even though discrepancies were sometimes found between different observations, and controversies also occurred. Inevitably, studies involved subtle or significant variations in experimental setup, lipid composition, sample preparation or biophysical technique used. A systematic study of gp41-membrane interactions using strict controls may provide a useful way to consolidate our understanding of the mechanism of gp41 mediated virus-cell fusion.
It is clear that all gp41 functional domains show binding to certain lipids. The binding is enhanced when negatively charged lipids or cholesterol are included in the liposomes. FP and MPER are the main fusogenic functional domains of gp41; NHR or CHR can significantly enhance the fusogenic activity of FP and MPER, respectively, when they are covalent linked; lipid conjugation can rescue or improve the membrane binding and fusogenic activity of NHR and CHR. A summary of gp41 domain – membrane interactions is presented in Table 7. Secondary structure and stability changes of gp41 functional domains are associated with lipid membrane interaction; generally, β-structure is induced for FP, helix structure is induced for MPER, and dissociation or destabilization are observed for NHR, CHR or the combined ectodomain.
Table 7.
Summary of gp41-lipid interactions
| Zwitteronic liposomes | Negative charged liposomes | Cholesterol containing liposomes/living cell membrane | ||||
|---|---|---|---|---|---|---|
| Gp41 Domain | Binding | fusogenicity | Binding | fusogenicity | Binding | fusogenicity |
| FP | n.d. | − | ++ | + | ++ | + |
| NHR | ++ | − | ++ | + | ++ | + |
| Lipo-NHR | ++ | + | ++ | n.d. | ++ | n.d. |
| CHR | +/++ | n.d. | ++ | n.d. | ++ | + |
| Loop | + | − | ++ | + | n.d. | n.d. |
| MPER | +++ | n.d. | n.d. | n.d. | +++ | ++ |
| FP+NHR | ++ | + | n.d. | ++ | n.d. | n.d. |
| CHR+MPER | n.d. | + | n.d. | n.d. | n.d. | ++ |
| ectodomain | +++ | − | n.d. | +++ | n.d. | n.d. |
n.d. not determined; − no binding or activity; + weak; ++ medium; +++ strong
Lipid interacting domains as models or targets for fusion inhibitor design
The crystal structure of gp41 NHR/CHR 6HB verified that gp41 was a suitable target for fusion inhibitor design, but the membrane context was not considered. The hydrophobic gp41-membrane interaction provides an additional dimension towards fully understanding the mechanism of HIV-1 cell fusion and inhibition. The importance of hydrophobic interactions in HIV-1 fusion inhibitor design were highlighted by design of a lipopeptide which contained a CHR peptide and a cholesterol tail, showing improved antiretroviral profile compare with available fusion inhibitors [55]. We also observed that certain surfactants containing a charged polar head group and a simple fatty acid chain could reach low micromolar fusion inhibitory activity, close to the most potent small molecule fusion inhibitors designed recently [153]. These observations suggest that understanding the hydrophobicity of gp41 interactions is key to designing and improving the fusion inhibitors that target 6HB.
Beyond the 6HB, more hydrophobic parts of gp41, especially FP and MPER, are also potential targets for new fusion inhibitor design, notwithstanding their importance in vaccine design. Less exploited parts of gp41 such as the loop region and CT, also play important roles in gp41 – membrane interaction. Peptides from specific gp41 functional domains showed various interactions with membrane and membrane mimetics. FP and MPER, the gp41 sequences that function by inserting into the target cellular and viral membranes, showed preferential interaction with negatively charged liposomes, a property of both viral and cell membranes. The CHR peptides showed a preference for binding to rigid membranes or lipid rafts where the cellular receptors and co-receptors used by HIV-1 envelope protein for entry are located.
The importance of gp41-membrane interaction outside of the 6HB sequence is evident from several inhibition studies. VIRIP [59] and short D-peptides [82] inhibit HIV-1 replication by targeting FP of gp41. FP-68742, a low molecular-weight entry inhibitor, can induce drug resistant HIV-1 mutations at both FP and the loop area [61]. Amphotericin B methyl ester (AME), a cholesterol binding compound that blocks HIV-1 entry, induced a drug resistant HIV-1 strain by deleting the CT part of gp41 [154–156]. A recently reported crystallographic structure showed that a cavity in MPER may be a binding site for small molecular-weight fusion inhibitors [109]. Systematic study of these areas in the context of gp41-membrane interactions may shed new light on future fusion inhibitor design.
Table 6.
Peptide-liposome binding affinity
| Peptide | Liposome | Kd/μM | ΔGm/kcal/mol | Partition coefficient/M−1 | Ref |
|---|---|---|---|---|---|
| FP16 (2) | PC/PE | 10 | [73] | ||
| FP23 (3) | PC/PE | 11.6±0.2, 12 | [72], [73] | ||
| PC/PE/10% KC | 9.5±0.61 | [72] | |||
| PC/PE/15% KC | 6.2±0.02 | [72] | |||
| PC/PE/30% KC | 5.2±0.32 | [72] | |||
| PC/PE/33% Ch | 9.24±0.06 | [72] | |||
| PC/PE/40% Ch | 5.3±0.4 | [72] | |||
| FP23-8W (3c) | DOPC-DOPE-Ch | 3.0×104 | [68] | ||
| FP23-H (3h) | PC/PE | 10 | [73] | ||
| FP33 (5) | PC:Ch (10:1) SUV | −9.3 | [94] | ||
| IFFA (5b) | PC:Ch (10:1) SUV | −8.3 | [94] | ||
| N54 (9) | PC SUV | (1.3±0.03)×104 | [69] | ||
| pFP23 (11) | EPC:Ch (5:1) | 22.1 | [74] | ||
| EPC:BPS:Ch(5:4:1) | 7.39 | [74] | |||
| complex membrane | 7.64 | [74] | |||
| pFP15 (14) | EPC:Chol (5:1) | 38.2 | [74] | ||
| EPC:BPS:Ch(5:4:1) | 9.67 | [74] | |||
| complex membrane | 18.4 | [74] | |||
| N36 (15) | PC/Ch | (1.2±0.2)×104 | [75] | ||
| PC/PS/Ch | (1.3±0.1)×104 | [75] | |||
| PS/PC | −8.5 | 3.3×104 | [71] | ||
| PC | −9.1 | 1×105, | [71] | ||
| PC SUV | (1.1±0.4)×104 | [69] | |||
| C16–N36 (15b) | PC SUV | (1.7±0.01)×104 | [69] | ||
| N36I62D (33c) | PC/Ch | (1.1±0.1)×104 | [75] | ||
| PC/PS/Ch | 0.7±0.1×104 | [75] | |||
