Abstract
Polyomavirus BK is a recognized cause of nephropathy and hemorrhagic cystitis in kidney or allogeneic hematopoietic stem cell transplant recipients. This study explored a role of genetic variations in capsid protein VP-1 gene as a factor in viral pathogenesis. VP-1 was amplified from 7 healthy subjects with viruria, 7 transplant patients with viruria, and 11 patients with viremia or nephropathy. PCR products were cloned and a total of 558 clonal sequences were subjected to phylogenetic analysis using standard methods. VP-1 quasispecies were found in 25/25 and coinfection with different genotypes in 12/ 25 subjects. Genotype II was found as an unexpected minority species in 5/25 individuals. Recombinant strains of uncertain biologic significance, which frequently contained genotype II and IV sequences were identified in 9/25 subjects. Viremia/nephropathy group was characterized by (a) greater sequence complexity in whole VP-1 versus BC loop and BC loop compared to the HI loop, (b) greater intra-strain genetic diversity in the BC loop compared to whole VP-1 protein and HI loop, (c) more non-synonymous substitutions (dN) in the BC loop compared to whole VP-1 and HI loop, (e) fewer synonymous substitutions (dS) compared to healthy-viruria group, and (f) selection pressure (dN/dS >1.0) exerted on VP-1. In conclusion, this study documents frequent occurrence of quasispecies in a host DNA polymerase dependent virus which is theoretically expected to show high replication fidelity. Quasispecies occur even in healthy subjects with viruria, but evolutionary selection pressure directed at the viral capsid protein VP-1 is seen only in patients with viremia.
Keywords: BK virus, viremia, polyomavirus, nephropathy, quasispecies
INTRODUCTION
Polyomavirus BK (BKV) is ubiquitous in man. Initial asymptomatic infection usually occurs in childhood. This is followed by latency within the urogenital tract [Chesters et al., 1983; Knowles, 2001]. Reactivation of infection occurs under conditions of immunosuppression, particularly renal transplantation. The initial manifestation is asymptomatic viruria. This is followed by viremia, and overt nephropathy, which may culminate in graft loss [Brennan et al., 2005; Brinkert et al., 2010; Hirsch and Randhawa, 2009; Low et al., 2004; O’Donnell et al., 2009; Randhawa and Demetris, 2000; Thomas et al., 2009]. BKV has also been associated with hemorrhagic cystitis in bone marrow and hematopoietic stem cell transplant patients and cancer patients treated with cyclophosphamide. Whereas viruria is generally considered to be insignificant, viremia and nephropathy trigger reduction in immunosuppression and anti-viral therapy.
The reason why only 1–10% of patients develop symptomatic disease is not clearly understood. The intensity of immunosuppression is one recognized variable. There is also evidence that viral genetic factors are important [Carr et al., 2006; Dugan et al., 2007; Gosert et al., 2008; Hirsch et al., 2003; Randhawa et al., 2002]. Genetic mutations and rearrangement are most extensively described in the non-coding control region of BKV [Sharma et al., 2007]. There are relatively few studies that have focused on the coding regions of the viral genome. The viral capsid protein 1 (VP-1) of BKV is responsible for virus assembly and maintaining the viral structural integrity. It is also essential as a receptor site for the infection of host cells. In mouse polyomavirus, small plaque and large plaque forming viral strains differ at a single amino acid located at position 92 of VP-1 [Freund et al., 1991]. There are also reports of increased mutations of the VP-1 gene in patients with BK nephropathy [Boldorini et al., 2009; Bolen et al., 1981; Jin and Gibson, 1996; Randhawa et al., 2002]. Thus, genetic alterations in VP-1 may be important with regards to (a) modulation of tissue injury, (b) evasion of host neutralizing antibodies, and (c) escape from VP-1 specific T-cell responses [Binggeli et al., 2007]. Accordingly, this study has characterized BKV sequence diversity and strain complexity in samples obtained from a broad spectrum of patients. The data highlights the frequent occurrence of mixed infections and presence of quasispecies in both immunosuppressed and healthy individuals. These observations have implications for design of diagnostic assays, effective vaccines and anti-microbial agents against BKV.
PATIENTS AND METHODS
Clinical material
Twenty eight urine samples were studied from the following three groups:
14 samples from 11 kidney transplant patients with BK viruria >6.46E+06 genomic equivalents per ml and viremia >3.21E+03 genomic equivalents per ml. Four of these patients had allograft nephropathy proven by a biopsy that had been performed within 0–45 days of urine collection.
7 samples from 7 kidney transplant patients with BK viruria <6.43E+05 genomic equivalents per ml/ml. These patients did not demonstrate viremia or active nephropathy within a 1 year interval of sample collection.
7 samples from healthy blood donors who had viruria but no evidence of viremia or active nephropathy.
Kidney transplant patients were recruited at the University of Pittsburgh Medical Center (UPMC) after informed consent (Institutional Review Board protocol #000586). Urine samples from healthy donors were provided by donors recruited at the University of Pittsburgh (n=2) or from the Blood Donation Center in Basel, Switzerland, as described previously [Egli et al., 2009]. All experiments are in accordance with ethical standards of the Declaration of Helsinki. Viral load measurements were performed using published quantitative PCR methods [Hirsch et al., 2002; Randhawa et al., 2005]. To confirm the diagnosis of nephropathy, in-situ hybridization for viral DNA was performed by the Clinical In-situ Hybridization Laboratory at UPMC.
DNA extraction and PCR amplification
Low molecular weight DNA was extracted from 560 μl urine using the QIAamp® Viral RNA Kit (Catalog 52906, QIAGen). Two 5 ml samples with low level viruria were processed using the QIAamp DNA Blood Maxi Kit (Catalog 51194, QIAGen). DNA was eluted in a final volume of 40 μl of buffer supplied by the manufacturer, and stored at −80 °C before downstream applications. The entire BKV VP-1 gene was amplified using published primers designated as SeqB7 (nucleotides 1484–1505, Dun numbering) and SeqB15 (nucleotides 2861-2839, Dun numbering) [Sharma et al., 2006b]. Amplification was carried out on an Applied Biosystem thermal cycler using rTth PCR Kit (Catalog 808-0187, Applied Biosystem), with rTth Taq polymerase 0.01 U/μl, dNTP (Catalog 200415, Stratagene) 0.1 mmol/L each, MgCl2 2.5 mmol/L, primers 50 nmol/L each, genomic DNA 3 μl, and a total reaction volume 20 μl. The thermal cycling program consisted of the following steps: 95 °C 4 min; 95 °C 15 sec, 58 °C 40 sec, and 72 °C 90 sec. After 40 cycles of amplification the PCR product was incubated at 72 °C for 15 min. Eight PCR reactions were pooled for each sample to generate enough DNA for cloning. In 5 samples where the initial amplification generated no visible product under agarose gel electrophoresis, the PCR reaction was diluted 10 times and amplified further for 30 cycles using a pair of nested PCR primers: Forward BKVP-1_FN 5′ and Reverse BKVP-1RN described in the section on sequencing. The reaction volume for the nested PCR was 100 μl. The sensitivity of this is 1000 copies/ml in plasma and 200 copies/ml in urine.
