Abstract
The use of microarray technology to measure gene expression on a genome-wide scale has been well established for more than a decade. Methods to process and analyse the vast quantity of expression data generated by a typical microarray experiment are similarly well-established. The Affymetrix Exon 1.0 ST array is a relatively new type of array, which has the capability to assess expression at the individual exon level. This allows a more comprehensive analysis of the transcriptome, and in particular enables the study of alternative splicing, a gene regulation mechanism important in both normal conditions and in diseases. Some aspects of exon array data analysis are shared with those for standard gene expression data but others present new challenges that have required development of novel tools. Here, I will introduce the exon array and present a detailed example tutorial for analysis of data generated using this platform.
Keywords: exon array, affymetrix, alternative splicing, microarray, gene expression
INTRODUCTION
Affymetrix have recently developed ‘whole transcript’ arrays, which are fundamentally different to their traditional 3′ expression arrays [1]. Probes are designed along the entire length of the gene as opposed to just the 3′-end. Consequently, data from all parts of the gene are available which avoids the need to infer expression of the entire gene based on measurements made at the 3′-end. As well as giving a better overall estimate of gene expression, it has the added advantage so that expression of individual exons can also be estimated.
The exon array consists of ∼1.4 million probesets and >5 million individual probes. Each probeset comprises four individual probes and usually corresponds to a single exon (longer exons may have more than one probeset designed to them). Again, there are differences to the design of traditional 3′ expression arrays, which have 11 probes per probeset, and make use of ‘mismatch probes’, with the central base mutated and intended to measure non-specific hybridization. The exon array contains only perfect match probes and non-specific hybridization is measured through two sets of negative control probes; one set, referred to as ‘antigenomic background probes’, is based on sequences having no match in the human, mouse or rat genomes. The second set, termed genomic background probes, are based on probes designed to Genscan Suboptimal exon predictions (unlikely to be transcribed) from an old version of the human genome (NCBI Build 31). In each set, there are generally 1000 probes per GC-content count (0–25 bases in the probe). This allows a GC-based background correction to be performed by matching the background signal to the GC content of each experimental probe, which is important as the hybridization signal observed from control probes tends to increase with GC content.
Probesets targeting individual exons can be further grouped into ‘transcript clusters’ and gene-level expression estimates can be obtained by considering all probes comprising a transcript cluster. The content of the exon array includes well-characterized genes as well as more speculative regions of transcription. Thus, it can potentially provide evidence for novel transcripts in more exploratory experimental designs. Depending on the level of evidence supporting the existence of a particular exon, the probesets are categorized as ‘core’ (annotated by RefSeq), ‘extended’ (mRNA evidence) or ‘full’ (bioinformatic prediction). The list of extended probesets comprises those annotated as ‘core’ or ‘extended’ and the list of full probesets includes ‘core’, ‘extended’ and ‘full’. The analysis at exon or gene level can be restricted to any level of evidence as appropriate for the study.
While the exon array can be used to analyse differential gene expressions in exactly the same way as traditional Affymetrix arrays, it has the advantage of simultaneously providing data at the exon level. Exon-level data can be used to investigate splicing events, which determine how individual exons are joined together to form a mature mRNA transcript. Many genes exist as different isoforms, where the exons at the gene locus are combined in different ways to generate multiple forms of the gene. For example, a cassette exon can give rise to two possible transcripts—one form including the exon and the other where it is ‘spliced out’ or skipped. There are many variations of splicing events and they can be combined together to generate a diverse set of transcripts. Alternative usage of 3′ sites, in particular, has implications for the earlier 3′ array design as shorter isoforms of a gene may not be detected, resulting in an incomplete estimate of gene expression.
Splicing is a highly regulated process and the idea that aberrant splicing could underlie many diseases is a very active area of research. The ability to investigate splicing on a genome-wide scale using the exon array is an exciting prospect. However, as is often the case with such developments, new challenges in terms of data analysis and interpretation can also arise. I will demonstrate the use of Affymetrix power tools (APT) [2] and R statistical software [3] to process and analyse data from the exon array platform. In particular, I will focus on data processing and filtering steps necessary before running a splicing analysis and briefly discuss ways to visualize and interpret the results. It is important to note that the methods presented here are just one way of approaching exon array data and many other tools and software packages are available or are under development in this fast-moving field.
