Summary
Pasteurella multocida toxin (PMT) is an AB toxin that causes pleiotropic effects in targeted host cells. The N-terminus of PMT (PMT-N) is believed to harbor the membrane receptor binding and translocation domains responsible for mediating cellular entry and delivery of the C-terminal catalytic domain into the host cytosol. Previous studies have implicated gangliosides as the host receptors for PMT binding. To gain further insight into the binding interactions involved in PMT binding to cell membranes, we explored the role of various membrane components in PMT binding, utilizing four different approaches: TLC-overlay binding experiments with 125I-labeled PMT, PMT-N or PMT-C; pull-down experiments using reconstituted membrane liposomes with full-length PMT, surface plasmon resonance (SPR) analysis of PMT-N binding to reconstituted membrane liposomes, and SPR analysis of PMT-N binding to HEK-293T cell membranes without or with sphingomyelinase, phospholipase D, or trypsin treatment. Results revealed that, in our experimental system, full-length PMT and PMT-N did not bind to gangliosides, including GM1, GM2 or GM3, but instead bound to membrane phospholipids, primarily the abundant sphingophospholipid sphingomyelin (SM) or phosphatidylcholine (PC) with other lipid components. Collectively, these studies demonstrate the importance of SM for PMT binding to membranes and suggest the involvement of a protein co-receptor.
Keywords: dermonecrotic toxin, sphingomyelin, surface plasmon resonance, thin layer chromatography, ganglioside, phosphatidylcholine
Introduction
Pasteurella multocida toxin (PMT) is a 1,285-amino acid AB toxin that is both a potent virulence factor associated with atrophic rhinitis and pasteurellosis [1–3] and a tool for understanding G-protein signaling [4, 5]. PMT is one of the most potent known mitogens and triggers mitosis in fibroblastic and osteoblastic cells [6–8] by activating heterotrimeric G proteins of the Gq, G13, and Gi families through deamidation of a critical active site glutamine residue [9–13]. As a consequence, PMT interferes with numerous cellular processes in targeted host cells.
The crystal structure of the PMT carboxyl-terminus (C-terminus; residues 575–1285) has been elucidated [14]. The C-terminus of PMT (PMT-C) consists of three domains, C1–C3, where C1 is the intracellular membrane-targeting domain [15], C2 is a domain of unknown function, and C3 is the minimal functional catalytic domain containing the active site C1165-H1205-D1220 catalytic triad responsible for deamidation of the target G proteins [16]. The amino-terminus (N-terminus) of PMT (PMT-N) is believed to be the binding domain and contains a putative membrane insertion motif (residues 370–470) believed to be involved in membrane translocation into the cytosol [17, 18].
PMT-N shares significant sequence similarity with the N-terminal regions of the cytotoxic necrotizing factors of E. coli (CNF1-3) [19–21] and Yersinia pseudotuberculosis (CNFY) [22]. These shared regions are believed to be important for toxin binding and translocation [17, 18, 23–26]. There is evidence for the involvement of the p37 laminin receptor precursor (LRP) in CNF1 binding [23–25] and heparin sulfate proteoglycan (HSPG) in both CNF1 and CNFY binding [23]. It has been suggested that gangliosides play an important role in PMT binding [27, 28], and there is no current experimental evidence for the necessity of a protein receptor for PMT.
AB toxins are known to employ various forms of receptors to gain entrance into cells, including gangliosides alone as for cholera toxin (CT) [29], proteins alone as for diphtheria toxin (DT) [30], or both proteins and gangliosides as for the tetanus neurotoxin (TeNT) and botulinum neurotoxins (BoNTs) [31, 32]. This raises the question as to whether binding gangliosides alone is sufficient for cellular entry of PMT, or gangliosides in conjunction with other cellular protein components might also be required for productive docking and endocytosis.
We used thin layer chromatography (TLC) overlay, liposome binding, and surface plasmon resonance (SPR) methods to further characterize the nature of the membrane receptors for PMT and to study the interaction properties of PMT on reconstituted membranes or membrane ghosts generated from lysed HEK-293T cells. We found that contrary to previous reports PMT did not bind sialogangliosides on TLC plates. Instead, our results implicate interplay among SM and PC membrane lipid components, as well as unidentified proteins in the binding of PMT to cells.
Results
Binding of PMT to lipid extract components from mammalian cells
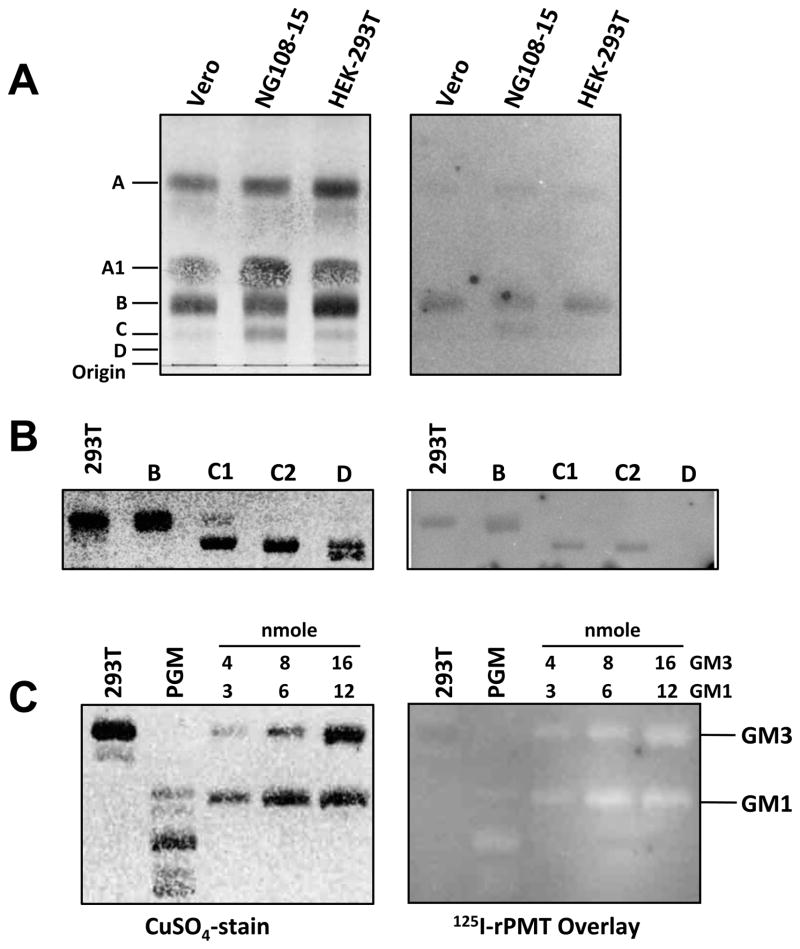
TLC in combination with overlaying labeled proteins is a valid method for determining protein-ligand interactions and has been used extensively for confirming protein toxin utilization of gangliosides and/or phospholipids as receptors for gaining entry into target cells, including cholera toxin [33], shiga-like toxins [34], Bacillus thuringiensis crystal toxin [35], TeNT [36] and BoNTs [37]. To test whether PMT can directly bind to membrane lipid components, we employed an overlay technique in which purified lipids or crude lipid extracts from PMT-susceptible Vero, HEK-293T and NG108-15 cells were resolved on TLC plates using appropriate solvent systems. After blocking with BSA, plates were overlayed for 1–2 h with 125I-rPMT in PBS, pH 7.4, at 37°C. After washing away unbound toxin, 125I-rPMT bound to lipids was detected using a phosphorimager. We found that 125I-rPMT was able to bind to a number of lipid components, with the three major binding components present in all three cell lines assigned as A, B, and C (Figure 1A, 1B and Supplementary Figure S1). However, there was an absence of 125I-rPMT binding to certain other lipid components, such as A1 and D, on the same TLC plate.
Figure 1. Binding of PMT to cellular membrane components.
(A) Total cell membrane extracts (50 nmol phosphate equivalent) developed in a solvent system of 75:25:4 (see experimental section). (B) Purified cell membrane components (10 nmol phosphate equivalent) developed in a solvent system of 75:25:4. (C) Purified gangliosides and porcine ganglioside mixture (PGM, 20 μg) developed in a solvent system of 25:20:3. Right panels are images of overlays with the indicated radiolabeled proteins recorded by phosphorimager. Left panels are images of the corresponding LC plates subsequently stained with CuSO4.
PMT does not bind GM1, GM2, GM3 or mixtures of porcine gangliosides (PGM) on TLC plates
Although previous reports suggested a role of gangliosides, namely GM1, GM2, and GM3, in cellular uptake of PMT [27, 28], 125I-rPMT did not bind these three mono-sialogangliosides or other di- or tri-sialogangliosides deposited on TLC plates at 37°C (Figure 1C and Supplementary Figure S2). Although prolonged exposure of 125I-rPMT at low temperature (16 hr, 4°C) showed 125I-rPMT binding to GM3, the asialoganglioside, lactosylceramide (LacCer), bound 125I-rPMT much more efficiently, even at 37°C (Supplementary Figure S2).