Cloning of PCR products and DNA sequencing
PCR products were purified using QIAquick Gel Extraction Kit (Catalog 28704, QIAGen). Briefly, the PCR product was first mixed with clean 6x loading buffer, and loaded to an agarose gel containing ethidium bromide. Gel electrophoresis was performed at 100 volts for 2 hours. A small slice of gel was cutoff vertically and examined under UV light. The DNA band position was marked by a thin tip. The gel slice was then aligned with the main gel. Following this, the target DNA band was cut off without ultraviolet exposure for gel extraction. Purified DNA was diluted into 20 μl ddH2O and cloned using pGEM®-T Easy Vector Systems (Catalog #A1360, Promega). The ligation system included 5 μl of 2x Rapid Ligation Buffer, 0.5 μl pGEM®-T Easy Vector, 3.5 μl purified BKV VP-1 gene PCR product and 1μl T4 DNA Ligase, with a total volume of 10 μl. Ligation was carried out by incubating at 4 °C overnight. Four microliters of ligation product was used to transform OneShot® TOP10 competent cells (Catalog C4040-10, Invitrogen). Plasmid DNA was extracted using QIAprep Spin Miniprep Kit (Catalog 27106, QIAGen) and subjected to EcoRI enzyme digestion. Typically, twenty clones with inserts of the correct size were picked for sequencing.
Approximately 100 ng of cloned DNA was mixed with 3.2 pmol of sequencing primers in 6 μl ddH2O and submitted for Sanger sequencing to the University of Pittsburgh Genomics and Proteomics Core Laboratories. Primers SeqB7, SeqB15, BKV_FN or BKV_RN were used as forward and reverse sequencing primers as appropriate. SeqB7 and SeqB15 primers have been published [Sharma et al., 2006c] and are known to show no mismatch with 160 BKV whole genome sequences available publicly. The Forward BKVP-1_FN primer has the sequence 5′ – CTTCTAGGCCTGTACGGGACTG – 3′. The Reverse BKVP-1_RN primer sequence is 5′-CTCAGATACTTCAGCCCCTGCTG - 3′(corresponding to positions 1515–1536 and 2839-2817 on the VP-1 gene using the Dun numbering system). The latter two primers show nucleotide identity with 153 known BKV whole genome sequences.
Evaluation of amplification and sequencing fidelity
Sequencing projects can be compromised by artifacts introduced in DNA amplification during the PCR amplification and sequencing steps. To evaluate this problem, one urinary DNA sample was amplified by nested PCR in five parallel reactions. PCR products for direct sequencing were purified using Millipore PCR cleanup columns (UFC7PCR50, Millipore). Using BK VP-1_FN and BK VP-1_RN primers, all five amplified products were sequenced in duplicate to obtain 10 high quality forward and reverse sequences, respectively 539 and 757 bp in length (after trimming of ends). An error rate of 1 in 12960 bp (0.00008 nucleotide differences per site) was obtained, which could only account for 0.087 substitutions per 1089 bp VP-1 sequence amplified in this study. In a second quality assurance experiment, one sample of cloned VP-1 DNA was sequenced 10 times using the SeqB7 primer. Ten identical high quality sequences of at least 716 bp were obtained.
Analysis of nucleotide substitutions
Chromatogram files from the sequencing facility were imported into Sequencher 4.5®. The 5′ and 3′ ends were trimmed up to 25% until the terminal 25 bases contained less than 3 bases with confidences values below 20. The Dunlop VP-1 gene sequence was imported as a reference. All remaining ambiguous bases were manually assigned by checking the chromatograms. Forward and reverse sequences were assembled as appropriate. Assembled clone sequences were checked manually, exported, and aligned using ClustalX. Alignments were further viewed and adjusted using BioEdit (Tom Hall, Department of Microbiology, North Carolina State University, North Carolina, USA: http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Phylogenetic Analyses
BKV sequences of known genotype or subgroup were downloaded from public sources [Luo et al., 2009]. Sequence alignments were done by ClustalX with default parameters [Thompson et al., 1994]. Neighbor Joining trees were constructed in Mega4.1 using the Kimura’s two-parameter method and viewed using Tree explorer [Kumar et al., 2008]. A bootstrap test with 1,000 replicates was used to estimate the confidence of branching patterns in the trees. Genotypes of experimental sequences were assigned by phylogenetic clustering with known reference sequences. BC, DE & HI loops were localized using published literature [Liddington et al., 1991; Murata et al., 2008]. Nucleotide diversity for sequence sets containing the predominant clones from each of the three subject categories was measured. This was done by computing the overall mean distance between all possible comparisons of two sequences in the sequence set of interest. These measurements reflect the mean number of nucleotide differences per site. Calculations are based on the nucleotide substitution model with a Jukes Cantor correction for multiple substitutions (Mega 4.1). The parameters dS and dN were respectively obtained by performing distance calculations on all potential synonymous site (dS) and nonsynonymous (dN) sites using the Nei-Gojobori method with Jukes and Cantor correction. A dN/dS ratio >1.0 was considered to be indicative of positive selection, while a ratio of <1.0 indicated purifying selection. Nucleotide and amino acid complexity were assessed for all clonal sequences in any given sample by first calculating Shannon entropy using the formula S = −Σi(pilnpi), where pi is the frequency of each sequence in the viral quasispecies. Then, the normalized entropy, Sn, was calculated using the formula Sn = S/lnN, where N is the total number of sequences analyzed [Minosse et al., 2006]. This quantity can vary from 0 (no diversity) to 1.0 (maximum diversity).
Analysis of Recombination Events
Recombinant clones and their putative parent strains were identified provisionally by inspection of sequence alignments. All potential recombinants were further validated by SimPlot software [Lole et al., 1999]. SimPlot bootscanning was done using default parameters with a window width of 140 bp and step size of 10 bp. VP-1 sequences of known strains different from the parental strains were used as an outgroup. The reference sequences used in this analysis were Dunlop (Accession No. V01108) for subgroup Ia, JL (Accession No. AB211370) for subgroup Ib, TW-3 (Accession No. AB211391) for genotype IV, and ETH-3 (Accession No. AB263916) for genotype II. The cross-over point in the bootscan plot was regarded as the approximate breakpoint region, which was further narrowed down to the extent possible by re-inspecting the sequence alignments.
Statistical analysis
Results of continuous variables were expressed as means with standard errors indicated. Comparisons between different subject groups were performed using unpaired Student’s t test in Microsoft Office Excel 2007.
RESULTS
1. Virologic parameters
The clinical and virologic parameters of the subjects studied are summarized in Table 1. Viremia was never seen in healthy subjects. In kidney transplant patients, viremia was associated with a circulating viral load that was at least two logs lower than the urinary viral load.
Table 1.
