Abstract
Targeted intracellular degradation provides a method to study the biological function of proteins and has numerous applications in biotechnology. One promising approach uses adaptor proteins to target substrates with genetically encoded degradation tags for proteolysis. Here, we describe an engineered split-adaptor system, in which adaptor assembly and delivery of substrates to the ClpXP protease depends on a small molecule (rapamycin). This degradation system does not require modification of endogenous proteases, functions robustly over a wide range of adaptor concentrations, and does not require new synthesis of adaptors or proteases to initiate degradation. We demonstrate the efficacy of this system in E. coli by degrading tagged variants of LacI repressor and FtsA, an essential cell-division protein. In the latter case, addition of rapamycin causes pronounced filamentation because daughter cells cannot divide. Strikingly, washing rapamycin away reverses this phenotype. Our system is highly modular, with clearly-defined interfaces for substrate binding, protease binding, and adaptor assembly, providing a clear path to extend this system to other degradation tags, proteases, or induction systems. Together, these new reagents should be useful in controlling protein degradation in bacteria.
Keywords: regulated protein degradation, small-molecule control of proteolysis, split adaptor, ClpXP protease, chemical genetics, cell division
INTRODUCTION
Targeted proteolysis provides a mechanism to control intracellular protein levels in a dynamic fashion and has applications in basic science and biological engineering (1-6). Indeed, the perturbation of protein stability has proven critical in the generation of synthetic cellular circuits and has been utilized to investigate loss-of-function phenotypes (6-10), providing an alternative and often complementary mechanism to transcriptional knockdowns. A proteolysis-based approach is particularly valuable when pre-existing proteins are inherently long-lived, as terminating the synthesis of such molecules results in relatively slow elimination of the gene product (11-16). In these cases, proteins are diluted via cell growth and division, often requiring many cell cycles to reach levels that are low enough to eliminate function. During this time period, cells may up-regulate compensatory pathways or acquire suppressor mutations, often obscuring the phenotype of the original perturbation.
When combined with temporal control, targeted proteolysis can be used to investigate essential proteins under control of their native transcriptional and translational control elements. Temperature-sensitive (ts) alleles share many of the advantages of targeted proteolysis, but tightly regulated ts-alleles are difficult to isolate for many genes. Furthermore, a temperature shift can result in unintended changes in cellular physiology, complicating interpretation. In yeast, for example, shifting the temperature form 25 to 37 °C alters the expression of nearly 1000 genes, half of which have unknown function (17). Small-molecule inhibitors/activators allow perturbations of native gene function with rapid temporal control under many environmental conditions (13). Unfortunately, the identification of highly specific, cell-permeable inhibitors is not trivial, and conclusively ruling out “off-target” effects is a substantial challenge.
In Escherichia coli and most bacteria, processive intracellular proteolysis is mediated by a group of energy-dependent AAA+ proteases, including ClpXP (18). The ClpXP enzyme degrades substrates via a multi-step process, which begins with the recognition of an ssrA tag or other short peptide sequences, which are exposed in native protein substrates (19,20). After ClpX binds a substrate, cycles of ATP hydrolysis drive translocation of the degradation tag through a narrow axial pore, producing a transient unfolding force once the native protein collides with the pore entrance (21-23). These pulling events eventually result in global substrate unfolding, allowing translocation of the denatured polypeptide into the lumen of ClpP, where it is cleaved into short peptide fragments (24,25). Although degradation rates differ between substrates, ClpX can unfold proteins with a wide range of thermodynamic stabilities and appears to have little sequence specificity in terms of substrate translocation (24,26,27). Thus, substrate selectivity is determined by the efficiency of the initial binding event, which can be modulated by accessory factors called adaptors. For example, the SspB adaptor improves ClpXP degradation of ssrA-tagged substrates by binding both to the protease and to a portion of the ssrA tag, thereby increasing the effective concentration of the substrate relative to the enzyme (28-31). Indeed, using synthetic degradation components, we found that tethering alone is sufficient for efficient substrate delivery by SspB (2).
Adaptor proteins have proven useful in engineering controlled degradation systems. For example, McGinness et al. (2006) showed that ClpXP degradation became almost entirely dependent on SspB when the C-terminal residues of the ssrA tag were mutated from LAA to DAS. Importantly, model substrates bearing DAS tags were selectively degraded in cells when transcription of the sspB gene from a lac promoter was induced using IPTG (4,6). Combining a genetically encoded degradation tag and small-molecule inducer has many of the advantages of classical genetics and pharmacology and can be applied to almost any protein target with temporal control provided by the presence or absence of the small molecule.
In the work reported here, we have engineered and characterized a new targeted degradation system in which the assembly and thus activity of a split adaptor is controlled by the small-molecule rapamycin. Experiments with purified components in vitro and with multiple protein substrates in E. coli show that ClpXP-mediated degradation of appropriately tagged proteins can be controlled in a rapamycin-dependent manner. This system is simple, generally applicable, requires few genomic modifications, and degrades substrates without the need for new protein synthesis. Moreover, this system should be relatively straightforward to port to other ClpXP-containing bacteria such as Caulobacter crescentus, Bacillus subtilis, and Mycobacterium tuberculosis (6,10,32).