Tools for exon array data analysis
APT is a suite of tools developed by Affymetrix for processing and analysing data from any GeneChip® array and can be obtained from http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx#1_2.
Standard processing of exon array data can be performed in APT with basic command line knowledge but there is also scope for more advanced users to adjust various parameters as well.
The R Statistical Software Package is a powerful, open-source environment for statistical data analysis and visualisation. It is available from http://www.r-project.org/.
To demonstrate the use of APT and R to analyse exon array data, I will use a publicly available data set that used this platform to investigate splicing in high and low hypoxia cancer samples [4]. The data are available from the Gene expression omnibus (GEO) [5] website http://www.ncbi.nlm.nih.gov/geo/ by searching for the data set accession number (GSE18300)—the record gives full information on the study and access to the raw data files, which can be downloaded and used to try out the methods presented. APT has a web forum for users and there is extensive documentation as well as a mailing list for R—these are very useful places to search if encountering any problems using the software.
Processing exon array data using APT
As with other Affymetrix arrays, raw signal intensity data are provided in .CEL files, each containing probe-level intensities from a single array (sample). These files can be processed in APT (as well as other software packages) to generate exon- and gene-level intensity estimates. As described above, probesets on the exon array consist of four individual probes and usually target a particular exon of a particular gene. Thus, exon-level intensity estimates correspond to the probeset-level estimates. Probesets are further grouped into transcript clusters enabling a gene-level estimate to be computed by summarizing data from all probes within the transcript cluster. These two values—the exon- and gene-level signal intensity estimates—form the basis of splicing analysis (discussed in more detail below). First, we present methods to process the CEL files to generate normalized expression data using APT and further processing in R.
The ‘apt-probeset-summarize’ command reads in raw CEL files and generates normalized exon- or gene-level signal intensities, depending on the specified arguments [6]. To run the command, a number of files providing information on array design, known as library files, are required. Affymetrix provide up-to-date versions of supporting files (library files, annotation files, etc.) for each type of array they manufacture.
The relevant files can be obtained from the Affymetrix website (www.affymetrix.com) after free registration for a username and password. At the time of writing, the library files for the Human Exon 1.0 ST array could be downloaded as a zip file by locating the array name in the list of products on the support page and checking the ‘library files’ box. However, note that the organization of the website and/or exact filenames may change over time. Click the ‘Human Exon 1.0 ST Array Analysis’ link to download the zip file. Unzip and check that the following files are among those available:
HuEx-1_0-st-v2.r2.pgf
HuEx-1_0-st-v2.r2.clf
HuEx-1_0-st-v2.r2.antigenomic.bgp
HuEx-1_0-st-v2.r2.genomic.bgp
HuEx-1_0-st-v2.r2.qcc
The probe group file (.pgf) and cel layout file (.clf) specify which probes belong to a given probeset and their location in the cel file respectively. These two files are used in place of the chip description file (CDF) provided for other Affymetrix arrays. The .bgp files contain information on background control probes and the .qcc file has details of all control probes. The library file package also includes a file named HuEx-1_0-st-v2.r2.all.ps, which, when specified in the analysis, produces summary estimates for all experimental probesets on the array. However, to perform a gene-level analysis, or to restrict the analysis to a certain annotation level (core, extended or full), additional files are needed. These can also be obtained from the library files page, under the ‘Human Exon 1.0 ST Array Probeset and Meta Probeset Files’ link.
The following six files should now be available:
HuEx-1_0-st-v2.r2.core.ps; HuEx-1_0-st-v2.r2.extended.ps; HuEx-1_0-st-v2.r2.full.ps
HuEx-1_0-st-v2.r2.core.mps; HuEx-1_0-st-v2.r2.extended.mps; HuEx-1_0-st-v2.r2.full.mps
Exon-level analysis
To process the data at the exon (or probeset) level, one of the .ps files should be specified; these simply list the probeset IDs annotated at the chosen level and the output contains summary estimates for the specified probesets only. Various summary methods are available within APT, including the RMA (Robust multi-array average) [7] and PLIER (Probe logarithmic intensity error) [8] algorithms, which are both widely accepted methods for processing Affymetrix microarray data. The processing steps include background correction, normalization and probeset summarization. RMA and PLIER are both model-based methods, which aim to generate robust signal estimates by down-weighing poorly performing probes. For a variety of reasons, poor probes would usually have low signal relative to others in the probeset. Although alternatively spliced exons could also have low signal (and therefore be indistinguishable from a poor probe) RMA and PLIER should be robust to this, specifically when splicing is limited to a small proportion of exons within the gene.