Virologic Parameters of Subjects Studied*
| Group | Sample | Day post-transplant | Viral load copies/ml | Total Clones | Unique Clones | Dominant Genotype | Coinfection | Recombinants | |
|---|---|---|---|---|---|---|---|---|---|
| Plasma | Urine | ||||||||
| Viremia/nephropathy | 1a | 868 | 8.13E+03 | 1.50E+09 | 20 | 17 | Ia | none | |
| 1b | 418 | 1.37E+05 | 3.90E+08 | 20 | 12 | Ia | none | ||
| 2a | 192 | 1.12E+04 | 3.83E+09 | 20 | 3 | Ib2 | none | ||
| 2b | 450 | 3.21E+03 | 6.28E+07 | 20 | 4 | Ia | none | ||
| 3a | 81 | 1.03E+05 | 6.79E+09 | 20 | 6 | II | none | ||
| 3b | 70 | 1.53E+05 | 3.78E+09 | 22 | 10 | II/20 | none | I-II/1, II-I/1 | |
| 4 | 316 | 3.27E+03 | 7.30E+07 | 20 | 12 | Ib2 | none | ||
| 5 | 167 | 5.26E+03 | 1.02E+08 | 20 | 7 | Ib1 | none | ||
| 6 | 257 | 4.66E+05 | 3.20E+09 | 20 | 17 | Ib2 | none | ||
| 7 | 175 | 8.02E+04 | 3.86E+08 | 20 | 9 | Ib1 | none | ||
| 8** | 161 | 9.52E+03 | 1.42E+08 | 21 | 13 | II/20 | 1b1/1 | ||
| 9** | 208 | 3.31E+03 | 1.05E+09 | 20 | 4 | Ib2 | none | ||
| 10** | 196 | 1.72E+04 | 6.46E+06 | 20 | 11 | Ib2 | none | ||
| 11** | 914 | 1.26E+05 | 1.06E+10 | 20 | 7 | Ib2 | none | ||
| Transplanted-viruria | 12 | 13 | 0 | 6.43E+05 | 22 | 19 | Ib1/20 | Ia/2 | |
| 13 | 40 | 0 | 2.50E+04 | 20 | 10 | II/13 | Ib2/2, Ib1/4, Ia/1 | ||
| 14 | 2737 | 0 | 2.20E+04 | 19 | 17 | Ib2/9 | Ia/1, Ib1/3, II/4 | II-I-II/1, II-I/1 | |
| 15 | 20 | 0 | 1.98E+03 | 20 | 16 | Ib2/14 | II/2, 1b1/1 | II-I/1, I-II/1, IV-I/1 | |
| 16 | 11 | 0 | 1.08E+03 | 18 | 13 | II/16 | 1b2/1 | II-I/1 | |
| 17 | 483 | 0 | 3.68E+03 | 20 | 15 | IV | none | ||
| 18 | 328 | 0 | 6.45E+03 | 20 | 16 | Ia/18 | II/1 | I-II/1 | |
| Healthy-viruria | 19 | N/A | 0 | 9.90E+03 | 20 | 8 | Ib2 | none | |
| 20 | N/A | 0 | 4.40E+03 | 20 | 12 | IV/17 | Ia/1 | IV-I/2 | |
| 21 | N/A | 0 | 3.22E+04 | 20 | 15 | II/14 | none | I-II/1, IV-II/1, II-I/1, II-IV/2, IV-II/1 | |
| 22 | N/A | 0 | 3.78E+03 | 20 | 16 | Ib2 | none | ||
| 23 | N/A | 0 | 1.86E+03 | 20 | 15 | IV/17 | II/1 | II-IV/2 | |
| 24 | N/A | 0 | 2.78E+03 | 20 | 16 | Ib2 | none | ||
| 25 | N/A | 0 | 1.01E+04 | 16 | 15 | IV/4 | 1b2/1, II/1 | II-IV/3, IV-II/3, I-IV/1, I-II-I/1, I-II/1, I-II-IV/1 |
The number of clones corresponding to each dominant, co-infecting or recobminant genotype is indicated after the slash, wherever this number is not identical to the total number of clones sequenced. Parent sequences contributing to recombination are separated by a dash and are listed in the order in which they are recombined. The number after the slash indicates the number of recombinants found (totals 29).
Biopsy proven polyomavirus nephropathy.
A total of 558 clones were obtained and sequenced from 25 subjects (28 urine samples). Taking into account only the predominant viral population the samples could be categorized by genotype as follows: 4 (14.3%) subgroup Ia, 3 (10.7%) subgroup Ib1, 11 (39.3%) subgroup Ib2, 6 (21.4%) genotype II and 4 (14.2%) genotype IV. Considering all 558 clone sequences, the distribution of genotypes was quite similar, namely Ia: 83 (14.8%), Ib1: 69 (12.4%), Ib2: 207 (37.1%), II: 112 (20.1%); IV: 58 (10.4%), recombinants: 29 (5.2%). Several subjects had co-infection with multiple genotypes, including 2 of 11 viremia/nephropathy subjects, 6 of 7 transplanted-viruria subjects, and 4 of 7 healthy-viruria subjects. A dominant genotype, defined as a viral population constituting more than 50% of the clones analyzed, was frequently present (Table 1).
2. Sequence Complexity
All subjects had multiple closely related viral populations that are referred to as viral quasispecies. It is believed that members of a quasispecies are related to each other by similar mutations and compete with each other in a mutagenicenvironment. The number of unique variants per sample in the study ranged from 3 to 19 (mean 12 ± 4.57, median 12.5). The mean complexity score was lower in viremia/nephropathy group compared with transplanted-viruria and healthy-viruria groups (0.54 ± 0.26 versus 0.81 ± 0.1 versus 0.79 ± 0.18, p<0.05, See Table 2). When different parts of the VP-1 protein were compared, complexity measured over the entire length of the VP-1 protein was higher than measurements made in the BC, DE, and HI loops. This was true in all three subject categories studied, and likely reflects differences in length amongst the polypeptides being compared. Within the viremia/nephropathy group, the complexity parameter was higher in the BC loop and DE loops compared to the HI loop. This relationship did not reach statistical significance in the transplanted-viruria and healthy-viruria subjects.
Table 2.
Quasispecies Complexity and Diversity as a Function of Clinical Status*
| Region | Complexity | Diversity | ||||
|---|---|---|---|---|---|---|
| Viremia/nephropathy | Transplanted-viruria | Healthy-viruria | Viremia/nephropathy | Transplanted-viruria | Healthy-viruria | |
| Whole VP1 | 0.54±0.261,a (0.17 – 0.93) | 0.81±0.11,c (0.59–0.89) | 0.79 ± 0.181,d (0.44–0.97) | 0.0015 ± 0.001e (0.0004–0.04) | 0.002±0.0011 (0.0007–0.0043) | 0.0047±0.0048 (0.0007–0.0152) |
| BC Loop | 0.20 ± 0.18a,b (0–0.59) | 0.27 ± 0.18c (0–0.48) | 0.31 ± 0.20d (0–0.57) | 0.006 ± 0.0072,e (0–0.0253) | 0.0019 ± 0.0015 (0–0.0041) | 0.0009 ± 0.0222 (0–0.06) |
| DE Loop | 0.06 ± 0.073,a,b (0–0.22) | 0.21 ± 0.063,c (0.1–0.26) | 0.19 ± 0.16d (0.0002–0.42) | 0.002 ± 0.002 (0–0.0078) | 0.0022 ± 0.0029 (0–0.0071) | 0.003 ± 0.004 (0–0.91) |
| HI Loop | 0.03 ± 0.044,a,b (0–0.11) | 0.12 ± 0.084,c (0–0.22) | 0.13 ± 0.15d (0–0.42) | 0.0013 ± 0.0023e (0–0.0072) | 0.0011 ± 0.002 (0–0.0032) | 0.0012 ± 0.0021 (0–0.85) |
Data is presented as mean +/− sd with the range indicated in parentheses. Statistical comparsions between columns are designated by numerical superscripts, while alphabetical designations mark comparisons between rows.