RESULTS AND DISCUSSION
Rapamycin-dependent control of adaptor function in vitro
We used a DAS+4 ssrA tag (AANDENYSENYADAS) to target substrates for adaptor-mediated degradation by ClpXP (4). The underlined residues in this tag sequence bind SspBCORE (the folded domain of SspB), whereas the bold residues interact with ClpXP. We split the functionally important portions of SspB into two components, one consisting of SspBCORE fused to FRB, the rapamycin-binding domain of mTOR, and another consisting of the highly flexible ClpX-binding tail of SspB (XB) fused to the FKBP12 protein (Fig. 1). Normally, these split components (SspBCORE-FRB and FKBP12-SspBXB) should not interact or deliver substrates to ClpXP. However, rapamycin-dependent dimerization of the FRB and FKBP12 portions of these split proteins (33-35) should drive assembly of a functional adaptor that delivers DAS+4-tagged substrates to ClpXP for degradation. We verified these expectations by incubating a GFP-DAS+4 substrate with ClpXP protease, FKBP12-SspBXB, SspBCORE-FRB, and an ATP regenerating system either in the presence or absence of rapamycin. Importantly, no substantial degradation was observed without rapamycin but robust degradation occurred when rapamycin was present (Fig. 2A).
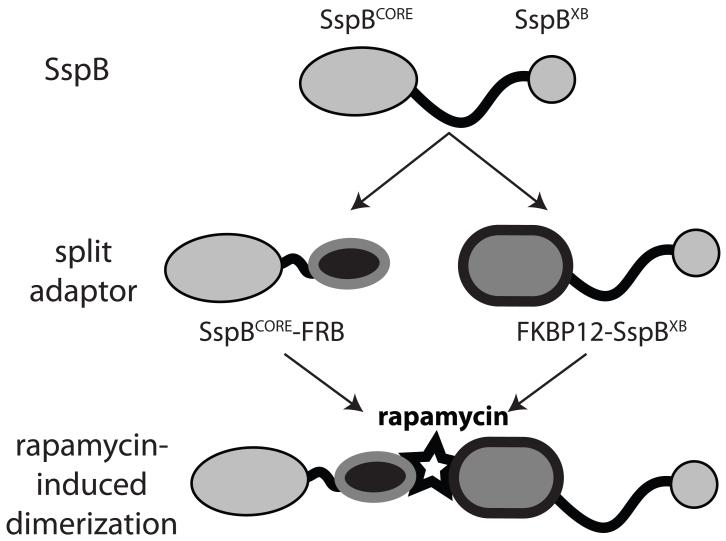
Figure 1. Cartoon of the split adaptor.
The SspB core domain (residues 1-113) was split from the ClpX-binding tail (SspBXB, residues 139-156). SspBCORE was fused to FRB, and FKBP12 was fused to SspBXB. Addition of rapamycin reconstitutes a complete adaptor.
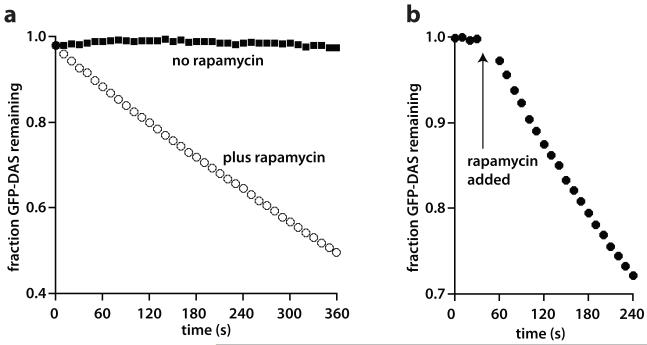
Figure 2. Rapamycin-dependent degradation.
(A) Incubation of GFP-DAS+4 (2 μM), ClpX 6 (0.3 μM), ClpP14 (0.9 μM), FKBP12-SspBXB (5 μM), SspBCORE-FRB (5 μM), and rapamcyin (8 μM) resulted in GFP-DAS+4 degradation at a rate of 0.58 min−1 enzyme−1. No degradation was observed in the absence of rapamycin. (B) Addition of rapamycin resulted in rapid assembly of the degradation complex. GFP-DAS+4 (2 μM), ClpX6 (0.3 μM), ClpP14 (0.9 μM), FKBP12-SspBXB (5 μM), SspBCORE-FRB (5 μM) were preincubated at 30 °C. Rapamycin (8 μM) was added at the time indicated by the arrow. Note that the y-axis does not extend to 0 in both panels.
Many molecules must interact for efficient degradation in our system, raising the possibility that slow assembly could limit the degradation rate. To test this possibility, we preincubated substrate, ClpXP, and the adapter components and monitored the kinetics of GFP-DAS+4 degradation before and after addition of rapamycin (Fig. 2B). After adding rapamycin, steady-state degradation was reached within 20 s, the dead-time of the experiment. Thus, rapamycin-dependent formation of the active proteolysis complex occurs on reasonably fast time scale.
Next, we determined how degradation efficiency varied with the stoichiometry of different components. First, we titrated a mixture containing equal amounts of the two split-adaptor components against a fixed concentration of protease, substrate, and rapamycin. Degradation increased in a roughly hyperbolic fashion (Fig. 3A), with half-maximal stimulation at ~0.5 μM SspBCORE-FRB and FKBP12-XB. Second, we added increasing concentrations of FKBP12-XB to fixed quantities of all other components and observed half-maximal stimulation at 2-4 μM FKBP12-XB (Fig. 3B). Third, we titrated increasing SspBCORE-FRB against fixed concentrations of the other components (Fig. 3C). Degradation increased initially and then was inhibited at high SspBCORE-FRB concentrations (Fig. 3C), probably because free SspBCORE-FRB competes with the limited number of SspBCORE-FRB•rapamycin•FKBP12-XB complexes for substrate binding. Finally, we measured degradation as the substrate concentration was varied in the presence or absence of rapamycin using fixed concentration of protease and the split-adaptor proteins (Fig. 3D). As expected, slow degradation was observed in the absence of rapamycin and robust degradation was observed with rapamycin present. In combination, these results show that many enzyme and adaptor concentrations mediate efficient rapamycin-dependent degradation. For maximum efficiency, however, the ratio of FKBP12-XB to SspBCORE-FRB should be equal to or greater than 1.