The following APT command can be used to generate exon-level intensity estimates for core probesets using RMA:
> apt-probeset-summarize -a rma-sketch –p HuEx-1_0-st-v2.r2.pgf -c HuEx-1_0-st-v2.r2.clf -s HuEx-1_0-st-v2.r2.core.ps – qc-probesets HuEx-1_0-st-v2.r2.qcc -o OUT_EXON *.CEL
A detailed description of each argument and notes can be found in Table 1.
Table 1:
Using Affymetrix power tools to process raw data from the exon array and generate exon or gene-level signal estimates
| Example argument to apt-probeset- summarize command | Notes |
|---|---|
| -a rma-sketch | Specifies the analysis to be performed. ‘rma-sketch’ (RMA using a subset of probes for memory efficiency) is one of the standard options. Other options, such as plier-sketch and plier-gcbg-sketch (incorporating a gc-based background correction) can be specified instead. It is possible to perform multiple analyses simultaneously by including more than one -a argument. |
| –p HuEx-1_0-st-v2.r2.pgf -c HuEx-1_0-st-v2.r2.clf | Specify the library files, which give information on probeset groups and the array layout. The –p *.pgf and –c *.clf arguments can be replaced with -d *.cdf if the user wishes to use a custom CDF with alternative probeset definitions [24]. |
| -s HuEx-1_0-st-v2.r2.core.ps | If a .ps file is specified with the –s argument, an exon-level analysis will be performed. To run a gene-level analysis, a .mps file is specified with the –m argument instead. In either case, analysis can be restricted to probes annotated as core, extended or full by specifying the relevant file. |
| --qc-probesets HuEx-1_0-st-v2.r2.qcc | Specify the .qcc file to process the control probesets on the array and check quality of data. |
| -o output_exon | Specify a directory to write the output files with the –o argument. This will be created in the current working directory unless a path to another location is given. Output files are named according to the analysis method (e.g. rma-sketch.summary.txt) and are given the same name for both exon and gene-level analyses; they will be over-written if another analysis is run with the same output folder specified. Therefore, it is useful to write files to a new folder for each analysis. It is also helpful to re-name immediately with an informative name to include the dataset, analysis method and whether it is an exon or gene-level analysis to enable easy identification of the data in the file. |
| *.CEL | Specify the .CEL files to be processed. If they are contained in the current working directory, *.CEL will suffice, but a path to the files can be given if required. There will be some differences between Windows and Linux systems regarding syntax for path names and use of the wildcard (*) character—Windows users will need to specify each .CEL file to be analysed individually. |
Gene-level analysis
To generate gene-level signal intensity estimates, one of the *.mps files is specified in place of the *.ps file.
> apt-probeset-summarize –a rma-sketch –p HuEx-1_0-st-v2.r2.pgf -c HuEx-1_0-st-v2.r2.clf -m HuEx-1_0-st-v2.r2.core.mps –qc-probesets HuEx-1_0-st-v2.r2.qcc-o OUT_GENE *.CEL
Each *.mps file defines which probesets are associated with each transcript cluster. Only probesets classified as mapping uniquely to the genome are listed, to avoid including signal from potentially cross-hybridizing probes in the computation of the gene-level signal estimate. It is worth noting that, in the gene level, output from apt-probeset-summarize, the column containing transcript cluster IDs is still named probeset_id, but is not to be confused with the exon level probeset_id. One decision regarding the analysis workflow is whether to use core, extended or full probesets for the calculation of exon/gene intensities. The more speculative content of the array tends to introduce a lot of low intensity noise from probes designed to regions that may not be transcribed. Restricting to core probesets should generate more reliable signal for the well-characterized content and is particularly recommended for the gene-level estimate.