| gp41579–601(17) | POPC/DMPG | 310±100 | [152] | ||
| --- | DMPG | 1410±380 | [152] | ||
| Loop-2 (18) | EPC | 190 ± 39 | [80] | ||
| --- | EPA | 1.56±0.4×103 | [80] | ||
| --- | EPG | 1.55±0.6×103 | [80] | ||
| --- | BPS | 3.2 ±1.4×103 | [80] | ||
| Sifuvirtide (21) | PC | 1.2 × 102 | [147] | ||
| POPC:EPOPC | (2.2±0.3)× 103 | [146] | |||
| T1249 (22) | POPC | −7.0 | (5.1±0.7)×103 | [139] | |
| POPC/Chol (82:18) | 3.1×103 | [139] | |||
| POPC/Chol (75:25) | 4.6×103 | [139] | |||
| C34 (23) | PS/PC | −7.1 | 3×103 | [71] | |
| PC | −7.4 | 5×103 | [71] | ||
| T20 (29) | POPC LUV | 5.41 × 104 | [65] | ||
| POPC | 1.6 × 103 | [139] | |||
| SIV T20 (29a) | PC/SM/PE/Ch (4.5:4.5:1:1) | (1.7±0.2)×104 | [66] | ||
| C8-SIV T20 (29b) | --- (same as above) | (1.2±0.3)×105 | [66] | ||
| SIV T20-C8 (29c) | --- | (3.2±0.7)×104 | [66] | ||
| Mut T20 (29d) | --- | (1.9±0.3)×104 | [66] | ||
| C8-MT20 (29e) | --- | (0.5±0.1)×104 | [66] | ||
| Mut T20-C8 (29f) | --- | (1.9±0.6)×104 | [66] | ||
| CHR5 (30) | POPC LUV | 1.66×105 | [65] | ||
| CTP (33) | POPC | −8.7 | (53±8.5)×103 | [139] | |
| MP20 (39) | DOPC-DOPE-Ch | 7.3×105 | [68] | ||
| SIV e-gp41 (50) | PC SUV | −10.1 | 4.33×105 | [91] |
Acknowledgments
The authors are grateful for support from the Natural Science Foundation of China (Grant 81072581) and intramural funding from Beijing Institute of Pharmacology & Toxicology to L. C., and National Institute of Health (NIH, USA) Grants GM087998 and NS059403 to M. G.
Abbreviations
- 2OH-β-CD
2-hydoxypropyl-β-cyclodextrin
- 6HB
six helix bundle
- AFM
atomic force microscopy
- AIS
amphipathic-at-interface sequence
- ANTS
8-Aminonaphthalene-1,3,6-trisulfonic acid sodium salt
- ATR-FTIR
attenuated total reflection Fourier transform infrared spectroscopy
- BPI
bovine brain L-α-phosphatidylinositol
- BPS
bovine brain L-α-phosphatidylserine
- CD
circular dichroism
- Ch
cholesterol
- CF
Carboxyfluorescein
- CHR
C-terminal heptad repeat
- CMTMR
((4-chloromethyl)benzoyl)amino)) tetramythyl rhodamine
- di-8-ANEPPS
1-(3-sulfonatopropyl)-4-[â-[2-(di-n-octylamino)-6-naphthyl]vinyl]pyridinium betaine
- CT
Cytoplasmic tail
- DiI
1,10-diocta decyl-3,3,30,30-tetramethyl indocarbocyanine perchlorate
- DiO
3,3′-dioctadecyl oxacarbocyanineperchlorate
- DLS
dynamic light scattering
- DNS-PE
N-(5-dimethylaminonaphthalene-1-sulfonyl) -sn-glycero-3-phosphoethanolamine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine
- DOPC
1,2-dioleoyl -sn-glycero-3-phosphocholine
- DOPE
1,2-dioleoyl -sn-glycerol-3-phosphoethanolamine
- DOPG
1,2-dioleoylphosphatidylglycerol
- DPC
dodecylphosphocholine
- DPPS
1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine
- DPX
p-xylyl enebis[pyridinium] bromide
- DXSA
12-doxyl-stearic acid
- EPA
egg L-α-phosphatidic acid
- EPC
egg L-R-phosphatidylcholine
- EPR
electron paramagnetic resonance
- FP
fusion peptide
- FPE
fluorescein-phosphatidylethanolamine
- FRET
Förster resonance energy-transfer
- gp41
41,000-Da transmembrane subunit glycoprotein of HIV-1
- HIV-1
human immuno deficiency virus, type 1
- HPTS
8-hydroxypyrene- 1,3,6-trisulfonic acid
- IFP
internal fusion peptide
- KC
6-ketocholestanol
- LPC
lysophosphatidyl choline
- LUV
large unilamellar vesicles
- MCC-DOPE
1,2-dioleoyl -sn-glycerol-3-phosphoethanolamine -N-[4- (pmaleimido methyl) cyclohexanecarboxamide]
- MPER
Membrane-proximal external region
- NBD
4- fluoro-7- nitrobenz-2-oxa-1,3-diazole
- NHR
N-terminal heptad repeat
- NNRTI
Non-Nucleotide Reverse Transcriptase Inhibitor
- NRTI
Nucleotide Reverse Transcriptase Inhibitor
- PA-DPH
1,6-diphenyl-1,3,5-hexatrienepropionic acid
- PBS
phosphate-buffered saline
- PC
phosphatidyl choline
- PE
dioleoylphosphatidylethanol amine
- PI
Protease Inhibitor
- POPC
1-palmitoyl-2-oleyl-sn- glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoylphosphatidylglycerol
- POPS
1-palmitoyl-2-oleoyl phosphatidylserine
- PS
phosphatidyl serine
- R18
Octadecylrhodamine Chloride B
- Rho
rhodamine
- Rho-PE
phosphatidyl-ethanol amine-rhodamine
- SM
sphingomyelin
- SOPC
1-stearoyl-2-oleoyl phosphatidylcholine
- TM
Transmembrane domain
- TMA-DPH
1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene
- TPE
egg trans-esterified L-R-phosphatidylethanolamine
References
- 1.McEnery R. Update on pandemic shows new HIV infections steadily declining. IAVI Rep. 2009;13:17. [PubMed] [Google Scholar]
- 2.Dorrucci M. Epidemiology of HIV update. Recent Prog Med. 2010;101:12–15. [PubMed] [Google Scholar]
- 3.Steffen I, Pohlmann S. Peptide-based inhibitors of the HIV envelope protein and other class I viral fusion proteins. Curr Pharm Design. 2010;16:1143–1158. doi: 10.2174/138161210790963751. [DOI] [PubMed] [Google Scholar]
- 4.Granich R, Crowley S, Vitoria M, Smyth C, Kahn JG, Bennett R, Lo YR, Souteyrand Y, Williams B. Highly active antiretroviral treatment as prevention of HIV transmission: Review of scientific evidence and update. Curr Opin HIV AIDS. 2010;5:298–304. doi: 10.1097/COH.0b013e32833a6c32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarchoan R, Broder S. Development of antiretroviral therapy for the acquired-immunodeficiency-syndrome and related disorders - a progress report. New Eng J Med. 1987;316:557–564. doi: 10.1056/NEJM198702263160925. [DOI] [PubMed] [Google Scholar]
- 6.Furman PA, Fyfe JA, Stclair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H, Barry DW. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human-immunodeficiency-virus reverse-transcriptase. Proc Natl Acad Sci USA. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merluzzi VJ, Hargrave KD, Labadia M, Grozinger K, Skoog M, Wu JC, Shih CK, Eckner K, Hattox S, Adams J, Rosehthal AS, Faanes R, Eckner RJ, Koup RA, Sullivan JL. Inhibition of HIV-1 replication by a nonnucleoside reverse-transcriptase inhibitor. Science. 1990;250:1411–1413. doi: 10.1126/science.1701568. [DOI] [PubMed] [Google Scholar]