Figure 3. Dependence of GFP-DAS+4 degradation on adaptor or substrate concentration.
(A) Degradation in the presence of increasing equimolar concentrations of FKBP12-SspBXB and SspBCORE-FRB. The solid line is a hyperbolic fit with an apparent binding constant of 0.51 μM.
| (1) |
(C) Degradation with a fixed concentration of FKBP12-SspBXB (5 μM) and increasing concentrations of SspBCORE-FRB. Data were fit using equation 1 with parameters: k1 = 1.2 min−1 enz−1, Ka = 1.3 μM, and Kb = 3.9 μM.
(D) Substrate dependence of degradation in the presence of SspBCORE-FRB (5 μM) and FKBP12-SspBXB (5 μM). Fitting the rapamycin data to the Michaelis-Menten equation gave KM = 1.3 μM, VMAX = 0.92 min−1 enz−1. In all panels, the concentrations of ClpX6 and ClpP14 were 0.3 and 0.9 μM, respectively. In panels A-C and the “plus rapamycin” experiment of panel D, the rapamycin concentration was 12 μM. In panels A-C, the GFP-DAS+4 concentration was 2 μM.
Controlled degradation of a transcriptional repressor
Given the promising results in vitro, we developed an assay for rapamycin-dependent degradation in vivo. To prevent uncontrolled substrate delivery and degradation, the sspB gene was deleted from the chromosome of E. coli strain W3110. Next, we introduced a DAS+4 tag at the C-terminus of the lacI transcriptional repressor, using a scar-less λ-RED-based recombination technique, and transformed this strain with a plasmid (pJD427) bearing constitutive promoters driving production of FKBP12-XB and SspBCORE-FRB (36). We expected LacI-DAS+4 to repress lacZ transcription and thus production of β-galactosidase in the absence of rapamycin (Fig. 4A), with addition of rapamycin resulting in LacI-DAS+4 degradation and induction of lacZ expression.
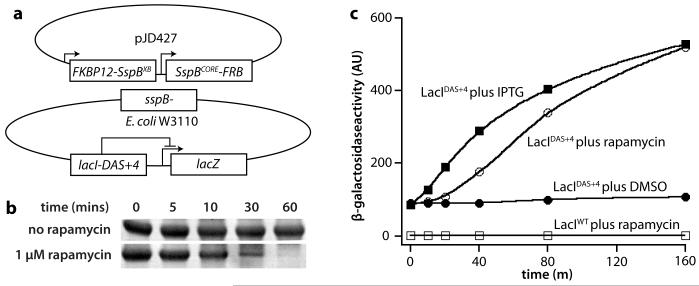
Figure 4. Degradation of LacI-DAS+4.
(A) Assay strains contained an sspB deletion and a DAS+4 tag at the C-terminus of LacI, which represses transcription of lacZ. Production of the split-adaptor components is mediated by plasmid-borne constitutive promoters. (B) Time course of degradation of LacI-DAS+4 (8 μM) by ClpX6 (0.3 μM), ClpP (0.9 μM), SspBCORE-FRB (5 μM), FKBP12-SspBXB (5 μM) and 10 μM rapamycin (when present). (C) β-galactosidase activities were measured in strains containing LacI-DAS+4 following addition of rapamycin (10 μM), IPTG (5 mM), or DMSO. An isogenic strain expressing wild-type LacI showed no response to rapamycin. In the absence of inducer, the LacI-DAS+4 strain exhibited increased basal β-galactosidase activity relative to the wild-type LacI strain. Further experiments indicated that the observed de-repression is not solely due to rapamycin-independent LacI-DAS+4 degradation or SspBCORE-FRB mediated inhibition of LacI activity (37).
In vitro, we observed rapamycin-dependent degradation of LacI-DAS+4 using ClpXP protease, FKBP12-SspBXB, SspBCORE-FRB, and an ATP regenerating system (Fig. 4B). To assay for LacI-DAS+4 degradation in vivo, we grew strain JD704 (W3110 lacI-DAS+4 sspB−/pJD427) in M9 medium at 37 °C to mid-log phase and added either rapamycin (growth of the W3110 parent strain was unchanged from 0 to 50 M rapamycin; not shown), IPTG as a positive control for induction, or an equal volume of DMSO (the solvent for rapamycin and IPTG) as a negative control. At various times, samples were taken and β-galactosidase activity was assayed and normalized to OD600. Addition of either rapamycin or IPTG led to increased β-galactosidase activity but DMSO had no effect (Fig. 4C). Rapamycin-dependent induction of lacZ mRNA levels in the LacI-DAS+4 strain was also observed by RT-qPCR, confirming a direct effect on transcription (data not shown). Compared with IPTG induction, rapamycin induction in the LacI-DAS+4 strain showed a lag and somewhat slower kinetics (Fig. 4C). This lag may result from slow diffusion of the drug into the cell or from relatively slow ClpXP-mediated degradation. We note, however, that rapamycin-dependent induction still occurred on the time scale of many biological processes. Relative to wild-type strains, LacI-DAS+4 strains exhibited elevated β-galactosidase activity, even in the absence of rapamycin. Control experiments revealed that this de-repression resulted from a combination of effects including disruption of the lac operon by introduction of the tag-coding sequence, as well as by adaptor- and rapamycin-independent effects on LacI-DAS+4 repressor activity (37).