Quality control of exon array data
The qcc argument to apt-probeset-summarize results in the generation of a file containing summary measures for each array, which can be assessed to check the quality of the data prior to analysis. As with other types of microarray data, deciding whether to exclude a sample as an outlier is dependent on a number of factors; however, any sample behaving differently to others in the experiment should be considered carefully, specifically if flagged by multiple quality control (QC) measures. Full details on the various metrics and control probes available on the exon array and their interpretation is given in the Affymetrix white paper on quality assessment [9]. To give a couple of examples, the pm_mean value is the mean raw intensity for all probes on the array—unusually bright or dim samples (high or low pm_mean values respectively) should be handled by normalization but this should be checked with probeset-level metrics as well. The mean absolute deviation (MAD) of the residuals for each chip (from the RMA or PLIER model fit) compared to the median for all chips is an useful metric for most studies. An unusually high value for this metric can suggest a problem with the given chip. Other useful functions for assessing chip quality, including identification of spatial artefacts, are available in the ‘affyPLM’ package [10] from BioConductor [11] (note that the CEL files will need to be processed directly in R to create the required objects).
If the number of samples is small, the data in the QC output file can be inspected by eye or otherwise easily plotted for visualization in R:
> qc <- read.table("./OUT_EXON/rma-sketch_core_exon.report.txt", sep="\t", header=T)
> plot(1:10, qc$pm_mean, ylim=c(0,1000), xlab="Array", ylab="Signal Intensity", main="Average Raw Intensity Signal")
> plot(1:10, qc$all_probeset_mad_residual_mean, ylim=c(0,0.3), xlab="Array", ylab="Mean absolute deviation", main="Deviation of Residuals from Median")
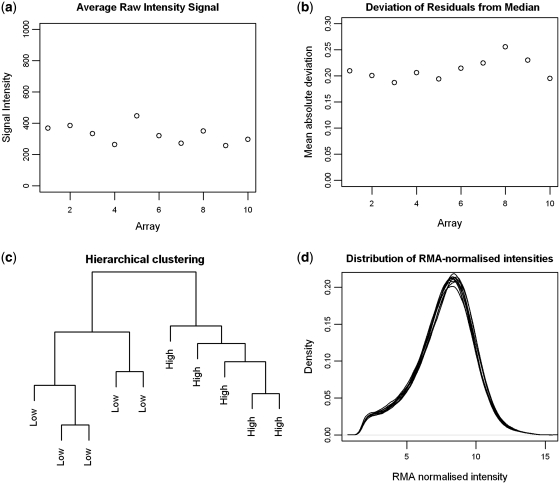
Chip 8 has the highest MAD value (Figure 1) but as it does not seem excessively high we would not exclude this array unless further checks gave more cause for concern. It is usually preferable to keep arrays unless there is very good reason to exclude them, as it reduces sample size, which is typically small in the first place.
Figure 1:
Example quality control plots for the 10 arrays in the example data set (GSE18300) showing (a) average raw signal intensity; (b) mean residual deviation; (c) hierarchical clustering and (d) distribution of normalized intensities.
Visualizing the normalized data using hierarchical clustering or density plots, e.g., can also be helpful to check for outlier samples.
> d.exon <- read.table("./OUT_EXON/rma-sketch_core_exon.summary.txt", sep="\t", header=T, row.names=1)
> d.t <- dist(t(d.exon))
> plot(hclust(d.t), main="Hierarchical clustering", labels=c(rep("Low", 5), rep("High", 5)))
> plot(density(d.exon[,1]), main="Distribution of RMA-normalised intensities", xlab="RMA normalised intensity")
> for(i in 2:ncol(d.exon)) {lines (density(d.exon[,i]))}
The output from these plots is shown in Figure 1 and suggests high-quality data from all arrays. Furthermore, the low and high hypoxia samples cluster into two groups, suggesting substantial differential expression at the gene and/or exon level.
At this point, it is useful to run another APT command to generate detection of P-values for each probeset. ‘Detected above background’ or DABG can be run as an analysis option to apt-probeset-summarize:
> apt-probeset-summarize -a dabg -p HuEx-1_0-st-v2.r2.pgf -c HuEx-1_0-st-v2.r2.clf -b HuEx-1_0-st-v2.r2.antigenomic.bgp-o ./OUT_DABG *.CEL
The signal intensity estimate obtained for each probeset is compared to the signal distribution from the set of anti-genomic probes (negative controls). The degree of overlap is used to compute a detection P-value, with P < 0.05, the usual threshold to consider a probeset detected. The matrix of detection P-values is written to a file called ‘dabg.summary.txt’ by default. This will be used later to filter for detected exons and genes.