- 8.Vacca JP, Dorsey BD, Schleif WA, Levin RB, McDaniel SL, Darke PL, Zugay J, Quintero JC, Blahy OM, Roth E, Sardana VV, Schlabach AJ, Graham PI, Condra JH, Gotlib L, Holloway MK, Lin J, Chen IW, Vastag K, Ostovic D, Anderson PS, Emini EA, Huff JR. L-735,524 - an orally bioavailable human-immunodeficiency-virus type-1 protease inhibitor. Proc Natl Acad Sci USA. 1994;91:4096–4100. doi: 10.1073/pnas.91.9.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeks SG, Smith M, Holodniy M, Kahn JO. HIV-1 protease inhibitors - a review for clinicians. JAMA-J Am Med Assoc. 1997;277:145–153. [PubMed] [Google Scholar]
- 10.Walmsley S, Henry K, Katlama C, Nelson M, Castagna A, Reynes J, Clotet B, Hui J, Salgo M, DeMasi R, Delehanty J. Enfuvirtide (T-20) cross-reactive glycoprotein 41 antibody does not impair the efficacy or safety of enfuvirtide. J Infect Diseases. 2003;188:1827–1833. doi: 10.1086/379810. [DOI] [PubMed] [Google Scholar]
- 11.Hardy N, Skolnik PR. Enfuvirtide, a new fusion inhibitor for therapy of human immunodeficiency virus infection. Pharmacotherapy. 2004;24:198–211. doi: 10.1592/phco.24.2.198.33141. [DOI] [PubMed] [Google Scholar]
- 12.Baldanti F, Paolucci S, Gulminetti R, Brandolini M, Barbarini G, Maserati R. Early emergence of raltegravir resistance mutations in patients receiving haart salvage regimens. J Med Virol. 2010;82:116–122. doi: 10.1002/jmv.21651. [DOI] [PubMed] [Google Scholar]
- 13.Autran B, Carcelain G, Li TS, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4(+) T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 14.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 15.Williams AB. New horizons: Antiretroviral therapy in 1997. J Assoc Nurses AIDS Care. 1997;8:26–38. doi: 10.1016/S1055-3290(97)80011-8. [DOI] [PubMed] [Google Scholar]
- 16.Yeni P. Update on HAART in HIV. J Hepatol. 2006;44:S100–S103. doi: 10.1016/j.jhep.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–45. [PubMed] [Google Scholar]
- 18.Aghokeng AF, Ayouba A, Mpoudi-Ngole E, Loul S, Liegeois F, Delaporte E, Peeters M. Extensive survey on the prevalence and genetic diversity of sivs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Inf Gen Evol. 2010;10:386–396. doi: 10.1016/j.meegid.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Ann Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 20.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 22.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: Discovery of d-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 23.Liu SW, Wu SG, Jiang SB. HIV entry inhibitors targeting gp41: From polypeptides to small-molecule compounds. Curr Pharm Design. 2007;13:143–162. doi: 10.2174/138161207779313722. [DOI] [PubMed] [Google Scholar]
- 24.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human-immunodeficiency-virus replication - correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang SB, Lin K, Strick N, Neurath AR. HIV-1 inhibition by a peptide. Nature. 1993;365:113–113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 26.Jiang SB, Lin K, Strick N, Neurath AR. Inhibition of HIV-1 infection by a fusion domain binding peptide from the HIV-1 envelope glycoprotein-gp41. Biochem Biophys Res Comm. 1993;195:533–538. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- 27.Wild C, Dubay JW, Greenwell T, Baird T, Oas TG, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human-immunodeficiency-virus type-1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 29.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 30.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 31.Tan KM, Liu JH, Wang JH, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Mathews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 34.Lazzarin A, Clotet B, Cooper D, Reynes J, Arasteh K, Nelson M, Katlama C, Stellbrink H, Delfraissy J, Lange J, Huson L, DeMasi R, Wat C, Delehanty J, Drobnes C, Salgo M, Grp TS. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. New Eng J Med. 2003;348:2186–2195. doi: 10.1056/NEJMoa035211. [DOI] [PubMed] [Google Scholar]
- 35.Zhou GY, Wu D, Hermel E, Balogh E, Gochin M. Design, synthesis, and evaluation of indole compounds as novel inhibitors targeting gp41. Bioorg Med Chem Lett. 2010;20:1500–1503. doi: 10.1016/j.bmcl.2010.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie D, Yao C, Wang L, Min WJ, Xu JH, Xiao JH, Huang MX, Chen B, Liu B, Li XL, Jiang H. An albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-life. Antimicrob Agents Chemother. 2010;54:191–196. doi: 10.1128/AAC.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welch BD, Francis JN, Redman JS, Paul S, Weinstock MT, Reeves JD, Lie YS, Whitby FG, Eckert DM, Hill CP, Root MJ, Kay MS. Design of a potent d-peptide HIV-1 entry inhibitor with a strong barrier to resistance. J Virol. 2010;84:11235–11244. doi: 10.1128/JVI.01339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Lu H, Zhu Q, Jiang SB, Liao Y. Structure-based design, synthesis and biological evaluation of new N-carboxyphenylpyrrole derivatives as HIV fusion inhibitors targeting gp41. Bioorg Med Chem Lett. 2010;20:189–192. doi: 10.1016/j.bmcl.2009.10.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Lu L, Qi Z, Lu H, Wang J, Yu XX, Chen YH, Jiang SB. Novel recombinant engineered gp41 n-terminal heptad repeat trimers and their potential as anti-HIV-1 therapeutics or microbicides. J Biol Chem. 2010;285:25506–25515. doi: 10.1074/jbc.M110.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai L, Jiang S. Development of peptide and small-molecule HIV-1 fusion inhibitors that target gp41. Chemmedchem. 2010;5:1813–24. doi: 10.1002/cmdc.201000289. [DOI] [PubMed] [Google Scholar]
- 41.Bianchi E.Anonymous, Pessi A, Geleziunas R, Bramhill D. US 07811577. Covalently stabilized chimeric coiled-coil HIV gp41 N-peptides with improved antiviral activity. 2010
- 42.Jiang SB, Zhao Q, Debnath AK. Peptide and non-peptide HIV fusion inhibitors. Curr Pharm Design. 2002;8:563–580. doi: 10.2174/1381612024607180. [DOI] [PubMed] [Google Scholar]