Putative BC loop (nucleotides 169 – 267, Dun numbering, Acc. V01108), DE loop (# 382 – 438), and HI loop (# 805 – 835) were identified using the published literature (40, 41)
p<0.05, complexity, Viremia/nephropathy vs both viruria groups in whole VP1 gene.
p<0.05, Diversity is significantly higher of BC Loop region in Viremia/nephropathy group than Healthy-viruria Group.
p<0.05, Complexity is significantly higher of DE Loop region in Transplanted-viruria group than Viremia/nephropathy group.
p<0.05, complexity of the HI Loop is significantly higher in the Transplanted-viruria group than the Viremia/nephropathy group.
p<0.05, Complexity, Whole VP1 gene region vs BC Loop, DE Loop, HI Loop of Viremia/nephropathy group.
p<0.05 Complexity, BC or DI Loops vs HI Loop in Viremia/nephropathy group.
p<0.05, Complexity, whole VP1 gene region vs BC Loop, DE Loop, HI Loop of Transplanted-viruria group.
p<0.05, Complexity, whole VP1 gene region vs BC Loop, DE Loop, HI Loop of Healthy-viruria group.
p<0.05, Diversity, BC Loop vs whole VP1 and HI Loop in Viremia/nephropathy group.
3. Sequence Diversity
Considering the whole VP-1 protein, overall sequence diversity expressed as the mean number of substitutions per site showed no statistically significant differences between the three study groups (0.0015 ± 0.001 viremia/nephropathy group versus 0.002 ± 0.0011 transplanted-viruria group versus 0.0055 ± 0.0045 healthy-viruria group). However, BC loop diversity in the viremia/nephropathy group was higher than the healthy-viruria group (0.006 ± 0.007 versus 0.0019 ± 0.015). Amongst the subjects with viremia or nephropathy, diversity was greater in the BC loop (0.006 ± 0.007) compared to the HI loop (0.0013 ± 0.002) and the whole VP-1 protein (0.0015 ± 0.001).
dN, a measure of non-synonymous substitutions per non-synonymous site, was similar in the three major subject categories (0.0015 ± 0.0012 viremia/nephropathy group versus 0.0014 ± 0.0004 transplanted-viruria group versus 0.002 ± 0.001 healthy-viruria group subjects). Looking at the individual loops with in the VP-1 protein, 10/28 BC loop, 14/28 DE loop, and 22/28 HI loop sequence sets had dN values of zero (indicating identical sequences in all clones analyzed). Nevertheless, the mean dN of the BC loop in the viremia/nephropathy group (0.008 ± 0.009) was higher compared with the whole VP1 gene 0.0015 ± 0.0012 (p<0.05) as well as the DE and HI loops (although the difference with respect to DE loop did not reach statistical significance). These differences were not noted in kidney transplant and healthy subjects with viruria..
In the whole VP-1 protein, dS, an index reflecting synonymous substitution per synonymous site, was lower in patients with viremia/nephropathy group compared to transplanted-viruria group and healthy-viruria groups (0.001 ± 0.0007 viremia/nephropathy group versus 0.0046 ± 0.0043 transplanted-viruria group versus 0.005 ± 0.003 healthy-viruria subjects). The difference between the viremia/nephropathy group and transplanted-viruria group did not reach statistical significance (p=0.064). Within the receptor binding loops, more than half of the patients analyzed in each of the three groups had dS values of zero. The mean dS of the BC Loop region in the viremia/nephropathy group (0.0004±0.0014) appeared to be lower compared with the whole VP1 gene region (0.001 ± 0.0007), but the difference did not reach statistical significance (p=0.17).
In the full length VP-1 sequence, the ratio dN/dS, which is a measure of evolutionary selection pressure, was assigned a higher value in viremia/nephropathy group compared to the transplanted-viruria and healthy-viruria groups (1.5 ± 1.08 versus 0.4 ± 0.16 versus 0.45 ± 0.16 respectively). Within individual VP-1 loops, the dN/dS ratio could frequently not be calculated since the denominator was zero, and the complete absence of synonymous mutations generated a quotient approaching infinity. The number of subjects in which a finite ratio could be obtained is noted in Table 3. This ratio exceeded 1.0 only in the BC loop of a patient with viremia/nephropathy (dN/dS=2.58).
Table 3.
Distribution of Synonymous and Non-synonymous Nucleotide Substitutions in Different Subject Categories
| Region | dN | dS | dN:dS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Viremia/nephropathy | Transplanted-viruria | Healthy-viruria | Viremia/nephropathy | Transplanted-viruria | Healthy-viruria | Viremia/nephropathy | Transplanted-viruria | Healthy-viruria | |
| Whole VP1 | 0.0015±0.00121 (0.0002–0.0044) | 0.0014±0.0004 (0.0006–0.0019) | 0.002±0.001 (0.001–0.67) | 0.001 ± 0.00072 (0–0.0024) | 0.0046±0.0043 (0.0012–0.014) | 0.005 ± 0.0032 (0.0023–0.064) | 1.5 ± 1.083 (0.125–3.375) (n=12) |
0.4±0.163 (0.11–0.59) (n=7) |
0.45±0.163 (0.005–0.62) (n=7) |
| BC Loop | 0.008 ± 0.0091 (0–0.0317) | 0.0021±0.0015 (0–0.0036) | 0.005±0.009 (0–0.0234) | 0.0004±0.0014 (0–0.053) | 0.0036±0.0063 (0–0.0139) | 0.005±0.009 (0–0.0238) | 2.58 (n=1) | 0.25±0.014 (0.24–0.26) (n=2) |
0.7±0.36 (0.47–0.98) (n=2) |
| DE Loop | 0.0024±0.0032 (0–0.0101) | 0.0014±0.0019 (0–0.0046) | 0.0038±0.0045 (0–0.0115) | 0.0011±0.0029 (0–0.0081) | 0.005±0.007 (0–0.0167) | 0.0037±0.008 (0–0.018) | 0.41±0.19 (0.28–0.54) (n=2) |
0.28±0.006 (0.28–0.29) (n=3) |
0.43 (n=1) |
| HI Loop | 0.0011 ± 0.00291 (0–0.0102) | 0.0016 ± 0.002 (0–0.0059) | 0.0017 ± 0.003 (0–0.0063) | 0.002 ± 0.005 (0–0.0151) | 0.013 ± 0.034 (0–0.0167) | 0 (0) | 0 (0) (n=2) | 0 (n=1) | N/A (dS=0 for all sequences) |
Data is presented as mean +/− sd with the range indicated in parentheses.