PMT binds positively charged phospholipid and sphingophospholipid headgroups
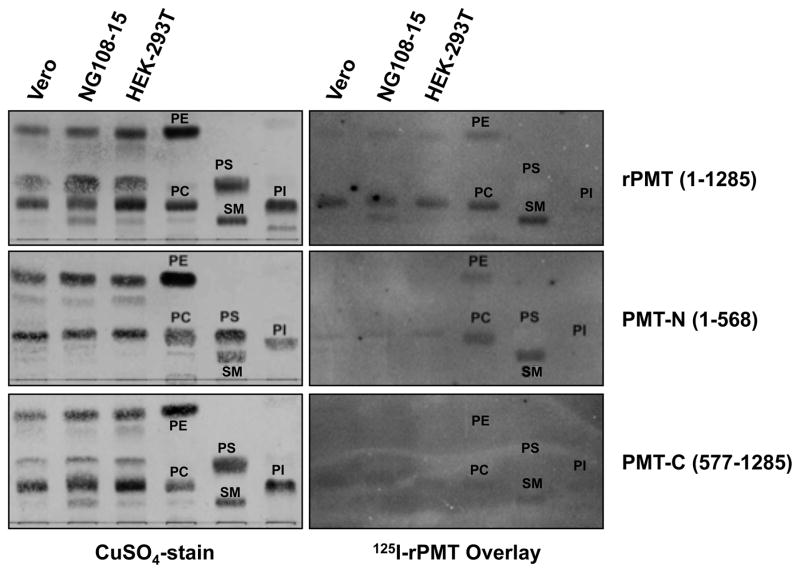
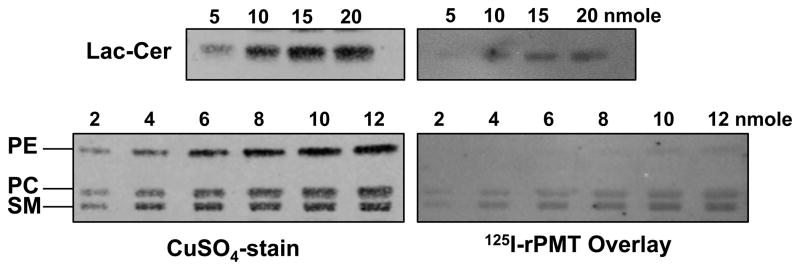
Among the four major phospholipids found in cell membranes, phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylserine (PS), and phosphatidylinositol (PI), and the abundant sphingophospholipid sphingomyelin (SM), the most efficient binding of 125I-rPMT was observed for PC and SM, while PE and PI showed only weak binding and PS showed no detectable binding (Figure 2). Studies using 125I-PMT-N and 125I-PMT-C revealed that the N terminus contributed to the strong binding observed for full-length 125I-rPMT to PC and SM and the weaker binding to PE, while the C terminus contributed only partially to the binding of PC and SM and was primarily responsible for the weak binding to PI (Figure 2). Semi-quantitative analysis of 125I-rPMT binding to the three positively charged phospholipid head groups clearly showed a binding preference of SM > PC ≫ PE (Figure 3). The asialoganglioside LacCer was also found to bind 125I-rPMT in a manner comparable to that of PC but less than SM (Figure 3).
Figure 2. Binding of PMT and PMT fragments to phospholipids.
Images of TLC plates loaded with cell membrane lipids (50 nmol phosphate equivalent) and purified phospholipids (10 nmol phosphate equivalent each), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS) and sphingomyelin (SM) and developed in a solvent system of 75:25:4 (see experimental section). Right panels are images of overlays with the indicated radiolabeled proteins recorded by phosphorimager. Left panels are images of the corresponding LC plates subsequently stained with CuSO4.
Figure 3. Relative affinity of PMT binding to phospholipids and lactosylceramide.
Images of TLC plates loaded with incremental amounts of lactosylceramide (LacCer) and developed in a solvent system of 85:15:1 (see experimental section) or with phosphatidylethanolamine (PE), phosphatidylcholine (PC) and sphingomyelin (SM) and developed in a solvent system of 75:25:4. Right panels are images of overlays with the indicated radiolabeled proteins recorded by phosphorimager. Left panels are images of the corresponding LC plates subsequently stained with CuSO4.
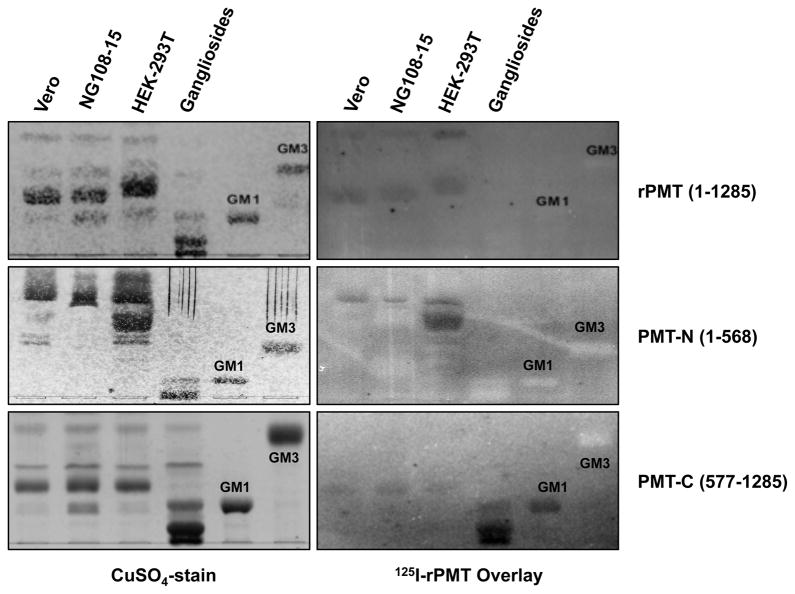
PMT-C uniquely binds highly polar gangliosides
Although full-length 125I-rPMT and 125I-PMT-N showed no affinity for sialogangliosides, 125I-PMT-C surprisingly showed affinity for GM1 and more polar di- and trisialogangliosides in mixtures of porcine brain gangliosides (Figure 4). This suggests that the binding characteristics of 125I-PMT-C are different than those of the full-length toxin.
Figure 4. Binding of PMT and PMT fragments to gangliosides.
Shown are representative images of TLC plates: (lanes 1–3) cell membrane lipid extracts as indicated (50 nmol phosphate equivalent); (lane 4) a ganglioside mixture (20 μg) containing mostly GM1, GD1a, GD1b, GT1b and lesser amounts of GM2 and GM3; (lane 5) purified GM1 (10μg); and (lane 6) purified GM3 (10μg). The labeled band in the three lanes for cellular membrane lipids corresponds to the lipid component B (PC). Only PMT-C bound GM1 and other more polar gangliosides. All TLC plates were developed in a solvent system of 25:20:3 (see experimental section). Right panels are images of overlays with the indicated radiolabeled proteins recorded by phosphorimager. Left panels are images of the corresponding TLC plates subsequently stained with CuSO4.
Other lipid components showed affinity for PMT on TLC
It was noted that full-length 125I-rPMT bound the more polar components in cellular lipid extracts (Supplementary Figure S1). Further TLC overlay analysis showed that 125I-rPMT also bound to cholesterol, stearic acid, and mono- and di-glycerides (Supplementary Figure S3). In addition to LacCer, 125I-rPMT also bound other asialogangliosides, including GalCer, GlcCer, CerI, and CerII (Supplementary Figure S4). However, with these less polar lipid compounds, specific binding to the N terminus or C terminus of PMT was not apparent.
Binding of PMT to liposomes
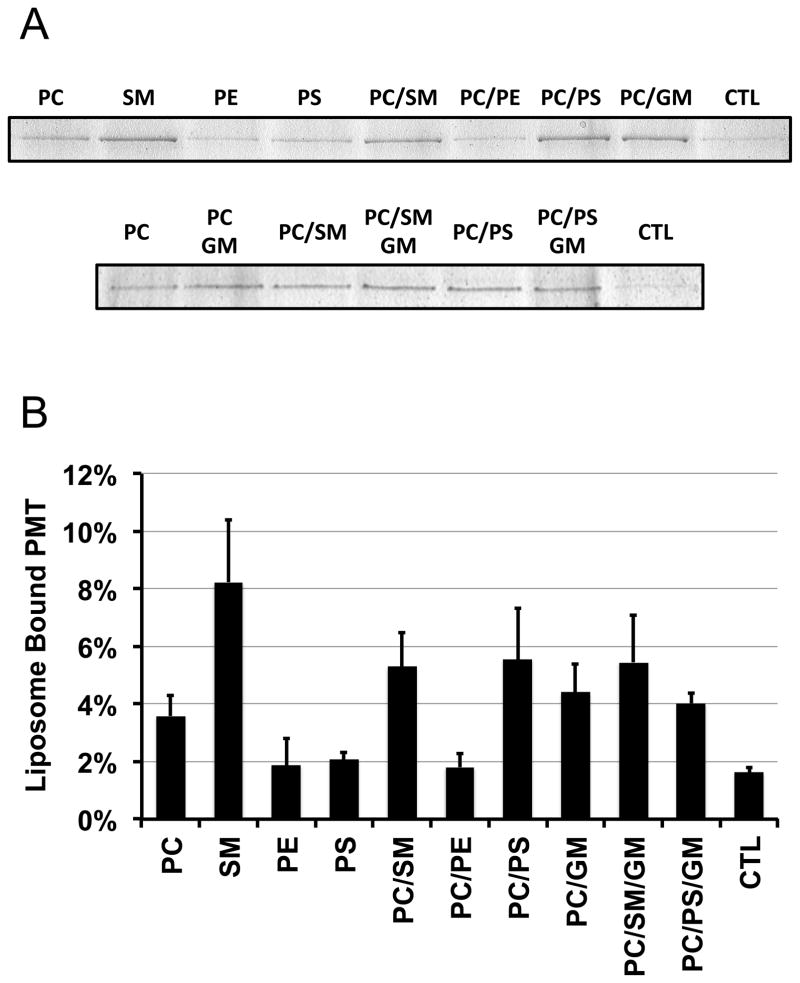
The importance of SM in PMT binding to membranes was further studied by PMT pull-down assay using liposomes containing a mixture of cholesterol with various purified phospholipids (Figure 5). The amount of rPMT found in the pellet of liposomes containing only PS, only PE or a mixture of PC and PE was similar to that of controls. PC-containing liposomes pulled down 2–3-fold more rPMT than control (p < 0.001), while SM-containing liposomes were the most effective at pulling down rPMT (4–5-fold versus control, p < 0.001). Although PS-containing liposomes were poor at pulling down rPMT, addition of PS to PC-containing liposomes enhanced the pull-down of rPMT over PC alone (p = 0.03). Likewise, addition of ganglioside mixtures (GM) to PC-containing liposomes also enhanced binding of rPMT to liposomes (p = 0.08). However, addition of GM to PC/SM or PC/PS liposomes did not enhance the binding of rPMT (p = 0.87 and 0.20, respectively).