- 43.Jiang SB, Lu H, Liu SW, Zhao Q, He YX, Debnath AK. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob Agents Chemother. 2004;48:4349–4359. doi: 10.1128/AAC.48.11.4349-4359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bianchi E, Finotto M, Ingallinella P, Hrin R, Carella AV, Hou XS, Schleif WA, Miller MD, Geleziunas R, Pessi A. Covalent stabilization of coiled coils of the HIV gp41 N region yields extremely potent and broad inhibitors of viral infection. Proc Natl Acad Sci USA. 2005;102:12903–12908. doi: 10.1073/pnas.0502449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lalezari JP, Bellos NC, Sathasivam K, Richmond GJ, Cohen CJ, Myers RA, Henry DH, Raskino C, Melby T, Murchison H, Zhang Y, Spence R, Greenberg ML, DeMasi RA, Miralles GD, Grp TS. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J Infect Diseases. 2005;191:1155–1163. doi: 10.1086/427993. [DOI] [PubMed] [Google Scholar]
- 46.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci USA. 2006;103:13938–13943. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai LF, Gochin M. A novel fluorescence intensity screening assay identifies new low-molecular-weight inhibitors of the gp41 coiled-coil domain of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2007;51:2388–2395. doi: 10.1128/AAC.00150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, Tvermoes NA, Matthews TJ, Greenberg ML, Delmedico MK. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc Natl Acad Sci USA. 2007;104:12772–12777. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He YX, Cheng JW, Lu H, Li JJ, Hu J, Qi Z, Liu ZH, Jiang SB, Dai QY. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc Natl Acad Sci USA. 2008;105:16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He YX, Xiao YH, Song HF, Liang Q, Ju D, Chen X, Lu H, Jing WG, Jiang SB, Zhang LQ. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem. 2008;283:11126–11134. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 51.Oishi S, Ito S, Nishikawa H, Watanabe K, Tanaka M, Ohno H, Izumi K, Sakagami Y, Kodama E, Matsuoka M, Fujii N. Design of a novel HIV-1 fusion inhibitor that displays a minimal interface for binding affinity. J Med Chem. 2008;51:388–391. doi: 10.1021/jm701109d. [DOI] [PubMed] [Google Scholar]
- 52.Qi Z, Shi WG, Xue N, Pan CG, Jing WG, Liu KL, Jiang SB. Rationally designed anti-HIV peptides containing multifunctional domains as molecule probes for studying the mechanisms of action of the first and second generation HIV fusion inhibitors. J Biol Chem. 2008;283:30376–30384. doi: 10.1074/jbc.M804672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoddart CA, Nault G, Galkina SA, Thibaudeau K, Bakis P, Bousquet-Gagnon N, Robitaille M, Bellomo M, Paradis V, Liscourt P, Lobach A, Rivard ME, Ptak RG, Mankowski MK, Bridon D, Quraishi O. Albumin-conjugated C34 peptide HIV-1 fusion inhibitor equipotent to C34 and T-20 in vitro with sustained activity in SCID-Hu Thy/Liv mice. J Biol Chem. 2008;283:34045–34052. doi: 10.1074/jbc.M805536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai LF, Balogh E, Gochin M. Stable extended human immunodeficiency virus type 1 gp41 coiled coil as an effective target in an assay for high-affinity fusion inhibitors. Antimicrob Agents Chemother. 2009;53:2444–2449. doi: 10.1128/AAC.00150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingallinella P, Bianchi E, Ladwa NA, Wang YJ, Hrin R, Veneziano M, Bonelli F, Ketas TJ, Moore JP, Miller MD, Pessi A. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc Natl Acad Sci USA. 2009;106:5801–5806. doi: 10.1073/pnas.0901007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katritzky AR, Tala SR, Lu H, Vakulenko AV, Chen QY, Sivapackiam J, Pandya K, Jiang SB, Debnath AK. Design, synthesis, and structure-activity relationship of a novel series of 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl)furans as HIV-1 entry inhibitors. J Med Chem. 2009;52:7631–7639. doi: 10.1021/jm900450n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naito T, Izumi K, Kodama E, Sakagami Y, Kajiwara K, Nishikawa H, Watanabe K, Sarafianos SG, Oishi S, Fujii N, Matsuoka M. SC29EK, a peptide fusion inhibitor with enhanced alpha-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide. Antimicrob Agents Chemother. 2009;53:1013–1018. doi: 10.1128/AAC.01211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang RR, Yang LM, Wang YH, Pang W, Tam SC, Tien P, Zheng YT. Sifuvirtide, a potent HIV fusion inhibitor peptide. Biochem Biophys Res Comm. 2009;382:540–544. doi: 10.1016/j.bbrc.2009.03.057. [DOI] [PubMed] [Google Scholar]
- 59.Munch J, Standker L, Adermann K, Schuz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing WG, Jiang SB, Forssmann WG, Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 60.Liu J, Deng YQ, Li QN, Dey AK, Moore JP, Lu M. Role of a putative gp41 dimerization domain in human immunodeficiency virus type 1 membrane fusion. J Virol. 2010;84:201–209. doi: 10.1128/JVI.01558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murray EJ, Leaman DP, Pawa N, Perkins H, Pickford C, Perros M, Zwick MB, Butler SL. A low-molecular-weight entry inhibitor of both CCR5-and CXCR4-tropic strains of human immunodeficiency virus type 1 targets a novel site on gp41. J Virol. 2010;84:7288–7299. doi: 10.1128/JVI.00535-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wexler-Cohen Y, Ashkenazi A, Viard M, Blumenthal R, Shai Y. Virus-cell and cell-cell fusion mediated by the HIV-1 envelope glycoprotein is inhibited by short gp41 N-terminal membrane-anchored peptides lacking the critical pocket domain. Faseb J. 2010;24:4196–4202. doi: 10.1096/fj.09-151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wexler-Cohen Y, Shai Y. Demonstrating the c-terminal boundary of the HIV 1 fusion conformation in a dynamic ongoing fusion process and implication for fusion inhibition. Faseb J. 2007;21:3677–3684. doi: 10.1096/fj.07-8582com. [DOI] [PubMed] [Google Scholar]