N/A: not applicable
P<0.05, dN, BC Loop vs whole VP1 region and HI Loop in Viremia/nephropathy group.
P<0.05, dS Viremia/nephropathy vs Healthy-viruria in whole VP1 gene.
P<0.05, dN/dS, Viremia/nephropathy vs Transplanted-viruria & Healthy-viruria groups in whole VP1 gene.
Putative BC loop (nucleotides 169 – 267, Dun numbering, Acc. V01108), DE loop (# 382 – 438), and HI loop (# 805 – 835) were identified using the published literature: (40, 41)
4. Distribution of nucleotide substitutions within the VP-1 gene
Nucleotide substitutions were noted throughout the VP-1 gene but most frequently affected the loop regions. ‘Hot spots’ defined as sites altered in at least 3 samples were seen at nucleotide positions 178, 217, 229, 244, 246, 577, 673, 687 (non-synonymous alterations) and 72, 678, 867, 888, 891, and 906 (synonymous alterations). Five (5) out of 8 non-synonymous hot spots were located with in the BC Loop. Furthermore, all samples that showed non-synonymous substitutions localizing to BC loop ‘hot spots’ belonged to the viremia/nephropathy group. The BC loop region is important because it interacts with cellular receptors, and facilitates BKV entry into the host cell. Occasional clones showed non-synonymous nucleotide substitutions at other functionally important sites outside of the receptor loops. These sites included the VP-1 nuclear localization signal site, disulfide binding sites related to capsid assembly (Cys10, Cys50, Cys88, Cys105, Cys208, Cys255, Cys268; BKV Dun numbering) [Ishizu et al., 2001; Kawano et al., 2009], calcium binding sites that help maintain capsid shape (Glu330, Glu331, Asp346) [Ishizu et al., 2001; Kawano et al., 2009], amino acids identified as important for BKV viability and growth [Dugan et al., 2007], and two immunologic epitopes recognized by T-cells, namely, LLMWEAVTV and AITEVECFL [Krymskaya et al., 2005; Sharma et al., 2006a].
5. Recombination events
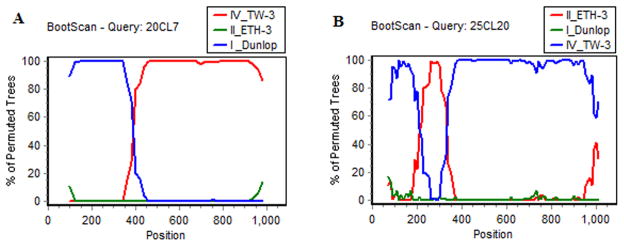
Viral genotype could not be assigned to 29/558 (5.2%) of clones which were recombinants of other known genotypes. These recombinant strains were derived from 9 subjects (1 of viremia/nephropathy group, 4 of transplanted-viruria group, 4 of healthy-viruria group). Considering only these nine subjects recombinant clones constituted 29/175 (16.6%) of the total clones analyzed. Twenty five (25/29, 86.2%) recombinants obtained from 8 patients had a parent strain of genotype II. Likewise, 17/29 (58.6%) recombinants obtained from 5 subjects had a parent strain of genotype IV (Table 1). Four of the latter subjects belonged to the healthy-viruria group, 3 were from Europe, and 2 had genotype IV as the predominant viral species in the urine. The breakpoints that generated the recombinant events were mapped and found to be concentrated between positions 184–285 (5 events), 296–399 (7 events), 415–504 (5 events), 540–579 (3 events), 628–807 (15 events), 828–981 (6 events) and 1020–1058 (1 event). Ten clones showed two separate breakpoints in the same sequence. Bootscans of representative recombinant sequences are shown in Figure 1. Phylogenetic trees of recombinant sequences confirmed assignment of parental sequences to the genotype level.
Figure 1.
Representative recombinant sequences identified by bootscans implemented in Simplot. Scans were generated using a sliding window of 140 nucleotides, moving 10 nucleotides at a time. The x-axis shows nucleotide positions on the VP-1 gene sequence (Dun numbering). The y-axis depicts the percent agreement in the permuted trees. The query sequences are indicated in the title line starting with patient number followed by clone number. Thus, 20CL7 refers to clone #7 from patient #20. Accession numbers of published sequences used in this plot are: Dunlop (V01108), TW-3 (AB211391), and ETH-3 (AB263916).
Figure 1A illustrates a recombinant sequence that conforms to type I in the initial segment (blue line) and type IV in the final segment (red line). A type II outgroup sequence (II_ETH, green line) was used to generate the plot.
Figure 1B depicts a more complex recombinant sequence with an initial segment conforming to genotype IV (blue line), which is followed by a short genotype II sequence (red line), following which the sequence reverts to genotype IV (blue line). BKV strain Dun (Type I) was used as the outgroup sequence (green line).
DISCUSSION
Genetic heterogeneity in the VP-1 gene has been used to divide BKV into four major genotypes I, II, III, and IV. Genotype I is the most prevalent in all major geographic areas with a prevalence range of 46–82% [Luo et al., 2008]. Subgroups have been described within genotype 1. These appear to have predilection for specific geographic regions, such as subgroup 1a for Africa, 1b-1 for Southeast Asia, 1b-2 for Europe, and 1c for northeast Asia [Zheng et al., 2007]. Genotype IV is typically the second most prevalent type (incidence 12–54%). There are also differences in geographic distribution for its constituent subgroups [Baksh et al., 2001; Di Taranto et al., 1997; Jin, 1993] Thus, genotype IVa1 comprised 8/15 (53%) of genotype IV strains obtained from southeast Asia (Philippines, Vietnam, and Mynamar), while genotype IVb1 and IVb2 accounted for 40% and 55% respectively of Korean and Japanese isolates. In contrast, 21/26 (81%) of Chinese strains were genotype IVc1, and all 22 genotype IV strains from Europe were subgroup IVc2 [Nishimoto et al., 2007]. Based on serology studies, Genotypes II and III have been considered to be uncommon strains [Stolt et al., 2003]. However, clonal sequence data from this study suggests that genotype II is present as a minority species much more commonly than hitherto recognized. More convincing proof of this will require development of genotype II specific serologic assays.
The frequent presence of infection by more than one viral genotype is another notable finding in this study. The actual frequency of co-infection is likely even higher than that shown, since an analysis based on 20 clones may not detect viral species present at a frequency of less than 5%. Coinfection of the same healthy subject with multiple BKV strains could be the result of social contact with other subjects excreting virus in the urine. The presence of BKV DNA in metropolitan sewage suggests that feco-oral transmission may be yet another mode of viral transmission from one individual to another. In kidney allograft patients, the transplanted organ, and blood transfusions associated with dialysis may act as additional sources of a second infectious strain of virus.