Figure 5. Binding of rPMT to liposomes.
(A) Shown are representative images from more than 3 independent repeats of Coomassie Blue-stained 10% SDS-PAGE analyses of pellets from pull-down experiments using the indicated liposomes, as described under Methods. The control (CTL) was from the pellets of a mock treatment of PMT solution without liposomes. (B) Quantitative analysis of the amount of rPMT bound to the indicated liposome preparations from 3 independent repeats of pull-down experiments similar that in (A), as described under Methods.
Analysis of PMT-N interaction with reconstituted phospholipid membranes by surface plasmon resonance (SPR)
SPR is a well-established method for studying the kinetic and thermodynamic interactions of toxins with their respective ganglioside [38, 39] and/or protein receptors [40], including shiga-like toxins [38], E. coli heat-labile toxin [41], CT [39, 41], anthrax toxin [40], TeNT [41] and BoNTs [42]. Kinetic analysis of SPR data has been established using a Langmuir binding-dissociation model, as described by O’Shannessy et al. [43]. In practice, most binding sensorgrams appear to show a biphasic behavior. The portion of the sensorgram showing rapid change is often attributed to a “bulk shift” or other nonspecific phenomenon and is generally ignored for calculations. However, our methodology does not ignore this phase. There have been attempts to analyze the dissociation curve as two distinct phases [39] or to treat it as a transitional behavior during binding [42]. For simplification, we treated the biphasic behavior as a sum of two separate events, a rapid binding component and a slow tight binding component. We found our approach fitted well across a wide range of analyte concentrations in both the association phase and the dissociation phase. Our model does not exclude the possibility that the rapid binding event will transition into the slower tight binding event, i.e., a conformational transition model.
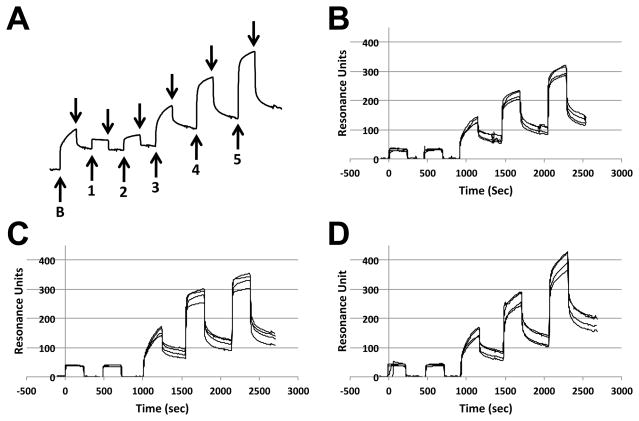
SPR sensorgrams of PMT-N binding to the three reconstituted membranes (PC only, PC/PS, or PC/SM) on an L1 Biacore chip clearly showed a rapid and a slow component to both the binding and dissociation events (Figure 6A). PC/SM showed the highest RU at high PMT-N concentrations and appeared to be far from saturation even at 7.5 μM PMT-N, suggesting a greater loading capacity for PC/SM membranes (compare Figure 6B, 6C and 6D). PC/PS appeared to be near saturation at 7.5 μM PMT-N, suggesting that PC/PS membranes contained the fewest number of binding sites among the three membranes (Figure 6C).
Figure 6. SPR traces of PMT binding to liposomes.
Example of traces generated using SPR from reconstituted lipids. Membrane vesicles were generated and loaded onto a L1 chip, as described in the experimental section. (A) Shown is an SPR trace for PC vesicles with initial injection of 3.75 μM BSA (labeled as B) followed by dissociation (indicated by downward arrows) and injections of PMT (labeled as 1–5) at concentrations of 7.5 nM, 75 nM, 750 nM, 3.75 μM, and 7.5 μM punctuated with dissociation events. Also shown are example traces from all four channels of the L1 chip preloaded with (B) PC, (C) PC/PS or (D) PC/SM vesicles, followed by 5 serial PMT-N injection-dissociation events, similar to the sequence shown in (A).
The sensorgrams were further analyzed using a bi-phasic exponential binding model (Equations 1–3) coupled with a bi-phasic exponential dissociation model (Equation 4), as defined in the experimental section. The best-fit kinetic parameters, using a least-squares method according to this model, were used to construct a theoretical resonance signal for the total resonance signals, consisting of the rapid and slow component resonance signals of PMT-N binding (Supplementary Figure S5). There was a general agreement between the calculated curves and the corresponding experimental sensorgrams. The calculated parameters showed highest binding capacity (Bmax) in PC/SM membranes for both rapid and slow components (Table 1). Although the dissociation rates (koff) were similar among the three types of membranes, PC/SM appeared to have a lower association rate (kon) for the slow components. Both the slower association rate (kon) and the higher Bmax calculated for PC/SM membraneswere consistent with the observed sensorgrams. For all three reconstituted membranes, rapid binding was detected at concentrations as low as 7.5 nM, but slow binding became more evident at concentrations of 750 nM or higher.
Table 1. PMT binding characteristics on various reconstituted membranes or natural membranes determined by SPR analysis.
The untreated and enzyme-treated natural membranes are designated as Native, Trypsin, SMase, PLCCab, or PLCSc. T indicates additional trypsin treatment after lipase treatment. The number of repeats N is the repeat of a set consisting of 4 recording signals from 4 flow channels of the same L1 chip. T-test p values for comparison among three reconstituted membranes were tabulated in supplementary Table S1, and among seven natural membranes in supplementary Table S2.
| Rapid Component
|
Slow Component
|
N (x4) | ||||||
|---|---|---|---|---|---|---|---|---|
| * Bmax (RU) | * KDx106 (M) | koff x102 (s−1) | Bmax (RU) | KD x106 (M) | koff x104 (s−1) | kon x10−2 (M−1s−1) | ||
| Reconstituted membranes | ||||||||
| PC | 243±2 | 3.78±0.12 | 29.6±24.3 | 165±30 | 0.54±0.10 | 8.46±2.77 | 16.1±6.2 | 3 |
| PC/PS | 221±1 | 1.57±0.03 | 24.4±15.6 | 185±30 | 0.41±0.11 | 5.58±0.75 | 14.0±3.0 | 3 |
| PC/SM | 264±3 | 2.80±0.13 | 26.6±15.5 | 256±42 | 0.84±0.21 | 6.57±0.51 | 8.22±1.93 | 3 |
| HEK-293T cell membranes | ||||||||
| Native | 530±29 | 12.4±1.1 | 4.9±1.0 | 802±193 | 0.68±0.24 | 5.7±1.1 | 9.3±2.8 | 4 |
| Trypsin | 201±1 | 3.07±0.15 | 5.3±0.7 | 572±52 | 0.60±0.07 | 6.4±0.3 | 10.8±1.3 | 2 |
| SMase | 4785±99 | 38.3±1.4 | 15.7±6.6 | 525±82 | 1.60±0.76 | 5.2±2.4 | 3.3±0.4 | 2 |
| SMase/T | 5516±64 | 48.5±0.9 | 14.0±5.5 | 616±54 | 1.46±0.63 | 5.2±1.7 | 3.7±0.5 | 3 |
| PLDCab | 423±3 | 14.3±0.3 | 12.9±3.3 | 423±83 | 0.91±0.32 | 8.6±1.3 | 10.2±2.8 | 3 |
| PLDCab/T | 416±3 | 9.44±0.24 | 11.1±0.3 | 548±37 | 0.79±0.19 | 7.5±0.8 | 10.2±3.0 | 2 |
| PLDSc | 313±8 | 7.18±0.32 | 11.6±4.6 | 518±48 | 0.68±0.13 | 6.8±1.1 | 10.2±1.7 | 3 |
Calculated from the RU at equilibrium of the rapid component, fitting to equation 5 using least-square method with a weight proportional to the concentration of PMT-N.