- 64.Wexler-Cohen Y, Shai Y. Membrane-anchored HIV-1 N-heptad repeat peptides are highly potent cell fusion inhibitors via an altered mode of action. Plos Pathog. 2009:5. doi: 10.1371/journal.ppat.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu SW, Jing WG, Cheung B, Lu H, Sun J, Yan XX, Niu JK, Farmar J, Wu SG, Jiang SB. HIV gp41 C-terminal heptad repeat contains multifunctional domains - relation to mechanisms of action of anti-HIV peptides. J Biol Chem. 2007;282:9612–9620. doi: 10.1074/jbc.M609148200. [DOI] [PubMed] [Google Scholar]
- 66.Peisajovich SG, Gallo SA, Blumenthal R, Shai Y. C-terminal octylation rescues an inactive T20 mutant - implications for the mechanism of HIV/simian immunodeficiency virus-induced membrane fusion. J Biol Chem. 2003;278:21012–21017. doi: 10.1074/jbc.M212773200. [DOI] [PubMed] [Google Scholar]
- 67.Chang DK, Cheng SF, Chien WJ. The amino-terminal fusion domain peptide of human immunodeficiency virus type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as helix with a conserved glycine at the micelle-water interface. J Virol. 1997;71:6593–6602. doi: 10.1128/jvi.71.9.6593-6602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suarez T, Gallaher WR, Agirre A, Goni FM, Nieva JL. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: Putative role during viral fusion. J Virol. 2000;74:8038–8047. doi: 10.1128/jvi.74.17.8038-8047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lev N, Shai Y. Fatty acids can substitute the HIV fusion peptide in lipid merging and fusion: An analogy between viral and palmitoylated eukaryotic fusion proteins. J Mol Biol. 2007;374:220–230. doi: 10.1016/j.jmb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 70.Pritsker M, Jones P, Blumenthal R, Shai YC. A synthetic all d-amino acid peptide corresponding to the N-terminal sequence of HIV-1 gp41 recognizes the wild-type fusion peptide in the membrane and inhibits HIV-1 envelope glycoprotein-mediated cell fusion. Proc Natl Acad Sci USA. 1998;95:7287–7292. doi: 10.1073/pnas.95.13.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kliger Y, Peisajovich SG, Blumenthal R, Shai Y. Membrane-induced conformational change during the activation of HIV-1 gp41. J Mol Biol. 2000;301:905–914. doi: 10.1006/jmbi.2000.4004. [DOI] [PubMed] [Google Scholar]
- 72.Buzon V, Cladera J. Effect of cholesterol on the interaction of the HIV gp41 fusion peptide with model membranes. Importance of the membrane dipole potential. Biochemistry. 2006;45:15768–15775. doi: 10.1021/bi060622i. [DOI] [PubMed] [Google Scholar]
- 73.Buzon V, Padros E, Cladera J. Interaction of fusion peptides from HIV gp41 with membranes: A time-resolved membrane binding, lipid mixing, and structural study. Biochemistry. 2005;44:13354–13364. doi: 10.1021/bi050382r. [DOI] [PubMed] [Google Scholar]
- 74.Moreno MR, Guillen J, Perez-Berna AJ, Amoros D, Gomez AI, Bernabeu A, Villalain J. Characterization of the interaction of two peptides from the N terminus of the nhr domain of HIV-1 gp41 with phospholipid membranes. Biochemistry. 2007;46:10572–10584. doi: 10.1021/bi700911g. [DOI] [PubMed] [Google Scholar]
- 75.Korazim O, Sackett K, Shai Y. Functional and structural characterization of HIV-1 gp41 ectodomain regions in phospholipid membranes suggests that the fusion-active conformation is extended. J Mol Biol. 2006;364:1103–1117. doi: 10.1016/j.jmb.2006.08.091. [DOI] [PubMed] [Google Scholar]
- 76.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 77.Rafalski M, Lear JD, Degrado WF. Phospholipid interactions of synthetic peptides representing the n-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 78.Sackett K, Nethercott MJ, Shai Y, Weliky DP. Hairpin folding of HIV gp41 abrogates lipid mixing function at physiologic ph and inhibits lipid mixing by exposed gp41 constructs. Biochemistry. 2009;48:2714–2722. doi: 10.1021/bi8019492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lev N, Fridmann-Sirkis Y, Blank L, Bitler A, Epand RF, Epand RM, Shai Y. Conformational stability and membrane interaction of the full-length ectodomain of HIV-1 gp41: Implication for mode of action. Biochemistry. 2009;48:3166–3175. doi: 10.1021/bi802243j. [DOI] [PubMed] [Google Scholar]
- 80.Pascual R, Moreno MR, Villalain J. A peptide pertaining to the loop segment of human immunodeficiency virus gp41 binds and interacts with model biomembranes: Implications for the fusion mechanism. J Virol. 2005;79:5142–5152. doi: 10.1128/JVI.79.8.5142-5152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Charloteaux B, Lorin A, Crowet JM, Stroobant V, Lins L, Thomas A, Brasseur R. The N-terminal 12 residue long peptide of HIV gp41 is the minimal peptide sufficient to induce significant T-cell-like membrane destabilization in vitro. J Mol Biol. 2006;359:597–609. doi: 10.1016/j.jmb.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 82.Gomara MJ, Lorizate M, Huarte N, Mingarro I, Perez-Paya E, Nieva JL. Hexapeptides that interfere with HIV-1 fusion peptide activity in liposomes block gp41-mediated membrane fusion. FEBS Lett. 2006;580:2561–2566. doi: 10.1016/j.febslet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 83.Lorizate M, de la Arada I, Huarte N, Sanchez-Martinez S, de la Torre BG, Andreu D, Arrondo JLR, Nieva JL. Structural analysis and assembly of the HIV-1 gp41 amino-terminal fusion peptide and the pretransmembrane amphipathic-at-interface sequence. Biochemistry. 2006;45:14337–14346. doi: 10.1021/bi0612521. [DOI] [PubMed] [Google Scholar]
- 84.Sackett K, Shai Y. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochemistry. 2002;41:4678–4685. doi: 10.1021/bi0255322. [DOI] [PubMed] [Google Scholar]
- 85.Shnaper S, Sackett K, Gallo SA, Blumenthal R, Shai Y. The C- and the N-terminal regions of glycoprotein 41 ectodomain fuse membranes enriched and not enriched with cholesterol, respectively. J Biol Chem. 2004;279:18526–18534. doi: 10.1074/jbc.M304950200. [DOI] [PubMed] [Google Scholar]
