Abstract
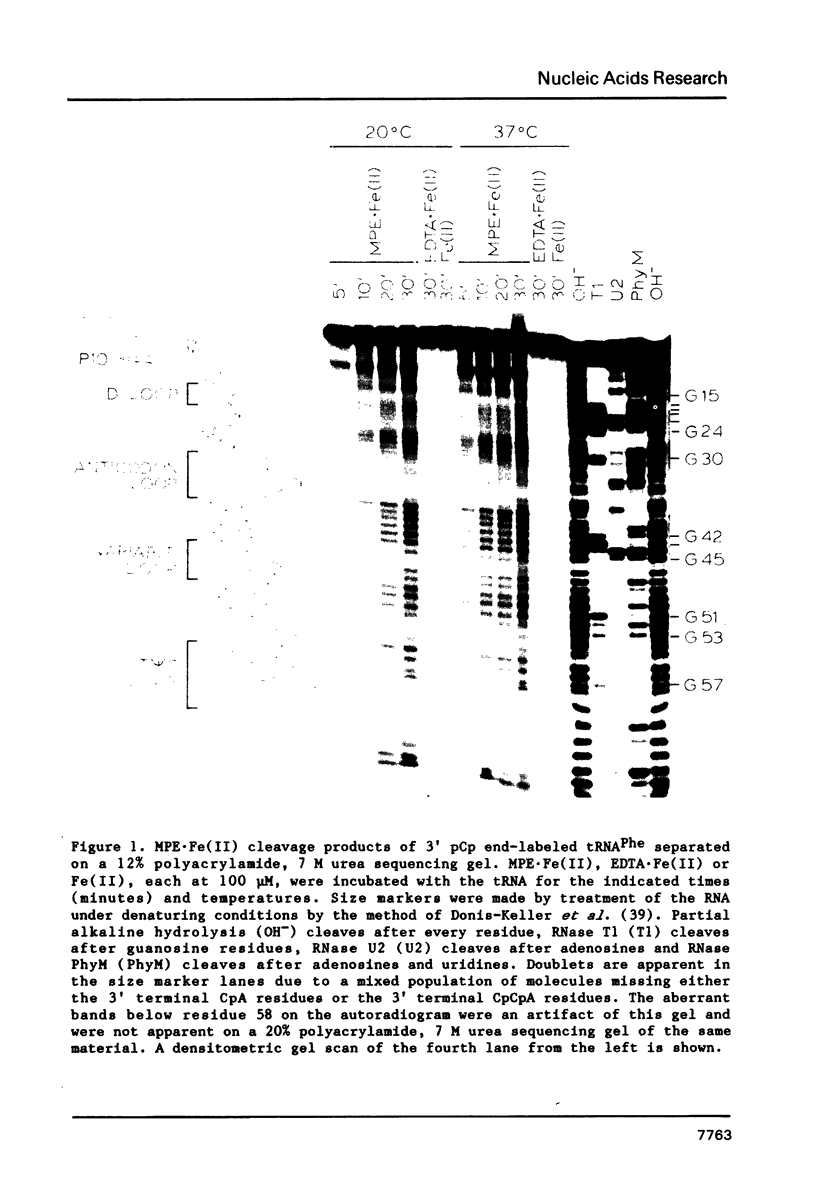
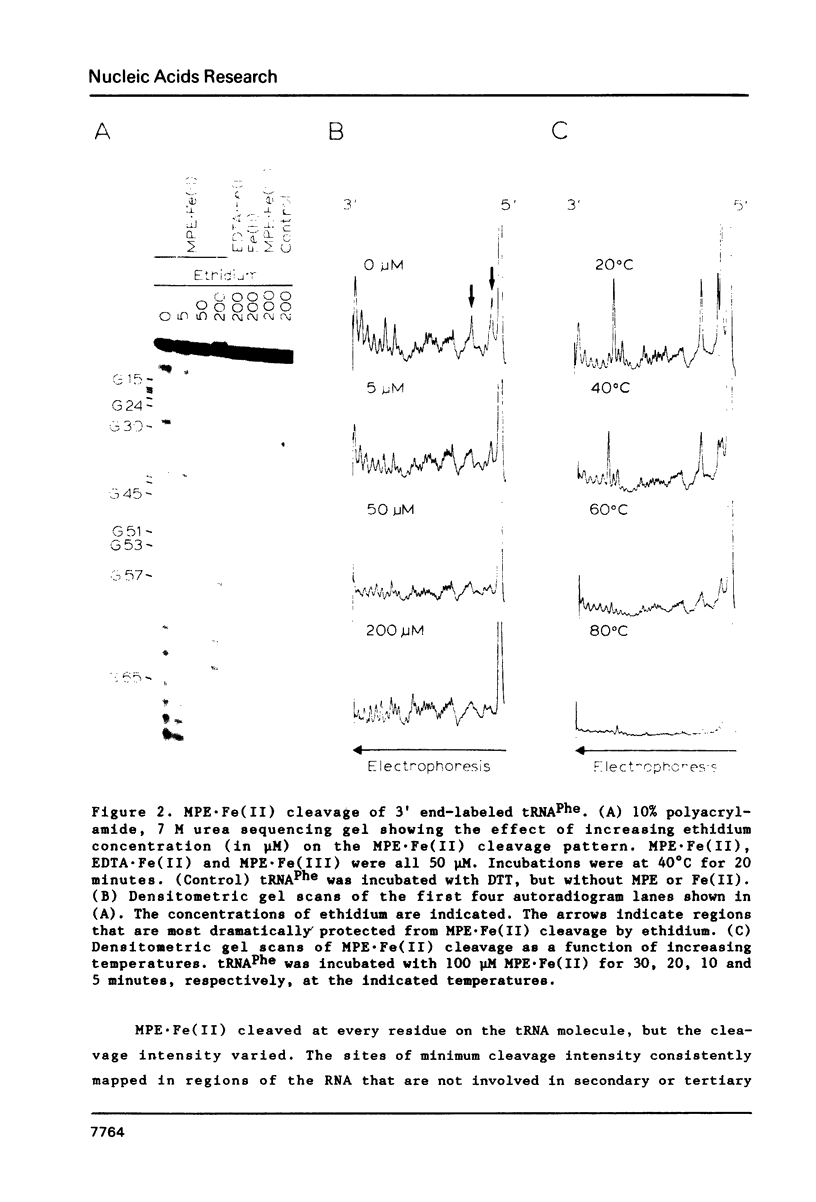
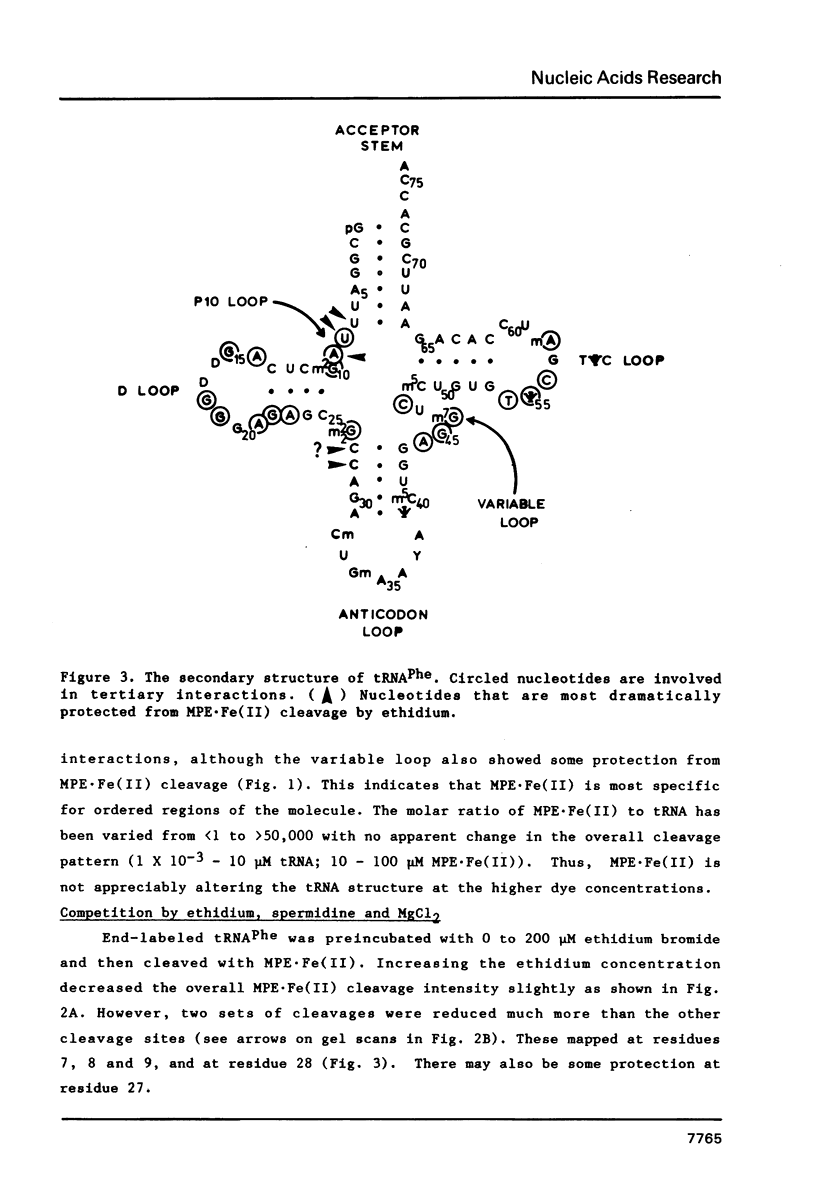
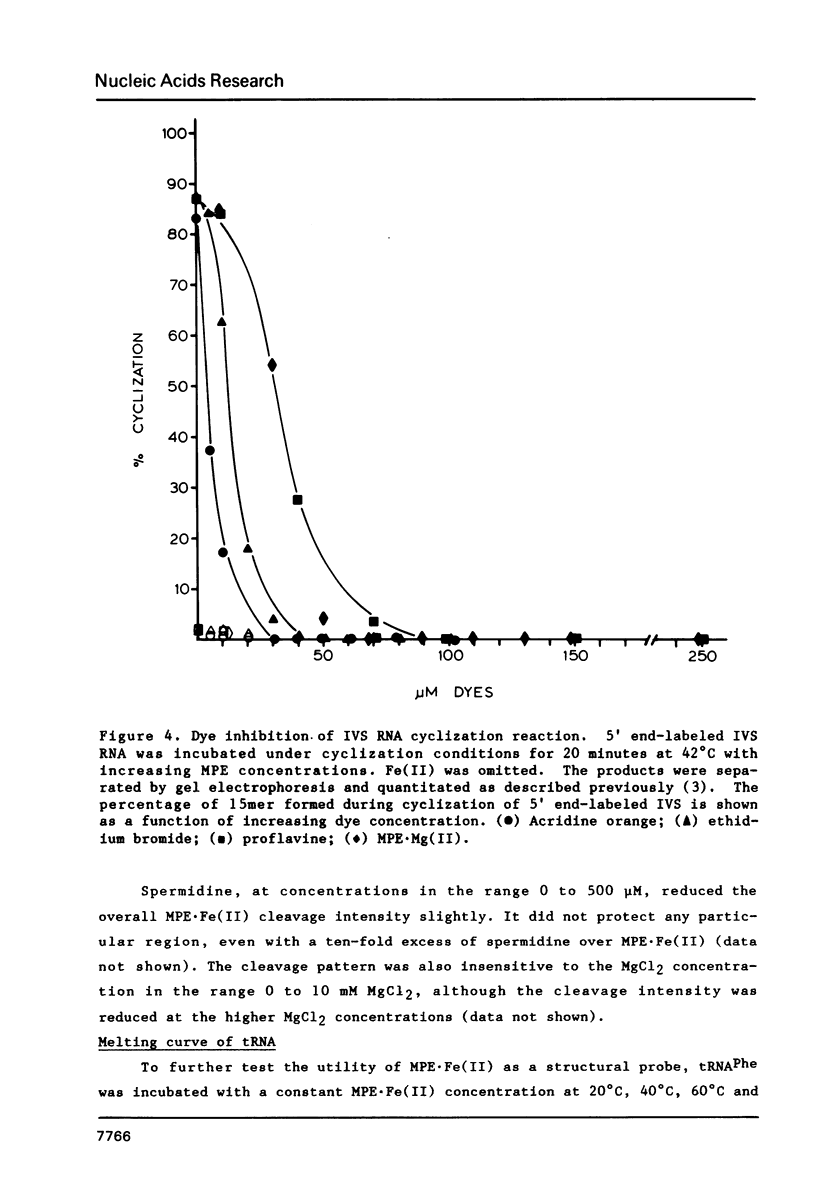
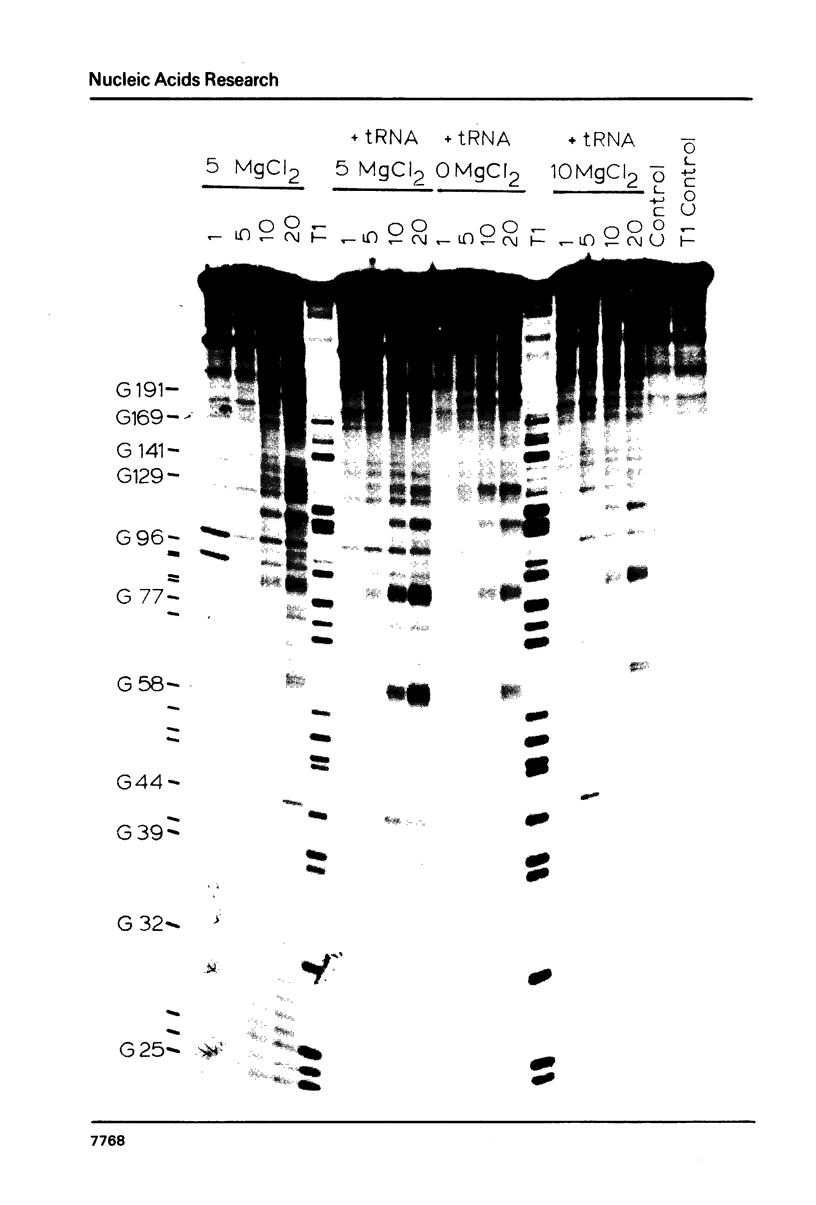
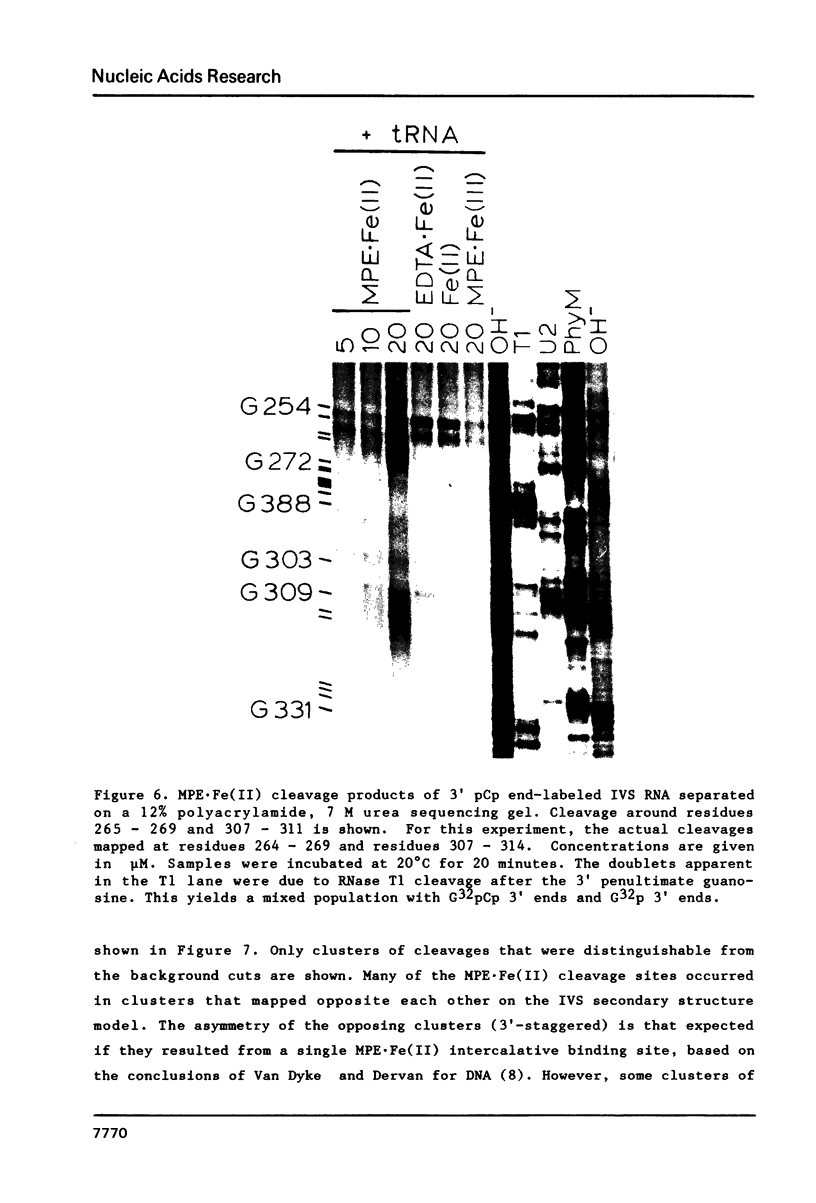
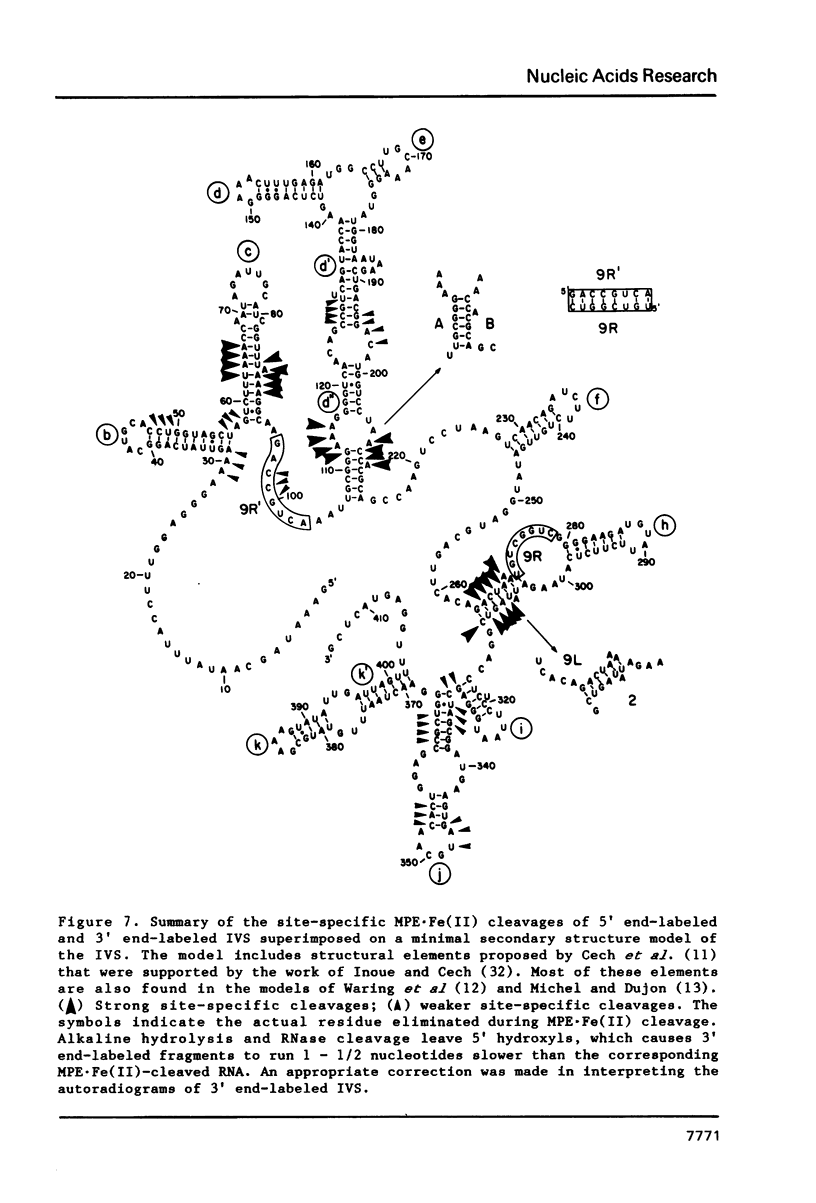
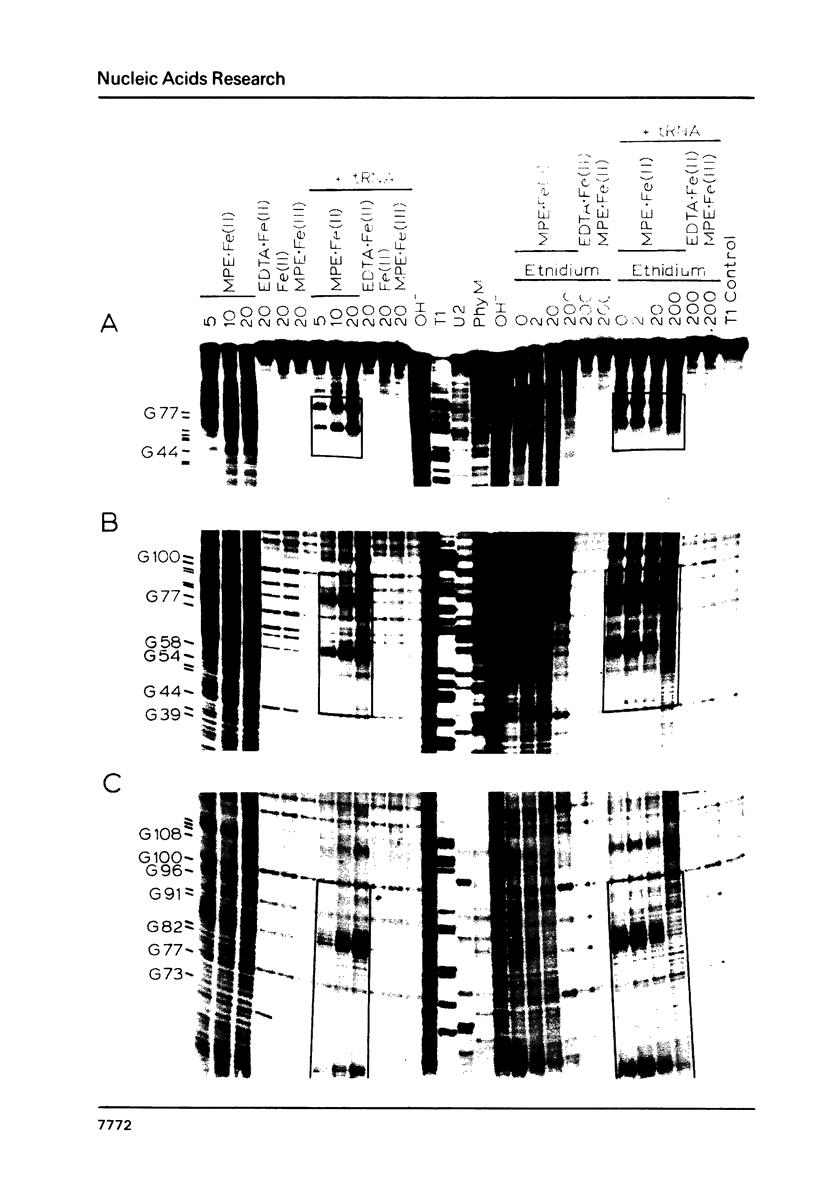
The intervening sequence (IVS) excised from the rRNA precursor of Tetrahymena thermophila is converted to a covalently closed circular RNA in the absence of proteins in vitro. This self-catalyzed cyclization reaction is inhibited by the intercalating dye methidiumpropyl.EDTA (MPE; R.P. Hertzberg and P.B. Dervan (1982) J. Am. Chem. Soc. 104, 313-315). The MPE binding sites have been localized by mapping the sites of MPE.Fe(II) cleavage of the IVS RNA. There are three major binding sites within the 414 nucleotide IVS