Once the data have been processed to generate exon- and gene-level intensity estimates, it is useful to import the data into the R statistical package for further processing (in particular, filtering) before any assessment of splicing. The exon-level data have already been read into R for the quality checks, so, now the gene-level data is needed:
> d.gene <- read.table("./OUT_GENE/rma-sketch.summary_core_gene.txt", sep="\t", header=T, row.names=1)
Signal intensity values from microarray experiments are usually log-transformed to make the data more appropriate for statistical analysis. The signal intensity estimates will already be on log2 scale if RMA was used to process the data but on the natural scale if PLIER was used. Since PLIER estimates can be close to zero, it is usual to add a small constant (e.g. 16) to all values to stabilize the variance, prior to log2 transforming the data.
## if processed with PLIER
> d.exon <- log2(d.exon+16)
> d.gene <- log2(d.gene+16)
The next step is to filter the data, which is critical in the case of exon array data to reduce the false-positive rate in the identification of potential splicing events [12]. The two major types of filter aim to remove probes with unusually low or high signal, which could be artefacts mistaken for splicing events. However, it is possible that stringent filtering will also remove some true splicing events (e.g. low signal intensity due to a poorly performing probeset is indistinguishable from the low signal due to a skipped exon). Thus, there is a balance between reducing the false-positive rate, while keeping the false-negative rate low as well. In practical terms, it is better to focus on reducing the false-positive rate to increase confidence in the splicing events that are identified. This maximizes the chance of successful validation (often required to confirm findings suggested by microarray data), which can be expensive and time-consuming.
Specific filtering steps recommended include:
Restrict analysis to core probesets
Remove undetected probesets
Remove potentially cross-hybridizing probesets
Remove genes undetected in both groups of samples
Each of these scenarios is discussed below, together with sample R code to perform the suggested filtering. The dimensions of various objects are given to illustrate the inclusion of some commands; the reader may find that their own data gives differing numbers and possibly that minor modifications to the code are needed if using the mouse/rat exon array or there are changes to the Affymetrix file formats in the future.
Filter for core probesets
As described earlier, core probesets are supported by RefSeq annotations and are expected to give the most reliable signal data. The analysis can easily be restricted to core probesets by specifying the core.ps and core.mps files in the exon- and gene-level processing in APT as described above.
Filter for undetected probesets
Following the filter outlined in the Affymetrix technical note ‘identifying and validating alternative splicing events’ [13], a probeset could be considered detected when the DABG P < 0.05 in ∼50% of the samples of at least one group. The rationale for ‘at least one group’ is that a skipped exon could be entirely unexpressed in one group but present in another.
> dabg <- read.table("./OUT_DABG/dabg.summary.txt", sep="\t", header=T, row.names=1)
> dim(dabg) # 1411399 10
> dabg.core <- dabg[match(row.names(d.exon), row.names(dabg)),]
> dim(dabg.core) # 287329 10
## define a function to count how many samples have a detection P < 0.05 and apply to each group separately
> count.det <- function(x){length (which(x<0.05))}
> group1.det <- apply(dabg.core[,1:5], 1, count.det)
> group2.det <- apply(dabg.core[,6:10], 1, count.det)
## retain probesets with P < 0.05 in three or more samples in at least one group
> x <- sort(union(which(group1.det>=3), which(group2.det>=3)))
> d.exon.fil <- d.exon[x,]
> dim(d.exon.fil) # 224600 10
Filter for cross-hybridizing probesets
Cross-hybridizing probesets may have artificially high signals due to more than one RNA product hybridizing to them. They are not included in the *.mps files to ensure that gene-level estimates are as reliable as possible, but are in the exon level *.ps files. Thus, exon-level signals from these probesets may suggest an increased rate of inclusion of that exon, but would be false positives if the additional signal comes from another RNA product. Such probesets can easily be filtered out using information in the exon array annotation file (current release is HuEx-1_0-st-v2.na30.hg19.probeset.csv).
The latest annotation file for the Human Exon 1.0 ST array can be obtained from the Affymetrix website by locating the product in the drop-down menu on the support page and checking the box for annotation files. Scroll down to find the files under the heading ‘Current NetAffx Annotation Files’ and choose the ‘HuEx-1_0-st-v2_Probeset_Annotations, CSV Format’ link.