Analysis of interaction between PMT-N and HEK-293T cell membranes by SPR
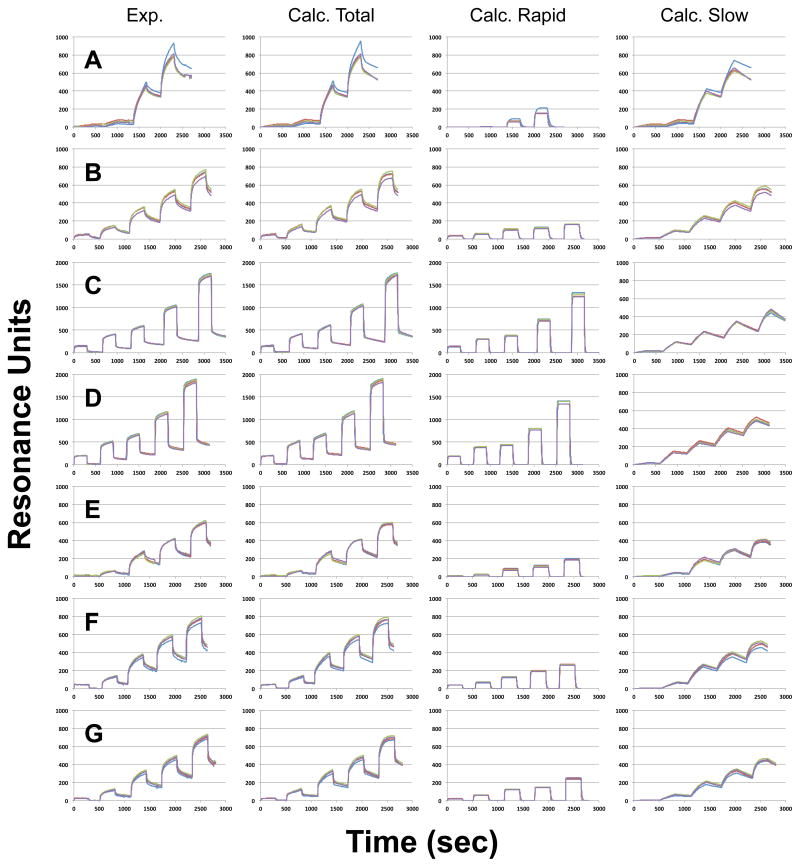
Liposomes prepared from membrane ghosts of HEK-293T cells were loaded onto an L-1 Biacore chip. SPR sensorgrams of PMT-N bound to these natural membrane liposomes showed concentration-dependent binding, with prominent binding observed at concentrations higher than 750 nM (Figure 7A), which was three times higher than that observed with reconstituted phospholipid membranes (Table 1). After fitting the sensorgrams to a two-component model, the calculated rapid binding component contributed less to the overall SPR signal for natural membranes than that for phospholipid membranes (20% versus 50%, respectively) (compare Figure 7A to Supplementary Figure S5). Even so, the sensorgrams for PMT-N binding to natural membranes did not fit well using a single step binding-dissociation model (Supplementary Figure S6).
Figure 7. SPR traces of PMT binding to cell membranes and enzyme-treated cell membranes.
Shown are representative sets of traces recorded from all four channels of the L1 chip, preloaded with (A) native membranes, or membranes treated with (B) trypsin, (C) SMase, (D) SMase and trypsin, (E) PLDCab, (F) PLDCab and trypsin, or (G) PLDSc. Four graphs shown in each row are the recorded traces (Exp.), the calculated curves (Calc. Total), the calculated curve for the rapid components (Calc. Rapid), and the calculated curves for the slow components (Calculated Slow), generated as described in the experimental section. For the native membranes, PMT-N at concentrations of 75 nM, 750 nM, 3.75 μM, or 7.5 μM were used (four binding-dissociation events). For all other membranes, an additional 15 μM concentration was used (5 binding-association events).
Effect of sphingomyelinase treatment on PMT-N binding to HEK-293T cell membranes
SMase cleaves the phosphocholine head group from SM and forms ceramide-rich domains in membranes [44]. To explore the importance of the head group in PMT-N interactions with membranes, HEK-293T cell membranes were treated with SMase on an L-1 Biacore chip. The results showed a noticeable decrease in the slow and tight binding component; however, the Bmax of the rapid and loose binding component increased by 10-fold (compare Figure 7C vs. 7A, Table 1). The KD for the rapid and slow binding components both increased by 3- and 2-fold, respectively. The changes in kinetic parameters upon SMase treatment were consistent with decreased PMT-N binding to membranes due to removal of the phosphocholine head group by SMase.
Effect of phospholipase D treatment on PMT-N binding to HEK-293T cell membranes
PLD cleaves the head group of phospholipids and forms phosphatidic acid. PLD from Streptomyces chromofuscus (PLDSc) has a substrate specificity for PC>PE≫PS, but it also has a weak contaminating SMase activity [45]. Cabbage phospholipase D (PLDCab) has a substrate preference for PC≈PE≈PG≫PI>PS [46]. Comparison between the effects of SMase vs. PLD could distinguish whether the choline head group alone or the unique structural features of sphingophospholipids contribute to the binding of PMT-N. PLDCab caused a 50% decrease in the slow binding Bmax, but only a 20% decrease in the rapid binding Bmax (Figure 7E vs. 7A, Table 1), whereas PLDSc decreased both the rapid and slow binding Bmax by 40% (Figure 7G vs. 7A, Table 1). The drastic changes seen with SMase were not detected with either PLD (Figure 7E vs. 7C and 7G vs. 7C), suggesting that the contribution of SM to PMT-N binding was due to the entire sphingophospholipid structure and not just the choline head group alone.
Effect of trypsin treatment on PMT-N binding to HEK-293T cell membranes
Trypsin treatment would remove some cell surface protein components; particularly those not present in microdomains or membrane lipid rafts. Trypsin treatment of native membranes deceased the slow binding Bmax, but showed little effect on kon or koff (Figure 7B vs. 7A, Table 1). The most prominent effects of trypsin were on the rapid binding component, where the Bmax decreased by more than half and became quickly saturable with a KD of less than one third of that for the untreated membranes. Trypsin treatment of the membranes that had already been exposed to SMase or PLDCab showed a small increase in Bmax for the slow binding component, but did not have an effect on other kinetic parameters of the rapid or slow binding components (Figure 7D vs. 7C, 7F vs. 7E, Table 1). These results implicate a protein component in natural membranes for both the rapid and slow binding components. However, removal of protein had minimal effect on membranes already depleted of choline or phosphocholine head groups, indicating that protein is only important for PMT binding when PMT is also interacting with PC and/or SM in the membrane.
Discussion
Two previous reports indicated that gangliosides, namely GM1, GM2 and GM3, were receptors for PMT [27, 28]. Our results using the TLC-overlay method, summarized in Table 2, indicate that gangliosides, including GM1, GM2 or GM3, were not the preferred binding targets for PMT in this experimental system. Instead, PMT bound well to asialogangliosides, such as LacCer, and in particular positively charged phospholipids (PC and PE) and sphingophospholipids (SM). We also found that addition of GM1, GM2 or GM3 to PC/SM- or PC/PS-containing liposomes did not enhance the binding of PMT in pull-down assays (Figure 5). It was noted that the ganglioside concentrations used in previous studies [27, 28] was above the critical micelle concentration, which might have interfered with the availability of PMT to gain access to the cell surface for optimal binding. In addition, the readout of the assays in one of the studies [27] relied on determination of the downstream mitogenic effect of the toxin on cells, which could have been skewed by indirect inhibitory effects on mitogenesis caused by the ganglioside mixtures [47–49].
Table 2.
Potential receptors for PMT screened by TLC-overlay method.
| Binding at 37°C | Binding at 4°C | Note | |
|---|---|---|---|
| Glycosphingolipids | |||
| GM1 | − | +/− | +/−, PMT-C only |
| GM2 | − | +/− | |
| GM3 | − | + | |
| GA1 | +/− | + | Asialo-form of GM1 |
| GA2 | − | + | Asialo-form of GM2 |
| GA3 (LacCer) | ++ | ++ | Asialo-form of GM3 |
| GD1a | − | − | +/−, PMT-C only |
| GD1b | − | − | +/−, PMT-C only |
| GT1b | − | − | +/−, PMT-C only |
| Trihexoside (Gb3) | − | + | |
| Globoside (Gb4) | − | − | |
| Ceramide I (Cer I) | + | ||
| Ceramide II (Cer II) | + | ||
| GalCer | + | ||
| GlcCer | + | ||
| Phospholipids | |||
| PC | ++ | +/−, PMT-C only | |
| PE | + | +, rPMT, PMT-N only | |
| PI | +/− | +/−, rPMT, PMT-C only | |
| PS | − | ||
| SM | +++ | +/−, PMT-C only | |
| Others | |||
| Cholesterol | + | ||
| Stearic acid | + | ||
| Monoglyceride | + | ||
| Diglyceride | + | ||
A qualitative score was given for lipids that always bound well (+, ++, or +++), that bound weakly or not always bound (+/−), or that never bound (−).
Although we did not observe binding of full-length PMT or PMT-N to gangliosides, we did observe binding of PMT-C to gangliosides, specifically GM1 and other more polar di- and trisialogangliosides (Figure 4). This suggests that removal of the N-terminus of PMT unmasks a ganglioside-binding site of the protein, which could potentially occur during the uptake, endosomal trafficking and/or the translocation process where the toxin presumably unfolds. It has been shown that the C1 domain itself can associate with the cell membrane [15]. Considering that the C1 domain alone (residues 575–719) has a calculated pI of 8.9, it is possible for PMT-C to have affinity for negatively charged gangliosides.
The finding that PMT and PMT-N bound to positively charged phospholipid or sphingophospholipid headgroups on TLC plates was not surprising considering that both have a pI of 5.1. Indeed, PE, PC and SM corresponded to the three major binding components A, B and C, respectively, present in cell membrane extracts (Figure 1 and Supplementary Figure S1). However, our finding that PMT-N bound specifically to the choline-containing PC and SM, but not as well to PE and not at all to PS, suggests that the recognition was not due solely to the positive charge of the head group. Additional binding studies using liposome pull-down assays with full-length PMT also supported these findings. Liposomes containing PS or PE alone did not bind PMT (Figure 5), but liposomes containing PC alone or especially SM alone were effective in pulling down PMT. Addition of certain lipid components to PC-containing liposomes enhanced binding, including SM, PS or a GM mixture (Figure 5), as well as LacCer and GM1 alone (data not shown), but PE did not. This suggests that the environment of the choline head group, such as that uniquely present in SM only liposomes, also contributes to recognition by PMT. It is possible that disruption of the uniform surface of PC-only liposomes could present a similarly favorable environment for binding.