Regulated proteolysis of an essential cell-division protein
Regulated proteolysis obviates the need to alter synthesis of a protein of interest. Unlike genetically-encoded knockouts, or techniques reliant on a regulated promoter, our approach leaves the natural regulatory components in place. This could be important for studying genes with complex regulatory patterns, such as those involved in cell-cycle progression or cell division (38-43).
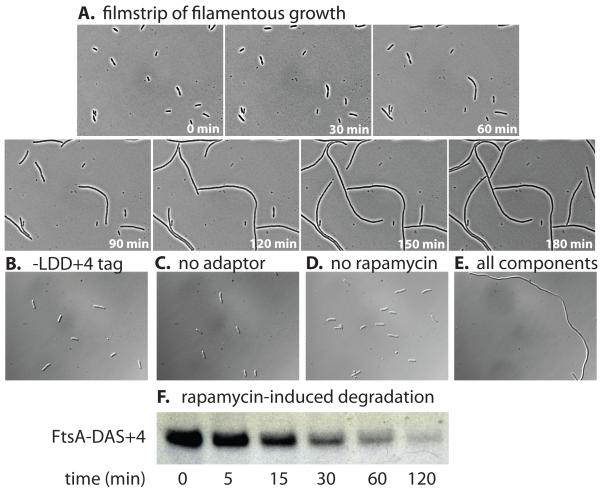
To assay the efficacy of our system in controlling a cell-cycle regulated protein, we appended a degradation tag to the C-terminus of the FtsA cell-division protein. FtsA is a membrane-associated ATP-binding protein that interacts with FtsZ (the Z-ring filament-forming protein) and recruits downstream cell-division proteins (44). Previous studies using depletion strains and temperature-sensitive ftsA mutants have shown that loss of FtsA function results in incomplete septation and long, filamentous cells (45-47). We integrated a sequence encoding a DAS+4 tag at the 3′ end of the chromosomal copy of ftsA in E. coli strain W3110 sspB− and transformed this strain with plasmid pJD427, encoding the split-adaptor proteins. For control experiments, we also constructed a strain in which FtsA had a C-terminal LDD+4 tag, which allows for SspBCORE-FRB binding but does not target proteins to ClpXP (4). After growing this strain in M9 medium, cells were spotted on an M9-agarose pad with 10 M rapamycin, pressed between two microscope slides, and then imaged as a function of time. As time progressed, cells formed long filaments, consistent with efficient proteolysis of FtsA-DAS+4 (Fig. 5A). To ensure that this phenotype depended on degradation, variants were grown in M9 medium to mid-log phase (O.D. 0.7), diluted 30-fold into the same medium with or without 10 μM rapamycin, and grown for an additional 2 h at 37 °C. At this time, cell morphology was observed using phase-contrast microscopy. Only strains grown in the presence of rapamycin, containing pJD427, and harboring a DAS+4 tagged copy of ftsA displayed a filamentation phenotype (Fig. 5B-E). To determine the efficiency of rapamycin-dependent degradation in vivo, we prepared cell lysates at various times after treatment with rapamycin and subjected them to SDS-PAGE and Western blotting using an anti-FtsA antibody (a generous gift of Dr. Miguel Vicente, Centro Nacional de Biotecnología, Madrid, Spain). Although protein synthesis continued, the steady-state level of FtsA decreased rapidly with a half-life of ~15 min (Fig. 5F). These results indicate that degradation is efficient and depends on the small-molecule inducer, the genetically-encoded degradation tag, and the split-adaptor system.
Figure 5. Rapamycin-dependent degradation of an essential cell division protein.
(A) E. coli W3110 sspB- bearing plasmid pJD427 was modified at the ftsA loci by addition of a DAS+4 tag fused to a kanamycin marker (JD784). This strain was grown in medium lacking rapamycin before cells were mounted on a microscope slide bearing LB/agar/rapamycin (10 μM) and imaged every 30 min. (B) E. coli W3110 sspB−/pJD427 with a non-degradable LDD+4 tag fused to FtsA exhibited normal cell morphology when grown in the presence of 10 μM rapamycin. Strain JD784 lacking the adaptor system (C) or grown in medium lacking rapamycin (D) also exhibited normal cell morphology. (E) Addition of both pJD427 and 10 μM rapamycin to strain JD784 resulted in a strong filamentation phenotype. (F) Western blotting shows intracellular degradation of FtsA-DAS+4 after addition of rapamycin (10 μM) in E. coli W3110 sspB−/pJD427. Protein synthesis was not blocked in this experiment. The sample volume in each lane was adjusted to contain the same amount of total cellular protein.
Because degradation depends on rapamycin, we tested if filamentation could be reversed by washing out the small molecule. Thus, we grew cells in M9 medium containing rapamycin to mid-log phase (Fig. 6A,B), harvested cells by centrifugation, washed with rapamycin-free medium, and then resuspended cells and diluted them 40-fold into rapamycin-free M9 medium. After growth for 3 h at 37 °C, cells appeared normal and were not filamentous (Fig. 6C). In principle, this rescue could arise from multiple rounds of division by the filamentous cells or by the death of these cells and growth of a minority of non-filamentous cells in the original culture. To distinguish between these possibilities, we imaged live cells after removal of rapamycin. Strain JD784 (W3110 sspB−, ftsA::ftsA-DAS+4, pJD427) was grown in M9 medium supplemented with rapamycin to OD 0.5. Cells were harvested, washed with medium lacking rapamycin, spotted on an M9-agarose pad, and placed between microscope slides for imaging. Importantly, this experiment revealed the formation of multiple septa throughout individual filamentous cells and consequent division, indicating that removal of rapamycin allowed these cells to accumulate FtsA-DAS+4 and to restart successful cell division within an hour of rapamycin removal (Fig. 6D).