Download and unzip the file, then read into R:
> annot <- read.table("HuEx-1_0-st-v2.na30.hg19.probeset.csv", sep=",", header=T)
> dim(annot) # 1422046 39
## reduce annotation table to core probesets passing the detection filter:
> annot.core <- annot[match(row.names(d.exon.fil), annot[,1]),]
> dim(annot.core) # 224600 39
> colnames(annot.core)
## keep only probesets with a value of 1 in the crosshyb_type column (map uniquely)
> keep <- which(annot.core$crosshyb_type==1) # rows containing non-cross-hybridizing probesets
> ids <- annot.core[keep,1] # extract corresponding probeset IDs
> d.exon.fil2 <- d.exon.fil[match(ids, row.names(d.exon.fil)),]
> dim(d.exon.fil2) # 179445 10
> write.table(d.exon.fil2, "./OUT_EXON/rma-sketch.summary_core_exon_filtered.txt", sep="\t", quote=F, row.names=T)
Filter for genes undetected in both groups
If a gene is not expressed overall in either of the groups being investigated, the concept of differential splicing becomes meaningless and it is therefore useful to remove any genes, as well as probesets, considered undetected. The DABG value is only appropriate at the probeset (exon) level, but simple criteria can be defined to decide if the gene should be considered expressed overall. For example, it might be reasonable to call a gene expressed in a particular sample if more than one-half its component exons were assigned a detection P < 0.05. To be considered expressed in a group overall, we might require that it is detected in more than one-half the samples of that group. This filter has a few more steps than the probeset detection filter but follows the same principles. First, use the annotation table to add transcript cluster ID as an extra column in the dabg.core detection matrix:
> dim(dabg.core) # 287329 11
> length(intersect(row.names(dabg.core), annot[,1]))# 286876 – 453 probeset IDs are missing from annotation file (likely to be control probes). Remove these to avoid NAs later on
> keep <- intersect(row.names(dabg.core), annot[,1])
> dabg.core2 <- dabg.core[match(keep, row.names(dabg.core)),]
> dim(dabg.core2) # 286876 10
> dabg.core2[,11] <- annot[match(row.names(dabg.core2), annot$probeset_id), 7] # look up transcript cluster IDs
> gene.ids <- unique(dabg.core2[,11])
> length(gene.ids) # 18705
Now, define a function to calculate the proportion of probesets with P < 0.05—this will be applied to each sample for each gene:
count.exon.det <- function(x){length (which(x<0.05))/length(x)}
Create an empty matrix to store results from applying the above function:
> gene.detection <- matrix(nrow=length(unique(dabg.core2[,11])), ncol=10)
> row.names (gene.detection) <- gene.ids
> colnames(gene.detection) <- colnames(d.gene)
Apply the function to each sample in turn:
> for(i in 1:10)
{
gene.detection[,i] <- tapply(dabg.core2[,i], dabg.core2[,11], count.exon.det)
}
Now, another function is needed to count how many samples in each group the gene was detected in (i.e. more than half the probesets were detected):
> count.gene.det <- function(x){length (which(x>=0.5))}
> genes.det.group1 <- apply(gene.detection[,1:5], 1, count.gene.det)
> genes.det.group2 <- apply(gene.detection[,6:10], 1, count.gene.det)
Keep genes where both counts are ≥3 (i.e. more than one-half the samples in the group):
> keep.genes <- which((genes.det.group1>=3)&(genes.det.group2>=3)) # row numbers
> length(keep.genes) # 13211
> keep.gene.ids <- row.names (gene.detection)[keep.genes]
## look up these gene ids in the gene summary matrix
> length(intersect(keep.gene.ids, row.names(d.gene))) # 12642 – this tells us that 569 genes are not in the gene summary output — they are missing because these genes have no core probesets that map uniquely —
> y <- match(keep.gene.ids, row.names(d.gene))
## remove the NAs
> y <- y[-which(is.na(y)=="TRUE")]
> d.gene.fil <- d.gene[y,]
> dim(d.gene.fil) # 12642 10
> write.table(d.gene.fil, "./OUT_GENE/rma-sketch.summary_core_gene_filtered.txt", sep="\t", quote=F, row.names=T)
Splicing analysis
To identify exons of a gene that are alternatively spliced, the concept of the splicing index has been introduced [14]. In the absence of alternative splicing, the expression level of each exon is expected to be similar and to the expression level of the gene overall (i.e. each and every exon is included in the gene product. The exon to gene ratio is therefore expected to be close to one. The splicing index is defined as the log2 ratio of exon to gene intensity. Under the condition of no splicing, this index would equal zero, while a skipped exon would generate a negative value and an exon included at a higher rate than others would generate a positive value. Usually, it is of interest to identify differential exon usage between two samples or groups of samples, as this could indicate preferential expression of one isoform over another with potential biological consequences. Several tools can be used to identify differential splicing, including the APT program MiDAS [15], the ‘limma’ package from BioConductor [16, 17], a specialised R package called ExonMap [18] and commercial software such as Partek Genomics Suite [19]. This is an active area of research but the basis of these tools is to use an ANOVA approach to identify exons behaving differently to others at the same gene locus. Another R package ‘aroma.affymetrix’ [20] has comprehensive methods for both processing exon array data and detection of alternative splicing (FIRMA: Finding isoforms using robust multichip analysis) [21].