Analysis of SPR sensorgrams with SM plus PC vesicles likewise showed a higher capacity for binding (Bmax for slow component) to PMT-N than PC-only vesicles. Natural membranes treated with SMase showed the most drastic changes in PMT-N binding, with 10-fold increase in the rapid binding Bmax and 2–3-fold increase in both the rapid and slow KD. The increase in KD for the tight, slow-binding component resulted primarily from a decrease in kon, while the increase in the KD for the rapid-binding component resulted primarily from an increase in koff. This change reflects that SM contributes to both rapid and tight binding of PMT-N to membranes. Although it is puzzling that loss of binding affinity is accompanied by a dramatic increase in binding capacity in the rapid phase, it is possible that the loss of the phosphocholine head group from SM might result in a reorganization of the membrane to expose more surface area and thereby expose more low-affinity sites for binding or it could distribute low-affinity binding sites to make them more available. Our TLC-overlay experiments also showed that PMT bound to both CerI and CerII. Removal of the phosphocholine head group is known to induce the resulting ceramide moieties to laterally segregate into microdomains [50]. This reorganized ceramide microdomains may be responsible for the observed enhanced binding. In contrast, there was a decrease in the slow component Bmax, indicating a loss in the number of tight-binding components available and further implicating the importance of SM for binding. On the other hand, treatment with either PLD did not show any drastic enhancement of rapid binding nor did it change the kinetic characteristics; however, a small decrease in Bmax for both the rapid and slow binding phases was observed.
SM has recently been implicated in the cellular binding and uptake of the vacuolating toxin VacA from Helicobacter pylori [51, 52]. In these studies, the presence of SM appears to be required for VacA entry, since SMase treatment abolished VacA activity while adding SM back to cells restored VacA activity. This is in contrast to our findings, which suggest that removing the phosphocholine head group of SM only altered the binding affinity (KD) of PMT by a factor of 3. Indeed, no single treatment of the membranes, by SMase, PLD or trypsin, resulted in complete abolishment of PMT binding, with each of the treatments resulting in a reduction of about a third of the slow-binding component. This points to the possibility of multiple co-receptors for PMT binding. However, it is not clear whether the observed reduction in binding, as evidenced by increase in KD, upon SMase treatment could lead to nonproductive, albeit tight binding that might impair the normal endocytic process.
We found that the kon observed for the slow component of PMT-N binding with the natural membranes was about 1,000-fold lower than that reported for CT binding to GM1 receptors (103 versus 106 M−1s−1), whereas koff was 100-fold higher, such that the overall KD is about 100,000-fold higher (10−7 M for PMT-N binding versus 10−12 M for CT-GM1 binding) [39, 41]. On the other hand, PMT-N binding to membranes is on the same order of that found for BoNT/A binding to GT1b [42]. BoNT/A is an example of a toxin known to bind to cells through multiple interactions with two different types of co-receptors, ganglioside and protein [31] (see Supplementary Table S3).
Considering that PMT-N shares significant sequence similarity with the N-terminal regions of the CNFs [3] and CNF1 and CNFY have been found to bind to protein (LRP) and heavily glycosylated protein (HSPG) receptors [23–25], it is not unreasonable to propose that PMT-N likewise might have multiple binding components that it interacts with during the internalization process. Such a cellular entry model for PMT might involve an initial nonspecific binding to lower affinity but more abundant components such as PC that would bring the protein to the cell surface. A more specific binding to SM would follow this, along with possible binding to a putative protein co-receptor that would then trigger endocytosis. In addition to PC, other membrane lipids might contribute to the initial binding, particularly since we observed that PMT bound to a number of other glycosphingolipids and phospholipids (see Table 2).
Although trypsin treatment did not completely abolish PMT binding interactions with membranes and did not affect the binding kinetics, this does not necessarily rule out the possibility of a protein co-receptor for PMT. It is possible that there may be a limitation of the number of critical receptors or events involved in toxin uptake, which might be mediated through a protein co-receptor. In summary, our data point to an important role of SM in PMT binding to membranes, as evidenced through four different methods, TLC-overlay, liposome pull-down, SPR of liposomes, and SPR of natural membranes. Based on our findings, we propose a model for PMT interaction with host cells that involves initial low-affinity nonspecific binding to cells, followed by a more specific, tight-binding interaction with SM and possibly other membrane components including a putative protein co-receptor.
Materials and Methods
Materials
African green monkey kidney (Vero) cells, HEK-293T cells, NG108-15 cells, and Swiss 3T3 cells were obtained from the ATCC. Dulbecco’s modified Eagle’s medium (DMEM, GIBCO), heat-inactivated bovine growth serum (BGS, HyClone), and tissue-culture-grade penicillin G and streptomycin were obtained from Fisher. Opti-prep® density gradient solution (60%) was obtained from VWR. Mouse anti-EEA1 (G-4, sc-137130) and goat anti-mouse IgG-HRP (sc-2005) antibodies from Santa Cruz. [125I] NaI was purchased from MP Biomedicals. Protease inhibitor cocktail (1X PI, P8849), L-α-phosphatidylcholine (PC), L-α-phosphatidylethanolamine (PE), L-α-phosphatidyl-L-serine (PS), sphingomyelin (SM), stearic acid (SA), 1-stearoyl glycerol (MG), 1,2-distearoyl glycerol (DG), Bacillus cereus sphingomyelinase (SMase), cabbage phospholipase D (PLDCab), Streptomyces chromofuscus phospholipase D (PLDSc), trypsin-EDTA (1X solution), and His-select® nickel affinity gel were obtained from Sigma. Cerebroside (GalCer), glucocerebroside (GlcCer), lactosyl ceramide (LacCer), monoasialogangliosides GM1, GM2 and GM3, a mixture of monoasialogangliosides (GM1, GM2 and GM3), Gb3, Gb4, GA1, GA2, GD1a, GD1b, GT1b, and porcine or bovine brain gangliosides (PGM or BGM, respectively) were obtained from Matreya Inc. Ceramide I (CerI) and Ceramide II (CerII) were obtained from Alexis Biochemicals. Trypsin (mass spec grade) was obtained from Promega. The aluminum-backed silica TLC plates were purchased from Whatman. Iodogen reagent and BCA protein assay kit were obtained from Pierce. HiTrap-ANX, PD-10, and L1 chip were purchased from GE Healthcare Sciences. All other reagents were obtained from Fisher or RPI, unless otherwise noted.
Preparation of recombinant PMT (rPMT)
rPMT (residues 1–1285) and PMT-C (residues 577–1285), each with an N-terminal His6-tag, respectively cloned into pTrcHis and pET33b vector, and PMT-N (residues 1–568) with a C-terminal His6-tag cloned into pET21 vector, were expressed and purified using previously published methods [8]. Briefly, proteins were expressed in E. coli under IPTG induction. Cells were harvested and lysed using sonication, and proteins were purified using a nickel affinity (His-Select) and anion exchange chromatography (ANX), and then desalted using a PD-10 column. Concentrations were measured via BCA assay and verified via densitometry on a 10% SDS-PAGE gel with BSA as reference.
Labeling of rPMT, PMT-N and PMT-C with [125I] sodium iodide
A PD-10 column was equilibrated by washing with 20 mL of PBS, blocked with BSA (a solution of 10 mg in 1 mL PBS), then washed again with 20 mL PBS. [125I]Sodium iodide (100 μCi) was added to a 2-mL screwed-capped centrifuge tube precoated with 50 μg of Iodogen, and the protein to be labeled (50 μg) was added, followed by PBS for a total volume of 100 μL. The tube was incubated at room temperature for 3 minutes, with occasional swirling. The contents of the tube were applied to the PD-10 column, and 10 μL of NaI (1 g/mL) was added quickly to the column to dilute any remaining free label. The labeled protein was eluted from the column in 12 × 0.5 mL fractions of PBS. Fractions containing the labeled protein (#6–9) were combined and mixed with BSA (10 mg in 1 mL of PBS) in a screw-capped 15-mL polypropylene tube. The labeled protein was stored at 4°C until use. Functional viability of 125I-rPMT to bind to and enter cells and traffic to endosomes was confirmed by subcellular fractionation (Supplementary Figure S7). Functional viability of 125I-PMT-N to bind membrane lipids on TLC plates was confirmed by competition with unlabeled PMT-N (Supplementary Figure S8).
Cell culture
Vero, HEK-293T, NG108-15 or Swiss 3T3 cells were cultured and maintained at 37°C and 5% CO2 in DMEM, pH 7.4, supplemented with 10% heat-inactivated BGS, 100 U/mL penicillin G, and 100 μg/mL streptomycin.
Subcellular fractionation
Swiss 3T3 cells, maintained as above, were plated onto 6-well plates at a density of 0.3 × 106 cells/well and incubated for two days prior to toxin treatment. For toxin treatment, medium was replaced with fresh, cold DMEM medium containing 125I-rPMT (50 μg/mL) and incubated on ice for 30 min, then at 37°C for 4 hrs. Cells were washed thrice with cold PBS, harvested by centrifugation at 4°C and 3,000 × g, followed by suspension in 0.5 mL cold sucrose buffer (250 mM sucrose, containing 0.5 mM EDTA and 1X PI) and homogenized by passing 30 times through a 26-gauge needle. The homogenate was centrifuged for 10 min at 800 × g and 4°C to separate nuclear pellet (P) and supernatant (S) fractions. The S fraction was then further fractionated by gradient centrifugation, starting with a mixture of the S fraction in 50% Opti-prep solution at the bottom and step-wise addition of 40%, 30%, 20% and 10% Opti-prep solutions (total volume of 12 mL), followed by centrifugation at 88,000 × g for 18 hrs at 4°C using a SW-40Ti rotor (Beckman). Fractions (1 mL) were collected and separated by SDS-PAGE and subjected to either Western blot analysis using anti-EEA1 as primary antibody or phosphorimager (Fuji FLA-3000) analysis using Fujifilm Image Gauge software to detect radiolabeled PMT.