Samples showing infection with a single genotype also showed multiple genetically related populations. The high fidelity of DNA polymerase under the conditions used indicates that these reflect BKV quasispecies rather than a laboratory artifact. Quasispecies are most widely recognized in RNA viruses, which have error prone mechanisms for replication of nucleic acids. In contrast, DNA viruses use the host replication system with high fidelity, good proof reading capacity, and efficient post replication repair capacity. Hence, the mutation rate of DNA viruses may be up to 100 times lower than RNA viruses [Drake and Holland, 1999]. BKV also uses host DNA polymerase for its replication, but agnoprotein coded by the viral genome inhibits double-stranded DNA break repair activity. This could potentially increase the error rate in BKV DNA replication [Darbinyan et al., 2004]. It is pertinent to mention that quasispecies have been described in another DNA virus, namely, canine parvovirus [Shackelton et al., 2005]. The work reported here documents that VP-1 quasispecies are common in the context of human BKV infection. This likely reflects the documented high turnover rate of this virus in the renal tubular epithelium: in kidney transplant patients, it has been estimated that between 1E+07 to 1E+09 virions can be produced and cleared per patient per day [Funk et al., 2006]. In healthy subjects BKV is typically latent, but approximately 5% do excrete virus asymptomatically, and the viral turnover in this small subset of healthy subjects with viruria appears to be sufficient to result in formation of quasispecies.
The viremia/nephropathy group had higher nucleotide substitution rate in the BC loop region compared to transplanted-viruria and healthy-viruria subjects. The BC loop region was also characterized by a high dN/dS ratio indicating that this part of the viral genome was under positive selection pressure in viremia/nephropathy subjects. Given the function of the BC loop in virus-host cell receptor interactions, this selection is likely driven by enhanced cellular entry of the virus into the cell. Less likely possibilities include replicative fitness, immune escape or better adaptability to the metabolic milieu of the cell. BC loop mutations are known to enhance the plaque forming ability of mouse polyomavirus [Bauer et al., 1995; Freund et al., 1991]. Recently, loop specific polymorphisms in polyomavirus JC virus have been associated with favorable prognosis in patients with progressive multifocal encephalopathy [Delbue et al., 2009]. Some of the adaptive mutations in these patients have been shown to be under selection pressure at the codon level [Sunyaev et al., 2009]. In contrast with the experience reported here, a prior study evaluating genetic distances between 176 publicly available polyomavirus sequences found no evidence of positive (diversifying) selection either at the whole genome or individual gene level [Mes et al., 2010]. Analysis of nucleotide substitutions at synonymous and non-synonymous sites suggested that BKV genes evolve under strong constraints that result in a net purifying selection at the population level. The present study differs from the aforementioned one in having analyzed intra-host (as opposed to between-host) nucleotide substitutions in the setting of post-kidney transplant viremia or nephropathy. The BKV genome has also been reported to be stable in molecular clones of BKV derived from six post-transplant patients with viruria and serum creatinine ranging from 1.1 to 1.7 mg/dl [Takasaka et al., 2006]. Plasma BKV status was not evaluated in the latter study. In a rare case of BKV associated capillary leak syndrome clones of infectious virus derived from the urine, muscle, and heart showed high intra-strain genetic diversity, but no positive selection as judged by the dN/dS ratio [Chen et al., 2004].
Recombination is an important mechanism of viral genetic variation which can alter virulence as well as host range. [Brown, 1997; Gibbs et al., 2001; Gibbs and Weiller, 1999; Malim and Emerman, 2001; Rest and Mindell, 2003]. Sequence rearrangements in the non-coding control region of BKV are well documented [Carr et al., 2006; Perets et al., 2009; Sharma et al., 2007]. An earlier study from this laboratory described two BKV strains with potential recombinations in the VP-1 gene [Luo et al., 2009]. In the current work, recombinants constitute approximately 5% of all clonal sequences examined. While recombinant sequences can be created as an in-vitro artifact, the frequent finding of genotype II footprints, presence of sequences not represented in the majority genotype, the non-random distribution of breakpoints, and the occurrence of multiple breakpoints, some of which were common to strains derived from different subjects, makes it unlikely that all of the observed recombinants are in-vitro artifacts. One could argue that horizontal exchange of genetic material is to be expected in individuals co-infected with multiple BK virus strains. Other investigators have accrued evidence that even BKV coding regions other than VP-1 can participate in genetic recombination events. Thus, it has been proposed that a region of the BKV genome upstream of the VP3 gene is derived from a recombination event with SV40 in the course of evolution [Yasunaga and Miyata, 1982]. Yogo et al identified a Non coding control region (NCCR)-Large T antigen recombinant JC virus in brain tissue derived from a patient with progressive multifocal encephalopathy. The NCCR in this viral strain had a small deletion as well as a 29 bp sequence originating from the early region of the JCV genome [Yogo et al., 1994]. Genetic recombination has been reported in culture systems when wild type BKV is introduced in permissive BSC-1 cells along with either early or late defective SV40 genome regions [O’Neill and Miller, 1985]. In experimentally constructed hybrid sequences, coding portions of BKV and polyomavirus SV40 genome can function if placed downstream of the regulatory region of polyomavirus JC [Chuke et al., 1986]. Notwithstanding these considerations, the clinical significance of the VP-1 recombinant BKV species observed by us is uncertain: these always represented a minor viral population in the samples studied.
In conclusion, this study has used clonal sequencing of BKV VP-1 to document frequent occurrence of mixed infections and recombinant strains. Genotype II was found as an unexpected minority in 25% of individuals and contributed to 55% of the recombinant sequences. VP-1 Quasispecies were found in the urine of both immunosuppressed and healthy individuals. Evidence of evolutionary selection pressure on quasispecies was demonstrable only in patients with viremia or nephropathy. In healthy subjects, generation of quasispecies may reflect an evolving attempt to achieve immune evasion. The viral fitness of the quasispecies was not assessed in this study. However, it has been shown by others changes in VP-1 as well as NCCR sequence can alter rates of viral replication [Gosert et al., 2008; Nukuzuma et al., 2006].
Acknowledgments
Supported by grants RO1 AI 51227 and AI 63360 to PR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Institute of Allergy and Infectious Disease. Jill March provided excellent secretarial assistance.