To continue the demonstration of APT for exon array data analysis, I will briefly outline an example splicing analysis using the MiDAS tool. MiDAS can be run directly on the exon and gene-level summaries generated by the apt-probeset-summarize command; however, as noted above, it is important to filter the files to remove potential artefacts before running any splicing analysis. The filtered files are then provided as input to MiDAS.
First, a text file describing the samples in your experiment containing two columns is required: the first column should have the header ‘cel_files’ and list the names of the .CEL files corresponding to the samples to be analysed (note that the filenames have to be exactly correct to be recognized—they will also be the column headers in the exon/gene summary files); the second column should have the header ‘group_id’—this can contain arbitrary names that describe the group each sample belongs to (samples assigned the same name will be considered as one group). MiDAS is an ANOVA-based test, so two or more groups can be analysed simultaneously and at least three samples per group are required to estimate the variance. Run MiDAS with the following command:
> apt-midas –cel-files cels.txt –g ./OUT_GENE/rma-sketch_core_gene.summary_filtered.txt –e ./OUT_EXON/rma-sketch_core_exon.summary_filtered.txt –m HuEx-1_0-st-v2.r2.core.mps –o ./OUT_MIDAS –nol
By default, the apt-midas command log transforms the data—this is only applicable to PLIER estimates and needs to be switched off if using RMA data (or if you have already log2 transformed PLIER estimates) with the –nol argument.
The results file generated by MiDAS contains a list of the probesets that have passed filtering and an associated raw P-value for differential splicing of the exon targeted by that probeset. It can be read into R and sorted on P-value to find the exons most likely to be interesting in terms of alternative splicing:
> midas <- read.table("midas.pvalues.txt", sep="\t", header=T)
> head(midas)
> o <- order(midas$pvalue)
> midas.ordered <- midas[o,]
Further filtering based on P-value and/or magnitude of the splicing index (analogous to fold change filter in standard gene expression analyses) can be applied to generate a shortlist. Currently, MiDAS doesn’t output the SI value, but it can be computed in R as the difference between the log scale exon and gene intensities. Manual inspection of the data can be very useful at this point to decide which genes to follow-up.
Interpreting the results from a splicing analysis can be the most challenging aspect of exon array analysis. In some situations, differential splicing can be dramatic; e.g. a large number of brain-specific isoforms have been found in tissue comparisons [22]. However, in many common analysis scenarios, there may only be a subtle change in the relative proportions of different isoforms between samples/groups, which can be difficult both to detect and interpret.
The first step in trying to understand the biology underlying an exon identified as differentially spliced is to place the data in the context of known isoforms of that gene. APT does not have any visualization capabilities but again, other tools are available for this purpose. In particular, the R package ExonMap [18] and commercial software Partek Genomics Suite [19] both perform analysis of splicing and provide excellent visualization functions. Another possibility is looking at the gene of interest in the UCSC Genome Browser [23], giving access to full details of known and predicted genes/mRNAs from a wide variety of sources. This can be very useful to pinpoint splicing that affects an exon that has been predicted but is not part of the RefSeq transcripts for example. Another useful feature of the UCSC Genome Browser is that a track showing the location of exon array probesets can also be displayed, allowing cross-referencing between the splicing results and gene structure information. Finally, with some R code, it is relatively straightforward to generate a graph of the data for a particular gene, which can be coloured for each group.
In summary, data generated with the exon array has the potential to give deeper biological insights into gene expression and regulation, particularly with regard to splice isoforms, than standard gene expression arrays. A range of tools are available to analyse these data, drawing on previous algorithms for microarray expression data or novel approaches specific for exon arrays. Similar analyses can now be performed with next-generation sequencing technology and this will undoubtedly become more routine with reducing costs, increased read counts and development of appropriate tools. However, analysis of alternative splicing and isoform representation is arguably one of the most challenging aspects of RNA-Seq data and the exon array will remain an extremely useful platform in this transition period.