Ames test for phosphate contents in lipid extracts
The phosphate content in phospholipids and lipid extracts was determined by using the Ames phosphate assay [53]. Briefly, 0.01–0.1 mL of sample was mixed with 0.03 mL of 10% Mg(NO3)2 solution in absolute ethanol in a 10-mL glass tube. The mixture was dried and ashed behind a safety shield in a fume hood by shaking the tube over a flame until brown fumes disappeared. After cooling the tubes to room temperature, 0.3 ml of 0.5 N HCl was added to each tube and incubated for 15 min at 95°C. The tubes were allowed to cool down to room temperature and 0.7 ml of reaction mixture (6:4:1 ratio of 0.42% ammonium molybdate:water:20% ascorbic acid) was added to each tube. The tubes were then incubated at 45°C for 20 min, and the absorbance was read at 820 nm. The concentrations of phosphate in the samples were determined using a standard curve generated from serial dilutions of a known phosphate standard.
Preparation of total cellular lipids for TLC analysis
Confluent monolayers of Vero, HEK-293T or NG108-15 cells, maintained as above, were collected, washed and resuspended in PBS (3 mL per 150 mm culture dish used). To 1 mL of resuspended cells, 1 ml of a 1:1 chloroform:methanol mixture was added, and the lipids were extracted from the cells by a combination of sonication, vortexing, and pipetting, followed by incubation at room temperature overnight. The mixture was centrifuged at 16,000 × g for 5 minutes, and the supernatant was transferred to a new tube, dried by SpeedVac, and stored at −20°C until use.
Thin layer chromatography for phospholipids and overlay with radio-labeled rPMT, PMT-N or PMT-C
Aluminum-backed silica-gel TLC plates (6.5 cm × 10 cm) were loaded with lipid samples (50 nmoles phosphate equivalent) and dried for 1 hr under vacuum in a desiccator. The plates were then developed in a TLC chamber with a solvent system of 80:20:2, 70:30:3,70:25:4, or 25:20:3 (v/v/v, chloroform/methanol/0.05% CaCl2 in distilled water). Plates were then dried and either stained with 10% cupric sulfate solution (in 8% phosphoric acid) or used for overlay experiments. For overlay experiments, the dried plates were blocked in 1% BSA in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 136 mM NaCl, 2.6 mM KCl). The BSA-blocked TLC plates were transferred into an overlay solution (125I-labeled rPMT, PMT-N, or PMT-C in 15 ml PBS) and incubated for 2 hr at 37°C with a gentle rocking. Plates were then washed four times with ice-cold PBS and dried for 1 hr by heating gently under a 40-Watt light bulb. The plates were then stored in phosphorimager cassettes overnight, and the image was visualized using phosphorimager analysis. After the radio image was acquired, all the lipid spots on the TLC plate were visualized by CuSO4 staining. The corresponding CuSO4-stained images were recorded with a desktop scanner and compared to the phosphorimager-generated images.
Purification of lipid components in cell membrane
Large-scale lipid purification was performed using silica-gel chromatography. Fresh bovine trachea (~300 g) were diced and placed in a blender with 700 mL distilled water. After blending the tissues, the mixture was centrifuged for 5 min at 2,500 × g. The supernatant was discarded. Four samples of 20 g each were resuspended in 100 ml of a 1:1 mixture of chloroform:methanol (v/v). After overnight extraction on a rocker at room temperature, the insoluble fractions were removed by centrifugation for 5 min at 2,500 × g. The extracts were then combined and evaporated under vacuum. The dried lipids were resuspended in 50 mL of a mixture of 1:1 chloroform:methanol (v/v) and loaded onto a column containing 40 ml of silica gel. Phospholipid components A, B, C and D were eluted from the column with a 10% stepwise chloroform/methanol gradient from 100:0 to 0:100 (v/v), and 10 ml fractions were pooled, according to TLC analysis, and allowed to evaporate in a fume hood. Some of the pooled fractions were further purified with appropriate chloroform/methanol mixtures to resolve into additional components.
Preparation of liposomes for solution binding
Liposomes were prepared by first mixing 0.1 mg of cholesterol and 0.9 mg of phospholipid, either PC, SM, PE, PS, PC/SM (3:1), PC/PE (3:1) or PC/SM (3:1) in the form of freshly prepared solutions in 50:50 chloroform/methanol (v/v) into a 2-mL microfuge tube. Some mixtures contained additional 0.1 mg of a mixture of GM1, GM2 and GM3. The mixture was dried under vacuum for 1 hr using a SpeedVac. The film of dried lipids was then rehydrated with 1 mL of ice-cold PBS for 1 hr and resuspended by vortexing (2 × 30 sec) and short pulses of ultrasonication in a bath (Branson). The opaque, large multi-lamellar vesicles (MLV) were stored at −80°C. Immediately before use, frozen liposomes were sonicated using a bath sonicator (Branson 1200) for 10 min to produce small uni-lamellar vesicles (SUV). The SUV suspension (100 μL) so prepared was then incubated with 10 μg of purified rPMT for 1 hr at 4°C and then centrifuged for 10 min at 30,000 × g. For quantitation, the pellets were resolubilized with 100 μL of SDS-PAGE sample buffer, and 20 μl of each was removed and analyzed by 10% SDS-PAGE, using 1% of the total supernatant from a mock treatment as a reference on the same gel. Images of the Coomassie Blue-stained gels were analyzed by using both Adobe ImageMeter and ImageJ software, where the correlation between the two methods was 0.86. The mean value of the two methods was used for analysis shown in Figure 5b.
Preparation of liposomes for surface plasmon resonance
Lipids, PC only, PC/PS (1:1) or PC/SM (1:1), in powder form were weighed and re-suspended in 10 mM Tris-HCl buffer, pH 7.4, to a final concentration of 20 mM of lipid. The flask was purged with nitrogen and sealed before sonication for 1 hr using a bath sonicator. The resulting vesicles were extruded 19 times through a 100-micron pore using the extruder kit from Avanti Polar Lipids. SUVs were aliquoted, flash frozen and stored at −20°C before use.
Isolation of membranes from HEK-293T cells
Membranes were purified using a modified protocol utilizing density centrifugation to isolate membrane fractions from whole cell lysates [54]. Briefly, HEK-293T cells grown to confluence in DMEM with 10% BGS under 5% CO2 at 37°C were harvested and centrifuged at 2,500 × g. A volume of 2 mL of cell pellets was resuspended in 20 mL of Tris-Mg2+ buffer (TM, 100 mM Tris-HCl, 10 mM MgSO4, pH 7.4) and homogenized using 10 strokes of a Dounce homogenizer, and converted into an 8.5% sucrose solution by addition of 4.5 mL of 60% sucrose. The membranes and whole cells were pelleted for 15 min at 2,000 × g and washed 3x with 40 mL of TM with 8.5% sucrose. Sucrose was added to the pellet to make a 45% sucrose solution. An equal volume of 40%, 35% and 8.5% sucrose solutions were overlayed on top in sequence. The solution was centrifuged for 10 min at 70,000 × g at 4°C using an SW-40Ti rotor. The band at the 35–40% interface was collected and diluted with two volumes of TM buffer, pH 8.6, and centrifuged at 2,500 × g for 20 min. The ghosts were resuspended in 6 mL of TM-sucrose solution. The membranes were sonicated briefly (10 sec at power level 5.5 using Sonifier Cell Disrupter, Model 185D, Heat Systems, Ultrasonics Inc.) to form vesicles. Residual amount of heavy organelles was pelleted by centrifugation at 2,000 × g for 15 minutes. The supernatant was collected, and the protein content was measured via BCA assay. Vesicles were flash frozen and stored at −20°C until use.
Surface plasmon resonance analysis using Biacore 3000
All four flow cells in the L1 chip were primed with 20 mM CHAPS, pH 7.4, between trials. All trials were run at 25°C. Membrane extracts were loaded by lowering the pH to ~4.5 using sodium acetate buffer and injecting at 2 μL/min in Biacore buffer (autoclaved, sterile filtered and degassed 50 mM Tris-HCl, 150 mM NaCl, pH 7.4). Reconstituted membrane vesicles were loaded at a concentration of 500 μM in Biacore buffer at 5 μL/min. The loading was continued until less than 1 RU/sec baseline increase was observed (~30–45 min). The surface of the membrane was then conditioned using 1 M NaCl (at a flow rate of 20 μL/min for 2 min). Non-specific binding sites were blocked with BSA (0.5 mg/mL, at a flow rate of 5 μL/min for 5 min). After re-equilibration of the L1 chip with buffer, increasing concentrations of PMT-N were added to the L1 chip. Concentrations of PMT-N injected onto the L1 chip surface were 7.5 nM, 75 nM, 750 nM, 3.75 μM, or 7.5 μM, and in some cases, where necessary, 15 μM at a flow rate of 5 μL/min for 4 min for all reconstituted vesicles and for 5 min for all cell membrane isolates (see Figure 6A). Dissociation for at least 2 min by washing with Biacore buffer without PMT occurred before injection of the next highest concentration of PMT. Chips were washed between experiments thrice with 20 mM CHAPS, pH 7.4, followed by 1X Trypsin-EDTA solution.
Treatment of membranes on L1 chip with enzymes
Membranes on the L1 chip were treated with 0.1 U/μL of SMase, PLDCab, or PLDSc or with 0.20 μg/μL of MS-grade trypsin in Biacore buffer by applying to the surface at a flow rate of 1 μL/min for 30 min. Enzymes were removed by washing with 1 M NaCl, followed by Biacore buffer. These enzyme-treated membranes were used for SPR experiments. In some instances, after sequential PMT-binding/dissociation events using SMase or PLDCab-treated membranes, the membrane were then treated with trypsin (0.2 μg/μL) at a flow rate of 1 μL/min in Biacore buffer for 30 min to remove residual PMT-N and cleavable protein components of the membrane, followed by washing with Biacore buffer. The resulting lipase/trypsin-treated membranes were then used for further SPR experiments for PMT-N binding.