- 86.Koenig BW, Ferretti JA, Gawrisch K. Site-specific deuterium order parameters and membrane-bound behavior of a peptide fragment from the intracellular domain of HIV-1 gp41. Biochemistry. 1999;38:6327–34. doi: 10.1021/bi982800g. [DOI] [PubMed] [Google Scholar]
- 87.Gordon LM, Curtain CC, McCloyn V, Kirkpatrick A, Mobley PW, Waring AJ. The amino-terminal peptide of HIV-1 gp41 interacts with human serum-albumin. AIDS Res Hum Retroviruses. 1993;9:1145–1156. doi: 10.1089/aid.1993.9.1145. [DOI] [PubMed] [Google Scholar]
- 88.Chang DK, Cheng SF, Trivedi VD, Yang SH. The amino-terminal region of the fusion peptide of influenza virus hemagglutinin HA2 inserts into sodium dodecyl sulfate micelle with residues 16–18 at the aqueous boundary at acidic pH - oligomerization and the conformational flexibility. J Biol Chem. 2000;275:19150–19158. doi: 10.1074/jbc.M907148199. [DOI] [PubMed] [Google Scholar]
- 89.Chien MP, Jiang SB, Chang DK. The function of coreceptor as a basis for the kinetic dissection of HIV type 1 envelope protein-mediated cell fusion. Faseb J. 2008;22:1179–1192. doi: 10.1096/fj.07-9576com. [DOI] [PubMed] [Google Scholar]
- 90.Bellamy-McIntyre AK, Bar S, Ludlow L, Drummer HE, Poumbourios P. Role for the disulfide-bonded region of human immunodeficiency virus type 1 gp41 in receptor-triggered activation of membrane fusion function. Biochem Biophys Res Comm. 2010;394:904–908. doi: 10.1016/j.bbrc.2010.03.071. [DOI] [PubMed] [Google Scholar]
- 91.Peisajovich SG, Blank L, Epand RF, Epand RM, Shai Y. On the interaction between gp41 and membranes: The immunodominant loop stabilizes gp41 helical hairpin conformation. J Mol Biol. 2003;326:1489–1501. doi: 10.1016/s0022-2836(03)00040-8. [DOI] [PubMed] [Google Scholar]
- 92.Sackett K, Shai Y. How structure correlates to function for membrane associated HIV-1 gp41 constructs corresponding to the N-terminal half of the ectodomain. J Mol Biol. 2003;333:47–58. doi: 10.1016/j.jmb.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 93.Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang JH, Wagner G, Reinherz EL. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 94.Gerber D, Pritsker M, Gunther-Ausborn S, Johnson B, Blumenthal R, Shai Y. Inhibition of HIV-1 envelope glycoprotein-mediated cell fusion by a dl-amino acid-containing fusion peptide - possible recognition of the fusion complex. J Biol Chem. 2004;279:48224–48230. doi: 10.1074/jbc.M403436200. [DOI] [PubMed] [Google Scholar]
- 95.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. Plos Pathog. 2010:6. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang G. NMR of membrane-associated peptides and proteins. Curr Protein Pep Sci. 2008;9:50–69. doi: 10.2174/138920308783565714. [DOI] [PubMed] [Google Scholar]
- 97.Tamm LK, Lai AL, Li Y. Combined NMR and EPR spectroscopy to determine structures of viral fusion domains in membranes. Biochim Biophys Acta. 2007;1768:3052–60. doi: 10.1016/j.bbamem.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Biron Z, Khare S, Quadt SR, Hayek Y, Naider F, Anglister J. The 2F5 epitope is helical in the HIV-1 entry inhibitor T-20. Biochemistry. 2005;44:13602–11. doi: 10.1021/bi0509245. [DOI] [PubMed] [Google Scholar]
- 99.Biron Z, Khare S, Samson AO, Hayek Y, Naider F, Anglister J. A monomeric 3(10)-helix is formed in water by a 13-residue peptide representing the neutralizing determinant of HIV-1 on gp41. Biochemistry. 2002;41:12687–96. doi: 10.1021/bi026261y. [DOI] [PubMed] [Google Scholar]
- 100.Schibli DJ, Montelaro RC, Vogel HJ. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–8. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 101.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 ENV peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2450–1. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 102.Balogh E, Wu D, Zhou G, Gochin M. NMR second site screening for structure determination of ligands bound in the hydrophobic pocket of HIV-1 gp41. J Am Chem Soc. 2009;131:2821–2823. doi: 10.1021/ja8094558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: Discovery of d-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99:103–15. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 104.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci U S A. 2006;103:13938–43. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stewart KD, Huth JR, Ng TI, McDaniel K, Hutchinson RN, Stoll VS, Mendoza RR, Matayoshi ED, Carrick R, Mo H, Severin J, Walter K, Richardson PL, Barrett LW, Meadows R, Anderson S, Kohlbrenner W, Maring C, Kempf DJ, Molla A, Olejniczak ET. Non-peptide entry inhibitors of HIV-1 that target the gp41 coiled coil pocket. Bioorg Med Chem Lett. 2010;20:612–7. doi: 10.1016/j.bmcl.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 106.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. Three-dimensional solution structure of the 44 kda ectodomain of SIV gp41. EMBO J. 1998;17:4572–84. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cole JL, Garsky VM. Thermodynamics of peptide inhibitor binding to HIV-1 gp41. Biochemistry. 2001;40:5633–5641. doi: 10.1021/bi010085w. [DOI] [PubMed] [Google Scholar]
- 108.Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Short constrained peptides that inhibit HIV-1 entry. Proc Natl Acad Sci USA. 2002;99:14664–14669. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu J, Deng YQ, Dey AK, Moore JP, Lu M. Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry. 2009;48:2915–2923. doi: 10.1021/bi802303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schuy S, Schafer E, Yoder NC, Hobe S, Kumar K, Vogel R, Janshoff A. Coiled-coil lipopeptides mimicking the prehairpin intermediate of glycoprotein gp41. Angew Chem-Int Ed. 2009;48:751–754. doi: 10.1002/anie.200803080. [DOI] [PubMed] [Google Scholar]