Key Points.
The exon array has the potential to give deeper biological insights into gene expression and regulation, particularly, alternative splicing, than standard gene expression arrays.
It presents new challenges in terms of data analysis and can produce a high rate of false positive differential splicing events without careful filtering of the data first.
A number of tools have been developed for analysis of exon array data—this review focuses on the suite of tools developed by Affymetrix and the statistical software package R.
The tutorial steps presented here represent just one of the many possible approaches to exon array data analysis.
FUNDING
Wellcome Trust Core Grant (grant 075491/Z/04).
Acknowledgements
I would like to thank Dr M.Wayland for helpful comments and suggestions to improve the manuscript.
Biography
Helen Lockstone is a Bioinformatician with more than six years experience in microarray data analysis. She is Head of Functional Data Analysis at the Wellcome Trust Centre for Human Genetics, University of Oxford.
References
- 1.Affymetrix Technical Note. Array Design for the Human Exon 1.0 ST Array. http://www.affymetrix.com/support/technical/technotesmain.affx (27 September 2010, date last accessed) [Google Scholar]
- 2.Affymetrix Power Tools. http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx (27 September 2010, date last accessed)
- 3.The R Project for Statistical Computing. http://www.r-project.org/ (27 September 2010, date last accessed)
- 4.Moller-Levet CS, Betts GNJ, Harris AL, et al. Exon array analysis of head and neck cancers identifies a hypoxia related splice variant of LAMA3 associated with a poor prognosis. PLoS Comput Biol. 2009;5:e1000571. doi: 10.1371/journal.pcbi.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/ (27 September 2010, date last accessed)
- 6.apt-probeset-summarize manual. http://www.affymetrix.com/support/developer/powertools/changelog/apt-probeset-summarize.html (27 September 2010, date last accessed)
- 7.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 8.Affymetrix Technical Note. Guide to Probe Logarithmic Intensity Error (PLIER) Estimation. http://www.affymetrix.com/support/technical/technotesmain.affx (27 September 2010, date last accessed) [Google Scholar]
- 9.Affymetrix White Paper. Quality Assessment of Exon and Gene 1.0 ST Arrays. http://www.affymetrix.com/support/technical/whitepapers.affx (27 September 2010, date last accessed) [Google Scholar]
- 10.Brettschneider J, Collin F, Bolstad BM, Speed TP. Quality assessment for short oligonucleotide microarray data. Technometrics. 2008;50:241–64. [Google Scholar]
- 11.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open source development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Della Beffa C, Cordero F, Calogero RA. Dissecting an alternative splicing analysis workflow for GeneChip® Exon 1.0 ST Affymetrix arrays. BMC Genomics. 2008;9:571. doi: 10.1186/1471-2164-9-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Affymetrix Technical Note. Identifying and Validating Alternative Splicing Events. http://www.affymetrix.com/support/technical/technotesmain.affx (27 September 2010, date last accessed) [Google Scholar]
- 14.Srinivasan K, Shiue L, Hayes JD, et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–59. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Affymetrix White Paper. Alternative Transcript Analysis Methods for Exon Arrays v1.1. http://www.affymetrix.com/support/technical/whitepapers.affx; Apt-Midas Manual. http://www.affymetrix.com/support/developer/powertools/changelog/apt-midas.html (27 September 2010, date last accessed) [Google Scholar]
- 16.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 17.Shah SH, Pallas JA. Identifying differential exon splicing using linear models and correlation coefficients. BMC Bioinformatics. 2009;10:26. doi: 10.1186/1471-2105-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okoniewski MJ, Yates T, Dibben S, et al. An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol. 2007;8:R79. doi: 10.1186/gb-2007-8-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partek Genomics Suite. http://www.partek.com/partekgs. (27 September 2010, date last accessed)
- 20.Bengtsson H, Simpson K, Bullard J, et al. Aroma.affymetrix: a generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Technical Report 745. Berkeley: University of California; 2008. [Google Scholar]
- 21.Purdom E, Simpson KM, Robinson MD, et al. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–14. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark TA, Schweitzer AC, Chen TX, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UCSC Genome Browser. http://genome.ucsc.edu/ (27 September 2010, date last accessed)
- 24.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]