Analysis of SPR sensorgrams
The sensorgram of each association (binding) phase was analyzed according to a two-component exponential curve model, where the resonance signal R(t) at time point t is
| (Eq. 1) |
One of these two components represents a rapid on-off binding process and the other a slow on-off binding. Each of these two components can be further resolved, according to the kinetic analysis of O’Shannessy et al. [43], where the resonance signal R(t) at time t is
| (Eq. 2) |
and where C is the concentration of ligand, Rmax is the maximum resonance signal corresponding to the maximal binding Bmax, and kon and koff are the association and dissociation rates, respectively. We added a ligand concentration-dependent correction term, ΔRc, for Rmax. For the slow on-off phase, bound ligand was not completely dissociated before a higher concentration of ligand was applied in sequence. To correct for this, a baseline adjustment was made. This correction was equal to the residual resonance R′min after treatment of ligand at the previous concentration C′, which was subtracted from the Rmax and added back into the total signal. For the rapid on-off component, an equilibrium was quickly reached, so R′min = 0. The detailed curve fitting equation was described as
| (Eq. 3) |
For the dissociation phase, the sensorgram was treated as a sum of two dissociation components and described as
| (Eq. 4) |
The segments from a sensorgram of 4 or 5 continuous association events were simultaneously fitted to equations 1, 3 and 4 with constant Rmax(A), kon(A), koff(A) and Rmax(B); kon(B), koff(B) and 5 concentration-dependent ΔRC values for Rmax(A) and for Rmax(B). The curve fitting was solved using a nonlinear least squares method calculated in Solver in Excel through minimization of the sum of the squares. For the rapid on-off component, the ligand concentration-dependent apparent maximum for each concentration at equilibrium, Rmax(C) = Rmax + ΔRC, was fitted to a thermodynamic equilibrium model, according to the Langmuir isotherm,
| (Eq. 5) |
where KD was the dissociation constant at thermodynamic equilibrium.
Supplementary Material
Supplementary Figure S1. Binding of PMT to lipid extracts from Vero cells.
Supplementary Figure S2. Binding of PMT to gangliosides.
Supplementary Figure S3. Binding of PMT and PMT fragments to nonpolar lipid components.
Supplementary Figure S4. Binding of PMT and PMT fragments to less polar ceramides.
Supplementary Figure S5. Calculated curves for SPR traces.
Supplementary Figure S6. SPR sensorgrams of PMT-N binding to cell membranes.
Supplementary Figure S7. Uptake of radiolabeled PMT into Swiss 3T3 cells.
Supplementary Figure S8. Competition between 125I-labeled and unlabeled PMT-N for binding to PC and SM in TLC-overlay assay.
Supplementary Table S1. P values calculated with the t-distribution for comparing PMT-binding characteristics on reconstituted lipids.
Supplementary Table S2. P values calculated with the t-distribution for comparing PMT-binding characteristics on native lipids and enzyme-treated lipids.
Supplementary Table S3. Cellular receptors for selected bacterial toxins.
Acknowledgments
This work was supported by NIH/NIAID grant AI038396 (to B.A.W), NIH/GM GM075937 (to C.M.R.), and the NIH/CBI Training Grant and DHS graduate fellowship (to M.C.B.). We thank Liping Wang for technical assistance with the Biacore facilities.
Abbreviations
- rPMT
recombinant Pasteurella multocida toxin
- PMT-N
N-terminal fragment of PMT (residues 1–568)
- PMT-C
C-terminal fragment of PMT (residues 577–1285)
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- PI
phosphatidylinositol
- PG
phosphatidylglycerol
- SM
sphingomyelin
- SMase
Bacillus cereus sphingomyelinase
- PLD
phospholipase D
- PLDSc
PLD from Streptomyces chromofuscus
- PLDCab
PLD from cabbage
- GM
monosialoganglioside
- PGM
porcine ganglioside mixture
- LacCer
lactosyl ceramide
- GalCer
galactosyl ceramide
- GlcCer
glucosyl ceramide
- CerI
ceramide I
- CerII
ceramide II
- CHL
cholesterol
- MG
monoglyceride
- DG
diglyceride
- SPR
surface plasmon resonance
- RU
SPR resonance units
- TLC
thin-layer chromatography
- T
trypsin
- DMEM
Dulbecco’s modified eagle medium
- BGS
bovine growth serum
- MLV
multilamellar vesicles
- SUV
small unilamellar vesicles
References
- 1.Foged NT. Pasteurella multocida toxin. The characterisation of the toxin and its significance in the diagnosis and prevention of progressive atrophic rhinitis in pigs. APMIS Suppl. 1992;25:1–56. [PubMed] [Google Scholar]
- 2.Pedersen KB, Barfod K. The aetiological significance of Bordetella bronchiseptica and Pasteurella multocida in atrophic rhinitis of swine. Nord Vet Med. 1981;33:513–522. [PubMed] [Google Scholar]
- 3.Wilson BA, Ho MH. Pasteurella multocida toxin. In: Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. Elsevier; Amsterdam; Boston: 2006. pp. 430–447. [Google Scholar]
- 4.Wilson BA, Ho M. Pasteurella multocida toxin as a tool for studying Gq signal transduction. Rev Physiol Biochem Pharmacol. 2004;152:93–109. doi: 10.1007/s10254-004-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson BA, Ho MF. Recent insights into Pasteurella multocida toxin and other G-protein-modulating bacterial toxins. Future Microbiology. 2010;5:1185–1201. doi: 10.2217/fmb.10.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullan PB, Lax AJ. Pasteurella multocida toxin is a mitogen for bone cells in primary culture. Infection and Immunity. 1996;64:959–965. doi: 10.1128/iai.64.3.959-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozengurt E, Higgins T, Chanter N, Lax AJ, Staddon JM. Pasteurella multocida toxin - Potent mitogen for cultured fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:123–127. doi: 10.1073/pnas.87.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson BA, Aminova LR, Ponferrada VG, Ho MF. Differential modulation and subsequent blockade of mitogenic signaling and cell cycle progression by Pasteurella multocida toxin. Infection and Immunity. 2000;68:4531–4538. doi: 10.1128/iai.68.8.4531-4538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orth JHC, Fester I, Preuss I, Agnoletto L, Wilson BA, Aktories K. Activation of G alpha(i) and subsequent uncoupling of receptor-G alpha(i) signaling by Pasteurella multocida toxin. Journal of Biological Chemistry. 2008;283:23288–23294. doi: 10.1074/jbc.M803435200. [DOI] [PubMed] [Google Scholar]
- 10.Orth JHC, Lang S, Taniguchi M, Aktories K. Pasteurella multocida toxin-induced activation of RhoA is mediated via two families of G alpha proteins, G alpha(q) and G alpha(12/13) Journal of Biological Chemistry. 2005;280:36701–36707. doi: 10.1074/jbc.M507203200. [DOI] [PubMed] [Google Scholar]
- 11.Orth JHC, Preuss I, Fester I, Schlosser A, Wilson BA, Aktories K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7179–7184. doi: 10.1073/pnas.0900160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson BA, Zhu X, Ho M, Lu L. Pasteurella multocida toxin activates the inositol triphosphate signaling pathway in Xenopus oocytes via G(q)alpha-coupled phospholipase C-beta1. J Biol Chem. 1997;272:1268–1275. doi: 10.1074/jbc.272.2.1268. [DOI] [PubMed] [Google Scholar]
- 13.Zywietz A, Gohla A, Schmelz M, Schultz G, Offermanns S. Pleiotropic effects of Pasteurella multocida toxin are mediated by G(q)-dependent and -independent mechanisms - Involvement of G(q) but not G(11) Journal of Biological Chemistry. 2001;276:3840–3845. doi: 10.1074/jbc.M007819200. [DOI] [PubMed] [Google Scholar]
- 14.Kitadokoro K, Kamitani S, Miyazawa M, Hanajima-Ozawa M, Fukui A, Miyake M, Horiguchi Y. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5139–5144. doi: 10.1073/pnas.0608197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamitani S, Kitadokoro K, Miyazawa M, Toshima H, Fukui A, Abe H, Miyake M, Horiguchi Y. Characterization of the membrane-targeting C1 domain in Pasteurella multocida toxin. Journal of Biological Chemistry. 2010;285:25467–25475. doi: 10.1074/jbc.M110.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aminova LR, Luo SH, Bannai Y, Ho MF, Wilson BA. The C3 domain of Pasteurella multocida toxin is the minimal domain responsible for activation of G(q)-dependent calcium and mitogenic signaling. Protein Science. 2008;17:945–949. doi: 10.1110/ps.083445408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin MR, Lakey JH, Lax AJ. Identification and characterization of the Pasteurella multocida toxin translocation domain. Molecular Microbiology. 2004;54:239–250. doi: 10.1111/j.1365-2958.2004.04264.x. [DOI] [PubMed] [Google Scholar]