Figure 6. Rapamycin-dependent filamentation is reversible.
(A) JD784 cells exhibit normal morphology before treatment with rapamycin. (B) After growth in the presence of 10 μM rapamycin, strong filamentation was observed. (C) Growth of filamentous cells in rapamycin-free medium results in the appearance of cells with normal morphology. (D) Filamentous cells grown in the presence of rapamycin were harvested and washed to remove rapamycin, immobilized in LB/agar on a microscope, slide and imaged every 10 min using an inverted microscope (the initial image was taken 10-15 min after beginning the washing procedure). Arrows indicate the formation of septa in dividing cells.
Discussion
Adaptor proteins must be capable of binding substrates and also tethering the bound substrate to a protease (Fig. 7A). In the wild-type SspB adaptor, these functions reside in a core substrate-binding domain and an unstructured C-terminal tail, respectively (29-31, 48). The results presented here show that these adaptor functions can reside in separate fusion proteins, with adaptor assembly and function depending on the presence of rapamycin, which mediates dimerization of the FRB and FKBP12 domains of the split components (Fig. 7B). This split-adaptor system allows rapamycin-dependent degradation of appropriately tagged substrates by the wild-type ClpXP protease both in vitro and in vivo. Our results also provide a framework for extending the system. For example, it should be straightforward to change substrate specificity by exchanging the substrate-binding domain from E. coli SspB with a different peptide-binding domain. The modularity of our adaptor design may also facilitate the incorporation of other dimerization domains. Indeed, recent reports have demonstrated spatio-temporal control of protein dimerization using light (49-51). Replacing the FKPB12 and FRB domains in our split adaptor system with such a pair (e.g. PhyB and PIF) could result in a light-induced protein degradation system affording both spatial and temporal control.
Figure 7. Split adaptor systems preserve critical protein-binding interfaces.
(A) Substrate delivery by wild-type SspB requires simultaneous binding between the XB tails of the adaptor and the N-domain of ClpX and between the substrate-binding domain of SspB and a ssrA tag on the substrate. (B) These critical interfaces are preserved in our engineered system but formation of a complete adaptor is under control of rapamycin, a small-molecule inducer of dimerization.
Critically, our split-adaptor system combines the advantages of small-molecule control with utilization of the endogenous ClpXP protease. Thus, there is no requirement for introduction of an exogenous protease, which might degrade non-intended substrates. Moreover, our system was effective over a wide-range of component stoichiometries, simplifying its implementation in vivo. ClpXP is present in many bacteria, and E. coli SspB has been shown to deliver substrates to ClpX homologs from C. crescentus, B. subtilis, and M. tuberculosis (6,10,32). Porting the split-adaptor system to these or other bacterial species should be as simple as introducing a plasmid expressing roughly comparable amounts of the two adaptor components and adding an appropriate degradation tag to the protein target of interest. We found that a split-adaptor system using some parts of C. crescentus SspB and modified C. crescentus degradation tags worked well in vitro, in a manner that was not affected by the presence of E. coli SspB (37). This ability to interchange modular components should prove useful in optimizing split-adaptor systems with altered degradation-tag specificity.
We have shown that modification of the wild-type LacI repressor by addition of a C-terminal DAS+4 tag allows rapamycin-dependent degradation and induction of β-galactosidase expression. The kinetics of β-galactosidase induction using rapamycin were slower than those observed using IPTG, a small molecule that binds directly to LacI repressor and reduces its affinity for operator DNA ~1000-fold (52). Rapamycin may enter cells more slowly than IPTG, or the rate of rapamycin-mediated induction may be limited by other factors, such as the rate of repressor dissociation from the operator or the rate of ClpXP degradation. Importantly, however, similar final levels of β-galactosidase expression were observed for both small-molecule inducers indicating near complete degradation of LacI repressor by our system. Other transcription factors may also be potential targets for rapamycin-dependent induction. For example, we found that modification of the tetracycline repressor (derived from transposon Tn10) with a C-terminal DAS+4 tag resulted in rapamycin-dependent ClpXP degradation in vitro (J.H. Davis, unpublished).
Finally, we have demonstrated that an essential cellular protein, which is subject to complex transcriptional and post-transcriptional regulation can be studied using our proteolysis-based knockdown system. After fusing the DAS+4 tag to the cell-division protein FtsA, we observed pronounced filamentation upon induction of the degradation system using rapamycin. Importantly, filamentation was completely dependent on the presence of the DAS+4 tag, the adaptor system, and rapamycin. Unlike a genetically-encoded knockout, our small-molecule dependent system is reversible. After removing rapamycin from the culture medium, we observed the division of filamentous cells, eventually returning to normal cell morphology.
Rapamycin-dependent assembly of split adaptors has many potential applications. Because this system does not require synthesis of new adaptor proteins (4,6,10), degradation could be initiated at the same time as inhibition of transcription or translation, allowing for faster clearance of target proteins from the cell. Additionally, this system, which relies on the pre-existing pool of adaptors, could be used to directly study proteins involved in transcription and translation. Indeed, Moore et al. (2008) demonstrated that ClpXP could forcefully extract a ribosomal protein from an intact 50S particle in vitro. Split-adaptor degradation systems might allow similar experiments in vivo, helping to elucidate the physiological function of essential genes required for transcription and translation.