- 111.Barz B, Wong TC, Kosztin I. Membrane curvature and surface area per lipid affect the conformation and oligomeric state of HIV-1 fusion peptide: A combined ftir and md simulation study. Biochim Biophys Acta-Biomem. 2008;1778:945–953. doi: 10.1016/j.bbamem.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 112.Liao ZH, Graham DR, Hildreth JEK. Lipid rafts and HIV pathogenesis: Virion-associated cholesterol is required for fusion and infection of susceptible cells. AIDS Res Hum Retroviruses. 2003;19:675–687. doi: 10.1089/088922203322280900. [DOI] [PubMed] [Google Scholar]
- 113.Harada S, Yusa K, Monde K, Akaike T, Maeda Y. Influence of membrane fluidity on human immunodeficiency virus type 1 entry. Biochem Biophys Res Comm. 2005;329:480–486. doi: 10.1016/j.bbrc.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 114.Quintana FJ, Gerber D, Kent SC, Cohen IR, Shai Y. HIV-1 fusion peptide targets the tcr and inhibits antigen-specific T cell activation. J Clin Invest. 2005;115:2149–2158. doi: 10.1172/JCI23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bloch I, Quintana FJ, Gerber D, Cohen T, Cohen IR, Shai Y. T-cell inactivation and immunosuppressive activity induced by HIV gp41 via novel interacting motif. Faseb J. 2007;21:393–401. doi: 10.1096/fj.06-7061com. [DOI] [PubMed] [Google Scholar]
- 116.Yang J, Gabrys CM, Weliky DP. Solid-state nuclear magnetic resonance evidence for an extended beta strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry. 2001;40:8126–37. doi: 10.1021/bi0100283. [DOI] [PubMed] [Google Scholar]
- 117.Li Y, Tamm LK. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys J. 2007;93:876–85. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol. 1997;4:732–8. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 119.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–80. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 120.Barz B, Wong TC, Kosztin I. Membrane curvature and surface area per lipid affect the conformation and oligomeric state of HIV-1 fusion peptide: A combined ftir and md simulation study. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 121.Gordon LM, Mobley PW, Pilpa R, Sherman MA, Waring AJ. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in membrane environments using C-13-enhanced fourier transform infrared spectroscopy. Biochim Biophy Acta-Biomem. 2002;1559:96–120. doi: 10.1016/s0005-2736(01)00443-6. [DOI] [PubMed] [Google Scholar]
- 122.Li YL, Tamm LK. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophy J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peisajovich SG, Epand RF, Pritsker M, Shai Y, Epand RM. The polar region consecutive to the HIV fusion peptide participates in membrane fusion. Biochemistry. 2000;39:1826–33. doi: 10.1021/bi991887i. [DOI] [PubMed] [Google Scholar]
- 124.Yang J, Parkanzky PD, Khunte BA, Canlas CG, Yang R, Gabrys CM, Weliky DP. Solid state NMR measurements of conformation and conformational distributions in the membrane-bound HIV-1 fusion peptide. J Mol Graph Model. 2001;19:129–35. doi: 10.1016/s1093-3263(00)00128-5. [DOI] [PubMed] [Google Scholar]
- 125.Qiang W, Bodner ML, Weliky DP. Solid-state nmr spectroscopy of human immunodeficiency virus fusion peptides associated with host-cell-like membranes: 2D correlation spectra and distance measurements support a fully extended conformation and models for specific antiparallel strand registries. J Am Chem Soc. 2008;130:5459–71. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sackett K, Nethercott MJ, Epand RF, Epand RM, Kindra DR, Shai Y, Weliky DP. Comparative analysis of membrane-associated fusion peptide secondary structure and lipid mixing function of HIV gp41 constructs that model the early pre-hairpin intermediate and final hairpin conformations. J Mol Biol. 2010;397:301–315. doi: 10.1016/j.jmb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Castano S, Desbat B. Structure and orientation study of fusion peptide FP23 of gp41 from HIV-1 alone or inserted into various lipid membrane models (mono-, bi- and multibi-layers) by FR-IR spectroscopies and brewster angle microscopy. Biochim Biophys Acta. 2005;1715:81–95. doi: 10.1016/j.bbamem.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 128.Jiang S, Lin K, Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu SW, Lu H, Niu J, Xu YJ, Wu SG, Jiang SB. Different from the HIV fusion inhibitor C34, the anti-HIV drug fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J Biol Chem. 2005;280:11259–11273. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 130.Rabenstein M, Shin YK. A peptide from the heptad repeat of human-immunodeficiency-virus gp41 shows both membrane-binding and coiled-coil formation. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 131.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human-immunodeficiency-virus type-1 gp41 are potent inhibitors of virus-infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chinnadurai R, Rajan D, Munch J, Kirchhoff F. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J Virol. 2007;81:6563–6572. doi: 10.1128/JVI.02546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eggink D, Baldwin CE, Deng YQ, Langedijk JPM, Lu M, Sanders RW, Berkhout B. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J Virol. 2008;82:6678–6688. doi: 10.1128/JVI.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eggink D, Langedijk JPM, Bonvin A, Deng YQ, Lu M, Berkhout B, Sanders RW. Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J Biol Chem. 2009;284:26941–26950. doi: 10.1074/jbc.M109.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Melby T, Demasi R, Cammack N, Miralles GD, Greenberg ML. Evolution of genotypic and phenotypic resistance during chronic treatment with the fusion inhibitor T-1249. AIDS Res Hum Retroviruses. 2007;23:1366–1373. doi: 10.1089/aid.2006.0277. [DOI] [PubMed] [Google Scholar]
- 136.Miralles GD, Melby T, DeMasi R, Zhang Y, Spence R, Cammack N, Matthews TJ, Greenberg M. Baseline and on-treatment gp41 genotype and susceptibility to enfuvirtide (enf) and T-1249 in a 10-day study of T-1249 in patients failing an ENF-containing regimen (T1249–102) Antiviral Ther. 2003;8:21. [Google Scholar]