- 18.Pullinger GD, Sowdhamini R, Lax AJ. Localization of functional domains of the mitogenic toxin of Pasteurella multocida. Infection and Immunity. 2001;69:7839–7850. doi: 10.1128/IAI.69.12.7839-7850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide-sequence of the gene encoding cytotoxic necrotizing factor-I of Escherichia coli. Infection and Immunity. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oswald E, Sugai M, Labigne A, Wu HC, Fiorentini C, Boquet P, Obrien AD. Cytotoxic necrotizing factor type-2 produced by virulent Escherichia coli modifies the small GTP-binding proteins-Rho involved in assembly of actin stress fibers. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoll T, Markwirth G, Reipschlager S, Schmidt G. A new member of a growing toxin family - Escherichia coli cytotoxic necrotizing factor 3 (CNF3) Toxicon. 2009;54:745–753. doi: 10.1016/j.toxicon.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 22.Lockman HA, Gillespie RA, Baker BD, Shakhnovich E. Yersinia pseudotuberculosis produces a cytotoxic necrotizing factor. Infection and Immunity. 2002;70:2708–2714. doi: 10.1128/IAI.70.5.2708-2714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blumenthal B, Hoffmann C, Aktories M, Backert S, Schmidt G. The cytotoxic necrotizing factors from Yersinia pseudotuberculosis and from Escherichia coli bind to different cellular receptors but take the same route to the cytosol. Infection and Immunity. 2007;75:3344–3353. doi: 10.1128/IAI.01937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung JW, Hong SJ, Kim KJ, Goti D, Stins MF, Shin S, Dawson VL, Dawson TM, Kim KS. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. Journal of Biological Chemistry. 2003;278:16857–16862. doi: 10.1074/jbc.M301028200. [DOI] [PubMed] [Google Scholar]
- 25.Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. Journal of Biological Chemistry. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- 26.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Molecular Microbiology. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 27.Dudet LI, Chailler P, Dubreuil JD, MartineauDoize B. Pasteurella multocida toxin stimulates mitogenesis and cytoskeleton reorganization in Swiss 3T3 fibroblasts. Journal of Cellular Physiology. 1996;168:173–182. doi: 10.1002/(SICI)1097-4652(199607)168:1<173::AID-JCP21>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Pettit RK, Ackermann MR, Rimler RB. Receptor-mediated binding of Pasteurella multocida dermonecrotic toxin to canine osteosarcoma and monkey kidney (Vero) cells. Laboratory Investigation. 1993;69:94–100. [PubMed] [Google Scholar]
- 29.Merritt EA, Sarfaty S, Vandenakker F, Lhoir C, Martial JA, Hol WGJ. Crystal structure of cholera toxin B-pentamer bound to receptor G(M1) pentasaccharide. Protein Science. 1994;3:166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naglich JG, Metherall JE, Russell DW, Eidels L. Expression cloning of a diphtheria-toxin receptor - Identity with a heparin-binding EGF-like growth-factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 31.Binz T, Rummel A. Cell entry strategy of clostridial neurotoxins. Journal of Neurochemistry. 2009;109:1584–1595. doi: 10.1111/j.1471-4159.2009.06093.x. [DOI] [PubMed] [Google Scholar]
- 32.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 33.Cumar FA, Maggio B, Caputto R. Ganglioside-cholera toxin interactions -a binding and lipid monolayer study. Molecular and Cellular Biochemistry. 1982;46:155–160. doi: 10.1007/BF00239664. [DOI] [PubMed] [Google Scholar]
- 34.Berenson CS, Nawar HF, Yohe HC, Castle SA, Ashline DJ, Reinhold VN, Hajishengallis G, Connell TD. Mammalian cell ganglioside-binding specificities of E. coli enterotoxins LT-IIb and variant LT-IIb(T13I) Glycobiology. 2010;20:41–54. doi: 10.1093/glycob/cwp141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffitts JS, Haslam SM, Yang T, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 2005;307:922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 36.Walton KM, Sandberg K, Rogers TB, Schnaar RL. Complex ganglioside expression and tetanus toxin binding by PC12 pheochromocytoma cells. J Biol Chem. 1988;263:2055–2063. [PubMed] [Google Scholar]
- 37.Takamizawa K, Iwamori M, Kozaki S, Sakaguchi G, Tanaka R, Takayama H, Nagai Y. TLC immunostaining characterization of Clostridium botulinum type A neurotoxin binding to gangliosides and free fatty acids. FEBS Lett. 1986;201:229–232. doi: 10.1016/0014-5793(86)80614-7. [DOI] [PubMed] [Google Scholar]
- 38.Flagler MJ, Mahajan SS, Kulkarni AA, Iyer SS, Weiss AA. Comparison of binding platforms yields insights into receptor binding differences between Shiga toxins 1 and 2. Biochemistry. 2010;49:1649–1657. doi: 10.1021/bi902084y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuziemko GM, Stroh M, Stevens RC. Cholera toxin binding affinity and specificity for gangliosides determined by surface plasmon resonance. Biochemistry. 1996;35:6375–6384. doi: 10.1021/bi952314i. [DOI] [PubMed] [Google Scholar]
- 40.Wigelsworth DJ, Krantz BA, Christensen KA, Lacy DB, Juris SJ, Collier RJ. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. Journal of Biological Chemistry. 2004;279:23349–23356. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- 41.MacKenzie CR, Hirama T, Lee KK, Altman E, Young NM. Quantitative analysis of bacterial toxin affinity and specificity for glycolipid receptors by surface plasmon resonance. J Biol Chem. 1997;272:5533–5538. doi: 10.1074/jbc.272.9.5533. [DOI] [PubMed] [Google Scholar]
- 42.Yowler BC, Schengrund CL. Botulinum neurotoxin A changes conformation upon binding to ganglioside GT1b. Biochemistry. 2004;43:9725–9731. doi: 10.1021/bi0494673. [DOI] [PubMed] [Google Scholar]
- 43.O’Shannessy DJ, Brighamburke M, Soneson KK, Hensley P, Brooks I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface-plasmon resonance - Use of nonlinear least-squares analysis-methods. Analytical Biochemistry. 1993;212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- 44.Oda M, Takahashi M, Matsuno T, Uoo K, Nagahama M, Sakurai J. Hemolysis induced by Bacillus cereus sphingomyelinase. Biochimica Et Biophysica Acta-Biomembranes. 2010;1798:1073–1080. doi: 10.1016/j.bbamem.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Yang HY, Roberts MF. Phosphohydrolase and transphosphatidylation reactions of two Streptomyces phospholipase D enzymes: Covalent versus noncovalent catalysis. Protein Science. 2003;12:2087–2098. doi: 10.1110/ps.03192503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dippe M, Ulbrich-Hofmann R. Substrate specificity in phospholipid transformations by plant phospholipase D isoenzymes. Phytochemistry. 2009;70:361–365. doi: 10.1016/j.phytochem.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Hashiramoto A, Mizukami H, Yamashita T. Ganglioside GM3 promotes cell migration by regulating MAPK and c-Fos/AP-1. Oncogene. 2006;25:3948–3955. doi: 10.1038/sj.onc.1209416. [DOI] [PubMed] [Google Scholar]
- 48.Rusnati M, Tanghetti E, Urbinati C, Tulipano G, Marchesini S, Ziche M, Presta M. Interaction of fibroblast growth factor-2 (FGF-2) with free gangliosides: biochemical characterization and biological consequences in endothelial cell cultures. Mol Biol Cell. 1999;10:313–327. doi: 10.1091/mbc.10.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates AJ, VanBrocklyn J, Saqr HE, Guan Z, Stokes BT, O’Dorisio MS. Mechanisms through which gangliosides inhibit PDGF-stimulated mitogenesis in intact Swiss 3T3 cells: receptor tyrosine phosphorylation, intracellular calcium, and receptor binding. Exp Cell Res. 1993;204:38–45. doi: 10.1006/excr.1993.1006. [DOI] [PubMed] [Google Scholar]
- 50.Cremesti AE, Goni FM, Kolesnick R. Role of sphingomyelinase and ceramide in modulating rafts: do biophysical properties determine biologic outcome? FEBS Lett. 2002;531:47–53. doi: 10.1016/s0014-5793(02)03489-0. [DOI] [PubMed] [Google Scholar]
- 51.Gupta VR, Patel HK, Kostolansky SS, Ballivian RA, Eichberg J, Blanke SR. Sphingomyelin functions as a novel receptor for Helicobacter pylori VacA. Plos Pathogens. 2008;4:e1000073. doi: 10.1371/journal.ppat.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta VR, Wilson BA, Blanke SR. Sphingomyelin is important for the cellular entry and intracellular localization of Helicobacter pylori VacA. Cell Microbiol. 2010;12:1517–1533. doi: 10.1111/j.1462-5822.2010.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatase. In: Neufeld E, Ginsburg V, editors. Methods in Enzymology. Academic Press; New York: 1966. pp. 115–118. [Google Scholar]
- 54.Boone CW, Ford LE, Bond HE, Stuart DC, Lorenz D. Isolation of plasma membrane fragments from Hela cells. Journal of Cell Biology. 1969;41:378. doi: 10.1083/jcb.41.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Binding of PMT to lipid extracts from Vero cells.
Supplementary Figure S2. Binding of PMT to gangliosides.
Supplementary Figure S3. Binding of PMT and PMT fragments to nonpolar lipid components.
Supplementary Figure S4. Binding of PMT and PMT fragments to less polar ceramides.
Supplementary Figure S5. Calculated curves for SPR traces.
Supplementary Figure S6. SPR sensorgrams of PMT-N binding to cell membranes.
Supplementary Figure S7. Uptake of radiolabeled PMT into Swiss 3T3 cells.
Supplementary Figure S8. Competition between 125I-labeled and unlabeled PMT-N for binding to PC and SM in TLC-overlay assay.
Supplementary Table S1. P values calculated with the t-distribution for comparing PMT-binding characteristics on reconstituted lipids.
Supplementary Table S2. P values calculated with the t-distribution for comparing PMT-binding characteristics on native lipids and enzyme-treated lipids.
Supplementary Table S3. Cellular receptors for selected bacterial toxins.