METHODS
Plasmids and strains
GFP variants used in this study contained the sequence H6-IDDLG at the N-terminus in addition to the mutations S2R, S65G, S72A, and M78R. The sequences of the DAS+4 (AANDENYSENYADAS) and LDD+4 (AANDENYSENYALDD) tags were appended directly to the C-terminus of substrates.
LacI degradation experiments were performed in E. coli strain W3110 sspB− with the degradation tag (DAS+4) introduced at the C-terminus of LacI repressor as described below. Strains also contained pJD427, which drives constitutive production of the split adaptor components, SspBCORE-FRB from the weak proB promoter and FKBP12-SspBXB from the strong proC promoter (36); these promoter strengths were chosen to increase the chance that FKBP12-SspBXB would be in excess of SspBCORE-FRB, a condition that results in robust delivery in vitro (Fig. 3B, 3C). The pJD427 plasmid bears a medium-copy p15a origin of replication and also expresses chloramphenicol acetyltransferase. For each split-adaptor gene, transcription was terminated using the Bba_B0011 element and translation was initiated using the Bba_B0032 ribosome-binding site (for descriptions of these sequences, see www.partsregistry.org).
FtsA-depletion studies were performed in E. coli W3110 sspB− bearing pJD427. A DAS+4 degradation tag fused to a kanamycin-resistance marker was introduced at the C-terminus of the FtsA coding sequence via λ-RED mediated recombination using primers ftsA-a, ftsA-b (Table 1).
Table 1. Recombineering primers.
Residues underlined are used to form duplexes with the target sequence. Residues in upper-case font are homologous to the appropriate E. coli chromosomal locus.
| sspB-a | AAGCAGAACGTGAAATGCGTCTGGGCCGGAGTTAATCTGTgaattcgcggccgcttctag |
| sspB-b | CATTAAAAAGACAAAACAGGCCGCCTGGGCCTGTTTTGTActgcagcggccgctactagt |
| lacI-a | GCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGgaattcgcggccgcttctag |
| lacI-a | TGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCctgcagcggccgctactagt |
| sspB-ko-1 | AAGCAGAACGTGAAATGCGTCTGGGCCGGAGTTAATCTGTactgatttgttgtgaagtaa |
| sspB-ko-2 | CATTAAAAAGACAAAACAGGCCGCCTGGGCCTGTTTTGTAttacttcacaacaaatcagt |
| lacI-DAS−1 | GCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGgcagctaacgatgaaaacta |
| lacI-DAS−2 | TGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCtcagctagcgtcagcatagt |
| ftsA-a | GTGGATCAAGCGACTCAATAGTTGGCTGCGAAAAGAGTTTgcagctaacgatgaaaacta |
| ftsA-b | CTGTCGCCTGAGGCCGTAATCATCGTCGGCCTCATAAAAAttattagaaaaactcatcga |
Scar-less λ-red mediated chromosomal manipulation
The λ-red-mediated recombineering machinery, which is encoded on the plasmid vector pSIM5, was used to integrate degradation tags into the chromosome and to knock-out sspB (53). Scar-less genomic manipulation was achieved by first introducing a cassette encoding both selectable and counter-selectable markers. Successful recombinants were identified using the selectable marker, the entire cassette was then targeted for replacement, and recombinants were identified using the second counter-selectable marker.
Our targeting cassette contained a constitutive promoter, (Bba_J23116) and ribosome-binding site (Bba_B0032; www.partsregistry.org), used to drive production of our counter-selectable marker, mPheS as well as a kanamycin-resistance marker (Bba_P1003). mPheS, a mutant variant of E. coli phenylalanine tRNA synthetase (mPheS), incorporates ρ-chlorophenylalanine (ρ-Cl-Phe) into cellular tRNA and proteins, and is lethal when cells are grown in the presence of ρ-Cl-Phe (54-57).
The mPheS-kanR cassette was PCR amplified from plasmid pJD141, using primers with 20 bp of homology to the cassette and 40 bp of homology to target either the sspB ORF (primers sspB-a, sspB-b, Table 1) or the 3′ terminus of lacI (primers lacI-a, lacI-b, Table 1). Plasmid DNA was removed by restriction digestion with DpnI, followed by gel purification of the PCR product.
After induction of the λ-red recombination proteins by heat shock for 15 min at 42 °C, PCR products (100 ng) were electroporated into cells, which were then allowed to recover for 6 h at 30 °C, before plating on LB/kanamycin (20 μg/mL) at 30 °C. Successful recombinants exhibited resistance to 20 μg/mL kanamycin and sensitivity to 16 mM ρ-Cl-Phe and were verified by colony-PCR. dsDNA cassettes bearing the desired insertion sequence (flanked by the same 40 bp overhangs described above) were prepared by PCR using primers sspB-ko-1, sspB-ko-2 (generating sspB−) or lacI-DAS−1, lacI-DAS−2 (resulting in lacI-DAS+4). Cassettes were electroporated into cells prepared for recombination, and were allowed to recover at 30 °C for 6 h before plating on YEG-agar/16 mM ρ-Cl-Phe at 30 °C (YEG-agar consists of 0.5% yeast extract, 1% NaCl, 0.4% glucose, 1.5% agar). Successful replacement of the mPheS-kanR cassette was verified by PCR amplification and sequencing of the region of interest. pSIM5 was cured from the cells via serial dilution and growth at 30 °C under non-selective conditions.