- 137.Pan CG, Cai LF, Lu H, Qi Z, Jiang SB. Combinations of the first and next generations of human immunodeficiency virus (HIV) fusion inhibitors exhibit a highly potent synergistic effect against enfuvirtide-sensitive and -resistant HIV type 1 strains. J Virol. 2009;83:7862–7872. doi: 10.1128/JVI.00168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rusconi S, Scozzafava A, Mastrolorenzo A, Supuran CT. An update in the development of HIV entry inhibitors. Curr Top Med Chem. 2007;7:1273–1289. doi: 10.2174/156802607781212239. [DOI] [PubMed] [Google Scholar]
- 139.Veiga AS, Santos NC, Loura LMS, Fedorov A, Castanho M. HIV fusion inhibitor peptide T-1249 is able to insert or adsorb to lipidic bilayers. Putative correlation with improved efficiency. J Am Chem Soc. 2004;126:14758–14763. doi: 10.1021/ja0459882. [DOI] [PubMed] [Google Scholar]
- 140.Dai SJ, Song HF, Dou GF, Qian XH, Zhang YJ, Cai Y, Liu XW, Tang ZM. Quantification of sifuvirtide in monkey plasma by an on-line solid-phase extraction procedure combined with liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Comm Mass Spec. 2005;19:1273–1282. doi: 10.1002/rcm.1917. [DOI] [PubMed] [Google Scholar]
- 141.Liang Q, Mao YC, Yang S, Fang SS, Tan JX, Liao YP, Hu ZH. Drug-drug interactions of sifuvirtide with cytochrome p450 1A2, 2A6, 2D6, and 3A4 by using pooled human liver microsomes and primary human hepatocytes. Drug Metab Rev. 2006;38:93–93. [Google Scholar]
- 142.Che J, Meng Q, Chen Z, Hou Y, Shan C, Cheng Y. Quantitative analysis of a novel HIV fusion inhibitor (sifuvirtide) in HIV infected human plasma using high-performance liquid chromatography-electrospray ionization tandem mass spectrometry. J Pharmaceut Biomed Anal. 2010;51:927–933. doi: 10.1016/j.jpba.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 143.Covens K, Megens S, Dekeersmaeker N, Kabeya K, Balzarini J, De Wit S, Vandamme AM, Van Laethem K. The rare HIV-1 gp41 mutations 43T and 50V elevate enfuvirtide resistance levels of common enfuvirtide resistance mutations that did not impact susceptibility to sifuvirtide. Antiviral Res. 2010;86:253–260. doi: 10.1016/j.antiviral.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 144.Pan C, Lu H, Qi Z, Jiang SB. Synergistic efficacy of combination of enfuvirtide and sifuvirtide, the first- and next-generation HIV-fusion inhibitors. AIDS. 2009;23:639–641. doi: 10.1097/QAD.0b013e328325a4cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang QQ, Xiang SS, Jia YB, Ou L, Chen F, Song HF, Liang Q, Ju D. An improved on-line solid phase extraction coupled hplc-ms/ms system for quantification of sifuvirtide in human plasma. J Chromatog B-Anal Technol Biomed Life Sci. 2010;878:1893–1898. doi: 10.1016/j.jchromb.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 146.Franquelim HG, Loura LMS, Santos NC, Castanho M. Sifuvirtide screens rigid membrane surfaces. Establishment of a correlation between efficacy and membrane domain selectivity among HIV fusion inhibitor peptides. J Am Chem Soc. 2008;130:6215–6223. doi: 10.1021/ja711247n. [DOI] [PubMed] [Google Scholar]
- 147.Franquelim HG, Veiga AS, Weissmuller G, Santos NC, Castanho M. Unravelling the molecular basis of the selectivity of the HIV-1 fusion inhibitor sifuvirtide towards phosphatidylcholine-rich rigid membranes. Biochim Biophys Acta-Biomem. 2010;1798:1234–1243. doi: 10.1016/j.bbamem.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 148.Naider F, Anglister J. Peptides in the treatment of AIDS. Curr Opin Struct Biol. 2009;19:473–482. doi: 10.1016/j.sbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vishwanathan SA, Thomas A, Brasseur R, Epand RF, Hunter E, Epand RM. Hydrophobic substitutions in the first residue of the crac segment of the gp41 protein of HIV. Biochemistry. 2008;47:124–130. doi: 10.1021/bi7018892. [DOI] [PubMed] [Google Scholar]
- 150.Vishwanathan SA, Thomas A, Brasseur R, Epand RF, Hunter E, Epand RM. Large changes in the crac segment of gp41 of HIV do not destroy fusion activity if the segment interacts with cholesterol. Biochemistry. 2008;47:11869–11876. doi: 10.1021/bi8014828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Song L, Sun ZY, Coleman KE, Zwick MB, Gach JS, Wang JH, Reinherz EL, Wagner G, Kim M. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci USA. 2009;106:9057–62. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Santos NC, Prieto M, Castanho M. Interaction of the major epitope region of HIV protein gp41 with membrane model systems. A fluorescence spectroscopy study. Biochemistry. 1998;37:8674–8682. doi: 10.1021/bi9803933. [DOI] [PubMed] [Google Scholar]
- 153.Gochin M, Cai LF. The role of amphiphilicity and negative charge in glycoprotein 41 interactions in the hydrophobic pocket. J Med Chem. 2009;52:4338–4344. doi: 10.1021/jm900190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Waheed AA, Ablan SD, Sowder RC, Roser JD, Schaffner CP, Chertova E, Freed EO. Effect of mutations in the human immunodeficiency virus type 1 protease on cleavage of the gp41 cytoplasmic tail. J Virol. 2010;84:3121–3126. doi: 10.1128/JVI.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Waheed AA, Ablan SD, Roser JD, Sowder RC, Schaffner CP, Chertova E, Freed EO. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc Natl Acad Sci USA. 2007;104:8467–8471. doi: 10.1073/pnas.0701443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Waheed AA, Ablan SD, Mankowski MK, Cummins JE, Ptak RG, Schaffner CP, Freed EO. Inhibition of HIV-1 replication by amphotericin B methyl ester - selection for resistant variants. J Biol Chem. 2006;281:28699–28711. doi: 10.1074/jbc.M603609200. [DOI] [PubMed] [Google Scholar]
- 157.Epand RF, Sayer BG, Epand RM. The tryptophan-rich region of HIV gp41 and the promotion of cholesterol-rich domains. Biochemistry. 2005;44:5525–5531. doi: 10.1021/bi0500224. [DOI] [PubMed] [Google Scholar]