RNA. Two of these sites coincide with the A.B and 9L.2 pairings. These are structural elements that are conserved in all group I introns and are implicated as being functionally important for splicing. We propose that interaction of MPE with these sites is responsible for dye inhibition of cyclization. The reactions of MPE.Fe(II) with an RNA of known structure, tRNAPhe, and with the IVS RNA were studied as a function of temperature, ionic strength and ethidium concentration. Based on the comparison of the reaction with these two RNAs, we conclude that the dye is a very useful probe for structural regions of large RNAs, while it provides more limited structural information about the small, compact tRNA molecule.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Cech T. R. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell. 1984 Sep;38(2):501–510. doi: 10.1016/0092-8674(84)90505-1. [DOI] [PubMed] [Google Scholar]
- Helfgott D. C., Kallenbach N. R. Increased binding of ethidium bromide to polynucleotide duplexes containing mismatched bases. Nucleic Acids Res. 1979 Oct 25;7(4):1011–1017. doi: 10.1093/nar/7.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. R., Bolton P. H., Kearns D. R. Ethidium bromide binding to transfer RNA: transfer RNA as a model system for studying drug-RNA interactions. Biochemistry. 1978 Feb 21;17(4):601–607. doi: 10.1021/bi00597a007. [DOI] [PubMed] [Google Scholar]
- Jones C. R., Kearns D. R. Identification of a unique ethidium bromide binding site on yeast tRNAPhe by high resolution (300 MHz) nuclear magnetic resonance. Biochemistry. 1975 Jun 17;14(12):2660–2665. doi: 10.1021/bi00683a016. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Tinoco I., Jr Mutagen--oligonucleotide complexes with a bulged base as models for frameshift mutation. Nature. 1978 Aug 10;274(5671):609–610. doi: 10.1038/274609a0. [DOI] [PubMed] [Google Scholar]
- Liebman M., Rubin J., Sundaralingam M. Nonintercalative binding of ethidium bromide to nucleic acids: crystal structure of an ethidium--tRNA molecular complex. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4821–4825. doi: 10.1073/pnas.74.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Price J. V., Cech T. R. Coupling of Tetrahymena ribosomal RNA splicing to beta-galactosidase expression in Escherichia coli. Science. 1985 May 10;228(4700):719–722. doi: 10.1126/science.2986286. [DOI] [PubMed] [Google Scholar]
- Price J. V., Kieft G. L., Kent J. R., Sievers E. L., Cech T. R. Sequence requirements for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA. Nucleic Acids Res. 1985 Mar 25;13(6):1871–1889. doi: 10.1093/nar/13.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P. L., Filimonov V. V. Thermodynamic analysis of transfer RNA unfolding. J Mol Biol. 1978 Jul 15;122(4):447–464. doi: 10.1016/0022-2836(78)90421-7. [DOI] [PubMed] [Google Scholar]
- Sakai T. T., Cohen S. S. Effects of polyamines on the structure and reactivity of tRNA. Prog Nucleic Acid Res Mol Biol. 1976;17:15–42. doi: 10.1016/s0079-6603(08)60064-1. [DOI] [PubMed] [Google Scholar]
- Sakai T. T., Torget R., I J., Freda C. E., Cohen S. S. The binding of polyamines and of ethidium bromide to tRNA. Nucleic Acids Res. 1975 Jul;2(7):1005–1022. doi: 10.1093/nar/2.7.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M., Sakore T. D., Jain S. C., Banerjee A., Bhandary K. K., Reddy B. S., Lozansky E. D. beta-kinked DNA--a structure