Protein purification
E. coli ClpX, E. coli ClpP, E. coli SspB, and GFP-DAS+4 were expressed and purified as described (2,4,58). FKBP12-SspBXB was expressed with an N-terminal thrombin-cleavable His6 tag from a pET28 vector in E. coli BLR (F−, ompT, gal, dcm, lon, hsdSB(rB− mB−), λ(DE3), recA−). Cells were grown at room temperature in 1.5xYT broth (1.3% tryptone, 0.75% yeast extract, and 0.75% NaCl, [pH 7.0]) to OD600 0.7, induced with 1 mM IPTG, and harvested by centrifugation 4 h after induction. Cell pellets were resuspended in LB1 buffer (20 mM HEPES [pH 8.0], 400 mM NaCl, 100 mM KCl, 20 mM imidazole, 10% glycerol, and 10 mM 2-mercaptoethanol) and lysed by addition of 1 mg/mL lysozyme followed by sonication. Benzonase was added to lysates for 30 min prior to centrifugation at 8000 rpm in a Sorvall SA800 rotor. The supernatant was applied to a Ni2+-NTA affinity column (Qiagen), washed with 50 mL of LB1 buffer, and eluted with LB1 buffer supplemented with 190 mM imidazole. Fractions containing FKBP12-SspBXB were pooled and chromatographed on a Sephacryl S-100 gel filtration column in GF-1 buffer (50 mM Tris-HCl [pH 7.6], 1 mM dithiothreitol, 300 mM NaCl, 0.1 mM EDTA, and 10% glycerol). Fractions were analyzed by SDS-PAGE, concentrated to 1 mL, and incubated with thrombin overnight at room temperature. Cleaved FKBP12-SspBXB was purified away from thrombin using a S100 gel-filtration column. Analysis by SDS-PAGE confirmed complete cleavage. FKBP12-SspBXB was concentrated and stored at −80 °C.
SspBCORE-FRB contained an internal His6 tag separating the two domains and was expressed from a pACYC-derived vector in E. coli BLR as described for FKBP12-SspBXB. Harvested cells were resuspended in LB2 buffer (100 mM NaH2PO4 [pH 8.0], 10 mM Tris-HCl, 6 M GuHCl, 300 mM NaCl and 10 mM imidazole) and stored at −80 °C prior to purification. Cells were lysed by rapidly thawing the cell pellet, followed by centrifugation as described above. The supernatant was applied to a Ni2+-NTA column, washed with 50 mL LB2 buffer, and eluted with LB2 buffer supplemented with 240 mM imidazole. After overnight dialysis against GF2 buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 25 mM NaCl, 25 mM KCl, 1 mM dithiothreitol), soluble protein was chromatographed on a Sephacryl S100 gel-filtration column. Fractions were analyzed by SDS-PAGE, concentrated, and stored at −80 °C. LacI-DAS+4 containing a cleavable His6 tag was purified by Ni2+-NTA affinity chromatography, tag cleavage, and gel-filtration chromatography as described above.
β-galactosidase activity assays
To measure β-galactosidase activity, strains were grown in 1 mL of supplemented M9 medium (M9 salts, 1 mM thiamine hydrochloride, 0.2% casamino acids, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glycerol, and 35 μg/mL chloramphenicol when appropriate) at 37 °C in aerated culture vials. At mid-log phase, the OD600 was measured, an aliquot (20 μL) was taken for a Miller assay, and the cells were treated either with rapamycin (10 μM), IPTG (5 mM), or an equal volume of DMSO (the solvent for both IPTG and rapamycin). The final DMSO concentration in each sample was 0.5%. Growth of this strain was unaffected by DMSO concentrations up to 4% (data not shown). At different times after treatment, OD600 was measured (150 μL) in a SpectraMax plate reader (Molecular Devices) and samples (20 μL) were quenched by adding 80 μL Z-lysis buffer (B-PERII (Pierce) supplemented with 200 μg/mL spectinomycin and 1 mM PMSF). One mL of Z-assay buffer (66 mM Na2PO4 [pH 7.4], 6 mM KCl, 700 μM MgCl2, 1 mM DTT, and 0.67 mg/mL o-nitrophenyl β-d-galactoside) was then added to each sample, before aliquoting samples (150 μL) into a 96-well plate and measuring absorbance at 420 nm as a function of time using a SpectraMax plate reader (Molecular Devices). Sample OD600 was corrected for path-length using the equation,
For each sample and time point, β-galactosidase activity was reported as
Degradation assays
Degradation assays in vitro were performed in PD buffer (25 mM HEPES KOH [pH 7.6], 5 mM MgCl2, 10% glycerol, and 200 mM KCl) at 30 °C. GFP fluorescence was monitored by exciting with 467 nm light and measuring emission at 511 nm using a SpectraMax M5 96-well fluorescence plate reader (Molecular Devices) or a spectrofluorimeter (Photon Technology International). Each degradation reaction contained an ATP-regeneration mix, consisting of 4 mM ATP, 16 mM creatine phosphate, and 0.32 mg/mL creatine kinase (58). Degradation reactions of LacI-DAS+4 in vitro were quenched by boiling in GB buffer (50 mM Tris-HCl [pH 6.8], 75 mM dithiothreitol, 35 mM 2-mercaptoethanol, 0.003% bromophenol blue, 0.015% sodium dodecyl sulfate, 8% glycerol) and resolved by SDS-PAGE. Degradation assays in vivo were performed by centrifuging 1 mL of 0.5 OD600 cultures, resuspending the pellet in 50 μL WB (8 M urea, 50 mM Tris-HCL [pH 7.6]), normalizing each sample by total protein content using a Bradford assay, running SDS-PAGE, transferring by electro-blotting to a PVDF membrane, and probing using the polyclonal anti-FtsA antibody, MVM1 (a gift from Dr. Miguel Vicente, Centro Nacional de Biotecnología, Madrid, Spain).