References
- Baksh FK, Finkelstein SD, Swalsky PA, Stoner GL, Ryschkewitsch CF, Randhawa P. Molecular genotyping of BK and JC viruses in human polyomavirus-associated interstitial nephritis after renal transplantation. Am J Kidney Dis. 2001;38:354–365. doi: 10.1053/ajkd.2001.26101. [DOI] [PubMed] [Google Scholar]
- Bauer PH, Bronson RT, Fung SC, Freund R, Stehle T, Harrison SC, Benjamin TL. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J Virol. 1995;69:7925–7931. doi: 10.1128/jvi.69.12.7925-7931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binggeli S, Egli A, Schaub S, Binet I, Mayr M, Steiger J, Hirsch HH. Polyomavirus BK-specific cellular immune response to VP1 and large T-antigen in kidney transplant recipients. Am J Transplant. 2007;7:1131–1139. doi: 10.1111/j.1600-6143.2007.01754.x. [DOI] [PubMed] [Google Scholar]
- Boldorini R, Allegrini S, Miglio U, Paganotti A, Veggiani C, Mischitelli M, Monga G, Pietropaolo V. Genomic mutations of viral protein 1 and BK virus nephropathy in kidney transplant recipients. J Med Virol. 2009;81:1385–1393. doi: 10.1002/jmv.21520. [DOI] [PubMed] [Google Scholar]
- Bolen JB, Anders DG, Trempy J, Consigli RA. Differences in the subpopulations of the structural proteins of polyoma virions and capsids: biological functions of the multiple VP1 species. J Virol. 1981;37:80–91. doi: 10.1128/jvi.37.1.80-91.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan DC, Agha I, Bohl DL, Schnitzler MA, Hardinger KL, Lockwood M, Torrence S, Schuessler R, Roby T, Gaudreault-Keener M, Storch GA. Incidence of BK with tacrolimus versus cyclosporine and impact of preemptive immunosuppression reduction. Am J Transplant. 2005;5:582–594. doi: 10.1111/j.1600-6143.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- Brinkert F, Briem-Richter A, Ilchmann C, Kemper MJ, Ganschow R. Prevalence of polyomavirus viruria (JC virus/BK virus) in children following liver transplantation. Pediatric Transplantation. 2010;14:105–108. doi: 10.1111/j.1399-3046.2009.01139.x. [DOI] [PubMed] [Google Scholar]
- Brown DW. Threat to Humans from Virus Infections of Non-human Primates. Rev Med Virol. 1997;7:239–246. doi: 10.1002/(sici)1099-1654(199712)7:4<239::aid-rmv210>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Carr MJ, McCormack GP, Mutton KJ, Crowley B. Unique BK virus non-coding control region (NCCR) variants in hematopoietic stem cell transplant recipients with and without hemorrhagic cystitis. J Med Virol. 2006;78:485–493. doi: 10.1002/jmv.20566. [DOI] [PubMed] [Google Scholar]
- Chen YP, Sharp PM, Fowkes M, Kocher O, Joseph JT, Koralnik IJ. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J Gen Virol. 2004;85:2651–2663. doi: 10.1099/vir.0.79920-0. [DOI] [PubMed] [Google Scholar]
- Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- Chuke WF, Walker DL, Peitzman LB, Frisque RJ. Construction and Characterization of Hybrid Polyomavirus Genomes. J Virol. 1986;60:960–971. doi: 10.1128/jvi.60.3.960-971.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinyan A, Siddiqui KM, Slonina D, Darbinian N, Amini S, White MK, Khalili K. Role of JC virus agnoprotein in DNA repair. J Virol. 2004;78:8593–8600. doi: 10.1128/JVI.78.16.8593-8600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbue S, Branchetti E, Bertolacci S, Tavazzi E, Marchioni E, Maserati R, Minnucci G, Tremolada S, Vago G, Ferrante P. JC virus VP1 loop-specific polymorphisms are associated with favorable prognosis for progressive multifocal leukoencephalopathy. J Neurovirol. 2009;15:51–56. doi: 10.1080/13550280802425467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Taranto C, Pietropaolo V, Orsi GB, Jin L, Sinibaldi L, Degener AM. Detection of BK polyomavirus genotypes in healthy and HIV-positive children. Eur J Epidemiol. 1997;13:653–657. doi: 10.1023/a:1007371320999. [DOI] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan AS, Gasparovic ML, Tsomaia N, Mierke DF, O’Hara BA, Manley K, Atwood WJ. Identification of amino acid residues in BK virus VP1 that are critical for viability and growth. J Virol. 2007;81:11798–11808. doi: 10.1128/JVI.01316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli A, Kohli S, Dickenmann M, Hirsch HH. Inhibition of polyomavirus BK-specific T-Cell responses by immunosuppressive drugs. Transplantation. 2009;88:1161–1168. doi: 10.1097/TP.0b013e3181bca422. [DOI] [PubMed] [Google Scholar]
- Freund R, Garcea RL, Sahli R, Benjamin TL. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J Virol. 1991;65:350–355. doi: 10.1128/jvi.65.1.350-355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk GA, Steiger J, Hirsch HH. Rapid dynamics of polyomavirus type BK in renal transplant recipients. J Infect Dis. 2006;193:80–87. doi: 10.1086/498530. [DOI] [PubMed] [Google Scholar]
- Gibbs MJ, Armstrong JS, Gibbs AJ. Recombination in the hemagglutinin gene of the 1918 “Spanish flu”. Science. 2001;293:1842–1845. doi: 10.1126/science.1061662. [DOI] [PubMed] [Google Scholar]
- Gibbs MJ, Weiller GF. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci U S A. 1999;96:8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Rinaldo CH, Funk GA, Egli A, Ramos E, Drachenberg CB, Hirsch HH. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med. 2008;205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HH, Knowles W, Dickenmann M, Passweg J, Klimkait T, Mihatsch MJ, Steiger J. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med. 2002;347:488–496. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- Hirsch HH, Randhawa P. BK virus in solid organ transplant recipients. Am J Transplant. 2009;9(Suppl 4):S136–146. doi: 10.1111/j.1600-6143.2009.02904.x. [DOI] [PubMed] [Google Scholar]
- Hirsch HH, Steiger J, Knowles W, Dickenmann M, Passweg J, Klimkait T, Mihatsch MJ, Mayr M, Nickeleit V. Polyomavirus BK. Lancet Infect Dis. 2003;3:611–623. doi: 10.1016/s1473-3099(03)00770-9. [DOI] [PubMed] [Google Scholar]
- Ishizu KI, Watanabe H, Han SI, Kanesashi SN, Hoque M, Yajima H, Kataoka K, Handa H. Roles of disulfide linkage and calcium ion-mediated interactions in assembly and disassembly of virus-like particles composed of simian virus 40 VP1 capsid protein. J Virol. 2001;75:61–72. doi: 10.1128/JVI.75.1.61-72.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L. Rapid genomic typing of BK virus directly from clinical specimens. Mol Cell Probes. 1993;7:331–334. doi: 10.1006/mcpr.1993.1047. [DOI] [PubMed] [Google Scholar]
- Jin L, Gibson PE. Genomic Function and Variation of Human Polyomavirus BK (BKV) Rev Med Virol. 1996;6:201–214. doi: 10.1002/(SICI)1099-1654(199612)6:4<201::AID-RMV177>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kawano MA, Xing L, Tsukamoto H, Inoue T, Handa H, Cheng RH. Calcium bridge triggers capsid disassembly in the cell entry process of simian virus 40. J Biol Chem. 2009;284(50):34703–34712. doi: 10.1074/jbc.M109.015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles WA. The epidemiology of BK virus and the occurrence of antigenic and genomic subtypes. In: Khalili K, Stoner GL, editors. Human polyomaviruses: molecular and clinical perspectives. New York: Wiley-Liss; 2001. pp. 527–560. [Google Scholar]