that gives rise to drug intercalation and DNA breathing--and its wider significance in determining the premelting and melting behavior of DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):293–314. doi: 10.1101/sqb.1983.047.01.035. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Luehrsen K. R., Woese C. R., Pace N. R. An unusual 5S rRNA, from Sulfolobus acidocaldarius, and its implications for a general 5S rRNA structure. Nucleic Acids Res. 1981 Nov 25;9(22):6129–6137. doi: 10.1093/nar/9.22.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Hearst J. E. Structure of E. coli 16S RNA elucidated by psoralen crosslinking. Cell. 1983 Apr;32(4):1355–1365. doi: 10.1016/0092-8674(83)90316-1. [DOI] [PubMed] [Google Scholar]
- Urbanke C., Römer R., Maass G. The binding of ethidium bromide to different conformations of tRNA. Unfolding of tertiary structure. Eur J Biochem. 1973 Mar 15;33(3):511–516. doi: 10.1111/j.1432-1033.1973.tb02710.x. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Footprinting with MPE.Fe(II). Complementary-strand analyses of distamycin- and actinomycin-binding sites on heterogeneous DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):347–353. doi: 10.1101/sqb.1983.047.01.040. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNA structure analysis using methidiumpropyl-EDTA.Fe(II): a base-pair-specific RNA structure probe. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6978–6982. doi: 10.1073/pnas.81.22.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W., Scazzocchio C., Brown T. A. Internal structure of a mitochondrial intron of Aspergillus nidulans. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6332–6336. doi: 10.1073/pnas.79.20.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Scazzocchio C., Brown T. A., Davies R. W. Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J Mol Biol. 1983 Jul 5;167(3):595–605. doi: 10.1016/s0022-2836(83)80100-4. [DOI] [PubMed] [Google Scholar]
- Wells B. D., Cantor C. R. A strong ethidium binding site in the acceptor stem of most or all transfer RNAs. Nucleic Acids Res. 1977;4(5):1667–1680. doi: 10.1093/nar/4.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Grabowski P. J., Cech T. R. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983 Feb 17;301(5901):578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- de Bruijn M. H., Klug A. A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire 'dihydrouridine' loop and stem. EMBO J. 1983;2(8):1309–1321. doi: 10.1002/j.1460-2075.1983.tb01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]