Microscopy
Live cells were spotted onto M9-1.5% agarose pads (supplemented with 10 μM rapamycin when applicable) and imaged using an Axiovert 200 microscope (Zeiss) with a 633/1.4 NA objective (Zeiss) fitted with an objective heater (Bioptechs) maintained at 37 °C.
ACKNOWLEDGEMENTS
We thank the M. Vicente, K. McGinness, C. Tsokos, M. Laub, and the entire Sauer, Baker and Laub labs for helpful discussions, equipment, and reagents. Supported by NIH grant AI-16892.
REFERENCES
- 1.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis JH, Baker TA, Sauer RT. Engineering synthetic adaptors and substrates for controlled ClpXP degradation. J Biol Chem. 2009;284:21848–21855. doi: 10.1074/jbc.M109.017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore SD, Baker TA, Sauer RT. Forced extraction of targeted components from complex macromolecular assemblies. Proc Natl Acad Sci U S A. 2008;105:11685–90. doi: 10.1073/pnas.0805633105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Taxis C, Stier G, Spadaccini R, Knop M. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol Syst Biol. 2009;5:267. doi: 10.1038/msb.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffith KL, Grossman AD. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol Microbiol. 2008;70:1012–1025. doi: 10.1111/j.1365-2958.2008.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto KM, Kim KB, Kumagi A, Mercurio F, Crews CM, Deshais RJ. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stricker J, Cookson S, Bennet MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JH, Wei JR, Wallach JB, Robbins RS, Rubin EJ, Schnappinger D. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic Acids Res. 2011;39:2210–2220. doi: 10.1093/nar/gkq1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 12.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Rappleye CA, Roth JR. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J Bacteriol. 1997;179:5827–5834. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren P, Woodnutt G, Burnham MK, Rosenbergy M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 16.Janse DM, Crosas B, Finley D, Church GM. Localization to the proteasome is sufficient for degradation. J Biol Chem. 2004;279:21415–420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 17.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, et al. Remodeling of the yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 20.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, et al. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crytal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 23.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 24.Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- 26.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci U S A. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 29.Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent tethering of SspB to ClpXP is required for efficient substrate deliver: a protein-design study. Mol Cell. 2004;13:443–449. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- 30.Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- 31.McGinness KE, Bolon DN, Kaganovich M, Baker TA, Sauer RT. Altered tethering of the SspB adaptor to the ClpXP protease causes changes in substrate delivery. J Biol Chem. 2007;282:11465–11473. doi: 10.1074/jbc.M610671200. [DOI] [PubMed] [Google Scholar]
- 32.Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci U S A. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Zheng XF, Brown EJ, Schreiber SJ. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Graffenried CL, Laughlin ST, Kohler JJ, Bertozzi CR. A small-molecule switch for Golgi sulfotransferases. Proc Natl Acad Sci U S A. 101:16715–16720. doi: 10.1073/pnas.0403681101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. Conditional Protein Splicing: A New Tool to Control Protein Structure and Function in Vitro and in Vivo. J Am Chem Soc. 2003;125:10561–10569. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 36.Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011;39:1131–1141. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JH. Ph. D Thesis. Massachusetts Institute of Technology; U.S.A.: 2010. Understanding and harnessing energy-dependent proteolysis for controlled protein degradation in bacteria. [Google Scholar]
- 38.Carballès F, Bertrand C, Bouchè JP, Cam K. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol microbiol. 1999;34:442–450. doi: 10.1046/j.1365-2958.1999.01605.x. [DOI] [PubMed] [Google Scholar]
- 39.Garrido T, Sanchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laub MT. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 41.Sitnikov DM, Schineller JB, Baldwin TO. Control of cell division in Escherichia coli: regulation of transcription of the ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci U S A. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vicente M, Gomez MJ, Ayala JA. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell Mol Life Sci. 1998;54:317–24. doi: 10.1007/s000180050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang XD, Boer P, Rothfield LI. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–93. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hale CA, de Boer PAJ. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 47.Walker JR, Kovarik A, Allen JS, Gustafson RA. Regulation of bacterial cell division: temperature-sensitive mutants of the Escherichia coli that are defective in septum formation. J Bacteriol. 1975;123:693–703. doi: 10.1128/jb.123.2.693-703.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 50.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signaling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 52.Riggs AD, Newby RF, Bourgeois S. Lac Repressor-operator interaction II: Effect of galactosides and other ligands. J Mol Biol. 1970;51:303–314. doi: 10.1016/0022-2836(70)90144-0. [DOI] [PubMed] [Google Scholar]
- 53.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 54.Cox MM, Layton SL, Jiang T, Cole K, Hargis BM, Berghman, et al. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotechnol. 2007;7:59. doi: 10.1186/1472-6750-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffith KL, Fitzpatrick MM, Keen EF, Wolf RE. Two functions of the C-terminal domain of Escherichia coli Rob: mediating “sequestration-dispersal” as a novel off-on switch for regulating Rob’s activity as a transcription activator and preventing degradation of Rob by Lon protease. J Mol Biol. 2009;338:415–430. doi: 10.1016/j.jmb.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kast P. pKSS—A second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene. 1994;138:109–114. doi: 10.1016/0378-1119(94)90790-0. [DOI] [PubMed] [Google Scholar]
- 57.Lalioti M, Heath J. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 2001;29:E14. doi: 10.1093/nar/29.3.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin Y, Davis JH, Brau RR, Martin A, Kenniston JA, Baker TA, Sauer RT, Lang MJ. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc Natl Acad Sci U S A. 2009;106:19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]