- Krymskaya L, Sharma MC, Maritnez J, Haq W, Huang EC, Limaye AP, Diamond DJ, Lacey SF. Cross reactivity of T lymphocytes recognizing a human cytotoxic T-lymphocyte epitope wtihin BK and JC virus VP1 polypeptides. J Virol. 2005;79:11170–11178. doi: 10.1128/JVI.79.17.11170-11178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J, Humes HD, Szczypka M, Imperiale M. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology. 2004;323:182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol. 2009;83:2285–2297. doi: 10.1128/JVI.02180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Bueno M, Kant J, Randhawa P. Biologic diversity of polyomavirus BK genomic sequences: Implications for molecular diagnostic laboratories. J Med Virol. 2008;80:1850–1857. doi: 10.1002/jmv.21281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 sequence variation: drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- Mes TH, van Doornum GJ, Schutten M. Population genetic tests suggestthat the epidemiologies of JCV and BKV are strikingly different. Infect Genet Evol. 2010;10:397–403. doi: 10.1016/j.meegid.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Minosse C, Calcaterra S, Abbate I, Selleri M, Zaniratti MS, Capobianchi MR. Possible compartmentalization of hepatitis C viral replication in the genital tract of HIV-1-coinfected women. J Infect Dis. 2006;194:1529–1536. doi: 10.1086/508889. [DOI] [PubMed] [Google Scholar]
- Murata H, Teferedegne B, Sheng L, Lewis AM, Jr, Peden K. Identification of a neutralization epitope in the VP1 capsid protein of SV40. Virology. 2008;381:116–122. doi: 10.1016/j.virol.2008.07.032. [DOI] [PubMed] [Google Scholar]
- Nishimoto Y, Zheng HY, Zhong S, Ikegaya H, Chen Q, Sugimoto C, Kitamura T, Yogo Y. An Asian origin for subtype IV BK virus based on phylogenetic analysis. J Mol Evol. 2007;65:103–111. doi: 10.1007/s00239-006-0269-6. [DOI] [PubMed] [Google Scholar]
- Nukuzuma S, Takasaka T, Zheng H-Y, Zhong S, Chen Q, Kitamura T, Yogo Y. Subtype I BK polyomavirusstrains grow more efficiently in human renal epithelial cells than subtype IV strains. J Gen Virol. 2006;87:1893–1901. doi: 10.1099/vir.0.81698-0. [DOI] [PubMed] [Google Scholar]
- O’Donnell PH, Swanson K, Josephson MA, Artz AS, Parsad SD, Ramaprasad C, Pursell K, Rich E, Stock W, van Besien K. BK virus infection isassociated with hematuria and renal impairment in recipients of allogeneic hematopoetic stem cell transplants. Biol Blood Marrow Transplant. 2009;15:1038–1048. e1031. doi: 10.1016/j.bbmt.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill FJ, Miller TH. Isolation of a papovavirus with a bipartite genome containing unlinked SV40 and BKV sequences. Virology. 1985;143:75–87. doi: 10.1016/0042-6822(85)90098-4. [DOI] [PubMed] [Google Scholar]
- Perets TT, Silberstein I, Rubinov J, Sarid R, Mendelson E, Shulman LM. High frequency and diversity of rearrangements in polyomavirus bk noncoding regulatory regions cloned from urine and plasma of Israeli renal transplant patients and evidence for a new genetic subtype. J Clin Microbiol. 2009;47:1402–1411. doi: 10.1128/JCM.02065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa P, Uhrmacher J, Pasculle W, Vats A, Shapiro R, Eghtsead B, Weck K. A comparative study of BK and JC virus infections in organ transplant recipients. J Med Virol. 2005;77:238–243. doi: 10.1002/jmv.20442. [DOI] [PubMed] [Google Scholar]
- Randhawa PS, Demetris AJ. Nephropathy due to polyomavirus type BK. N Engl J Med. 2000;342:1361–1363. doi: 10.1056/NEJM200005043421809. [DOI] [PubMed] [Google Scholar]
- Randhawa PS, Khaleel-Ur-Rehman K, Swalsky PA, Vats A, Scantlebury V, Shapiro R, Finkelstein S. DNA sequencing of viral capsid protein VP-1 region in patients with BK virus interstitial nephritis. Transplantation. 2002;73:1090–1094. doi: 10.1097/00007890-200204150-00013. [DOI] [PubMed] [Google Scholar]
- Rest JS, Mindell DP. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect Genet Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton LA, Parrish CR, Truyen U, Holmes EC. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci USA. 2005;102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MC, Zhou W, Martinez J, Krymskaya L, Srivastava T, Haq W, Diamond DJ, Lacey SF. Cross-reactive CTL recognizing two HLA-A*02-restricted epitopes within the BK virus and JC virus VP1 polypeptides are frequent in immunocompetent individuals. Virology. 2006a;350:128–136. doi: 10.1016/j.virol.2006.02.040. [DOI] [PubMed] [Google Scholar]
- Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. Phylogenetic analysis of polyomavirus BK sequences. J Virol. 2006b;80:8869–8879. doi: 10.1128/JVI.00510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. A phylogenetic analysis of polyomavirus BK sequences. J Virol. 2006c;80:8869–8879. doi: 10.1128/JVI.00510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa PS. Polyomavirus BK non-coding control region rearrangements in health and disease. J Med Virol. 2007;79:1199–1207. doi: 10.1002/jmv.20909. [DOI] [PubMed] [Google Scholar]
- Stolt A, Sasnauskas K, Koskela P, Lehtinen M, Dillner J. Seroepidemiology of the human polyomaviruses. J Gen Virol. 2003;84:1499–1504. doi: 10.1099/vir.0.18842-0. [DOI] [PubMed] [Google Scholar]
- Sunyaev SR, Lugovskoy A, Simon K, Gorelik L. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML) PLoS Genet. 2009;5:e1000368. doi: 10.1371/journal.pgen.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaka T, Goya N, Ishida H, Tanabe K, Toma H, Fujioka T, Omori S, Zheng HY, Chen Q, Nukuzuma S, Kitamura T, Yogo Y. Stability of the BK polyomavirus genome in renal-transplant patients without nephropathy. J Gen Virol. 2006;87:303–306. doi: 10.1099/vir.0.81368-0. [DOI] [PubMed] [Google Scholar]
- Thomas LD, Milstone AP, Vilchez RA, Zanwar P, Butel JS, Dummer JS. Polyomavirus infection and its impact on renal function and long-term outcomes after lung transplantation. Transplantation. 2009;88:360–366. doi: 10.1097/TP.0b013e3181ae5ff9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga T, Miyata T. Evolutionary changes of nucleotide sequences of papova viruses BKV and SV40: they are possibly hybrids. J Mol Evol. 1982;19:72–79. doi: 10.1007/BF02100225. [DOI] [PubMed] [Google Scholar]
- Yogo Y, Guo J, Iida T, Satoh K, Taguchi F, Takahashi H, Hall WW, Nagashima K. Occurrence of multiple JC virus variants with distinctive regulatory sequences in the brain of a single patient with progressive multifocal leukoencephalopathy. Virus Genes. 1994;8:99–105. doi: 10.1007/BF01703608. [DOI] [PubMed] [Google Scholar]
- Zheng HY, Nishimoto Y, Chen Q, Hasegawa M, Zhong S, Ikegaya H, Ohno N, Sugimoto C, Takasaka T, Kitamura T, Yogo Y. Relationships between BK virus lineages and human populations. Microbes Infect. 2007;9:204–213. doi: 10.1016/j.micinf.2006.11.008. [DOI] [PubMed] [Google Scholar]